Abstract
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease facilitated by aberrant immune responses directed against cells and tissues, resulting in inflammation and organ damage. In the majority of patients, genetic predisposition is accompanied by additional factors conferring disease expression. While the exact molecular mechanisms remain elusive, epigenetic alterations in immune cells have been demonstrated to play a key role in disease pathogenesis through the dysregulation of gene expression. Since epigenetic marks are dynamic, allowing cells and tissues to differentiate and adjust, they can be influenced by environmental factors and also be targeted in therapeutic interventions. Here, we summarize reports on DNA methylation patterns in SLE, underlying molecular defects and their effect on immune cell function. We discuss the potential of DNA methylation as biomarker or therapeutic target in SLE.
Keywords: : biomarker, chromatin, CREM, DNMT, epigenetics, inflammation, methylation, SLE, transcription factor, treatment
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease that can affect any organ of the human body. Disease expression is facilitated by aberrant immune responses directed against cells and tissues, resulting in inflammation and organ damage [1]. Despite intense scientific efforts targeting the pathophysiology of SLE, the exact molecular mechanisms remain elusive [1,2]. Provided familial clusters with several SLE patients among first degree relatives, and associations with nucleotide polymorphisms and copy number variants, genetic factors centrally contributing to the pathophysiology have been identified. However, mutations in single genes (e.g., complement factor genes C1q, C2 or C4) explain the onset of disease only in a very small number of patients. Furthermore, genetically identical monozygotic twins exhibit disease concordance in only 20–40%. Thus, combined hormonal and environmental factors are implicated in disease expression in genetically predisposed individuals [1–4].
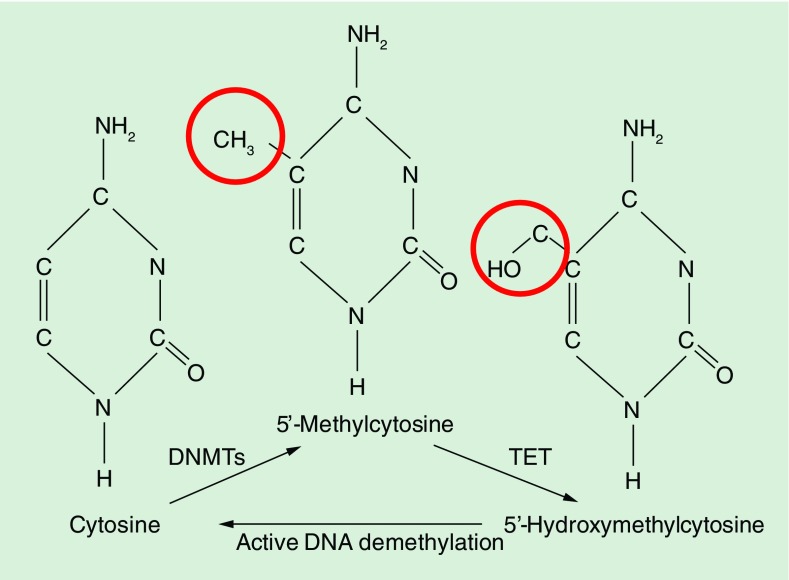
Over the past two decades, it has been convincingly documented that disrupted transcription factor networks and gene expression profiles in immune cells contribute to the pro-inflammatory phenotype in SLE [1,2,5–7]. However, the identification of primary and disease causing molecular pathomechanisms versus such phenomena secondary to chronic inflammation can be challenging. The expression of genes is regulated on multiple levels, including the transcription of DNA into RNA, translation of RNA into protein products, and post-translational inactivation and/or degradation [8–10]. In response to the recruitment of transcription regulatory factors, cis-DNA sequences control gene transcription [6–8,11]. The accessibility of DNA to transcription factors, however, can be altered by so-called epigenetic events that regulate chromatin accessibility without affecting the underlying DNA sequence. Covalent addition of a methyl group to the 5′ carbon position of cytosine within cytosine-phosphate-guanosine (CpG) dinucleotides is a highly efficient mechanism for preventing transcription factor recruitment (Figure 1). This process, referred to as DNA methylation, is mediated through enzymes, so-called DNA methyltransferases (DNMTs) [6–8,11]. DNMT1 is responsible for remethylation of hemimethylated DNA during cell division, restoring and conserving the methylation patterns of the paternal DNA strand, thus referred to as a maintenance methyltransferase. DNMTs 3a and 3b confer DNA methylation in newly replicated DNA, but may also methylate DNA regions that have not been methylated previously. Thus, they are summarized in the group of de novo methyltransferases. However, recent evidence suggests that the situation is more complicated and also DNMT1 can be involved in de novo methylation [12,13].
Figure 1. . Modifications to cytosine during DNA methylation.
Addition of a methyl group to the 5′ carbon position of cytosine within cytosine-phosphate-guanosine dinucleotides is a highly efficient mechanism for preventing transcription factor recruitment. DNA methylation is mediated through highly conserved enzymes, so-called DNA methyltransferases. DNA hydroxymethylation is the result of oxidation of methylated cytosine-phosphate-guanosine DNA by ten eleven translocation family enzymes, which makes it an intermediate product during active DNA demethylation processes. Hydroxymethylated cytosine may actively be removed by DNA repair pathways, suggesting a role of DNA hydroxymethylation during active DNA demethylation processes.
DNA methylation is not the only epigenetic event and cannot be addressed independently. The cell's genomic DNA is organized and packed as chromatin, a macromolecular complex containing DNA, basic histone proteins and nonhistone chromatin proteins, including DNA polymerases, RNA polymerases and transcription factors. A total of 147 base-pairs of genomic DNA are coiled around histone octamers, consisting of two copies of each histone H2A, H2B, H3 and H4, comprising the nucleosome core [6–8,10]. Epigenetic factors mediate the repositioning of nucleosomes, resulting in variable accessibility of DNA to transcription factors and RNA polymerases, regulating gene expression. Key epigenetic modifications in addition to the aforementioned DNA methylation are histone modifications (including methylation, acetylation, ubiquitination, phosphorylation, etc.). Of note, DNA methylation and histone modifications usually reflect and influence one another, thus cannot be seen as individual or even independent events [14]. Generally, high levels of DNA methylation and repressive histone modifications characterize inactive (hetero-)chromatin, while low levels of DNA methylation and permissive histone modifications characterize actively transcribed genes.
In immune cells from SLE patient, distinct modifications to the epigenome have been reported, and it became almost certain that some of these alterations directly contribute to disease expression and tissue damage rather than being a symptom of chronic inflammation and/or secondary to other molecular events [6,7,11,15]. Epigenetic marks are generally dynamic, allowing cells and tissues to differentiate and adjust to the (micro-)environment. Since epigenetic events regulate cell- and tissue-specific gene expression, their disruption can result in majorly affected tissue homeostasis and damage [6–8,11]. Conversely, the dynamic character of epigenetic marks also promises potential as a therapeutic target in SLE and other inflammatory conditions.
For this review, current literature on DNA methylation in SLE was identified in a PubMed search and from the authors’ collection of manuscripts on the topic. Relevant articles were selected by the authors, who, owing to space limitations, do not claim absoluteness, and apologize to colleagues whose important work may not have been included. The following will include discussion of altered DNA methylation patterns in immune cells from SLE patients, their contribution to inflammation and tissue damage, key mechanisms contributing to disturbed DNA methylation, their potential as disease biomarkers, and potential therapeutic interventions targeting DNA methylation in SLE. Interactions between DNA methylation and other epigenetic events are briefly discussed. A detailed discussion of involved mechanisms, however, is beyond the scope of this manuscript and reviewed elsewhere [16,17].
DNA methylation in the pathophsyiology of SLE
DNA methylation shaping the immune system
Closely controlled signal- and receptor-specific gene expression patterns define the phenotype of immune cells [7,18–20]. Lineage-defining proteins include transcription factors, cytokines and/or chemokines, signaling molecules and surface receptors [18,20]. Regulatory and effector T cells are central control elements of the immune system, providing inflammatory stimuli where appropriate and terminating them to prevent tissue damage. Regulatory and effector phenotypes exist in the cluster of differentiation (CD)4+, the CD8+ and the CD4-CD8- (so-called ‘double negative’; DN) T-cell compartments [18,20–22]. Most scientific data were generated for the differentiation of CD4+ T helper (Th) cell subsets. In human and murine Th1 cell differentiation, the IFN-γ (IFN) gene undergoes epigenetic remodeling including DNA demethylation allowing gene transcription, while lineage-inappropriate genes become methylated. For Th2 cells, reduced DNA methylation has been reported for the lineage-defining IL-4 (IL4) gene in humans and mice, while Th1 cytokine genes exhibit epigenetic silencing [6–8,11,18,20,23,24]. In concordance to these findings, Th17 cells in humans and mice also exhibit permissive DNA methylation patterns within the lineage-defining IL-17 (IL17) cytokine gene cluster [25–27]. Furthermore, DNA methylation is also suspected to be involved in the generation of murine and human DN T cells from CD8+ T lymphocytes in health and disease [28,29]. DN T cells comprise a more recently appreciated immune cell subset with incompletely understood function. Contrary to earlier hypotheses, DN T cells do not only exist in lymphoproliferative conditions in humans (autoimmune lymphoproliferative syndrome) and mice (e.g., Murphy Roths large mice homozygous for the lymphoproliferation spontaneous mutation [Faslpr]: MRL.lpr) [22], but rather exert regulatory and/or effector functions during immune responses [22,30,31].
Altered DNA methylation in SLE results in immune dysregulation
Altered epigenetic patterns are a hallmark of immune cells in SLE [6,7]. Global DNA methylation is reduced in lymphocytes from SLE patients and correlates with disease activity [6,7,11]. A number of cytokine genes are regulated by epigenetic remodeling and have been demonstrated to be hypomethylated in SLE, resulting in increased expression in CD4+ T cells (Table 1). Some of these cytokines contribute to tissue damage and/or (auto-)antibody production, including interleukin (IL-)4 [32], IL-6 [33], IL-10 [34,35], IL-13 [34] and IL-17A [6,25,26,36]. Increased expression of pro-inflammatory IL-6 is involved in broad immune activation through neutrophil differentiation, B- and T-cell activation, and the induction of immunoglobulin production in SLE [37]. The immune-modulatory cytokine IL-10 has anti-inflammatory effects on a variety of immune cells. However, IL-10 serum and tissue levels correlate with disease activity and tissue damage in SLE. This may be due to cell- and tissue-specific pro-inflammatory functions of IL-10, which is involved in the induction of B-cell differentiation, activation, and immunoglobulin class switch and increased IgG production [38]. Indeed, blockade of IL-10 with antibodies induced remission in a considerable subset of SLE patients [39]. The pro-inflammatory effector cytokine IL-17A instigates inflammation and promotes antibody production. Increased IL-17A expression was detected in T cells and in inflamed tissues from SLE patients [2,25,30,40,41].
Table 1. . Genes with reduced DNA methylation in systemic lupus erythematosus (in alphabetical order).
| Gene (protein) | (Mainly) expressed by | Physiological function | Proposed effects in SLE | Ref. |
|---|---|---|---|---|
| CD6 (Cluster of differentiation 6) | On the cell surface of T cells and some other immune cells | Important for continuation of T-cell activation | Enhanced T-cell activation | [57] |
| Promoter P1 of CREM (cAMP response element modulator α) | T cells, multiple other cells and tissues | Transcription factor, regulating multiple cellular functions, including cytokine expression in T cells | Involved in the generation of DN T cells, promotes effector phenotypes through the regulation of IL-2 and IL-17 in CD4+ T cells | [25,26,28,29,74,76,118] |
| ESR1 (estrogen receptor 1 or α) | Ubiquitously expressed | Nuclear receptor that is activated by estrogen | Increased estrogen signaling, contributing to immune activation (through CREM-α?) | [70,73] |
| Human endogenous retroviral elements (HERVs) | Generally all human cells, in SLE: B and T cells | None. HERVs are the remainders of ancient retroviral infections, and usually silenced by epigenetic events. | increased expression of interferon-related genes raised the possibility of a viral contribution to SLE; HERV protein products have been demonstrated to induce (auto-)antibody production | [45,50,53,55] |
| IFI44L (interferon-induced 44-like protein) | PBMCs | While the function of IFI44L is unknown, increased IFI44L expression is a component of the Type 1 interferon response signature and part of the cellular response to viral infection |
‘Interferon signature’ gene Global immune activation |
[43,185,186] |
| IKZF4 (IKAROS family zinc finger 4, encoding for Eos) | Lymphocytes | Member of the Ikaros family of transcription factors, implicated in the control of lymphoid development | ‘Interferon signature’ gene Global immune activation |
[42] |
| IL10 (Interleukin-10 gene, encoding for IL-10) | T cells, B cells, monocytes, others | Immune regulatory cytokine, inhibition of T-cell activation, B-cell differentiation, activation and immunoglobulin production | B-cell activation, (auto-) antibody production | [9,35,187,188] |
| IL17A (IL-17A gene, encoding for IL-17A) | T cells, NK cells, mast cells, neutrophils | Induction of chemokines, cytokines, recruitment of neutrophils: defense against bacteria and fungi | Induction of tissue damage in SLE | [26,27,40,41] |
| IL17F (IL-17F gene, encoding for IL-17F) | T cells, NK cells, mast cells, neutrophils | Induction of chemokines, cytokines, recruitment of neutrophils: defense against bacteria and fungi | Increased CREM-α-recruitment > reduced expression of IL-17F > increased IL-17A homodimers > enhanced inflammation | [27,40,41,118] |
| IRF7 (Interferon regulatory factor 7) | Constitutively expressed by lymphoid tissues, and plasmacytoid dendritic cells; inducible in many cells and tissues | Transcriptional activation of virus-inducible cellular genes, including type I interferon genes | ‘Interferon signature’ gene Global immune activation |
[42] |
| ITGAL (integrin alpha L gene, encoding for CD11A) | T cells | Cellular adhesion and costimulation | Increased T-cell-mediated inflammation | [189–192] |
| KIR2DL4 (killer cell immunoglobulin-like receptor 2DL4, encoding for KIR) | On the surface of NK cells and some T cells | Detection of virally infected cells or transformed cells | Increased expression of KIR on T cells, resulting in T-cell activation | [60,66,67] |
| MX1 (myxovirus resistance 1 gene, encoding for interferon-induced GTP-binding protein Mx1, or MxA) | Neutrophils, leukocytes | GTPase, mediates resistance against RNA viruses | ‘Interferon signature’ gene Global immune activation |
[43] |
| PP2A (serine/threonine protein phosphatase 2A) | Eukaryotic cells | Phosphatase with complex functions in many aspects of cell function | Increased PP2A expression in T cells from SLE patients mediates DNA demethylation through suppression of MAPK signaling pathways and induces epigenetic remodeling of the IL17 locus | [27,68,69] |
| PRF1 (Perforin) | CD8+ cytotoxic T cells and NK cells | Cytolytic protein | Perforin expression in CD4+ T cells, may contribute to T-cell-induced death of monocytes/macrophages in SLE | [64,65] |
| TNFSF5 (tumor necrosis factor ligand superfamily member 5, encoding for CD40L/CD154) | Activated T cells | Costimulatory molecule, B cell maturation and activation | Increased B cell costimulation and antibody production | [59,60,65,140,193] |
| TNFSF7 (TNF ligand superfamily member 7, encoding for CD70) | Activated T cells | B-cell activation, IgG synthesis, T-cell costimulation | Increased B cell activation, IgG synthesis, T-cell costimulation | [5–7,141] |
CREM-α: Cyclic adenosine-monophosphate response element modulator alpha; HERV: Human endogenous retroviral element; PBMC: Peripheral blood mononuclear cell; SLE: Systemic lupus erythematosus.
Most recently, reduced methylation of interferon-associated genes has been documented in CD4+ T cells (IKAROS family zinc finger 4: IKZF4, interferon regulatory factor 7: IRF7) and neutrophils (myxovirus resistance 1 gene: MX1, interferon-induced 44-like gene: IFI44L, interferon-induced transmembrane protein 1: IFITM1, poly adenosine-diphosphate ribose polymerase 9: PARP9, interferon-induced protein with tetratricopeptide repeats 3: IFIT3, DEAD (Asp-Glu-Ala-Asp) box polypeptide 60: DDX60, lymphocyte antigen 6E: LY6E, interferon-stimulated gene 15: ISG15) from SLE patients [42,43] (Table 1). Type 1 interferons play a pivotal role in the pathophysiology of SLE and associated disorders, and entertain the so-called interferon signature, increased expression of a set of interferon-associated and -dependent genes [44].
Another indication that altered DNA methylation and gene expression contribute to the pathophysiology of SLE is the reduced methylation of the human endogenous retroviral elements. Elevated serum levels of type I interferon and increased expression of interferon-related genes raised the possibility of a viral contribution to SLE [45]. Indeed, retroviral infections with human T lymphotropic virus type I or HIV-1 resemble symptoms of SLE [46]. However, retroviruses have not been isolated from patients with SLE. The study of lupus-prone New Zealand mice offered an alternative explanation for the aforementioned findings: activation of endogenous retroviral sequences (ERVS) [47]. In humans and mice, ERVS were integrated into the genome during evolutionary history through retroviral infections. In modern humans, ERVS account for approximately 8% of the entire genome [7,48]. Indeed, abnormal expression of an ERVS was reported in lupus-prone mice [48,49] and in patients with SLE [50]. Furthermore, ERV protein products have been demonstrated to induce (auto-)antibody production [50]. Increased transcription of ERVS has been linked to two mechanisms: increased transcriptional activity through permissive promoter haplotypes within the HRES-1 locus [51,52] and reduced DNA methylation within human endogenous retroviral elements of B and T cells [53], resulting in a general disturbance of gene expression, particularly of genes contributing to the development of SLE [6]. Furthermore, stimulation of human B lymphocytes with anti-IgM antibodies increased HRES methylation, which was reversed in the presence of IL-6 [54]. Without B-cell receptor engagement, B lymphocytes from SLE patients are hypomethylated as a result of reduced ERK/DNMT1 signaling [55]. Thus, it is generally accepted that HRES-1 is the target of epigenetic regulation in SLE, contributing to disease expression through immune dysregulation and autoantibody production.
In addition to cytokines, intra- and extracellular signaling molecules can be regulated through DNA methylation, some of which exhibit alterations in T and B cells from SLE patients (Table 1). Reduced methylation of regulatory regions of genes encoding for costimulatory molecules, including TNF ligand superfamily member 7: TNFSF7 (CD70/CD62L) [56], cluster of differentiation 6: CD6 [57], integrin α L: ITGAL (CD11A) [58], TNF ligand superfamily member 5: TNFSF5 (CD40L/CD154) [59,60] in T cells, and cluster of differentiation 5: CD5 [54] in B cells has been documented [6,7,11,61–63]. Reduced DNA methylation in SLE T cells results in expression of usually CD8+ T cell- and NK cell-specific perforin (PRF1 gene) in CD4+ T cells, likely contributing to increased T cell mediated cell death of monocytes and macrophages [64,65]. Increased expression of the stimulatory and inhibitory killer-cell immunoglobulin-like receptor on the surface of T cells in SLE patients, resulting in T-cell activation, was linked to demethylation of the killer cell immunoglobulin-like receptor 2DL4 (KIR2DL4) gene [60,66,67]. Lastly, T cells from SLE patients are characterized by increased expression of the serine/threonine protein phosphatase 2A (PP2A), which has also been linked to reduced methylation of CpG elements within the proximal promoter of the PP2A gene [68,69]. More detailed information on the function and effects of hypomethylated genes in immune cells from SLE patients are provided in Table 1.
Female predominance is a key characteristic of ‘classical’ SLE during the reproductive phase (also see later). Evidence suggests that this may (at least partially) be due to increased estrogen levels in women during the reproductive phase [70–72]. Recently, Liu et al. demonstrated reduced DNA methylation within the proximal promoter of the estrogen receptor 1 (ESR1) gene, encoding for the ubiquitously expressed estrogen receptor α, in women with SLE and in rheumatoid arthritis patients when compared with healthy controls, suggesting the involvement of reduced DNA methylation and increased estrogen signaling in the pathophysiology of SLE [73].
More recently, it became apparent that DNA methylation patterns in T cells from SLE patients are even more complex. While the aforementioned cytokine and signaling molecule genes are characterized by reduced DNA methylation, other genes are hypermethylated in SLE T cells favoring reduced expression, including IL-2 [7,25,28,74], the regulatory T-cell signature transcription factor Forkhead-Box-Protein P3 (Foxp3) [33,75], the surface co-receptors CD8A and CD8B [2] and Notch 1 [76], as well as the ubiquitously expressed glucocorticoid receptor (GR) [77] (Table 2).
Table 2. . Genes with increased DNA methylation in systemic lupus erythematosus (in alphabetical order).
| Gene (protein) | (Mainly) expressed by | Physiological function | Proposed effects in SLE | Ref. |
|---|---|---|---|---|
| CD8A, CD8B (Cluster of differentiation genes 8A and 8B, encoding for CD8A, CD8B) | CD8+ T cells | Co-receptor to the CD3/T cell receptor complex | Generation of effector CD3+CD4-CD8- DN T cells | [28,78] |
| IL2 (IL-2 gene, encoding for IL-2) | T cells | Proliferation and activation of T cells | Impaired generation of regulatory T cells, reduced activation-induced cell death and longer survival of autoreactive T cells, impaired function of cytotoxic CD8+ T cells, effector CD4+ T-cell differentiation and cytokine expression | [25,30,41,74] |
| Foxp3 (Forkhead-Box-Protein P3) | Regulatory T cells | Transcription factor, controlling regulatory T-cell functions | Reduced number and altered function of regulatory T cells | [33,75] |
| NOTCH1 (Notch-1 trans-membrane receptor) | T cells | Role during T-cell lineage determination, e.g., polarization of T helper cells | T-cell activation, increased IL-17A expression | [76] |
| NR3C1 (nuclear receptor subfamily 3 group C member 1, encoding for glucocorticoid receptor) | Multiple cells and tissues | Regulates development, metabolisms, immune responses, etc. Exerts pleiotropic effects in different cells and tissues | Unknown, potentially increased immune activation | [77] |
SLE: Systemic lupus erythematosus.
In contrast to the previously discussed pro-inflammatory effector cytokines, IL-2 fails to be expressed in T cells of SLE patients, which is (at least partially) the result of increased DNA methylation of IL2 regulatory regions. Altered IL-2 expression contributes to reduced numbers and impaired function of regulatory T cells in SLE, impaired activation-induced cell death, and altered cytotoxic CD8+ T cell function [6]. Regulatory T cells are key mediators of peripheral self-tolerance that can actively suppress effector T cells, inhibit inflammation, and prevent or terminate autoimmune processes. Several studies demonstrated increased DNA methylation of regulatory regions of the Foxp3 gene in T cells from SLE patients, which may contribute to reduced gene expression and reduced number and/or altered function on regulatory T cells in SLE [33,75]. During the generation of DN T cells from CD8+ T cells, DNA methylation plays a key role in silencing CD8A and CD8B expression. Site-specific DNA methylation contributes to epigenetic remodeling of the CD8 gene cluster and subsequent down-regulation of CD8 co-receptor expression of the surface of T cells from SLE patients and lupus-prone MRL.lpr mice, contributing to expanded numbers of DN T cells in the peripheral blood and tissues [28,30,78]. DN T cells in SLE patients have been demonstrated to exhibit pro-inflammatory effector phenotypes that are reflected by a pro-inflammatory epigenetic signature, permitting expression of pro-inflammatory effector cytokines including IFN-γ, IL-17F, IL-12, IL-18 and others [78]. These findings suggest similar molecular mechanisms linking T cell subset determination and lineage-specific cytokine expression, for example, through the modulation of DNA methylation.
The Notch family of trans-membrane coreceptors represents evolutionarily conserved pathways that transduce signals between neighboring cells and determines decisions during cell proliferation, survival, differentiation and lineage determination. T cells from SLE patients display defective upregulation of Notch-1 receptor expression in response to T-cell receptor activation. Decreased Notch-1 expression in T cells from active SLE patients was linked with enhanced histone H3 methylation and CpG DNA methylation of the human NOTCH1 promoter. Of note, decreased Notch-1 expression was associated with increased IL-17A expression, suggesting a key role for Notch-1 in SLE immunopathogenesis [36,76].
The GR is ubiquitously expressed in most cells and tissues and mediates multiple effects. Several studies reported reduced expression of the GR on peripheral blood mononuclear cells (PBMCs) from SLE patients, reflecting disease activity and tissue damage [79–81]. Differential GR expression has been linked with genetic and splice variants. Most recently, increased DNA methylation at the promoter region of the (GR encoding) nuclear receptor subfamily 3 group C member 1 (NR3C1) gene in PBMCs from SLE patients was linked with reduced gene expression, offering another example for complex methylation patterns in SLE [77].
DNA hydroxymethylation in SLE
DNA hydroxymethylation is a recently appreciated epigenetic modification. It appears likely that genomic DNA hydroxymethylation can act either as an intermediary in the delicate balance between DNA methylation and demethylation, or it can act as relatively stable component of genomic DNA [82–85]. Currently, our understanding of its exact function and impact of gene regulation is somewhat incomplete. Various cells and tissues exhibit positive correlation between gene expression and DNA hydroxymethylation [82,85,86]. In the context of SLE, DNA hydroxymethylation is under investigation for its role in gene regulation, which has been suggested by studies focusing on stem cell biology, cancer development and neurological disease [87]. DNA hydroxymethylation is the result of oxidation of methylated cytosines within CpG dinucleotides by hydroxytransferase ten eleven translocation (TET) family enzymes [88,89], which makes it an intermediate product during active DNA demethylation processes [90–92] (Figure 1). Furthermore, lower affinity of methyl-binding proteins to hydroxymethylated DNA as compared with methylated DNA, and the potential of transcription factor recruitment to hydroxymethylated regions suggest distinct permissive functions during the regulation of gene expression [92,93]. Recently, increased TET family mRNA expression and DNA hydroxymethylation have been reported in CD4+ T cells from SLE patients as compared with healthy controls [94]. However, in concordance to DNA methylation, DNA hydroxymethylation patterns in SLE appear complex with areas of increased (2748 genes) and decreased (47 genes) hydroxymethylation.
Mediators of epigenetic alterations in SLE
DNMTs
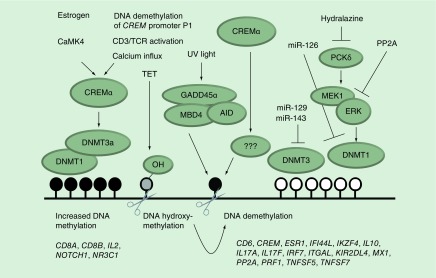
As mentioned before, DNMTs have historically been classified as maintenance (DNMT1) or de novo (DNMT3a, DNMT3b) enzymes. More recently it has become clear that DNMT1 can also be involved in de novo gene methylation, suggesting that the historic view is probably an oversimplification of the in vivo situation in complex biological systems [6–8,11–13]. In some studies, CD4+ T cells from SLE patients exhibited reduced expression of DNMT1 and DNMT3a when compared with controls [95,96]. However, conflicting results also exist, not showing significant differences between SLE patients and controls [97]. Differences in DNMT mRNA expression patterns between studies may be due to variable disease activity between groups, and possible discrepancies between DNMT mRNA expression and protein levels or activity. Furthermore, DNA methylation through DNMTs is signal-, target-, cell- and tissue-specific and not solely dependent on DNMT mRNA expression levels [6,7]. Indeed, DNMT1 activity largely depends on the MAP kinase activity that is altered in SLE T cells (see below) [27,68,69] (Figure 2).
Figure 2. . Selected molecular mechanisms contributing to altered DNA methylation in systemic lupus erythematosus.
Cyclic adenosine-monophosphate response element modulator (CREM) α is expressed at increased levels in T cells from systemic lupus erythematosus (SLE) patients. Estrogen receptor signaling, increased CaMK4 activity, CD3/TCR stimulation, increased cellular calcium influx and also reduced DNA methylation of the CREM promoter P1 increase CREM-α expression. CREM-α contributes to regionally increased DNA methylation in SLE T cells through its interactions with DNMT1, and DNMT3a. Conversely, CREM-α reduces DNA methylation of the IL17 gene in a yet to be determined manner. Another DNA demethylation mechanism involves TET proteins in T cells from SLE patients. TET proteins mediate DNA hydroxymethylation (-OH), an intermediate of several active demethylation pathways. Growth arrest and DNA damage-inducible protein alpha (GADD45-α) is overexpressed in SLE T cells and further inducible by UV irradiation. GADD45α in conjunction with activation-induced deaminase, and methyl–cytosine-phosphate-guanosine-binding domain 4 related G:T glycosylase mediates DNA demethylation. Mitogen activated protein kinas (extracellular signal-regulated kinase) activation through the protein kinase C (PKC)-δ can be inhibited by hydralazine, or PP2A (which is increased in T cells from SLE patients). Impaired extracellular signal-regulated kinase activation results in reduced activity of DNMT1. Furthermore, DNMT expression and activity can be altered by miRNAs (miR-126, -129, -143) which are expressed at increased levels in SLE T cells. Examples of affected genes are provided below the depiction of DNA methylation patterns observed in SLE. The above-mentioned molecular mechanisms contributing to altered DNA methylation, however, have not been shown for all genes affected by DNA dysmethylation (also see Tables 1 & 2).
AID: Activation-induced deaminase; CREM-α: Cyclic adenosine-monophosphate response element modulator alpha; DNMT: DNA methyltransferase; ERK: Extracellular signal-regulated kinase; MBD4: Methyl-CpG-binding domain 4; TET: Ten eleven translocation.
Micro-RNAs
Micro-RNAs (miR) are 21–23 base pair spanning noncoding RNAs that function as posttranscriptional regulators of gene expression. They derive from larger transcripts, which are usually derived from noncoding intergenic DNA, cleaved by nuclear ribonucleases and then exported to the cytoplasm [98,99]. In the cytoplasm, the enzyme Dicer processes the transcripts into mature miRNAs [100,101], which can interfere with gene expression through duplex formation with target genes or transcripts, usually at the 3′ untranslated region (3′UTR), inducing translational repression, mRNA cleavage, or translational arrest [77,102,103]. Micro-RNA expression can be either the result or the cause of altered DNA methylation, and several interconnections between miRNA expression and epigenetic regulation of cellular functions have been reported. Micro-RNA129 and miR143 influence DNA methylation through the regulation of DNMT3a and DNMT3b [104–106]. Recently, miR126 has been linked with reduced DNMT1 expression in CD4+ T cells from SLE patients and subsequently increased expression of CD11A and CD70, contributing to increased B- and T-cell activity [107] (Figure 2).
Mitogen activated protein kinases
Hypomethylation of DNA in T cells from patients with SLE has been linked to altered mitogen activated protein kinase (MAPK) signaling pathways. In SLE patients with active disease, reduced activation of extracellular signal-regulated kinases (ERKs) has been linked with impaired DNMT1 activity, resulting in gradual DNA demethylation and increased expression of the previously mentioned genes encoding for CD11A, CD70, CD40L and IL-17A, several interferon-regulated genes and the development of anti-DNA antibodies [27,108,109]. Reduced activity of ERK kinases has been linked to impaired activity of the protein kinase C (PKC)δ [27,108,109]. Recently, Sunahori et al. demonstrated that increased expression of PP2A results in DNA demethylation through suppression of ERK signaling pathways and reduced activity of DNMT1 [27,68,69] (Figure 2).
DNA-interacting proteins GADD45α, AID & MBD4
The growth arrest and DNA damage inducible protein (GADD45)α is expressed at increased levels in T cells from SLE patients [110]. GADD45α was shown to mediate active DNA demethylation through its interaction with activation-induced deaminase and methyl-CpG-binding domain (MBD) 4 in a complicated mechanism involving 5-methyl-cytosine-deaminase and G:T mismatch-specific thymine glycosylase [7,111] (Figure 2). GADD45α interacts with additional proteins, including the chromatin regulatory protein high mobility group box (HMGB)1. The expression of HMGB1 was increased in T cells from SLE patients. Of note, HMGB1 interacts with MeCP2, a protein that centrally participates in the recognition of methylated CpG islands, subsequently directing DNA demethylation in SLE signature genes, including ITGAL (CD11A), and TNFSF7 (CD70) [112].
Transcription factor: DNA interactions promoting alterations in DNA methylation
Recent large-scale studies targeted the interplay between genetic predisposition, for example, through single nucleotide polymorphisms or copy number variants, and epigenetic mechanisms during disease expression. In the encyclopedia of DNA elements project, transcriptionally active genomic regions were systemically mapped to transcription factor recruitment patterns and chromatin structure [113]. Encyclopedia of DNA elements data for the first time demonstrated on a large scale that epigenetic marks, including DNA methylation and histone acetylation occur over relatively long ranges, allowing for chromatin ‘opening’ and accessibility to transcription factors. In this context it is of special interest that the recruitment of the histone acetyltranferase p300 maps to unmethylated CpG islands, further underscoring the previously discussed interplay between the various epigenetic mechanisms. Transcription factor recruitment on a genome-wide scale, however, is highly targeted and mostly to 5′ regulatory promoter regions [113]. The long-suspected impact of genomic variants on transcription factor recruitment, epigenetic modifications and subsequent RNA transcription has been targeted and proven by combining high-density genotyping and integrated chromatin and transcription factor mapping. As previously suspected, genomic variants with a particularly high impact on gene expression were mostly located in or around recruitment domains for master regulatory factors of immune cell differentiation and stimulus-dependent gene activation [114,115].
Understanding the exact contribution of transcription factor recruitment to subsequently induced epigenetic events may offer broad implications for the diagnosis, treatment monitoring and target-directed treatment of autoimmune disorders. In the context of SLE, several studies demonstrated connections between transcription factor binding, DNA methylation and histone modifications, which even correlated with disease phenotypes [116].
Recently, transcription factor regulatory factor box 1 has been shown to recruit to the promoter regions of ITGAL (CD11A), and TNFSF7 (CD70), resulting in corecruitment of histone methyltransferase suppressor of variegation 3–9 homolog 1 (SUV39H1), histone deacetylase (HDAC)1 and DNMT1, subsequently inhibiting gene expression. Since regulatory factor box 1 expression is reduced in T cells from SLE patients, these observations offer an interesting molecular pathomechanisms in SLE that may hold potential for future therapeutic interventions [117].
The transcription factor cyclic adenosine-monophosphate (cAMP) response element regulator (CREM)α has been demonstrated to be involved in the antithetic regulation of IL-2 and IL-17A expression in CD4+ T cells from SLE patients, favoring effector T-cell phenotypes [25,26,74,118]. CREM-α trans-represses IL2 and induced epigenetic remodeling through DNMT1 and DNMT3a recruitment, resulting in increased DNA methylation of the IL2 promoter. Conversely, at the IL17A promoter, CREM-α acts as trans-activator and (in a yet to be determined fashion) fails to mediate DNMT recruitment and induces DNA demethylation [25,26,74]. Numbers of the aforementioned CD3+TCR+CD4-CD8- (DN) T cells are expanded in the peripheral blood and inflamed tissues of SLE patients [30]. DN T cells in SLE exhibit effector phenotypes with increased IL-17A expression. Of note, CREM-α contributes to epigenetic remodeling of the CD8 genes in CD8+ T cells, promoting downregulation of CD8A and CD8B surface coreceptors and the generation of DN T cells [28,29]. Though not experimentally proven yet, it appears likely that CREM-α may also have similar effects on the IL2 and IL17 genes in DN T cells and CD4+ T cells of SLE patients (Figure 2).
The expression of the transcriptional regulator CREM-α in T cells is regulated by various factors, including CD3/T cell receptor (TCR) activation, calcium influx, and estrogen receptor signaling [36,70,71]. CREM-α expression is controlled by the CREM promoter P1 that becomes activated by recruitment of the transcription factor signaling protein (Sp)-1 [119]. In response to estrogen receptor activation, Sp-1 expression is induced, which may in turn (through the induction of CREM-α expression) contribute to the female predominance in SLE [70]. Furthermore, the PP2A, which is expressed in greater amounts in T cells from SLE patients, contributes to CREM-α expression through specific dephosphorylation and activation of the transcription factor Sp-1 at serine residue 59 [119,120]. Another inducer of CREM-α is the calcium/calmodulin-dependent protein kinase 4, a multifunctional serine/threonine kinase that regulates various cellular processes, including gene expression [121–126]. Calmodulin-dependent protein kinase 4 expression is pathologically increased in T cells from patients with SLE and in lupus-prone MRL.lpr mice [121,123,127,128], where it favors effector phenotypes with reduced IL-2 and increased IL-17 expression. This is mediated through the activation of CREM-α that has been demonstrated to trans-regulate IL2 and IL17 in a diametric fashion and induce epigenetic remodeling through its interaction with epigenetic modifiers [25,26,74]. Lastly, the expression of CREM-α itself is controlled by DNA methylation. Effector memory CD4+ T cells from SLE patients exhibit significantly reduced DNA methylation of the CREM promoter P1, contributing to increased CREM-α mRNA expression [25].
Observations from genome-wide approaches delivered additional interactions between transcription factors, epigenetic- and transcriptional events. These include tissue-specific long range interactions between regulatory regions of physically distant but coregulated genes, the transcription of noncoding RNAs and activation-dependent and highly specific histone modifications, indicating the complexity and beauty of epigenetic regulation of chromatin conformation and gene expression [113]. However, to our knowledge, such complex epigenetic events have not been tested for their involvement in SLE and other autoimmune disorders yet.
TET proteins & DNA hydroxymethylation
TET methylcytosine dioxygenase proteins can oxidize methylated cytosine within CpG DNA into 5-hydroxyl-methyl cytosine, carboxyl-methyl cytosine or formyl-methyl-cytosine [89,129–131]. During cell division, DNMT1 exhibit greater affinity to hemi-methylated DNA as compared with hemihydroxymethylated DNA, while both DNMT3a and DNMT3b have approximately equal activity on all three DNA substrates. Binding of MBD family proteins to methylated DNA inhibits Tet activity, suggesting that MBD binding may play a role in regulating DNA hydroxymethylation [132]. Furthermore, hydroxymethylated cytosine may actively be removed by DNA repair pathways, suggesting a role of DNA hydroxymethylation during active DNA demethylation processes [87,133] (Figure 2).
Demographic factors: gender & age
Female predominance exists in most autoimmune disorders and is particularly pronounced in SLE (f:m = 9–10:1). Since SLE in prepubertal children is about equally rare in girls and boys, ovarian hormones appear central in the pathophysiology of SLE. In the context of autoimmunity, estrogens have been most widely studied and have been demonstrated to be involved in T cell subset differentiation and distribution [134,135]. Since Th cell determination and cytokine expression are controlled by epigenetic mechanisms, it appears likely that estrogen affects chromatin composition [5–7,70,71,136]. Recently, Moulton et al. demonstrated that estrogen receptor signaling enhances the expression of the transcription factor CREM-α. As mentioned above, CREM-α largely contributes to the differentiation of effector CD4+ T-cell phenotypes and the generation of DN T cells in SLE [6,7,25,26,28,36,74,118]. Another difference between genders is the presence of a second X chromosome in women. Most X-linked genes are not gender-specific, however, exhibit equal expression patterns between them. Comparable gene expression is achieved by a complex epigenetic event referred to as X chromosome inactivation. It includes DNA methylation, histone modifications and miRNA expression. X-linked genes that require silencing can potentially contribute to the pathophysiology of SLE. Indeed, several genes have been linked with SLE [6,7,137]. Reduced DNA methylation of the X chromosomal CD40L gene contributes to female predominance of SLE and other autoimmune disorders [59,138–140]. Along these lines, women who lack one X chromosome (Turner syndrome: 45, X0) exhibit lower incidences of SLE that are comparable to male cohorts, while individuals with an additional X chromosome (Klinefelter's syndrome: 47, XXY) are at an increased risk [6,7,63,72].
Disrupted epigenetic patterns accumulate over time, contributing to increased incidence of autoimmune/inflammatory disorders with age. The activity of DNMTs, particularly DNMT1, is decreased in the elderly [2,6,7,141]. Indeed, elderly men exhibit greater SLE incidence when compared with elderly women, and with increasing age epigenetic events appear to have an impact even more substantial than genetic predisposition [65]. One result of cumulative DNA demethylation is the generation and accumulation of so-called ‘senescent’ T cells. Senescent T cells are characterized by reduced CD28 expression, shortened telomere length and increased expression of the aforementioned proteins KIR, perforin and CD70, contributing to immune activation in SLE [6,15,32,66]. Together, these observations suggest that, after hormonal influences become weaker, cumulative age-related (likely epigenetic) events may be a key contributor to disease expression. Indeed, the degree of DNA methylation can be used as a biomarker for life expectancy [142].
Exposure to environmental factors
Most autoimmune/inflammatory disorders, including SLE, only cosegregate to a certain extent in first-degree relatives or monozygotic (identical) twins. Thus, it is almost certain that additional, for example, environmental factors contribute to disease expression in genetically predisposed individuals [1,2,6,7].
Drug-induced DNA hypomethylation represents a well-accepted environmental trigger for environmentally caused inflammation. The nucleoside analog 5-azacytidine irreversibly reduces the activity of DNMT1. The maintenance enzyme DNMT1 recognizes DNA hemimethylation during mitotic cell division, securing stable DNA methylation patterns in daughter cells by catalyzing the transfer of S-adenosin methionine (SAM) to the cytosine residue of the unmethylated DNA along the paternal ‘blue print’. Cell metabolism and alterations secondary to diet and/or oxidative stress are more recently appreciated modifiers of DNA methylation and inflammatory responses [143]. Modifications to DNA and/or histone proteins depend on substrates derived directly from diet or products of intermediary metabolism. Through methionine adenosyltransferase, a redox-sensitive enzyme in the so-called SAM cycle, SAM derives from adenosine triphosphate (ATP) and methionine [143]. Thus, the availability of B vitamins and methionine directly regulate SAM generation. Global DNA methylation is reduced in SLE patients and in the elderly, who exhibit an increased incidence of autoimmune disorders, suggesting alterations to the SAM cycle and/or DNMT activity as likely contributors to DNA demethylation in both instances [5,6,144]. Particularly in older individuals with reduced DNMT1 activity, nutritional intake of SAM may be another environmental factor contributing to DNA demethylation [144]. Furthermore, hydralazine (a smooth muscle relaxant used to treat hypertension that has been linked with drug-induced lupus) inhibits the activity of PKC-δ, resulting in impaired ERK kinase activation and subsequently altered activity of DNMT1 [108,109,145] (Figure 2).
Sunlight exposure can induce flares in SLE patients, and the photosensitivity which is reported by more than 70% of patients is a classification criterion for SLE [146,147]. Recently, it was demonstrated that UV irradiation of T cells from SLE patients resulted in reduced DNMT1 mRNA expression and reduced DNA methylation [147]. This may be triggered by the induction of GADD45α through UV exposure. As mentioned previously, GADD45α in conjunction with activation-induced deaminase and MBD4 promotes DNA demethylation in T cells from SLE patients [7,110,111] (Figure 2).
As discussed previously, demethylation of methylation-sensitive genes can result in increased expression of pro-inflammatory and/or immune activating genes, including intracellular signaling molecules, cytokines, chemokines and cell surface receptors. Furthermore, DNA demethylation by 5-azacytidine mediates immune activation and antibody formation through the increased immunogenicity of hypo- or demethylated DNA [1,2,6,7,141].
Mitochondria & the epigenome
As mentioned above, cell metabolism and oxidative stress are more recently appreciated modifiers of epigenetic patterns and cell function [143]. In addition to dietary and age-related effects of SAM metabolsim or DNA methylation, mitochondria play a central role during epigenetic remodeling, and metabolic alterations. Indeed, metabolic disturbances are another example of the tight interconnections between epigenetic events beyond DNA methylation. The generation of acetyl coenzyme A conveys carbon atoms within acetyl groups to the citric acid cycle to be the oxidized for energy production. Another important, however less appreciated function of acetyl coenzyme A is the delivery of acetyl groups, which are incorporated into lysine residues of histone during posttranslational modifications catalyzed by acetyltransferases [143,148].
Furthermore, mitochondria are responsible for the de novo synthesis of flavin adenine dinucleotides, which are essential during histone demethylation through lysine-specific demethylases [149,150]. Of note, H2O2 is a side product of this reaction, resulting in oxidative stress [151]. Lastly, mitochondrial oxidative phosphorylation is the dominant source of ATP. Depletion of ATP in T cells from SLE patients may affect the activity of both the aforementioned methionine adenosyltransferase and adenosine monophosphate activated protein kinase (AMPK) [152,153]. Of note, AMPK phosphorylates histone proteins and inhibits the mechanistic target of rapamycin (mTOR). The mTOR complex functions as nutrient/energy/redox sensor and controls protein synthesis and function. Recently, mTOR has been demonstrated to be involved in altered epigenetic patterns in SLE, including methylation, demethylation and histone phosphorylation, resulting in enhanced T-cell activation [143,154].
Epigenetic alterations as biomarkers for SLE
Currently available biomarkers for the diagnosis and activity assessment of SLE are somewhat limited. While some autoantibodies (particularly anti-nuclear antibodies) are highly sensitive, they lack disease specificity [155]. Others are highly specific (double-stranded DNA: dsDNA) but not very sensitive [156]. Furthermore, monitoring of disease activity can be challenging, since medication and infections can affect laboratory tests, such as lymphocyte counts or serum complement levels [157].
Epigenetic biomarkers, particularly DNA methylation, have been intensively studied in other disorders, including malignancies, and promise potential in the search for diagnostic and prognostic biomarkers also in autoimmune disease, such as SLE [158]. To date, we are only at the very beginning of understanding the impact and pattern of epigenetic changes during the onset and course of autoimmune disorders. A major obstacle in the search for epigenetic biomarkers is the fact that disease-specific molecular events are frequently accompanied by strong, yet unspecific inflammatory responses [159,160].
Most of the currently applied epigenetic disease biomarkers come from the cancer field. DNA methylation is most commonly used, since it is the best studied and most stable epigenetic modification. It can be measured in cells not specifically pretreated, and from extracellular DNA, for example, from body fluids [161,162]. As mentioned above, region-specific DNA methylation patterns have been demonstrated in immune cells from SLE patients. However, provided relatively small sample sizes and variable experimental settings, a correlation of epigenetic changes with disease activity or their usability as biomarkers for the diagnosis of SLE as compared with other autoimmune/inflammatory conditions has not been convincingly tested. Furthermore, the question of whether epigenetic alterations are causes or consequences of chronic systemic inflammation has not been fully answered for most genes [6,160]. A recent study suggested differential methylation of the interferon-induced protein 44-like (IFI44L) gene promoter as epigenetic biomarker for the diagnosis (but currently not the disease activity) of SLE. Differential hypomethylation of CG elements within the promotor region of PBMCs allows differentiation between Asian and European SLE patients from other autoimmune disorders and healthy controls [163]. Though the exact function of the IFI44L protein is currently unknown, increased expression is a component of type-1 interferon responses to viral infections [164]. Since the so-called interferon signature is a hallmark of immune cells and tissues in SLE, and provided that the addition of type-1 interferon did not result in reduced IFI44L methlylation in PBMCs, DNA methylation in this region may not be the result but rather the origin of inflammation and a direct indicator of disease activity in SLE [2,163]. With the increasingly broad availability of high-throughput technologies, new biomarkers will be identified and introduced into the clinical care of patients with autoimmune/inflammatory disorders beyond SLE.
Targeting epigenetic patterns in therapeutic interventions
SLE is a highly variable disease, involving complex pathomechanisms that can vary significantly between individual patients. One increasingly important focus of research is the identification and introduction of pharmacological treatment tailored to the individual patient. Studying epigenetic patterns in particular may provide targets for future therapeutic interventions. While for some epigenetic alterations (particularly histone modifications and miRNAs) epigenetic treatment appears within reach, targeted alteration of DNA methylation appears even more complex [165]. However, epigenetic events cannot easily be dissected into DNA methylation, histone modifications and miRNA expression, since epigenetic events affect one another and epigenetic patterns can be ‘translated’ [14,16,17].
Fragments of genomic DNA can act as antigens and can be detected in the peripheral blood of patients with SLE. Accumulation of single- and/or double-stranded DNA in the extracellular space has been explained by increased apoptosis, and defective clearance of apoptotic material and cell debris in SLE patients. As mentioned above, global DNA methylation is reduced in lymphocytes from SLE patients. Similar to microbial DNA, hypomethylated human DNA fragments may have increased tolerogenic potential, thus contributing to anti-DNA antibody generation, which can be detected years before the onset of disease. Provided disease-specificity of some of the epigenetic alterations detected in SLE, they may prove useful not only as disease biomarkers, but also as therapeutic targets [6,7,11,63,166].
Currently available treatment regimens in SLE include antimalarial medication (chloroquine and hydroxy-chloroquine), corticosteroids and immune-modulators/-suppressants (methotrexate, cyclophosphamide, mycophenolate mofetil, etc.). Antimalarial agents are effective for the prevention of flares in SLE patients. Though most of their effects are currently not understood, they influence micro-RNA expression in lupus-prone mice, suggesting some incompletely understood epigenetic effects [165,167]. Methotrexate inhibits DNA synthesis through inhibiting dihydrofolate reductase activity, which is the effect of high-dose methotrexate treatment used in cancer. Furthermore, it may block DNMT activity by depleting s-adenosyl methionine (SAM), the substrate to DNMT during DNA remethylation, thus mediating gradual DNA demethylation [5,168,169]. Most recently, mycophenolate mofetil, a selective inhibitor of inosine 5-monophosphate dehydrogenase frequently used in patients with lupus nephritis, has been shown to influence histone modifications – another key epigenetic mechanism – in SLE T cells, while not affecting DNA methylation [170]. Cyclophosphamide is an alkylating agent used in cancer treatment and severe vasculitis. It exerts its effects mainly through inhibiting DNA replication. The precise mechanism of cyclophosphamide in SLE and vasculitis treatment are poorly understood. Recently, it has been shown to increase DNA methylation through the induction of DNMT1 activity [5,171]. Although diametric, the epigenetic effects of methotrexate and cyclophosphamide on DNA methylation may partially explain their effectiveness in SLE and underscore the complexity of region-, cell- and tissue-specific DNA methylation patterns.
DNA methylation may be altered with already available ‘epigenetic drugs’, such as DNMT inhibitors azacytidine or decitabine. While not standard in the field of autoimmune/inflammatory diseases, epigenetic interventions have already made their way into cancer treatment [172–174]. Unfortunately, currently available drugs act in an untargeted way on the entire genome and may therefore result in deregulation of previously unaffected genes. Furthermore, DNA hypermethylation (reversible by these drugs) only occurs at a relatively small number of genes in lymphocytes from SLE patients, which tend to be hypomethylated [6,7,11,15,61–63]. Target-directed and gene-specific epigenetic modifiers are currently unavailable, but may be required to reverse disease-causing or disease-modifying epigenetic alterations in SLE.
Another potential (but currently hypothetical) intervention may be the induction of tolerance to extracellular chromatin components through desensitization approaches. Furthermore, correcting globally altered epigenetic marks may reduce immunogenicity of extracellular nucleic acids and reduce autoantibody generation [6].
Mitochondria cause oxidative stress, activating the enzyme mTOR, thus indirectly regulating the activity of histone and DNA-modifying enzymes, including the inactivation of DNMT1 [109]. Of note, treatment with the antioxidant acetylcysteine reduces oxidative stress and mTOR activity, resulting in reduced disease activity in SLE [175]. Since mTOR influences epigenetic events (including DNA and histone methylation and demethylation, as well as histone phosphorylation), thus mediating enhanced T-cell activation in SLE, anti-oxidants or inhibition of mTOR may be candidates in the search for individualized epigenetic treatment in SLE [175–177].
Blockade of transcription factors that are increased in T cells from SLE patients, such as CREM-α [36] or Stat3 [35], or preventing their activation may be promising epigenetic interventions mediating target-directed changes. As discussed above, CREM-α reciprocally mediates epigenetic silencing of some genes (IL2, CD8, and NOTCH1 [7,28,29,74,76]) while conferring epigenetic ‘opening’ to exert others (IL17A [26,118], IL10 [35]). To date, blockade of Stat transcription factor activation through Janus kinase (JAK) inhibitors is generally achievable, but not widely available for clinical use in SLE [178]. Furthermore, epigenetic alterations mediated through JAK/Stat inhibition have (to our knowledge) not been experimentally tested.
Genome-wide hypoacetylation is a hallmark of immune cells in SLE. In the context of animal models and inflammatory conditions, HDAC inhibitors are reported to have anti-inflammatory effects, and have thus been proposed as potentially useful in autoimmune-inflammatory disorders, including SLE [179–182]. Though histone modifications are beyond the scope of this manuscript, altering histone acetylation may indirectly affect DNA methylation and shall therefore be briefly mentioned here [7,13–15]. The already available HDAC inhibitor trichostatin A is one promising candidate for SLE treatment, given its ability to modify gene expression of SLE signature genes, including CD154, and IL-10 in T cells, and type I interferons in activated plasmacytoid DCs, and increase the expression of Foxp3 in regulatory T cells, thereby promoting immune-regulatory responses in humans and mice [117,165,183,184]. However, underlying mechanisms remain unclear and currently available epigenetic interventions are not target-directed. Thus, a general recommendation for the treatment of SLE cannot be given, since off-target effects may cause additional symptoms.
Conclusion & future perspective
The central contribution of epigenetic alterations, particularly DNA methylation in immune cells of SLE patients, to systemic inflammation and tissue damage is widely accepted. The challenge is in distinguishing between primary, disease-causing alterations and those secondary to systemic inflammation and global immune activation. However, recent reports link defined molecular alterations to epigenetic remodeling and gene dysregulation in T cells from SLE patients, suggesting a causative involvement in the pathophysiology. We are just beginning to understand the molecular events contributing to disease expression in genetically predisposed individuals and future studies are warranted. High-throughput applications will allow rapid and comprehensive DNA methylation mapping in immune cells and tissues from SLE patients during early and late disease stages and with variable outcomes. Thus, epigenetic alterations have potential as biomarkers for diagnosis and outcome assessment, as well as objects for targeted and individualized interventions in SLE.
Executive summary.
Current state
The diagnosis systemic lupus erythematosus (SLE) is based on clinical symptoms in conjunction with several sometimes limited laboratory tests.
Genetic predisposition plays a central role for the pathogenesis of SLE.
In most cases, genetic variants in single genes are not strong enough to confer disease.
Additional variables, including epigenetic alterations are necessary for disease expression.
Molecular mechanisms that alter the expression of key genes in SLE are under investigation, including epigenetic mechanisms.
The heterogeneity of clinical symptoms and organ involvement together with low penetrance of genetic associations complicate the interpretation of exome-wide genetic and epigenetic approaches.
Another obstacle is the differentiation between disease-causing alterations, and such that are the result of global immune activation and chronic inflammation in SLE.
Currently available treatment options are (mostly) not target-directed and induce a wide range of intended and unintended events (toxicity and side-effects).
DNA methylation & immune regulation
DNA methylation is a potent epigenetic mechanism, regulating gene expression through DNA accessibility to transcription factors and RNA polymerases.
Demographic factors (gender, age) and the environment (UV light, medication, infections, etc.) can modify DNA methylation patterns, some of which are heritable and induce more or less stabile alterations to gene expression.
DNA hydroxymethylation is a recently identified modification of cytosine with yet to be determined function.
Both DNA methylation and DNA hydroxymethylation have central effects on gene expression in the immune system and influence immune responses.
Altered DNA methylation in SLE
DNA methylation patterns in SLE are complex with regions of increased or reduced DNA methylation.
Altered DNA methylation patterns contribute to immune dysregulation and inflammation in SLE.
DNA hydroxymethylation is a recently appreciated epigenetic mark potentially influencing gene expression in SLE.
A manifold of molecular defects contribute to altered DNA methylation in SLE.
Future perspective
Provided its stability and (at least in some genes) disease-specific patterns, DNA methylation promises potential as biomarker for the diagnosis and risk-assessment in individual SLE patients.
Identifying genetic variants as risk factors for SLE, combining them with cell-specific epigenetic profiles and monitoring alterations over time, will provide novel tools for early risk-assessment, timely diagnosis and treatment initiation, or even preventive measures.
Either reverting disease-specific molecular pathomechanisms conferring epigenetic alterations in SLE (e.g., PP2A, CaMK4, CREM-α, etc.), or applying targeted therapeutic manipulations to the epigenome may become individualized treatment options in SLE with reduced side-effects.
Acknowledgements
The authors thank C Hedrich for her English-language consultation on this manuscript.
Footnotes
Financial & competing interests disclosure
The work of CM Hedrich is supported by the intramural MeDDrive program, TU Dresden and the Fitz-Thyssen Foundation; GC Tsokos is supported by grants from the National Institutes of Health. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Tsokos GC. Systemic lupus erythematosus. N. Engl. J. Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Crispin JC, Hedrich CM, Tsokos GC. Gene-function studies in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2013;9(8):476–484. doi: 10.1038/nrrheum.2013.78. [DOI] [PubMed] [Google Scholar]
- 3.Javierre BM, Fernandez AF, Richter J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20(2):170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The influence of DNA methylation on systemic lupus erythematosus (SLE) disease expression is demonstrated in genetically identical but disease discordant twins.
- 4.Javierre BM, Hernando H, Ballestar E. Environmental triggers and epigenetic deregulation in autoimmune disease. Discov. Med. 2011;12(67):535–545. [PubMed] [Google Scholar]
- 5.Hedrich CM. Systemic Lupus Erythematosus. Elsevier; 2016. p. 255. [Google Scholar]
- 6.Hedrich CM, Crispin JC, Tsokos GC. Epigenetic regulation of cytokine expression in systemic lupus erythematosus with special focus on T cells. Autoimmunity. 2014;47(4):234–241. doi: 10.3109/08916934.2013.801462. [DOI] [PubMed] [Google Scholar]
- 7.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol. Med. 2011;17(12):714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballestar E. An introduction to epigenetics. Adv. Exp. Med. Biol. 2011;711:1–11. doi: 10.1007/978-1-4419-8216-2_1. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann SR, Rosen-Wolff A, Tsokos GC, Hedrich CM. Biological properties and regulation of IL-10 related cytokines and their contribution to autoimmune disease and tissue injury. Clin. Immunol. 2012;143(2):116–127. doi: 10.1016/j.clim.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Hedrich CM, Bream JH. Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol. Res. 2010;47(1–3):185–206. doi: 10.1007/s12026-009-8150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballestar E. Epigenetic alterations in autoimmune rheumatic diseases. Nat. Rev. Rheumatol. 2011;7(5):263–271. doi: 10.1038/nrrheum.2011.16. [DOI] [PubMed] [Google Scholar]
- 12.Ooi SK, O'donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J. Cell Sci. 2009;122(Pt 16):2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner C, Fuks F. DNA methyltransferases: facts, clues, mysteries. Curr. Top. Microbiol. Immunol. 2006;301:45–66. doi: 10.1007/3-540-31390-7_3. [DOI] [PubMed] [Google Scholar]
- 14.Brenner C, Fuks F. A methylation rendezvous: reader meets writers. Dev. Cell. 2007;12(6):843–844. doi: 10.1016/j.devcel.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Brooks WH, Le Dantec C, Pers JO, Youinou P, Renaudineau Y. Epigenetics and autoimmunity. J. Autoimmun. 2010;34(3):J207–J219. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Meroni PL, Penatti AE. Epigenetics and systemic lupus erythematosus: unmet needs. Clin. Rev. Allergy Immunol. 2016;50(3):367–376. doi: 10.1007/s12016-015-8497-4. [DOI] [PubMed] [Google Scholar]
- 17.Xiao G, Zuo X. Epigenetics in systemic lupus erythematosus. Biomed. Rep. 2016;4(2):135–139. doi: 10.3892/br.2015.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josefowicz SZ. Regulators of chromatin state and transcription in CD4 T-cell polarization. Immunology. 2013;139(3):299–308. doi: 10.1111/imm.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim PS, Shannon MF, Hardy K. Epigenetic control of inducible gene expression in the immune system. Epigenomics. 2010;2(6):775–795. doi: 10.2217/epi.10.55. [DOI] [PubMed] [Google Scholar]
- 20.Rothenberg EV. The chromatin landscape and transcription factors in T cell programming. Trends Immunol. 2014;35(5):195–204. doi: 10.1016/j.it.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillhouse EE, Lesage S. A comprehensive review of the phenotype and function of antigen-specific immunoregulatory double negative T cells. J. Autoimmun. 2013;40:58–65. doi: 10.1016/j.jaut.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Martina MN, Noel S, Saxena A, Rabb H, Hamad AR. Double negative (DN) alphabeta T cells: misperception and overdue recognition. Immunol. Cell Biol. 2015;93(3):305–310. doi: 10.1038/icb.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoenborn JR, Dorschner MO, Sekimata M, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat. Immunol. 2007;8(7):732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv. Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- 25.Hedrich CM, Crispin JC, Rauen T, et al. cAMP response element modulator alpha controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc. Natl Acad. Sci. USA. 2012;109(41):16606–16611. doi: 10.1073/pnas.1210129109. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Diametric effects of the transcription factor cyclic adenosine-monophosphate response element modulator α on DNA methylation patterns and gene expression of IL2 and IL17 are documented.
- 26.Rauen T, Hedrich CM, Juang YT, Tenbrock K, Tsokos GC. cAMP-responsive element modulator (CREM)alpha protein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. J. Biol. Chem. 2011;286(50):43437–43446. doi: 10.1074/jbc.M111.299313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolidis SA, Rauen T, Hedrich CM, Tsokos GC, Crispin JC. Protein phosphatase 2A enables expression of interleukin 17 (IL-17) through chromatin remodeling. J. Biol. Chem. 2013;288(37):26775–26784. doi: 10.1074/jbc.M113.483743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hedrich CM, Crispin JC, Rauen T, et al. cAMP responsive element modulator (CREM) alpha mediates chromatin remodeling of CD8 during the generation of CD3+ CD4- CD8- T cells. J. Biol. Chem. 2014;289(4):2361–2370. doi: 10.1074/jbc.M113.523605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedrich CM, Rauen T, Crispin JC, et al. cAMP-responsive element modulator alpha (CREMalpha) trans-represses the transmembrane glycoprotein CD8 and contributes to the generation of CD3+CD4-CD8- T cells in health and disease. J. Biol. Chem. 2013;288(44):31880–31887. doi: 10.1074/jbc.M113.508655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crispin JC, Oukka M, Bayliss G, et al. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J. Immunol. 2008;181(12):8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Rodriguez N, Apostolidis SA, Penaloza-Macmaster P, et al. Programmed cell death 1 and Helios distinguish TCR-alphabeta+ double-negative (CD4-CD8-) T cells that derive from self-reactive CD8 T cells. J. Immunol. 2015;194(9):4207–4214. doi: 10.4049/jimmunol.1402775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Chen Y, Richardson B. Decreased DNA methyltransferase levels contribute to abnormal gene expression in “senescent” CD4(+)CD28(-) T cells. Clin. Immunol. 2009;132(2):257–265. doi: 10.1016/j.clim.2009.03.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lal G, Zhang N, Van Der Touw W, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J. Immunol. 2009;182(1):259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao M, Tang J, Gao F, et al. Hypomethylation of IL10 and IL13 promoters in CD4+ T cells of patients with systemic lupus erythematosus. J. Biomed. Biotechnol. 2010;2010:931018. doi: 10.1155/2010/931018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedrich CM, Rauen T, Apostolidis SA, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc. Natl Acad. Sci. USA. 2014;111(37):13457–13462. doi: 10.1073/pnas.1408023111. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Interactions between Stat3 and the epigenetic modifier p300 is demonstrated and linked to increased gene expression in SLE.
- 36.Rauen T, Hedrich CM, Tenbrock K, Tsokos GC. cAMP responsive element modulator: a critical regulator of cytokine production. Trends Mol. Med. 2013;19(4):262–269. doi: 10.1016/j.molmed.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohl K, Tenbrock K. Inflammatory cytokines in systemic lupus erythematosus. J. Biomed. Biotechnol. 2011:432595. doi: 10.1155/2011/432595. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng H, Wang W, Zhou M, Li R, Pan HF, Ye DQ. Role of interleukin-10 and interleukin-10 receptor in systemic lupus erythematosus. Clin. Rheumatol. 2013;32(9):1255–1266. doi: 10.1007/s10067-013-2294-3. [DOI] [PubMed] [Google Scholar]
- 39.Llorente L, Richaud-Patin Y, Garcia-Padilla C, et al. Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum. 2000;43(8):1790–1800. doi: 10.1002/1529-0131(200008)43:8<1790::AID-ANR15>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 40.Apostolidis SA, Crispin JC, Tsokos GC. IL-17-producing T cells in lupus nephritis. Lupus. 2011;20(2):120–124. doi: 10.1177/0961203310389100. [DOI] [PubMed] [Google Scholar]
- 41.Apostolidis SA, Lieberman LA, Kis-Toth K, Crispin JC, Tsokos GC. The dysregulation of cytokine networks in systemic lupus erythematosus. J. Interferon Cytokine Res. 2011;31(10):769–779. doi: 10.1089/jir.2011.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Absher DM, Li X, Waite LL, et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9(8):e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coit P, Yalavarthi S, Ognenovski M, et al. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J. Autoimmun. 2015;58:59–66. doi: 10.1016/j.jaut.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crow MK, Olferiev M, Kirou KA. Targeting of type I interferon in systemic autoimmune diseases. Transl. Res. 2015;165(2):296–305. doi: 10.1016/j.trsl.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rich SA. Human lupus inclusions and interferon. Science. 1981;213(4509):772–775. doi: 10.1126/science.6166984. [DOI] [PubMed] [Google Scholar]
- 46.Perl A. Mechanisms of viral pathogenesis in rheumatic disease. Ann. Rheum. Dis. 1999;58(8):454–461. doi: 10.1136/ard.58.8.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshiki T, Mellors RC, Strand M, August JT. The viral envelope glycoprotein of murine leukemia virus and the pathogenesis of immune complex glomerulonephritis of New Zealand mice. J. Exp. Med. 1974;140(4):1011–1027. doi: 10.1084/jem.140.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krieg AM, Steinberg AD. Analysis of thymic endogenous retroviral expression in murine lupus. Genetic and immune studies. J. Clin. Invest. 1990;86(3):809–816. doi: 10.1172/JCI114778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caza TN, Fernandez DR, Talaber G, et al. HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann. Rheum. Dis. 2014;73(10):1888–1897. doi: 10.1136/annrheumdis-2013-203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perl A, Colombo E, Dai H, et al. Antibody reactivity to the HRES-1 endogenous retroviral element identifies a subset of patients with systemic lupus erythematosus and overlap syndromes. Correlation with antinuclear antibodies and HLA class II alleles. Arthritis Rheum. 1995;38(11):1660–1671. doi: 10.1002/art.1780381119. [DOI] [PubMed] [Google Scholar]
- 51.Magistrelli C, Samoilova E, Agarwal RK, et al. Polymorphic genotypes of the HRES-1 human endogenous retrovirus locus correlate with systemic lupus erythematosus and autoreactivity. Immunogenetics. 1999;49(10):829–834. doi: 10.1007/s002510050561. [DOI] [PubMed] [Google Scholar]
- 52.Pullmann R, Jr., Bonilla E, Phillips PE, Middleton FA, Perl A. Haplotypes of the HRES-1 endogenous retrovirus are associated with development and disease manifestations of systemic lupus erythematosus. Arthritis Rheum. 2008;58(2):532–540. doi: 10.1002/art.23161. [DOI] [PubMed] [Google Scholar]
- 53.Nakkuntod J, Avihingsanon Y, Mutirangura A, Hirankarn N. Hypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patients. Clin. Chim. Acta. 2011;412(15–16):1457–1461. doi: 10.1016/j.cca.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 54.Garaud S, Le Dantec C, Jousse-Joulin S, et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J. Immunol. 2009;182(9):5623–5632. doi: 10.4049/jimmunol.0802412. [DOI] [PubMed] [Google Scholar]
- 55.Fali T, Le Dantec C, Thabet Y, et al. DNA methylation modulates HRES1/p28 expression in B cells from patients with Lupus. Autoimmunity. 2014;47(4):265–271. doi: 10.3109/08916934.2013.826207. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The influence of altered DNA methylation on increased transcriptional activity of the HRES1 locus in SLE is demonstrated.
- 56.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J. Immunol. 2005;174(10):6212–6219. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 57.Singer NG, Richardson BC, Powers D, et al. Role of the CD6 glycoprotein in antigen-specific and autoreactive responses of cloned human T lymphocytes. Immunology. 1996;88(4):537–543. [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao M, Sun Y, Gao F, et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J. Autoimmun. 2010;35(1):58–69. doi: 10.1016/j.jaut.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 59.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 2007;179(9):6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 60.Strickland FM, Li Y, Johnson K, Sun Z, Richardson BC. CD4(+) T cells epigenetically modified by oxidative stress cause lupus-like autoimmunity in mice. J. Autoimmun. 2015;62:75–80. doi: 10.1016/j.jaut.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garaud S, Youinou P, Renaudineau Y. DNA methylation and B-cell autoreactivity. Adv. Exp. Med. Biol. 2011;711:50–60. doi: 10.1007/978-1-4419-8216-2_5. [DOI] [PubMed] [Google Scholar]
- 62.Renaudineau Y, Beauvillard D, Padelli M, Brooks WH, Youinou P. Epigenetic alterations and autoimmune disease. J. Dev. Orig. Health Dis. 2011;2(5):258–264. doi: 10.1017/S2040174411000353. [DOI] [PubMed] [Google Scholar]
- 63.Renaudineau Y, Youinou P. Epigenetics and autoimmunity, with special emphasis on methylation. Keio J. Med. 2011;60(1):10–16. doi: 10.2302/kjm.60.10. [DOI] [PubMed] [Google Scholar]
- 64.Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J. Immunol. 2004;172(6):3652–3661. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 65.Sawalha AH, Wang L, Nadig A, et al. Sex-specific differences in the relationship between genetic susceptibility, T cell DNA demethylation and lupus flare severity. J. Autoimmun. 2012;38(2–3):J216–222. doi: 10.1016/j.jaut.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin. Immunol. 2009;130(2):213–224. doi: 10.1016/j.clim.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Basu D, Liu Y, Wu A, et al. Stimulatory and inhibitory killer Ig-like receptor molecules are expressed and functional on lupus T cells. J. Immunol. 2009;183(5):3481–3487. doi: 10.4049/jimmunol.0900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sunahori K, Juang YT, Tsokos GC. Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity. J. Immunol. 2009;182(3):1500–1508. doi: 10.4049/jimmunol.182.3.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sunahori K, Nagpal K, Hedrich CM, Mizui M, Fitzgerald LM, Tsokos GC. The catalytic subunit of protein phosphatase 2A (PP2Ac) promotes DNA hypomethylation by suppressing the phosphorylated mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)/phosphorylated ERK/DNMT1 protein pathway in T-cells from controls and systemic lupus erythematosus patients. J. Biol. Chem. 2013;288(30):21936–21944. doi: 10.1074/jbc.M113.467266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moulton VR, Holcomb DR, Zajdel MC, Tsokos GC. Estrogen upregulates cyclic AMP response element modulator alpha expression and downregulates interleukin-2 production by human T lymphocytes. Mol. Med. 2012;18:370–378. doi: 10.2119/molmed.2011.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moulton VR, Tsokos GC. Why do women get lupus? Clin. Immunol. 2012;144(1):53–56. doi: 10.1016/j.clim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 72.Tedeschi SK, Bermas B, Costenbader KH. Sexual disparities in the incidence and course of SLE and RA. Clin. Immunol. 2013;149(2):211–218. doi: 10.1016/j.clim.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Liu HW, Lin HL, Yen JH, et al. Demethylation within the proximal promoter region of human estrogen receptor alpha gene correlates with its enhanced expression: Implications for female bias in lupus. Mol. Immunol. 2014;61(1):28–37. doi: 10.1016/j.molimm.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 74.Hedrich CM, Rauen T, Tsokos GC. cAMP-responsive element modulator (CREM)alpha protein signaling mediates epigenetic remodeling of the human interleukin-2 gene: implications in systemic lupus erythematosus. J. Biol. Chem. 2011;286(50):43429–43436. doi: 10.1074/jbc.M111.299339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ngalamika O, Liang G, Zhao M, et al. Peripheral whole blood FOXP3 TSDR methylation: a potential marker in severity assessment of autoimmune diseases and chronic infections. Immunol. Invest. 2015;44(2):126–136. doi: 10.3109/08820139.2014.938165. [DOI] [PubMed] [Google Scholar]
- 76.Rauen T, Grammatikos AP, Hedrich CM, et al. cAMP-responsive element modulator alpha (CREMalpha) contributes to decreased Notch-1 expression in T cells from patients with active systemic lupus erythematosus (SLE) J. Biol. Chem. 2012;287(51):42525–42532. doi: 10.1074/jbc.M112.425371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen H, Fan J, Shou Q, Zhang L, Ma H, Fan Y. Hypermethylation of glucocorticoid receptor gene promoter results in glucocorticoid receptor gene low expression in peripheral blood mononuclear cells of patients with systemic lupus erythematosus. Rheumatol. Int. 2015;35(8):1335–1342. doi: 10.1007/s00296-015-3266-5. [DOI] [PubMed] [Google Scholar]
- 78.Renauer PA, Coit P, Sawalha AH. The DNA methylation signature of human TCRalphabeta+CD4-CD8- double negative T cells reveals CG demethylation and a unique epigenetic architecture permissive to a broad stimulatory immune response. Clin. Immunol. 2015;156(1):19–27. doi: 10.1016/j.clim.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andreae J, Tripmacher R, Weltrich R, et al. Effect of glucocorticoid therapy on glucocorticoid receptors in children with autoimmune diseases. Pediatr. Res. 2001;49(1):130–135. doi: 10.1203/00006450-200101000-00025. [DOI] [PubMed] [Google Scholar]
- 80.Jiang T, Liu S, Tan M, et al. The phase-shift mutation in the glucocorticoid receptor gene: potential etiologic significance of neuroendocrine mechanisms in lupus nephritis. Clin. Chim. Acta. 2001;313(1–2):113–117. doi: 10.1016/s0009-8981(01)00661-1. [DOI] [PubMed] [Google Scholar]
- 81.Piotrowski P, Burzynski M, Lianeri M, et al. Glucocorticoid receptor beta splice variant expression in patients with high and low activity of systemic lupus erythematosus. Folia Histochem. Cytobiol. 2007;45(4):339–342. [PubMed] [Google Scholar]
- 82.Ehrlich M, Ehrlich KC. DNA cytosine methylation and hydroxymethylation at the borders. Epigenomics. 2014;6(6):563–566. doi: 10.2217/epi.14.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hackett JA, Sengupta R, Zylicz JJ, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339(6118):448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hahn MA, Qiu R, Wu X, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3(2):291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neri F, Incarnato D, Krepelova A, et al. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 2013;14(8):R91. doi: 10.1186/gb-2013-14-8-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song CX, Szulwach KE, Fu Y, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shukla A, Sehgal M, Singh TR. Hydroxymethylation and its potential implication in DNA repair system: a review and future perspectives. Gene. 2015;564(2):109–118. doi: 10.1016/j.gene.2015.03.075. [DOI] [PubMed] [Google Scholar]
- 88.Schomacher L. Mammalian DNA demethylation: multiple faces and upstream regulation. Epigenetics. 2013;8(7):679–684. doi: 10.4161/epi.24977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao Y, Chen J, Li K, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12(4):453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 91.Sui W, Tan Q, Yang M, et al. Genome-wide analysis of 5-hmC in the peripheral blood of systemic lupus erythematosus patients using an hMeDIP-chip. Int. J. Mol. Med. 2015;35(5):1467–1479. doi: 10.3892/ijmm.2015.2149. [DOI] [PubMed] [Google Scholar]
- 92.Xu Y, Wu F, Tan L, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Straussman R, Nejman D, Roberts D, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 2009;16(5):564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]; • The potential influence of DNA hydroxymethylation on altered gene expression in SLE is demonstrated.
- 94.Zhao M, Wang J, Liao W, et al. Increased 5-hydroxymethylcytosine in CD4(+) T cells in systemic lupus erythematosus. J. Autoimmun. 2016;69:64–73. doi: 10.1016/j.jaut.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 95.Januchowski R, Wudarski M, Chwalinska-Sadowska H, Jagodzinski PP. Prevalence of ZAP-70, LAT, SLP-76, and DNA methyltransferase 1 expression in CD4+ T cells of patients with systemic lupus erythematosus. Clin. Rheumatol. 2008;27(1):21–27. doi: 10.1007/s10067-007-0644-8. [DOI] [PubMed] [Google Scholar]
- 96.Lei W, Luo Y, Lei W, et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand. J. Rheumatol. 2009;38(5):369–374. doi: 10.1080/03009740902758875. [DOI] [PubMed] [Google Scholar]
- 97.Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Rosa-Leyva M, Vilardell-Tarres M. Transcript levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T cells from patients with systemic lupus erythematosus. Immunology. 2008;124(3):339–347. doi: 10.1111/j.1365-2567.2007.02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 99.Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 100.Grishok A, Pasquinelli AE, Conte D, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 101.Hutvagner G, Mclachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 102.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 103.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17(3):118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 104.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garzon R, Liu S, Fabbri M, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ng EK, Tsang WP, Ng SS, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br. J. Cancer. 2009;101(4):699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao S, Wang Y, Liang Y, et al. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63(5):1376–1386. doi: 10.1002/art.30196. [DOI] [PubMed] [Google Scholar]; • Reduced MAPK activity in T cells is linked to altered DNA methylation and gene expression in SLE.
- 108.Gorelik G, Sawalha AH, Patel D, Johnson K, Richardson B. T cell PKCdelta kinase inactivation induces lupus-like autoimmunity in mice. Clin. Immunol. 2015;158(2):193–203. doi: 10.1016/j.clim.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gorelik GJ, Yarlagadda S, Patel DR, Richardson BC. Protein kinase Cdelta oxidation contributes to ERK inactivation in lupus T cells. Arthritis Rheum. 2012;64(9):2964–2974. doi: 10.1002/art.34503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y, Zhao M, Yin H, et al. Overexpression of the growth arrest and DNA damage-induced 45alpha gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. 2010;62(5):1438–1447. doi: 10.1002/art.27363. [DOI] [PubMed] [Google Scholar]
- 111.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Y, Huang C, Zhao M, et al. A possible role of HMGB1 in DNA demethylation in CD4+ T cells from patients with systemic lupus erythematosus. Clin. Dev. Immunol. 2013:206298. doi: 10.1155/2013/206298. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Applying whole-genome mapping of transcription factor recruitment, transcriptional activity, DNA methylation and histone modification patterns, the authors provide evidence for interconnections between these mechanisms.
- 113.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Farh KK, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roadmap Epigenomics C, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Interactions between the transcription factor RFX1 and the epigenetic modifiers SUV39H1, HDAC1 and DNMT1 is demonstrated and linked to altered gene expression in SLE.
- 116.Zhao M, Liu S, Luo S, et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J. Autoimmun. 2014;54:127–136. doi: 10.1016/j.jaut.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 117.Zhao M, Wu X, Zhang Q, et al. RFX1 regulates CD70 and CD11a expression in lupus T cells by recruiting the histone methyltransferase SUV39H1. Arthritis Res. Ther. 2010;12(6):R227. doi: 10.1186/ar3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hedrich CM, Rauen T, Kis-Toth K, Kyttaris VC, Tsokos GC. cAMP-responsive element modulator alpha (CREMalpha) suppresses IL-17F protein expression in T lymphocytes from patients with systemic lupus erythematosus (SLE) J. Biol. Chem. 2012;287(7):4715–4725. doi: 10.1074/jbc.M111.323261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Juang YT, Rauen T, Wang Y, et al. Transcriptional activation of the cAMP-responsive modulator promoter in human T cells is regulated by protein phosphatase 2A-mediated dephosphorylation of SP-1 and reflects disease activity in patients with systemic lupus erythematosus. J. Biol. Chem. 2011;286(3):1795–1801. doi: 10.1074/jbc.M110.166785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vicart A, Lefebvre T, Imbert J, Fernandez A, Kahn-Perles B. Increased chromatin association of Sp1 in interphase cells by PP2A-mediated dephosphorylations. J. Mol. Biol. 2006;364(5):897–908. doi: 10.1016/j.jmb.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 121.Ichinose K, Rauen T, Juang YT, et al. Cutting edge: calcium/calmodulin-dependent protein kinase type IV is essential for mesangial cell proliferation and lupus nephritis. J. Immunol. 2011;187(11):5500–5504. doi: 10.4049/jimmunol.1102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ichinose K, Ushigusa T, Nishino A, et al. Lupus nephritis IgG induction of calcium/calmodulin-dependent protein kinase IV expression in podocytes and alteration of their function. Arthritis Rheumatol. 2016;68(4):944–952. doi: 10.1002/art.39499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Koga T, Ichinose K, Mizui M, Crispin JC, Tsokos GC. Calcium/calmodulin-dependent protein kinase IV suppresses IL-2 production and regulatory T cell activity in lupus. J. Immunol. 2012;189(7):3490–3496. doi: 10.4049/jimmunol.1201785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koga T, Mizui M, Yoshida N, et al. KN-93, an inhibitor of calcium/calmodulin-dependent protein kinase IV, promotes generation and function of Foxp3(+) regulatory T cells in MRL/lpr mice. Autoimmunity. 2014;47(7):445–450. doi: 10.3109/08916934.2014.915954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Koga T, Otomo K, Mizui M, et al. Calcium/calmodulin-dependent kinase IV facilitates the recruitment of interleukin-17-producing cells to target organs through the CCR6/CCL20 axis in Th17 cell-driven inflammatory diseases. Arthritis Rheumatol. 2016;68(8):1981–1988. doi: 10.1002/art.39665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Racioppi L, Means AR. Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: novel routes for an ancient traveller. Trends Immunol. 2008;29(12):600–607. doi: 10.1016/j.it.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 127.Ichinose K, Juang YT, Crispin JC, Kis-Toth K, Tsokos GC. Suppression of autoimmunity and organ pathology in lupus-prone mice upon inhibition of calcium/calmodulin-dependent protein kinase type IV. Arthritis Rheum. 2011;63(2):523–529. doi: 10.1002/art.30085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Juang YT, Wang Y, Solomou EE, et al. Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J. Clin. Invest. 2005;115(4):996–1005. doi: 10.1172/JCI200522854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hashimoto H, Liu Y, Upadhyay AK, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40(11):4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ko M, An J, Rao A. DNA methylation and hydroxymethylation in hematologic differentiation and transformation. Curr. Opin. Cell Biol. 2015;37:91–101. doi: 10.1016/j.ceb.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pernis AB. Estrogen and CD4+ T cells. Curr. Opin. Rheumatol. 2007;19(5):414–420. doi: 10.1097/BOR.0b013e328277ef2a. [DOI] [PubMed] [Google Scholar]
- 135.Tiniakou E, Costenbader KH, Kriegel MA. Sex-specific environmental influences on the development of autoimmune diseases. Clin. Immunol. 2013;149(2):182–191. doi: 10.1016/j.clim.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kanno Y, Vahedi G, Hirahara K, Singleton K, O'shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu. Rev. Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Invernizzi P, Pasini S, Selmi C, Miozzo M, Podda M. Skewing of X chromosome inactivation in autoimmunity. Autoimmunity. 2008;41(4):272–277. doi: 10.1080/08916930802024574. [DOI] [PubMed] [Google Scholar]
- 138.Lian X, Xiao R, Hu X, et al. DNA demethylation of CD40l in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum. 2012;64(7):2338–2345. doi: 10.1002/art.34376. [DOI] [PubMed] [Google Scholar]
- 139.Liao J, Liang G, Xie S, et al. CD40L demethylation in CD4(+) T cells from women with rheumatoid arthritis. Clin. Immunol. 2012;145(1):13–18. doi: 10.1016/j.clim.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 140.Zhou Y, Yuan J, Pan Y, et al. T cell CD40LG gene expression and the production of IgG by autologous B cells in systemic lupus erythematosus. Clin. Immunol. 2009;132(3):362–370. doi: 10.1016/j.clim.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+) T cells of systemic lupus erythematosus. J. Autoimmun. 2013;41:92–99. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 142.Jones MJ, Goodman SJ, Kobor MS. DNA methylation and healthy human aging. Aging cell. 2015;14(6):924–932. doi: 10.1111/acel.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The influence of diet and cell metabolism on epigenetic patterns, namely DNA methylation, is demonstrated.
- 143.Oaks Z, Perl A. Metabolic control of the epigenome in systemic lupus erythematosus. Autoimmunity. 2014;47(4):256–264. doi: 10.3109/08916934.2013.834495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Strickland FM, Hewagama A, Wu A, et al. Diet influences expression of autoimmune-associated genes and disease severity by epigenetic mechanisms in a transgenic mouse model of lupus. Arthritis Rheum. 2013;65(7):1872–1881. doi: 10.1002/art.37967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J. Immunol. 2007;179(8):5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 146.Wysenbeek AJ, Block DA, Fries JF. Prevalence and expression of photosensitivity in systemic lupus erythematosus. Ann. Rheum. Dis. 1989;48(6):461–463. doi: 10.1136/ard.48.6.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhu X, Li F, Yang B, Liang J, Qin H, Xu J. Effects of ultraviolet B exposure on DNA methylation in patients with systemic lupus erythematosus. Exp. Ther. Med. 2013;5(4):1219–1225. doi: 10.3892/etm.2013.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 2015;10(1):95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16(1):9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012;24(5):981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Perillo B, Ombra MN, Bertoni A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319(5860):202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 152.Gergely P, Jr, Grossman C, Niland B, et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46(1):175–190. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Griffey RH, Brown MS, Bankhurst AD, Sibbitt RR, Sibbitt WL., Jr Depletion of high-energy phosphates in the central nervous system of patients with systemic lupus erythematosus, as determined by phosphorus-31 nuclear magnetic resonance spectroscopy. Arthritis Rheum. 1990;33(6):827–833. doi: 10.1002/art.1780330609. [DOI] [PubMed] [Google Scholar]
- 154.Fernandez DR, Telarico T, Bonilla E, et al. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J. Immunol. 2009;182(4):2063–2073. doi: 10.4049/jimmunol.0803600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Solomon DH, Kavanaugh AJ, Schur PH American College of Rheumatology Ad Hoc Committee on Immunologic Testing G. Evidence-based guidelines for the use of immunologic tests: antinuclear antibody testing. Arthritis Rheum. 2002;47(4):434–444. doi: 10.1002/art.10561. [DOI] [PubMed] [Google Scholar]
- 156.Bizzaro N, Villalta D, Giavarina D, Tozzoli R. Are anti-nucleosome antibodies a better diagnostic marker than anti-dsDNA antibodies for systemic lupus erythematosus? A systematic review and a study of metanalysis. Autoimmun. Rev. 2012;12(2):97–106. doi: 10.1016/j.autrev.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 157.Biesen R, Rose T, Hoyer BF, Alexander T, Hiepe F. Autoantibodies, complement and type I interferon as biomarkers for personalized medicine in SLE. Lupus. 2016;25(8):823–829. doi: 10.1177/0961203316640922. [DOI] [PubMed] [Google Scholar]
- 158.Wu H, Zhao M, Tan L, Lu Q. The key culprit in the pathogenesis of systemic lupus erythematosus: Aberrant DNA methylation. Autoimmun. Rev. 2016;15(7):684–689. doi: 10.1016/j.autrev.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 159.Chechlinska M, Kowalewska M, Nowak R. Systemic inflammation as a confounding factor in cancer biomarker discovery and validation. Nat. Rev. Cancer. 2010;10(1):2–3. doi: 10.1038/nrc2782. [DOI] [PubMed] [Google Scholar]
- 160.Ospelt C. Epigenetic biomarkers in rheumatology – the future? Swiss Med. Wkly. 2016;146:w14312. doi: 10.4414/smw.2016.14312. [DOI] [PubMed] [Google Scholar]
- 161.Mikeska T, Craig JM. DNA methylation biomarkers: cancer and beyond. Genes. 2014;5(3):821–864. doi: 10.3390/genes5030821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 163.Zhao M, Zhou Y, Zhu B, et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 2016 doi: 10.1136/annrheumdis-2015-208410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zahoor MA, Xue G, Sato H, Murakami T, Takeshima SN, Aida Y. HIV-1 Vpr induces interferon-stimulated genes in human monocyte-derived macrophages. PLoS ONE. 2014;9(8):e106418. doi: 10.1371/journal.pone.0106418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guo Y, Sawalha AH, Lu Q. Epigenetics in the treatment of systemic lupus erythematosus: potential clinical application. Clin. Immunol. 2014;155(1):79–90. doi: 10.1016/j.clim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 166.Jonsen A, Bengtsson AA, Nived O, Truedsson L, Sturfelt G. Gene-environment interactions in the aetiology of systemic lupus erythematosus. Autoimmunity. 2007;40(8):613–617. doi: 10.1080/08916930701511051. [DOI] [PubMed] [Google Scholar]
- 167.Chafin CB, Regna NL, Hammond SE, Reilly CM. Cellular and urinary microRNA alterations in NZB/W mice with hydroxychloroquine or prednisone treatment. Int. Immunopharmacol. 2013;17(3):894–906. doi: 10.1016/j.intimp.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chan ES, Cronstein BN. Methotrexate – how does it really work? Nat. Rev. Rheumatol. 2010;6(3):175–178. doi: 10.1038/nrrheum.2010.5. [DOI] [PubMed] [Google Scholar]
- 169.Nihal M, Wu J, Wood GS. Methotrexate inhibits the viability of human melanoma cell lines and enhances Fas/Fas-ligand expression, apoptosis and response to interferon-alpha: rationale for its use in combination therapy. Arch. Biochem. Biophys. 2014;563:101–107. doi: 10.1016/j.abb.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang Y, Tang Q, Zhao M, et al. The effect of mycophenolic acid on epigenetic modifications in lupus CD4+T cells. Clin. Immunol. 2015;158(1):67–76. doi: 10.1016/j.clim.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 171.Zhang J, Yuan B, Zhang F, et al. Cyclophosphamide perturbs cytosine methylation in Jurkat-T cells through LSD1-mediated stabilization of DNMT1 protein. Chem. Res. Toxicol. 2011;24(11):2040–2043. doi: 10.1021/tx2003849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat. Rev. Genet. 2012;13(10):679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 173.Yamazaki J, Issa JP. Epigenetic aspects of MDS and its molecular targeted therapy. Int. J. Hematol. 2013;97(2):175–182. doi: 10.1007/s12185-012-1197-4. [DOI] [PubMed] [Google Scholar]
- 174.Nie J, Liu L, Li X, Han W. Decitabine, a new star in epigenetic therapy: the clinical application and biological mechanism in solid tumors. Cancer Lett. 2014;354(1):12–20. doi: 10.1016/j.canlet.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 175.Lai ZW, Hanczko R, Bonilla E, et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2012;64(9):2937–2946. doi: 10.1002/art.34502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gu Z, Tan W, Ji J, et al. Rapamycin reverses the senescent phenotype and improves immunoregulation of mesenchymal stem cells from MRL/lpr mice and systemic lupus erythematosus patients through inhibition of the mTOR signaling pathway. Aging (Albany NY) 2016;8(5):1102–1114. doi: 10.18632/aging.100925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Koga T, Hedrich CM, Mizui M, et al. CaMK4-dependent activation of AKT/mTOR and CREM-alpha underlies autoimmunity-associated Th17 imbalance. J. Clin. Invest. 2014;124(5):2234–2245. doi: 10.1172/JCI73411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Verstovsek S. Therapeutic potential of JAK2 inhibitors. Hematology Am. Soc. Hematol. Educ. Program. 2009;2009:636–642. doi: 10.1182/asheducation-2009.1.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Grabiec AM, Korchynskyi O, Tak PP, Reedquist KA. Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann. Rheum. Dis. 2012;71(3):424–431. doi: 10.1136/ard.2011.154211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grabiec AM, Krausz S, De Jager W, et al. Histone deacetylase inhibitors suppress inflammatory activation of rheumatoid arthritis patient synovial macrophages and tissue. J. Immunol. 2010;184(5):2718–2728. doi: 10.4049/jimmunol.0901467. [DOI] [PubMed] [Google Scholar]
- 181.Lewis EC, Blaabjerg L, Storling J, et al. The oral histone deacetylase inhibitor ITF2357 reduces cytokines and protects islet beta cells in vivo and in vitro . Mol. Med. 2011;17(5–6):369–377. doi: 10.2119/molmed.2010.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Munro SK, Mitchell MD, Ponnampalam AP. Histone deacetylase inhibition by trichostatin A mitigates LPS induced TNFalpha and IL-10 production in human placental explants. Placenta. 2013;34(7):567–573. doi: 10.1016/j.placenta.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 183.Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J. Clin. Invest. 2003;111(4):539–552. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Salvi V, Bosisio D, Mitola S, Andreoli L, Tincani A, Sozzani S. Trichostatin A blocks type I interferon production by activated plasmacytoid dendritic cells. Immunobiology. 2010;215(9–10):756–761. doi: 10.1016/j.imbio.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 185.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl Acad. Sci. USA. 2003;100(5):2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schoggins JW, Wilson SJ, Panis M, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hedrich CM, Ramakrishnan A, Dabitao D, Wang F, Ranatunga D, Bream JH. Dynamic DNA methylation patterns across the mouse and human IL10 genes during CD4+ T cell activation; influence of IL-27. Mol. Immunol. 2010;48(1–3):73–81. doi: 10.1016/j.molimm.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hofmann SR, Moller J, Rauen T, et al. Dynamic CpG-DNA methylation of Il10 and Il19 in CD4+ T lymphocytes and macrophages: effects on tissue-specific gene expression. Klin. Padiatr. 2012;224(2):53–60. doi: 10.1055/s-0031-1291359. [DOI] [PubMed] [Google Scholar]
- 189.Hogg N, Laschinger M, Giles K, Mcdowall A. T-cell integrins: more than just sticking points. J. Cell Sci. 2003;116(Pt 23):4695–4705. doi: 10.1242/jcs.00876. [DOI] [PubMed] [Google Scholar]
- 190.Quddus J, Johnson KJ, Gavalchin J, et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J. Clin. Invest. 1993;92(1):38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Richardson B, Powers D, Hooper F, Yung RL, O'rourke K. Lymphocyte function-associated antigen 1 overexpression and T cell autoreactivity. Arthritis Rheum. 1994;37(9):1363–1372. doi: 10.1002/art.1780370915. [DOI] [PubMed] [Google Scholar]
- 192.Yung R, Powers D, Johnson K, et al. Mechanisms of drug-induced lupus. II. T cells overexpressing lymphocyte function-associated antigen 1 become autoreactive and cause a lupuslike disease in syngeneic mice. J. Clin. Invest. 1996;97(12):2866–2871. doi: 10.1172/JCI118743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vinuesa CG, Linterman MA, Goodnow CC, Randall KL. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunol. Rev. 2010;237(1):72–89. doi: 10.1111/j.1600-065X.2010.00937.x. [DOI] [PubMed] [Google Scholar]