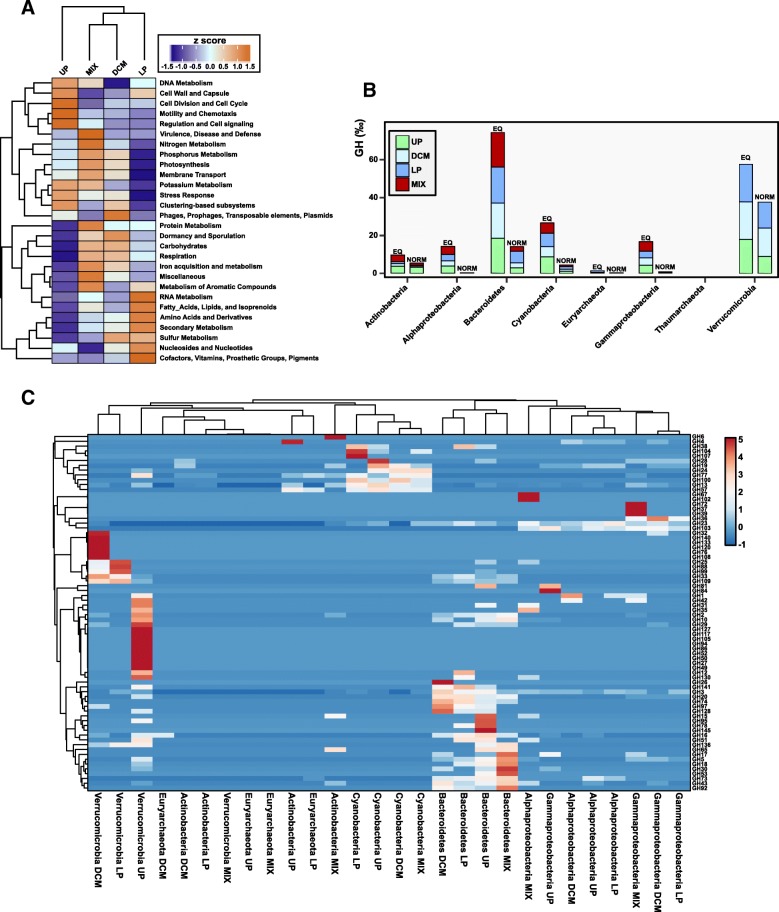
Fig. 5.
a SEED subsystems-based heatmap using the assembled coding sequences coming from contigs > 1 Kb. Proteins were grouped by depth (stratified samples UP, DCM, and LP) and season (MIX). For each one of the SEED categories, values were normalized by their standard score (z-score). b Number of glycoside hydrolases (GHs) detected in all the contigs > 5 Kb assigned the different phyla using the Carbohydrate-Active enZYmes (CAZy) database. EQ, number of GH per 1000 genes analyzed. NORM, number of GH per 1000 genes normalized by the percentage of 16S rRNA reads. c Heatmap of the different GH families. Abundance of GH was clustered by phylum and depth of the samples