Abstract
Aim:
African–Americans (AA) have increased prostate cancer risk and a greater mortality rate than European–Americans (EA). AA exhibit a high prevalence of vitamin D deficiency. We examined the global prostate transcriptome in AA and EA, and the effect of vitamin D3 supplementation.
Patients & methods:
Twenty-seven male subjects (ten AA and 17 EA), slated to undergo prostatectomy were enrolled in the study. Fourteen subjects received vitamin D3 (4000 IU daily) and 13 subjects received placebo for 2 months prior to surgery.
Results:
AA show higher expression of genes associated with immune response and inflammation.
Conclusion:
Systems level analyses support the concept that Inflammatory processes may contribute to disease progression in AA. These transcripts can be modulated by a short course of vitamin D3 supplementation.
Keywords: : African–American, health disparities, prostate, RNA-seq, transcription, vitamin D
There are considerable and persistent racial disparities in prostate cancer outcomes. Prostate cancer disproportionately affects African–American (AA) men in terms of incidence, morbidity, and mortality, even after adjustment for stage. AA men have a two- to three times increased risk of developing prostate cancer and have a greater mortality rate compared with European–American (EA) men. Reduced access to healthcare services contributes to racial disparities in prostate cancer outcomes, but even in equal access healthcare systems such as the Veterans Administration (VA), AA veterans have higher serum prostate-specific antigen (PSA) values and higher-grade tumors than EA veterans even when presenting at the same stage of disease [1,2]. Thus, access to healthcare is necessary but not sufficient for eliminating racial differences in prostate cancer outcomes. A better understanding of the biological mechanisms underlying these disparities is needed to develop strategies to overcome them.
Exposure of skin to sunlight in the ultraviolet B (UVB) range of the spectrum (290–315 nm) results in the photolytic conversion of 7-dehydrocholesterol to previtamin D3, which is transformed to vitamin D3 (cholecalciferol) by thermally induced isomerization [3,4]. Vitamin D3 can be obtained from the diet; however, it is distributed very poorly in natural foodstuffs. Dark skin pigmentation, due to increased melanin levels, likely evolved in equatorial regions to protect individuals from skin cancers. Increased skin pigmentation, however, limits one's ability to produce vitamin D3 [5,6]. Vitamin D deficiency occurs when serum levels of 25(OH)D are at <50 nmol (<20 ng/ml); as a result, a majority of AA are vitamin D deficient [6]. Until recently, higher dose vitamin D3 supplementation was not viewed as a viable treatment modality due to concerns about potential toxicity. However, Vieth et al. [7] examined the efficacy and safety of relatively high intakes of vitamin D3 by assessing the effects of 1000 and 4000 international units (IU) per day in 61 adults for up to 5 months. They found that vitamin D3 at a dose of 4000 IU/day was effective in elevating the serum 25(OH)D concentration to values ≥40 ng/ml of serum. Our own clinical experience with prolonged supplementation with 4000 IU/day for 12 months has demonstrated the safety of this regimen. We have observed that 4000 IU/day are extremely effective at raising circulating 25(OH)D across racial groups [8,9], to levels measured in athletes during summer training [10].
Prostate cells express the vitamin D receptor (VDR), vitamin D-25-hydroxylase, 25-hydroxy-vitamin D-1-alpha-hydroxylase and the 25-hydroxyvitamin D-24-hydroxylase [11–16]. Therefore, normal prostate cells can synthesize 25(OH)D3 (calcidiol) from vitamin D3 (cholecalciferol), and 1,25(OH)2D3 (calcitriol) from 25(OH)D3 [17,18]. 1,25(OH)2 D3 is the hormonal, most potent form of vitamin D and in prostate cells it acts in a paracrine/autocrine fashion.
Several mechanisms of vitamin D-mediated anticancer action have been identified [19]. Vitamin D suppresses the expression of cyclo-oxygenase-II, the key enzyme for the synthesis of prostaglandins, mediators of inflammation and thought to be important for cancer progression [20]; cyclo-oxygenase-II expression in biopsy cores and prostate cancer surgical specimen is an independent predictor of recurrence [21]. Furthermore, there is considerable evidence that calcitriol inhibits nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling, and decreases the levels of the angiogenic and pro-inflammatory cytokine IL-8 in prostate cancer cells [22]. NF-κB is a transcription factor that plays a central role in the control of inflammation and is expressed at high levels in prostate cancers with high Gleason scores [23]. This is only a very limited list of the many molecular pathways and mechanisms affected by vitamin D, as it is now well established that VDR may recognize cognate VDRE present within the regulatory sequences of hundreds of human genes, implicating vitamin D in a vast network of gene regulation, and underlying its broad physiological actions [24,25]. While it is well established that vitamin D and calcium are crucial for normal skeletal growth and for maintenance of the mechanical and structural integrity of the skeleton [26], the recent emphasis on nonskeletal functions of vitamin D has to do with the realization that vitamin D deficiency has major implications for human health in general, and cancer biology in particular [9].
Racial disparities in prostate cancer outcomes mirror racial differences in circulating levels of vitamin D [27]. Furthermore, about 60% of AA men have suboptimal levels of circulating 25(OH)D3 [28,29]. For this reason, vitamin D3 supplementation is likely to benefit these men. Vitamin D supplementation has no effect on free or total prostate-specific antigen (PSA) in AA men [30]. The effects of 25-OHD levels on the risk of total, low- and high-grade prostate cancer were examined in two separate studies, the SELECT [31] and the PCPT [32]. In the former, plasma 25-OHD levels were associated with a linear decrease in prostate cancer risk for high-grade cancers in AA men and an apparent ‘U’-shaped effect in other men reflecting detection bias. In the latter, which minimized detection bias, serum 25-OHD levels were associated with a linear decrease in the risk of high-grade prostate cancers. These data support the hypothesis that circulating levels of 25-OHD decrease the risk of clinically relevant prostate cancers and emphasize the need to further assess the influence of vitamin D supplementation on prostate cancer prevention.
Vitamin D promotes the differentiation of prostate cancer cells and maintains the differentiated phenotype of prostate epithelial cells, raising the possibility that long-term vitamin D deficiency may contribute to the progression from subclinical prostate cancer to clinical disease, especially among AA men [27]. Therefore, eliminating racial disparities in circulating levels of vitamin D could help reduce disparities in prostate cancer outcomes.
We completed an open-label clinical trial aimed at assessing the safety and potential efficacy of vitamin D3 supplementation at 4000 IU per day for one year in patients diagnosed with low-risk prostate cancer [33]. The combination of active surveillance and vitamin D3 supplementation resulted in a decreased number of positive cores at repeat biopsy in half of subjects enrolled in this trial, and a comparison between supplemented subjects and historical controls suggested that supplementation with vitamin D3 at 4000 IU per day may benefit patients with low-risk prostate cancer on active surveillance [33].
These observations prompted us to initiate a prospective clinical study aimed at examining the effects of vitamin D3 supplementation at 4000 IU per day for 2 months in male subjects who selected surgical removal of the prostate (prostatectomy) as a definitive treatment for their prostate cancer. According to current standard of care, a 2-month interval between biopsy and prostatectomy is recommended to resolve the inflammation due to the biopsy procedure. Moreover, we reported that the initial 2 months of vitamin D3 supplementation register the fastest raise in serum levels of 25(OH)D3 [8,9].
The primary goal of this study was to examine molecular differences in gene expression patterns relevant to prostate cancer disparities between AA and EA men, and investigate the global effects of vitamin D3 supplementation on the prostate transcriptome. To further this objective, we undertook a series of genome wide expression profiling experiments using high-throughput (HT) RNA sequencing. RNA was purified from prostate tissue specimens obtained at surgery from subjects enrolled in the study. Transcriptional profiles of each of the patient's tissue samples were generated and systems level analyses were performed.
Patients & methods
Human subjects
This human study was approved by the Institutional Review Board (IRB) of the Medical University of South Carolina (MUSC; SC, USA), and the Ralph H Johnson VA Medical Center (VAMC; SC, USA) and by the Research and Development (R&D) Committee of the VAMC. This interventional study was performed under investigational new drug (IND) 77839, granted to SGC by the US FDA. Male subjects enrolled in this study were diagnosed with localized prostate cancer. The study enrolled 27 subjects (ten AA and 17 EA men), who had selected surgical removal of the prostate (prostatectomy) as a definitive treatment for their prostate cancer. According to current standard of care, a 2-month interval between biopsy and prostatectomy is recommended to resolve the inflammation due to the biopsy procedure. Enrolled subjects were randomized to vitamin D3 (Carlson Laboratories, IL, USA) supplementation at 4000 IU per day or placebo for 2 months prior to surgery. Two blood samples were obtained from each subject (at enrollment and on the day of surgery) to measure serum levels of 25-hydroxyvitamin D3 (25[OH]D3) in nanograms (ng) per milliliter (ml). In total 14 subjects (five AA and nine EA men) took 4000 IU of vitamin D3 per day for 2 months prior to surgery; 13 subjects (five AA and eight EA men) received placebo for 2 months prior to surgery. Based on the serum levels of 25(OH)D at study exit, we concluded that there was a high level of compliance by all enrolled subjects.
Tissue sample procurement & RNA purification
Surgical specimens were received in the frozen section laboratory of the MUSC or the VAMC, depending on where the prostatectomy was performed. Nonmalignant tissue samples were excised from the peripheral zone of the prostate under the supervision of the attending pathologist, to ensure that the excision of tissue samples did not interfere with the diagnostic priorities of standard of care. Specifically, the attending pathologist identified for us nonmalignant tissue away from cancer lesions, based on his knowledge of the location, within the prostate, of cancer-positive biopsy cores prior to surgery. Tissue samples were transferred to sterile tubes, quick-frozen in liquid nitrogen and transported to the Hollings Cancer Center Genomics Core Facility (SC, USA). Total RNA from each de-identified tissue sample was purified on a Qiagen RNeasy column (Qiagen, CA, USA) according to manufacturer's instructions. RNA integrity was verified using RNA 6000 Nano Assay chips run in Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA).
RNA sequencing (RNA-seq) & analyses
Around 100–200 ng of total RNA was used to prepare RNA-Seq libraries using the TruSeq RNA Sample Prep Kit (Illumina, CA, USA), following the protocol described by the manufacturer. High-throughput sequencing (HTS) was performed using an Illumina HiSeq2500 with each sample sequenced to a minimum depth of ˜50 million reads. Data were subjected to Illumina quality control (QC) procedures (>80% of the data yielded a Phred score of 30). Secondary analysis was carried out on an OnRamp Bioinformatics Genomics Research Platform (OnRamp Bioinformatics, CA, USA). OnRamp's advanced Genomics Analysis Engine utilized an automated RNAseq workflow to process the data, including data validation and quality control, read alignment to the human genome (hg19) using TopHat2 [34], which revealed >93% mapping of the paired end reads, generation of gene-level count data with HTSeq and differential expression analysis with DEseq2 [35], which enabled the inference of differential signals with robust statistical power. (Genomics Research Platform with RNAseq workflow v1.0.1, including FastQValidator v0.1.1a, Fastqc v0.11.3, Bowtie2 v2.1.0, TopHat2 v2.0.9, HTSeq v0.6.0, DEseq v1.8.0).
The resulting SAM files were sorted and inputted into the Python package HTSeq to generate count data for gene-level differential expression analyses. In order to infer differential signal within the datasets with robust statistical power, we utilized DEseq2 [35]. Transcript count data from DESeq2 analysis of the samples were sorted according to their adjusted p-value or q-value, which is the smallest false discovery rate (FDR) at which a transcript is called significant. FDR adjustment is needed with large datasets such as RNAseq. FDR is the expected fraction of false-positive tests among significant tests and was calculated using the Benjamini–Hochberg multiple testing adjustment procedure. Statistical analysis of pathways and gene ontology (GO) terms was carried out using this sorted transcript list as described by us previously [36,37] and using Ingenuity Pathway Analysis (Qiagen) and the ToppGene Suite [38]. Area-proportional Venn diagrams were created using BioVenn [39].
Results
Patient cohort
The study enrolled 27 subjects (ten AA and 17 EA men), who had selected surgical removal of the prostate (prostatectomy) as a definitive treatment for their prostate cancer. Table 1 highlights the characteristics of enrolled subjects, their age and race distribution, disease stage and serum levels of 25(OH)D3. Baseline and exit values of serum levels of 25(OH)D3 are shown in ng/ml. Baseline Gleason grade refers to the pathology assessment of the prostate biopsy at diagnosis. Exit Gleason grade refers to the pathology assessment on the entire prostate after surgery, which may have resulted in upgrade, downgrade or no change of the pathology assessment. Overall, there was no significant change in pathology assessment on the prostate after surgery, compared with the previous biopsy, either by race or by supplementation. All 14 subjects receiving vitamin D3 supplementation had an increase in their serum concentration of 25(OH)D3. There were no significant changes in circulating levels of vitamin D in the 13 subjects receiving placebo. Differences in serum concentration of 25(OH)D3 measured at study entry between AA and EA subjects were erased after 2 months of supplementation (Supplementary Figure 1).
Table 1. . Characteristics of subjects enrolled in the prostatectomy study.
| Subect ID | Age (years) | Race | Baseline | Exit | Baseline | Exit | Randomized | Pathology |
|---|---|---|---|---|---|---|---|---|
| AA = 1; EA = 0 | 25(OH)D level | 25(OH)D level | Gleason score | Gleason score | D3 = 1 | Staging | ||
| 01 | 64 | 1 | 16.6 | 69.7 | 3 + 4 | 3 + 4 | 1 | pT2apN0 |
| 02 | 61 | 0 | 11.7 | 36.7 | 3 + 4 | 4 + 3 | 1 | pT2cpNX |
| 03 | 68 | 0 | 35.4 | 43.2 | 3 + 3 | 3 + 4 | 1 | pT2bpNX |
| 04 | 61 | 0 | 26.6 | 20.9 | 3 + 4 | 3 + 4 | 0 | pT2cpN0 |
| 05 | 66 | 0 | 21.3 | 19.3 | 3 + 3 | 3 + 4 | 0 | pT2cpN0 |
| 06 | 65 | 0 | 31.0 | 27.1 | 3 + 4 | 3 + 4 | 0 | pT3apN0 |
| 07 | 61 | 0 | 24.7 | 25.9 | 3 + 4 | 4 + 3 | 0 | pT3 pN0 |
| 08 | 62 | 0 | 36.4 | 55.0 | 4 + 4 | 3 + 4 | 1 | pT2cpN0 |
| 09 | 57 | 0 | 27.1 | 23.7 | 3 + 4 | 4 + 3 | 0 | pT2cpN0 |
| 10 | 60 | 0 | 46.9 | 58.2 | 4 + 3 | 3 + 4 | 1 | pT2cpN0 |
| 11 | 63 | 0 | 51.1 | 41.8 | 3 + 3 | 3 + 4 | 0 | pT2cpNX |
| 12 | 69 | 0 | 32.1 | 37.2 | 3 + 4 | 3 + 4 | 1 | pT2cpN0 |
| 13 | 58 | 0 | 39.7 | 36.8 | 3 + 3 | 3 + 3 | 0 | pT2cpNX |
| 14 | 50 | 0 | 56.9 | 41.3 | 3 + 4 | 3 + 3 | 0 | pT2cpN0 |
| 15 | 56 | 0 | 36.7 | 70.8 | 3 + 4 | 3 + 3 | 1 | pT2apN0 |
| 16 | 58 | 0 | 23.4 | 50.1 | 4 + 3 | 3 + 4 | 1 | pT2cpN0 |
| 17 | 67 | 1 | 32.7 | 34.1 | Intraductal | 4 + 3 | 0 | pT3bpN0 |
| 18 | 71 | 1 | 22.4 | 40.2 | 3 + 4 | 3 + 4 | 1 | pT2cpN0 |
| 19 | 70 | 1 | 30.3 | 39.5 | 3 + 4 | 3 + 4 | 1 | pT2cpN0 |
| 20 | 65 | 1 | 24.9 | 62.0 | 3 + 4 | 3 + 4 | 1 | pT3bpNX |
| 21 | 54 | 1 | 15.6 | 21.6 | 3 + 4 | 3 + 4 | 0 | pT2cpN0 |
| 22 | 61 | 1 | 14.1 | 32.8 | 3 + 4 | 3 + 4 | 1 | pT2c |
| 23 | 58 | 1 | 19.5 | 18.4 | 4 + 3 | 3 + 4 | 0 | pT2cpN0 |
| 24 | 63 | 0 | 19.9 | 35 | 3 + 3 | 3 + 3 | 1 | pT2cpN0 |
| 25 | 66 | 0 | 26.1 | 36.3 | 3 + 4 | 3 + 4 | 1 | pT2cpN0 |
| 26 | 62 | 1 | 33.7 | 29.4 | 3 + 3 | 3 + 3 | 0 | pT2cpNX |
| 27 | 62 | 1 | 28.8 | 20.9 | 3 + 4 | 3 + 3 | 0 | pT3apN0 |
Twenty-seven subjects completed the study. Age, race and randomization assignment for each enrolled subject are shown. Baseline and exit values of serum levels of 25(OH)D3 are shown in ng/ml. Baseline Gleason grade refers to the pathology assessment of the prostate biopsy preceding the surgery. Exit Gleason grade refers to the pathology assessment on the entire prostate after surgery, which may result in upgrade, downgrade or no change of the pathology assessment.
Differential prostate gene expression between AA and EA patients
We set EA subjects (17 samples) as the control and AA subjects (ten samples) as the test to uncover genes differentially expressed in AA. These cumulative patient datasets were analyzed to identify race-associated differences in prostate gene expression between samples from AA and EA subjects, as well as differences in molecular changes in the prostate associated with vitamin D3 supplementation. Fold-change (FC) estimation and hypothesis testing for differential expression were performed using the DESeq2 Bioconducter library [35,40–41]. For each gene, DESeq2 reported an estimated FC, and provided an adjusted p- or q-value equivalent to the smallest FDR incurred when declaring that test significant.
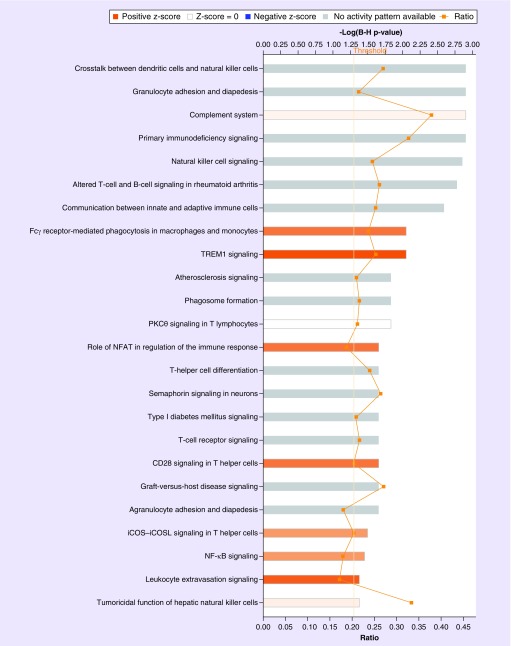
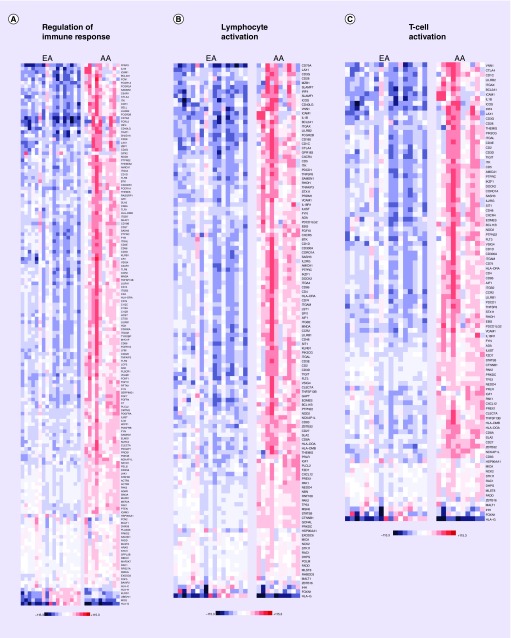
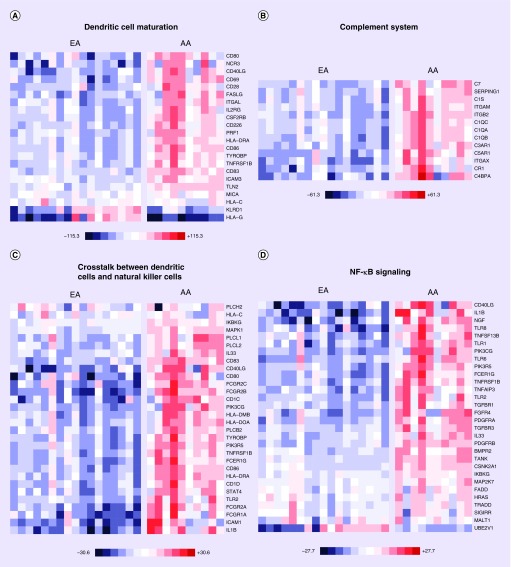
When we assessed differences in prostate gene expression between AA subjects (ten samples) compared with EA subjects (17 samples), this revealed that 3107 genes were differentially expressed between the two groups (q < 0.1). Pathway and GO analysis using (QIAGEN Ingenuity Pathway Analysis) with the 3107 differentially expressed genes uncovered major differences between the two groups (Supplementary Tables 1–3). The significant canonical pathways enriched in this dataset are presented with AA displaying elevated expression of transcripts related to immune response and inflammation (Figure 1 & Table 2; Supplementary Table 3). Examples of these canonical pathways include, ‘regulation of immune response’, ‘lymphocyte activation and T-cell activation’ (Figure 2) and ‘dendritic cell maturation’, ‘complement system’, ‘crosstalk between dendritic cells and natural killer cells’ and ‘NF-κB signaling’ (Figure 3).
Figure 1. . Immune and inflammatory canonical pathways enriched in prostate tissue specimens from African–American men compared with European–American men.
Analysis of the 3107 differentially expressed genes (q < 0.1) using Ingenuity Pathway Analysis (QIAGEN, CA, USA) uncovered altered immune system and inflammatory signatures between these two groups. Significant enriched canonical pathways are displayed along the y-axis. The x-axis (top) displays the -log of the p-value (calculated by Fisher's exact test right-tailed and adjusted for false discovery rate using Benjamini–Hochberg. Taller bars correspond to increased pathway significance. Orange colored bars indicate pathway activation in African–American relative to European–American. White bars indicate significant pathways in African–American compared with European–American that are neither activated nor inhibited. Gray bars indicate pathways that are significant but no prediction as to their activation or inhibition can be made. The orange points connected by a thin line represent the ratio (x-axis bottom). The ratio is calculated as follows: the number of genes in a particular pathway that are significantly enriched in the RNA-seq dataset are divided by the total number of genes that make up that pathway and are present in the reference gene set.
Table 2. . GO analysis of differences in prostate gene expression between African–American subjects and European–American subjects.
| ID | Name | q-value Bonferroni | Hit count in query list | Hit count in genome |
|---|---|---|---|---|
| GO:0002684 | Positive regulation of immune system process | 6.54E-17 | 172 | 732 |
| GO:0006955 | Immune response | 4.60E-16 | 276 | 1416 |
| GO:0045321 | Leukocyte activation | 2.20E-15 | 162 | 695 |
| GO:0001775 | Cell activation | 1.87E-14 | 195 | 916 |
| GO:0002682 | Regulation of immune system process | 4.74E-14 | 239 | 1212 |
The top 5 GO terms are presented. A more detailed analysis is presented in Supplementary Table 3.
Figure 2. . Heat map of gene expression changes in AA men compared with EA men involving transcripts involved in regulation of immune response, lymphocyte activation and T-cell activation.
Red and blue boxes colors depict relative over- and under-expression in AA relative to EA. The range of colors is between -115.3-fold and +115.3-fold and preserves qualitative relationships among individual values. All fold changes outside of this range have been truncated to ±115.3. Only transcripts found significant at the level q < 0.1 in the comparison, are shown.
AA: African–American; EA: European–American
Figure 3. . Heat map of gene expression changes in AA men compared with EA men involving transcripts involved in dendritic cell maturation, complement system, crosstalk between dendritic cells and natural killer cells and NF-κB signaling.
Red and blue boxes colors depict relative over- and under-expression in AA relative to EA. The range of colors is presented for each category and preserves qualitative relationships among individual values. All fold changes outside of these ranges have been truncated to ± the value noted. Only transcripts found significant at the level q < 0.1 in the comparison are shown.
AA: African–American; EA: European American.
Differential prostate gene expression between AA supplemented with vitamin D3 or placebo
We subsequently identified differentially expressed genes in prostate tissue specimens from five AA subjects supplemented with vitamin D3 at 4000 IU/day for 2 months compared with five AA subjects receiving placebo. 817 transcripts were significantly differentially expressed between these two groups (q < 0.4). GO analysis using the 817 differentially expressed genes revealed chemokine activity, chemokine receptor binding and G-protein coupled receptor binding as significantly enriched terms (Table 3, Supplementary Table 4). Pathway analysis indicated that transcripts belonging to the ‘calcium signaling’ (BioSystems: KEGG, 1.26E-05), and ‘chemokine receptors bind chemokines’ (BioSystems: REACTOME, 1.53E-02) pathways were significant in the vitamin D3 or placebo comparison. It must be noted that vitamin D3 supplementation in EA patients had no significant effect on gene expression (of the 17 samples examined, nine were supplemented with vitamin D3 and eight received placebo).
Table 3. . GO, pathway and co-expression analysis of differences in in prostate gene expression between African–Americans supplemented with vitamin D3 or placebo.
| Category | ID | Name | Source | q-value Bonferroni | Hit count in query list | Hit count in genome |
|---|---|---|---|---|---|---|
| GO: Molecular Function | GO:0008092 | Cytoskeletal protein binding | 5.46E-04 | 60 | 792 | |
| GO: Molecular Function | GO:0032403 | Protein complex binding | 1.16E-03 | 66 | 924 | |
| GO: Molecular Function | GO:0008009 | Chemokine activity | 1.31E-03 | 11 | 46 | |
| GO: Molecular Function | GO:0015631 | Tubulin binding | 6.18E-03 | 26 | 251 | |
| GO: Molecular Function | GO:0042379 | Chemokine receptor binding | 1.24E-02 | 11 | 57 | |
| Pathway | 83050 | Calcium signaling pathway | Bio-Systems: KEGG | 1.26E-05 | 26 | 181 |
| Pathway | 198906 | Calcium regulation in the cardiac cell | Bio-Systems: Wiki-Pathways | 2.40E-05 | 23 | 149 |
| Pathway | P00031 | Inflammation mediated by chemokine and cytokine signaling pathway | PantherDB | 3.96E-05 | 26 | 191 |
| Pathway | 154409 | Gastric acid secretion | Bio-Systems: KEGG | 1.70E-03 | 14 | 75 |
| Pathway | 908257 | Adrenergic signaling in cardiomyocytes | Bio-Systems: KEGG | 2.19E-03 | 20 | 149 |
| Coexpression | 17297478-SuppTable5 | Human Intestine_Vecchi07_1024genes | GeneSigDB | 2.50E-16 | 82 | 781 |
| Coexpression | 18498629-GeneList | Human Breast_Loi08_239genes | GeneSigDB | 1.90E-14 | 36 | 178 |
| Coexpression | M8124 | Genes upregulated in basal subtype of breast cancer samles | MSigDB C2: Broad Institute | 3.47E-14 | 70 | 647 |
| Coexpression | 20421987-TableS1 | Human Lung_Hou10_1067genes | GeneSigDB | 1.09E-12 | 72 | 724 |
| Coexpression | M19391 | Genes downregulated in prostate cancer samples | MSigDB C2: Broad Institute | 1.32E-11 | 55 | 480 |
The top five results for each are presented. A more detailed analysis is presented in Supplementary Table 4.
Comparison of transcripts regulated by vitamin D3 supplementation in AA with those differentially expressed between AA & EA
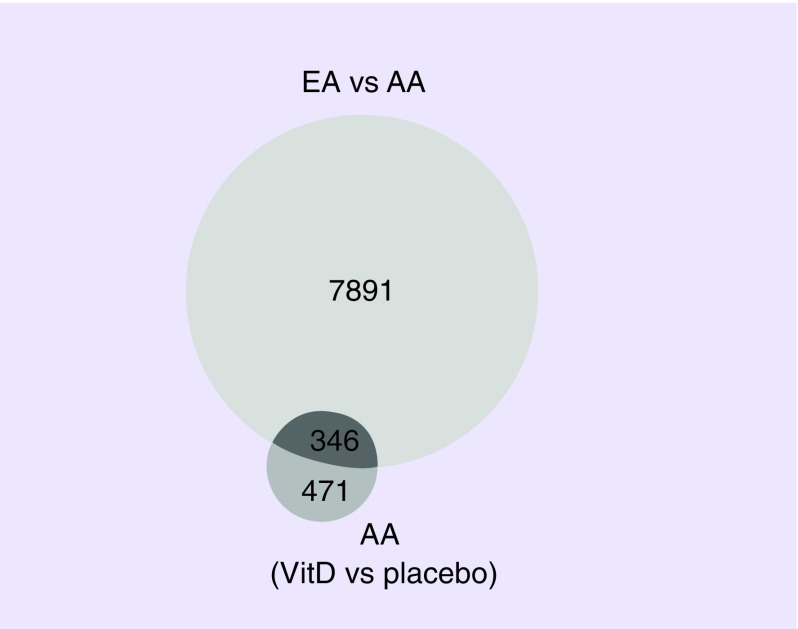
We examined the lists of transcripts regulated by vitamin D3 for overlap with those that were differentially expressed between AA and EA (Figure 4). Among those that overlapped were unc-5 netrin receptor C (UNC5C), fibroblast growth factor 10 (FGF10), a junctional protein associated with coronary artery disease (KIAA1462), ADAM-like decysin 1, (ADAMDEC1), vitrin (VIT), tachykinin receptor 2 (TACR2), FRMD6 antisense RNA 2 (FRMD6-AS2), adaptor related protein complex 1 sigma 3 subunit (AP1S3), pleckstrin homology domain containing N1 (PLEKHN1), coiled-coil domain containing 27 (CCDC27), FGF10 antisense RNA 1 (FGF10-AS1), myosin, heavy chain 6, cardiac muscle, alpha (MYH6), cingulin-like 1 (CGNL1), ventricular zone expressed PH domain containing 1 (VEPH1) and collagen, type IV, alpha 3 (COL4A3) (q < 0.1) in both comparisons.
Figure 4. . Area-proportional Venn diagram highlighting the overlap of differentially expressed transcripts by race and vitamin D3 supplementation.
Overlap between differentially expressed transcripts in prostate tissue specimens from AA subjects compared with EA subjects (8237 unique transcripts), and differentially expressed genes in prostate tissue specimens from AA subjects supplemented with vitamin D3 at 4000 IU per day for 2 months compared with AA subjects receiving placebo (817 unique transcripts).
AA: African–American; EA: European–American.
Furthermore, comparison of the 8238 transcripts that were differentially expressed between AA and EA subjects (q < 0.4) and the 817 genes that were differentially expressed in AA subjects supplemented with vitamin D3 compared with AA subjects receiving placebo (q < 0.4) revealed an overlap of 346 genes (Figure 4). This overlap suggested that a considerable number of genes that are differentially expressed between across racial groups, can be affected by a very short course of vitamin D3 supplementation in AA subjects.
GO and pathway analysis of these 346 genes using Toppfun revealed enriched terms generally related to development and differentiation (Supplementary Table 1). Co-expression analysis with this list of 346 revealed a signature related to ‘M19391; genes downregulated in prostate cancer samples’, MSigDB C2: Broad Institute (2.121E-9). GO and pathway analysis of the 471 genes that did not overlap revealed primarily immune signatures including ‘CXCR chemokine receptor binding’, ‘cytoskeletal protein binding’, ‘chemokine activity’ and ‘chemokine receptor binding’ (Supplementary Table 2).
Comparison of transcripts differentially expressed between AA & EA in patients who were treated with placebo versus those treated with vitamin D3.
We explored the lists of transcripts differentially regulated between AA and EA subjects who received placebo and observed that 1984 and 6896 transcripts were significantly differentially expressed at FDR values of <0.1 and <0.4, respectively. Subsequently we examined transcripts differentially regulated between AA and EA subjects who received vitamin D and noted that 2855 and 6383 transcripts were significantly differentially expressed at FDR values of <0.1 and <0.4, respectively. We next examined the overlap in transcripts between these two comparisons. 3701 transcripts were unique to the vitamin D treatment. A total of 4216 transcripts were unique to the placebo treatment. G0 and pathway analysis of the 3701 transcripts are presented in Supplementary Table 5. This revealed cytokine receptor activity (3.33E-03), immune response (8.87E-26), inflammatory response (7.51E-20), regulation of immune system process (1.05E-19), leukocyte aggregation (4.49E-17), leukocyte cell–cell adhesion (4.76E-17) and leukocyte activation (5.05E-17). Significantly enriched pathways included the TCR signaling pathway (2.96E-04), hematopoietic cell lineage (2.96E-04), cytokine-cytokine receptor interaction (2.96E-04), inflammation mediated by chemokine and cytokine signaling pathway (2.96E-04) and the B-cell receptor signaling pathway (2.96E-04). Examination of the 4216 transcripts unique to the placebo treatment did not reveal inflammatory signatures (Supplementary Table 6). Significantly enriched GO terms included cell projection organization (2.66E-12), neurogenesis (2.79E-06) and generation of neurons (4.71E-06). Pathways that were significantly enriched included Focal adhesion (1.02E-04), vascular smooth muscle contraction (1.02E-04) and nonintegrin membrane–extracellular matrix interactions (2.01E-04).
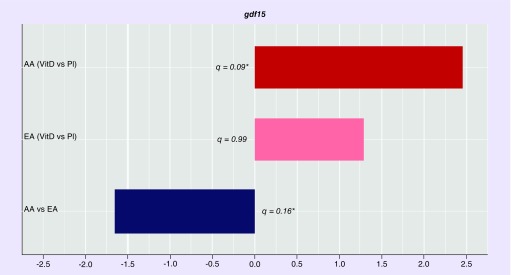
Analysis of the expression patterns of growth differentiation factor 15 (GDF15) mRNA.
GDF15 is one example of a vitamin D sensitive gene ‘captured’ by our research approach. Low expression of GDF15 is associated with prostate cancer progression [42,43]. We examined the expression pattern of GDF15 mRNA in our RNAseq datasets (Figure 5). GDF15 expression was significantly downregulated in from AA subjects compared with EA subjects (q = 0.16). In AA subjects receiving vitamin D3 supplementation, GDF15 mRNA expression was significantly upregulated relative to those receiving placebo (q = 0.09). In EA subjects receiving vitamin D3 supplementation, GDF15 mRNA expression was upregulated (although this was not statistically significant, q = 0.99) relative to those receiving placebo.
Figure 5. . Analysis of the expression patterns of GDF15 mRNA.
GDF15 mRNA expression was significantly downregulated investigated in from AA subjects compared with EA subjects. In AA subjects receiving vitamin D3 supplementation, GDF15 mRNA expression was significantly upregulated relative to those receiving Pl. In EA subjects receiving vitamin D3 supplementation, GDF15 mRNA expression was upregulated (although not significantly) relative to those receiving placebo.
AA: African–American; EA: European–American: PI: Placebo; VitD: Vitamin D.
Discussion
The objective of this clinical study was to investigate the molecular effects of vitamin D3 supplementation (4000 IU per day for 2 months) on prostate tissue specimens obtained at surgery, by means of HT RNA-sequencing, with special emphasis on differential gene expression patterns in AA men compared with EA men, and between supplemented and placebo groups in both. As we demonstrate in this study, advanced sequencing technologies and big data analytic tools, such as the OnRamp Genomic Research Platform can be easily applied to the analysis of prostate tissue samples obtained from prostate cancer patients undergoing prostatectomy as part of their standard medical care.
A major objective of this study was to investigate the molecular mechanisms relevant to prostate cancer disparities between AA and EA men, and explore the potentially beneficial effects of vitamin D3 supplementation on the prostate in AA men. We noted that the global transcriptomes of AA and EA men were considerably different. Our observations of increased inflammatory and immune signatures are consistent with a previous report by Wallace and colleagues (Supplementary Figures 2 & 3) [44]. The goal of this earlier study was to apply Affymetrix array based genome-wide gene expression profiling of prostate tumors to determine differences in tumor biology between 33 AA and 36 EA patients. Their analysis uncovered 162 significant genes (q < 0.05) that were differently expressed between AA and EA patients. Using a disease association-based approach analysis, a common theme among these transcripts was autoimmunity and inflammation, including ‘immune response’, ‘stress response’, ‘cytokine signaling’ and ‘chemotaxis’ pathways. The authors showed that metastasis-promoting genes, including autocrine mobility factor receptor, chemokine (C-X-C motif) receptor 4 (CXCR4) and matrix metalloproteinase 9 (MMP-9), were expressed at higher levels in AA relative to EA, highlighting the existence of a distinct tumor microenvironment in these two patient groups. The results of our transcriptomic analyses using a newer technology (RNA-seq) applied to prostate tissue samples acquired prospectively as part of a randomized, interventional clinical study further support the existence of considerable biological differences within the prostate between AA and EA men, and suggest that overexpression of genes linked to inflammatory processes likely contribute to the increased severity and faster progression of prostate cancer in AA even at the early stage of disease. Using IPA canonical pathway analyses, we noted activation of ‘FCγ receptor mediated phagocytosis in macrophages and mononcytes’, ‘TREM1 signaling’, ‘role of NFAT in the regulation of the immune response’, ‘iCOS–iCOSL signaling in T helper cells’, ‘NF-κB signaling and leukocyte-extravasation signaling’ (Figure 1) in AA subjects, all of which highlight differences in immune and inflammatory response. A general trend we observed with GO and pathway analyses were upregulation of transcripts in AA compared with EA, that were relevant to the immune system: segulation of immune response, lymphocyte activation and T-cell activation (Figure 2) and dendritic cell maturation and complement system activation (Figure 3).
We also identified differentially expressed genes in prostate tissue specimens from five AA subjects supplemented with vitamin D3 at 4000 IU/day for 2 months compared with five AA subjects receiving placebo. Expression of 124 genes was significantly different between these two groups with a stringent FDR <0.1 cut-off, while expression of 817 genes was significantly different between these two groups at a less stringent FDR <0.4 cut-off. These results highlight the impact that even a short period of vitamin D3 supplementation can have on gene expression within the prostate. Comparison of the 124 genes (FDR <0.1) affected by vitamin D3 supplementation with the 3107 genes (FDR <0.1) differentially expressed between AA and EA subjects revealed a 15 genes overlap: UNC5C, FGF10, KIAA1462, ADAMDEC1, VIT, TACR2, FRMD6-AS2, AP1S3, PLEKHN1, CCDC27, FGF10-AS1, MYH6, CGNL1, VEPH1 and COL4A3. We released the FDR stringency to explore overlap between the differentially expressed transcripts in prostate tissue specimens from AA subjects compared with EA subjects, i.e., 8237 unique transcripts (FDR <0.4), and differentially expressed genes in prostate tissue specimens from AA subjects supplemented with vitamin D3 compared with AA subjects receiving placebo, i.e., 817 unique transcripts (FDR <0.4). This analysis revealed an overlap of 346 genes (Figure 3) and suggested that a considerable number of genes that are differentially expressed between AA and EA subjects, can also be affected by a very short course of vitamin D3 supplementation. Of note, when we performed co-expression analysis with this short gene list, we uncovered a signature corresponding to genes downregulated in prostate cancer samples, further indicating that at a molecular level, vitamin D has potentially beneficial effects. Furthermore, vitamin D3 supplementation in EA patients had no significant effect on gene expression (of the 17 samples examined, nine were supplemented with vitamin D3 and eight received placebo), consistent with the concept that even a short course of supplementation will especially impact transcription in the prostate of AA men, possibly because of their pronounced vitamin D deficiency.
GDF15 is a protein belonging to the TGF-β superfamily. It functions in regulating inflammatory and apoptotic pathways in injured tissues and during disease processes [42]. We examined the expression pattern of GDF15 mRNA in our RNAseq datasets (Figure 5). GDF-15 is highly expressed in the prostate and has been associated with inflammation and tumorigenesis. In a recent study of prostatic inflammation, GDF-15 expression was determined via immunohistochemical staining of human prostatectomy specimens containing inflammation. Expression in luminal epithelial cells was found to be reduced with increasing inflammation severity, suggesting an inverse association between GDF-15 and inflammation [43].
In our patient cohort we observed that GDF-15 was down regulated in AA compared with EA (-1.65 fold and highly significant at an FDR = 0.16), suggesting increased prostatic inflammation in AA (Figure 5). Vitamin D3 supplementation in AA subjects resulted in upregulated GDF-15 expression, (2.45-fold in tissue samples from supplemented subjects relative to subjects receiving placebo, and highly significant at an FDR = 0.091). Based on the previously reported inverse association between GDF-15 and inflammation, a reduction in inflammatory processes would be expected in supplemented subjects. As accumulating evidence suggests that chronic prostatic inflammation may lead to prostate cancer development, vitamin D3 supplementation in AA is likely beneficial [43]. In supplemented EA subjects, we observed an upregulation of GDF-15 (1.28-fold but not statistically significant) compared with EA subjects receiving placebo.
It has recently been reported that in vitamin D deficient men initial biopsies are more likely to show prostate tumors with high Gleason grade and more advanced clinical stage than biopsies from men who are not vitamin D deficient [45]. Furthermore, this association was particularly strong for AA men who were vitamin D deficient [45], suggesting that vitamin D3 supplementation may prove helpful especially in the highest-risk group of AA men. Although there is some understanding of the vitamin D driven biochemical mechanisms and pathways affecting prostate cancer [46], the main objective of our research effort is to fill existing gaps in knowledge by identifying those mechanisms and pathways that are especially relevant to understand the effects of vitamin D on the prostate, as well as on prostate cancer disparities between AA and EA men. These data are needed to inform treatment recommendations for vitamin D3 supplementation and provide prescription guidelines to be used in the clinical setting as a treatment strategy for early-stage prostate cancer.
Finally we examined the expression patterns of vitamin D associated genes CYP27A1, GC (group-specific component [vitamin D binding protein]), CYP3A4, CYP2R1, DHCR7, NADSYN1, CYP27B1 and CYP24A1. We interrogated our datasets for any differences in expression patterns of these transcripts between supplemented and placebo-receiving AA subjects but noted that they were not differentially expressed. This result is not surprising because we have consistently observed that vitamin D3 supplementation normalizes all the vitamin D related biochemical parameters that we have measured in AA compared with EA. If there were physiologically relevant genetic differences mapped through single nucleotide variations associated with these genes, we would have expected transcriptomic differences.
Conclusion
This report represents an important first step in our effort to elucidate the molecular underpinnings of health disparities in prostate cancer. The results of our RNA-seq analyses highlight significant differences in the transcription profiles in prostate tissue samples between AA and EA men. Additional differences were observed between subjects supplemented with vitamin D3 and subjects receiving placebo, suggesting that even a short period of vitamin D3 supplementation can have a significant impact on prostate gene expression. In view of the widespread vitamin D deficiency among AA men and their increased risk of developing prostate cancer, a deeper understanding of race-based transcriptomic differences and vitamin D driven pathways in prostate tissue will allow us to better justify vitamin D3 supplementation as a therapeutic option for early-stage prostate cancer, especially in AA men.
We acknowledge that the sample size is a limitation of this study. Therefore, in future studies we plan to enlarge the enrollment of eligible subjects by expanding the scope of RNA-seq analyses to single-core prostate biopsy samples obtained prospectively. The results of the RNA-seq analyses reported here were obtained with tissue samples of <50 mg, equivalent to the weight of a single-core biopsy. These additional subjects will also be stratified according to race, serum levels of vitamin D, serum levels of PSA, Gleason score, and supplementation. These future clinical studies will allow us to validate the concept that the prostate appears to be, at the molecular level, a ‘sentinel’ organ for health disparities.
Executive Summary.
Prostate cancer disproportionately affects African–American (AA) men in terms of incidence, morbidity and mortality, even after adjusting for stage.
Racial disparities in prostate cancer outcomes mirror racial differences in circulating levels of vitamin D. AA men exhibit a high prevalence of vitamin D deficiency.
The first goal of this study was to determine whether there are significant differences in the transcription profile of prostate tissue specimens between AA and European–American (EA) men.
The second goal of this study was to determine whether vitamin D3 supplementation could affect these differences.
Twenty-seven subjects (ten AA and 17 EA men), slated to undergo prostatectomy, were enrolled in the study.
Fourteen of these subjects received vitamin D3 supplementation (4000 IU/day) and 13 subjects received placebo for 2 months before surgery.
RNA was purified from prostate tissue specimens obtained at surgery and RNA-seq analyses were performed on all samples.
A total of 3107 genes were differentially expressed (FDR <0.1). Pathway and GO analysis indicated that AA show higher expression of genes associated with immune response and inflammation.
A total of 817 genes were differentially expressed in AA subjects supplemented with vitamin D3 compared with those receiving placebo.
These results support the existence of fundamental biological differences within the prostate between AA and EA men and suggest that overexpression of genes linked to the inflammatory process may contribute to the increased severity and faster progression of prostate cancer in AA men.
These findings also suggest that a considerable number of genes that are differentially expressed in AA compared with EA subjects, can be affected by a short course of vitamin D3 supplementation.
The prostate appears to be, at the molecular level, a ‘sentinel’ organ for health disparities.
Supplementary Material
Acknowledgements
The authors thank Drs Thomas E Keane and Sandip M Prasad (Department of Urology, MUSC and VA Medical Center) for critical discussions and help with the recruitment of eligible subjects, and Thomas Nash (MUSC Bioinformatics Core, Center for Genomic Medicine, MUSC) for help with data analysis. We thank Dr Jeremy Davis-Turak and Mr Tim Wesselman, Onramp BioInformatics, Inc. for useful conversations on bioinformatics pipelines.
Footnotes
Financial & competing interests disclosure
This research was funded by Veterans Administration (VA) CSR&D Merit Award CX000753 and a Pilot Project grant from the MUSC Center for Genomic Medicine to SG-C and MUSC COM start-up funds to GTH. The authors also acknowledge support from the Genomics Shared Resource, Hollings Cancer Center, Medical University of South Carolina. This shared resource is supported in part by the Hollings Cancer Center, Medical University of South Carolina Support Grant (P30 CA 138313). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Freedland SJ, Amling CL, Dorey F, et al. Race as an outcome predictor after radical prostatectomy: results from the Shared Equal Access Regional Cancer Hospital (SEARCH) database. Urology. 2002;60(4):670–674. doi: 10.1016/s0090-4295(02)01847-2. [DOI] [PubMed] [Google Scholar]
- 2.Freedland SJ, Sutter ME, Naitoh J, Dorey F, Csathy GS, Aronson WJ. Clinical characteristics in black and white men with prostate cancer in an equal access medical center. Urology. 2000;55(3):387–390. doi: 10.1016/s0090-4295(99)00461-6. [DOI] [PubMed] [Google Scholar]
- 3.Esvelt RP, Schnoes HK, Deluca HF. Vitamin D3 from rat skins irradiated in vitro with ultraviolet light. Arch. Biochem. Biophys. 1978;188(2):282–286. doi: 10.1016/s0003-9861(78)80010-1. [DOI] [PubMed] [Google Scholar]
- 4.Maclaughlin JA, Holick MF. Mediation of cutaneous vitamin D3 synthesis by UV radiation. In: Goldsmith LA, editor. Biochemistry and Physiology of the Skin. Oxford University Press; Oxford, UK: 1983. [Google Scholar]
- 5.Matsuoka LY, Wortsman J, Haddad JG, Kolm P, Hollis BW. Racial pigmentation and the cutaneous synthesis of vitamin D. Arch. Dermatol. 1991;127(4):536–538. [PubMed] [Google Scholar]
- 6.Clemens TL, Adams JS, Henderson SL, Holick MF. Increased skin pigment reduces the capacity of skin to synthesise vitamin D3. Lancet (London, England) 1982;1(8263):74–76. doi: 10.1016/s0140-6736(82)90214-8. [DOI] [PubMed] [Google Scholar]
- 7.Vieth R, Chan PCR, MacFarlane GD. Efficacy and safety of vitamin D3 intake exceeding the lowest observed adverse effect level (LOAEL) Am. J. Clin. Nutr. 2001;73(2):288–294. doi: 10.1093/ajcn/73.2.288. [DOI] [PubMed] [Google Scholar]
- 8.Garrett-Mayer E, Wagner CL, Hollis BW, Kindy MS, Gattoni-Celli S. Vitamin D3 supplementation (4000 IU/d for 1 y) eliminates differences in circulating 25-hydroxyvitamin D between African American and white men. Am. J. Clin. Nutr. 2012;96(2):332–336. doi: 10.3945/ajcn.112.034256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollis BW, Marshall DT, Savage SJ, Garrett-Mayer E, Kindy MS, Gattoni-Celli S. Vitamin D3 supplementation, low-risk prostate cancer, and health disparities. J. Steroid Biochem. Mol. Biol. 2013;136:233–237. doi: 10.1016/j.jsbmb.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Halliday TM, Peterson NJ, Thomas JJ, Kleppinger K, Hollis BW, Larson-Meyer DE. Vitamin D status relative to diet, lifestyle, injury, and illness in college athletes. Med. Sci. Sports Exercise. 2011;43(2):335–343. doi: 10.1249/MSS.0b013e3181eb9d4d. [DOI] [PubMed] [Google Scholar]
- 11.Feldman D, Pike JW, Adams JS. Vitamin D (3rd Edition) Academic Press San Diego ix; CA, USA: 2011. [Google Scholar]
- 12.Miller GJ, Stapleton GE, Ferrara JA, et al. The human prostatic carcinoma cell line LNCaP expresses biologically active, specific receptors for 1 alpha,25-dihydroxyvitamin D3. Cancer Res. 1992;52(3):515–520. [PubMed] [Google Scholar]
- 13.Hendrickson WK, Flavin R, Kasperzyk JL, et al. Vitamin D receptor protein expression in tumor tissue and prostate cancer progression. J. Clin. Oncol. 2011;29(17):2378–2385. doi: 10.1200/JCO.2010.30.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellfolk M, Norlin M, Gyllensten K, Wikvall K. Regulation of human vitamin D(3) 25-hydroxylases in dermal fibroblasts and prostate cancer LNCaP cells. Mol. Pharmacol. 2009;75(6):1392–1399. doi: 10.1124/mol.108.053660. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz GG, Whitlatch LW, Chen TC, Lokeshwar BL, Holick MF. Human prostate cells synthesize 1,25-dihydroxyvitamin D3 from 25-hydroxyvitamin D3. Cancer Epidemiol. Biomarkers Prev. 1998;7(5):391–395. [PubMed] [Google Scholar]
- 16.Deeb KK, Luo W, Karpf AR, et al. Differential vitamin D 24-hydroxylase/CYP24A1 gene promoter methylation in endothelium from benign and malignant human prostate. Epigenetics. 2011;6(8):994–1000. doi: 10.4161/epi.6.8.16536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barreto AM, Schwartz GG, Woodruff R, Cramer SD. 25-Hydroxyvitamin D3, the prohormone of 1,25-dihydroxyvitamin D3, inhibits the proliferation of primary prostatic epithelial cells. Cancer Epidemiol. Biomarkers Prev. 2000;9(3):265–270. [PubMed] [Google Scholar]
- 18.Chen TC, Wang L, Whitlatch LW, Flanagan JN, Holick MF. Prostatic 25-hydroxyvitamin D-1alpha-hydroxylase and its implication in prostate cancer. J. Cell. Biochem. 2003;88(2):315–322. doi: 10.1002/jcb.10342. [DOI] [PubMed] [Google Scholar]
- 19.Krishnan AV, Feldman D. Vitamin D and prostate cancer (Chapter 86) In: Adams DFWPS, editor. Vitamin D (Third Edition) Academic Press; CA, USA: 2011. pp. 1675–1709. [Google Scholar]
- 20.Moreno J, Krishnan AV, Swami S, Nonn L, Peehl DM, Feldman D. Regulation of prostaglandin metabolism by calcitriol attenuates growth stimulation in prostate cancer cells. Cancer Res. 2005;65(17):7917–7925. doi: 10.1158/0008-5472.CAN-05-1435. [DOI] [PubMed] [Google Scholar]
- 21.Cohen BL, Gomez P, Omori Y, et al. Cyclooxygenase-2 (COX-2) expression is an independent predictor of prostate cancer recurrence. Int. J. Cancer. 2006;119(5):1082–1087. doi: 10.1002/ijc.21749. [DOI] [PubMed] [Google Scholar]
- 22.Bao BY, Yao J, Lee YF. 1alpha, 25-dihydroxyvitamin D3 suppresses interleukin-8-mediated prostate cancer cell angiogenesis. Carcinogenesis. 2006;27(9):1883–1893. doi: 10.1093/carcin/bgl041. [DOI] [PubMed] [Google Scholar]
- 23.Lessard L, Begin LR, Gleave ME, Mes-Masson AM, Saad F. Nuclear localisation of nuclear factor-kappaB transcription factors in prostate cancer: an immunohistochemical study. Br. J. Cancer. 2005;93(9):1019–1023. doi: 10.1038/sj.bjc.6602796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang TT, Tavera-Mendoza LE, Laperriere D, et al. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol. Endocrinol. 2005;19(11):2685–2695. doi: 10.1210/me.2005-0106. [DOI] [PubMed] [Google Scholar]
- 25.Ramagopalan SV, Heger A, Berlanga AJ, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20(10):1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gennari L, Merlotti D, De Paola V, Martini G, Nuti R. Update on the pharmacogenetics of the vitamin D receptor and osteoporosis. Pharmacogenomics. 2009;10(3):417–433. doi: 10.2217/14622416.10.3.417. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz GG. Vitamin D and the epidemiology of prostate cancer. Sem. Dialysis. 2005;18(4):276–289. doi: 10.1111/j.1525-139X.2005.18403.x. [DOI] [PubMed] [Google Scholar]
- 28.Nesby-O'Dell S, Scanlon KS, Cogswell ME, et al. Hypovitaminosis D prevalence and determinants among African American and white women of reproductive age: third National Health and Nutrition Examination Survey, 1988–1994. Am. J. Clin. Nutr. 2002;76(1):187–192. doi: 10.1093/ajcn/76.1.187. [DOI] [PubMed] [Google Scholar]
- 29.Looker AC, Pfeiffer CM, Lacher DA, Schleicher RL, Picciano MF, Yetley EA. Serum 25-hydroxyvitamin D status of the US population: 1988–1994 compared with 2000–2004. Am. J. Clin. Nutr. 2008;88(6):1519–1527. doi: 10.3945/ajcn.2008.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandler PD, Giovannucci EL, Scott JB, et al. Null association between vitamin D and PSA levels among black men in a vitamin D supplementation trial. Cancer Epidemiol. Biomarkers Prev. 2014;23(9):1944–1947. doi: 10.1158/1055-9965.EPI-14-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kristal AR, Till C, Song X, et al. Plasma vitamin D and prostate cancer risk: results from the Selenium and Vitamin E Cancer Prevention Trial. Cancer Epidemiol. Biomarkers Prev. 2014;23(8):1494–1504. doi: 10.1158/1055-9965.EPI-14-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schenk JM, Till CA, Tangen CM, et al. Serum 25-hydroxyvitamin D concentrations and risk of prostate cancer: results from the Prostate Cancer Prevention Trial. Cancer Epidemiol. Biomarkers Prev. 2014;23(8):1484–1493. doi: 10.1158/1055-9965.EPI-13-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall DT, Savage SJ, Garrett-Mayer E, et al. Vitamin D3 supplementation at 4000 international units per day for one year results in a decrease of positive cores at repeat biopsy in subjects with low-risk prostate cancer under active surveillance. J. Clin. Endocrinol. Metab. 2012;97(7):2315–2324. doi: 10.1210/jc.2012-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak I, Sasik R, Freeman WR, et al. A degenerative retinal process in HIV-associated non-infectious retinopathy. PLoS ONE. 2013;8(9):e74712. doi: 10.1371/journal.pone.0074712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paolini P, Pick D, Lapira J, et al. Developmental and extracellular matrix-remodeling processes in rosiglitazone-exposed neonatal rat cardiomyocytes. Pharmacogenomics. 2014;15(6):759–774. doi: 10.2217/pgs.14.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server Issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hulsen T, De Vlieg J, Alkema W. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Mccarthy DJ, Chen Y, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 42.Zimmers TA, Jin X, Hsiao EC, Mcgrath SA, Esquela AF, Koniaris LG. Growth differentiation factor-15/macrophage inhibitory cytokine-1 induction after kidney and lung injury. Shock (GA, USA) 2005;23(6):543–548. [PubMed] [Google Scholar]
- 43.Lambert JR, Whitson RJ, Iczkowski KA, et al. Reduced expression of GDF-15 is associated with atrophic inflammatory lesions of the prostate. Prostate. 2015;75(3):255–265. doi: 10.1002/pros.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallace TA, Prueitt RL, Yi M, et al. Tumor immunobiological differences in prostate cancer between African–American and European–American men. Cancer Res. 2008;68(3):927–936. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 45.Murphy AB, Nyame Y, Martin IK, et al. Vitamin D deficiency predicts prostate biopsy outcomes. Clin. Cancer Res. 2014;20(9):2289–2299. doi: 10.1158/1078-0432.CCR-13-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swami S, Krishnan AV, Wang JY, et al. Dietary vitamin D(3) and 1,25-dihydroxyvitamin D(3) (calcitriol) exhibit equivalent anticancer activity in mouse xenograft models of breast and prostate cancer. Endocrinology. 2012;153(6):2576–2587. doi: 10.1210/en.2011-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.