Abstract
Aim:
Long noncoding RNAs serve critical regulatory functions highly specific for a tissue and its developmental stage. Antisense long ncRNA (AS-lncRNA) methylation changes in acute lymphoblastic leukemia (ALL) versus normal pre-B-cell lymphoblasts were evaluated to identify potential differential methylation in this group of genes.
Materials & methods:
The methylome of ALL and normal lymphoblasts was examined by the methylated CpG island recovery assay followed by NGS.
Conclusion:
The potential effect of trans regulation by AS-lncRNA through DNA/RNA binding is significant as sequence alignment analysis of the 25 most differentially methylated AS-lncRNAs revealed 368 genes containing highly similar sequences with a median nucleotide identity of 90.8% and binding span of 122 base pairs. Regulation of biological processes and anatomical structure development were over represented. ALL classification schemes based on AS-lncRNA methylation can provide new insights into its pathogenesis and treatment.
Keywords: : DNA methylation, epigenetic regulations, lncRNA
Acute lymphoblastic leukemia (ALL) is the most common type of cancer in children. It is believed that ALL arises from a developmental abnormality within the pro-B or pre-B-cell lymphoblast stages that may be associated with genetic events such as chromosomal translocations producing fusion genes and ploidy changes. However, since the cellular developmental process is heavily regulated by epigenetic mechanisms, it is very likely that differential gene expression arising from DNA methylation may also play an etiologic role in this disease [1,2]. It is well recognized that DNA methylation and other epigenetic changes are also important in the pathogenesis of cancer in general [3]. Methylation of cytosine in CpG dinucleotides plays an essential role in regulating gene expression as appropriate for tissue and developmental stage and dysfunctional methylation may silence tumor suppressor genes, activate oncogenes or deregulate signaling pathways leading to tumorigenesis. Studies analyzing DNA methylation patterns in ALL have yielded new insights into the value of identifying epigenetic changes for the diagnosis and classification of this disease. Figueroa et al. [2]. have identified abnormal methylation patterns that are associated with distinct gene expression profiles in ALL suggesting that aberrant gene expression due to dysfunctional methylation is important in lymphoid malignant transformation. Nordlund et al. [4], in a study of over 500 B- and T-cell ALL samples, showed that DNA methylation patterns based on only 246 CpG sites could predict eight common leukemia subtypes with a sensitivity of 0.90 and specificity of 0.99.
Recently, attention has focused on the potential role of long noncoding RNAs (lncRNAs) in the pathogenesis of cancer [5,6]. Thousands of long noncoding RNAs are transcribed in the human genome, generally at much lower levels than protein coding transcripts. Although there is still controversy as to how many of these might represent inconsequential ‘transcriptional noise’, the functional role of many lncRNAs in the regulation of critical cellular developmental processes and disease pathogenesis, has been incontrovertibly confirmed [7,8]. There are several mechanisms by which lncRNAs may interact with their regulatory targets including the formation of RNA–RNA, RNA–DNA and RNA–DNA-protein complexes [9]. Bioinformatics analysis of the differential DNA methylation of noncoding RNA genes (ncRNAs) between normal precursor B-cell lymphoblasts and ALL cells as determined by next-generation sequencing (NGS) and has shown that a large number of ncRNAs are significantly differentially methylated including antisense, long intergenic/intronic, microRNA, and pseudogene classes [10].
The importance of the antisense subclass of lncRNAs (AS-lncRNAs) in the regulation of gene expression is being increasingly recognized [11]. As much as 70% of lncRNAs may be antisense to protein coding genes [12,13]. AS-lncRNAs utilize various mechanisms in their regulation of gene expression. AS-lncRNAs may act locally to control the expression of their sense protein-coding partners through post-transcriptional mRNA hybridization of the antisense and sense mRNAs or by interfering with sense transcription by RNA polymerase through promoter competition (RNA polymerase collision) [14]. However, they may also function distally genome-wide by recognizing reverse complementary sequences in RNA or DNA. Perhaps the most important consequence of such binding is the ability to modulate gene splicing, which controls transcription of alternative protein isoforms. This regulatory process may occur through chromatin remodeling, occlusion of intronic or exonic splice site enhancers or silencers, or recruitment of protein activators or repressors [15–17]. In addition, AS-lncRNAs appear to interact, in a counterbalancing fashion, with miRNAs to fine tune gene expression post transcriptionally [18].
LncRNAs, like protein coding genes, are subject to epigenetic regulation by DNA methylation and, therefore, aberrant methylation of lncRNAs may also result in adverse cellular effects. As pointed out by Huarte and Rinn [6], lncRNAs are capable of acting as tumor suppressor genes or oncogenes due to their roles in modulating critical cellular pathways that may lead to malignant transformation. This study has analyzed genome-wide NGS data interrogating the methylome of pre-B ALL versus normal pro- and pre-B-cell lymphoblasts to identify differential methylation of the subset of lncRNAs identified as antisense. We have identified epigenetic changes that may lead to novel approaches to classification and therapy and also provide new insights into the pathogenesis of ALL.
This paper is exclusively focused on methylation of AS-lncRNAs and does not attempt to evaluate their expression levels. This approach was taken because there is not a direct and consistent correlation between methylation and expression of any coding or noncoding genes due to the multiple possibilities for pre- and post-translational modification of expression by such mechanisms as methylation of alternative promoters or splice site inhibitors and activators, and competitive inhibitory effects of miRNAs. Therefore, there can be no predictable relationship between methylation and expression without an understanding of the many other factors regulating expression, which is well beyond the state of the art of molecular genetics at this time.
Materials & methods
Examination of the precursor B-cell methylome
A total of 20 de-identified patient samples and ten normal control umbilical cord blood samples were procured with the approval of the institutional review board of the University of Missouri. Lymphoid B-cell ontogeny begins with hematopoietic stem cells and flows through common lymphoid progenitor cells, progenitor B cells (pro-B), precursor B cells (pre-BI and pre-BII), immature (naive) B cells and, finally, mature B cells. In these studies, precursor cells were isolated from umbilical cord cells by flow cytometry using the following cell surface antigens: pro-B (CD34+, CD19+), pre-BI (CD34-, CD19+, CD45 low), pre-BII (CD34-, CD19+, CD45 medium) and naive B cells (CD34-, CD19+, CD45 high) as described by Almamun et al. [18]. We have analyzed the differential methylation of antisense-RNAs using the Methylated CpG Island Recovery Assay followed by NGS (MIRA-seq) technique [19].
Identification of differentially methylated antisense lncRNAs
Genomic coordinates for all lncRNAs annotated as ‘antisense’ (AS-lncRNAs) were obtained from the GRCh37 genome assembly through the Ensembl biomart. Our MIRA-seq alignment data included 5,216 (99.1%). Identification of methylation peaks within this population was performed using the Matlab® Bioinformatics Toolbox (The MathWorks Inc, Natick, MA, USA), which calculated the average number of reads per kilobase of transcript per million mapped reads (RPKM) for each nucleotide within the genomic limits of the AS-lncRNA gene for the ALL, pro-B, pre-BII and naive B-cell samples.
AS-lncRNAs showing low variance (standard deviation <0.2) across all samples were filtered out, which left a working dataset of 3120 genes with a mean and median of 2.16 RPKM and 1.17 RPKM per base pair.
Results & discussion
The results of t-test statistical comparison for sample types show that ALL, pre-BII and naive-B cells are significantly hypermethylated compared with pro-B cells (Table 1). Mean methylation per nucleotide in terms of RPKM are as follows: ALL, 2.175; pro-B, 2.067, pre-BII, 2.189 and naive B, 2.194.
Table 1. . p-values for significant differential methylation between sample subtypes.
| ALL | Pro-B | Pre-BII | Naive B | |
|---|---|---|---|---|
| ALL | – | 5.02 × 10-5 | 0.442647 | 0.578985 |
| Pro-B | – | 3.51 × 10-7 | 1.72 × 10-7 | |
| Pre-BII | – | 0.8161 | ||
| Naive B | – | |||
ALL: Acute lymphoblastic leukemia; Pre-B: Precursor B cell; Pro-B: Progenitor B cell.
Unsupervised hierarchical clustering
Unsupervised hierarchical clustering was performed using the clustergram function of the Matlab® Bioinformatics Toolbox with default settings except for implementing cosine rather than Euclidean as the distance measure. After correcting for multiple testing using the Bonferroni method, 139 of the 277 genes providing the most discrimination between ALL and normal lymphoblasts were statistically significantly differentially methylated (p < 0.05).
Identification of Genes with DNA sequences complementary to mRNA of AS-lncRNAs
DNA sequences complementary to the mRNA of AS-lncRNAs were identified with the UCSC blat sequence alignment tool. Output was parsed to identify genes located on the opposite strand using custom Python code yielding a total of 368 genes. All matches greater than 20 bp were retained. The median sequence identity was 90.8% and median match span was 122 bp. The vast majority of the matches were to protein coding genes (83%) with the remainder distributed between lincRNA (12.5%), antisense (5.7%), pseudogenes (1.9%) and processed transcripts (1.4%).
Evaluation of the biological function of the protein-coding genes potentially regulated by the AS-lncRNAs was performed using gene ontology (GO) enrichment algorithms from the Matlab® Bioinformatics toolbox. The reference gene group was all genes (20,314) labeled as ‘protein-coding’ in the Ensembl database. GO terms with a p < 0.5 were parsed, counted and categorized by the most specific, descriptive GO ancestor term.
ALL & normal B-cell precursor lymphoblast sub classification based on differential methylation of AS-lncRNAs
The sample size is small; however, these cases were carefully selected to include only those in the B-lymphoblastic leukemia, not otherwise specified category, thus excluding cases already falling into well-defined clinical categories with recurrent genetic abnormalities. Such cases are underrepresented in the literature and publically accessible databases; therefore it appears desirable to identify new methods for clinical stratification of these patients in order to define potential new diagnostic and treatment modalities. Our results indicate that consideration of AS-lncRNA methylation levels may provide new insights into the pathogenesis and treatment of B-cell ALL.
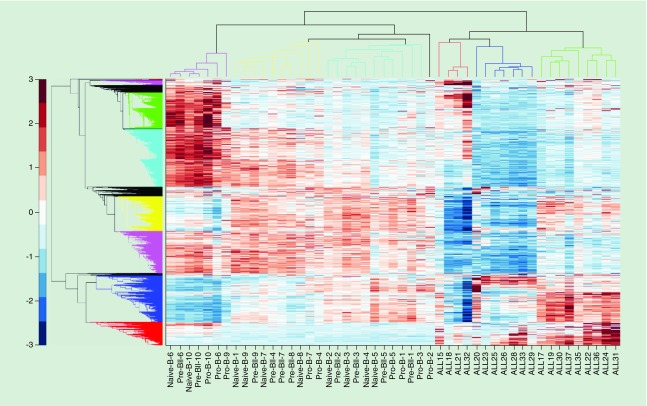
Hierarchical clustering cleanly segregates the ALL and normal lymphoblast sample populations and identifies three subclusters of both ALL and normal pre-B lymphoblasts based on average AS-lncRNA base pair methylation (Figure 1). While not statistically significant due to small sample size, the ALL cases display suggestive phenotypic characteristics: the middle ALL sub cluster (blue dendrogram) tended to have normal cytogenetic studies (five/six cases) and older age (mean: 90.9 months); the largest subgroup (green dendrogram) displayed more abnormal karyotypes including two cases with hyperdiploidy, two with deletion of chromosome 7 and one with duplication of chromosome 21 and intermediate age (mean: 60.7 months); and the smallest cluster (red dendrogram) contains examples of two translocations and one hyperdiploidy and is by far the youngest group (31.3 months) (Table 2).
Figure 1. . Unsupervised hierarchical clustering of 3120 antisense long ncRNA by mean reads per kilobase of transcript per million mapped reads per nucleotide.
The bottom gene subcluster (red gene dendrogram) contains the subset of antisense-RNA genes best discriminating between acute lymphoblastic leukemia; and normal precursor B cells and progenitor B-cell lymphoblasts.
Table 2. . Clinical phenotypes and karyotypes of patient samples.
| Group | Sample | Cytogenetics | Sex | Age (months) | Mean age (months) |
|---|---|---|---|---|---|
| Purple | A15 | Hyperdiploidy | M | 36 | |
| A18 | 15(der1)t(1,?), del(6)(q21) | F | 17 | ||
| A21 | t(3:19)(p25;p13) | F | 36 | ||
| A32 | None available | M | 36 | ||
| 31.3 | |||||
| Gold | A20 | 46 XY | M | 120 | |
| A23 | del(6)(q21;q27) | M | 180 | ||
| A25 | 46 XY | M | 48 | ||
| A26 | 47 XY? | M | 48 | ||
| A28 | None available | M | 36 | ||
| A29 | 46 XX | F | 24 | ||
| A33 | 46 XY | M | 180 | ||
| 90.9 | |||||
| Green | A17 | 46 XX | F | 72 | |
| A19 | Hyperdiploidy | M | 36 | ||
| A22 | 47 +21; 48, XX | F | 60 | ||
| A24 | 45, -7 -9 +der(9) t(8;9)(q112;p11) | M | 108 | ||
| A30 | 46 XX | F | 24 | ||
| A31 | 45, -7 XY | M | 132 | ||
| A35 | 46 XY | M | 22 | ||
| A36 | 46 XX | F | 72 | ||
| A37 | Hyperdiploidy | M | 20 | ||
| 60.7 | |||||
The set of 277 AS-lncRNA genes best discriminating between ALL and normal were further evaluated to characterize their relationship to protein-coding genes and their potential regulatory function. The 80 most statistically significant are listed in Table 3. Interestingly, it appears that there is a tendency for the normal lymphoblast samples to display similar distinct hypermethylation patterns within samples rather than across developmental stages, in other words, methylation varies more across subclasses than between stages of maturation. This suggests that methylation of normal lymphocytes may represent an inherent, inherited cellular feature rather than reflecting coordinated epigenetic control of gene expression critical to developmental maturation.
Table 3. . Top 80 most differentially hypermethylated antisense long noncoding RNAs and their neighboring sense genes.
| AS-lncRNA | p-value | Sense gene | AS-lncRNA | p-value | Sense gene |
|---|---|---|---|---|---|
| RP5-1024C24.1 | 8.06118 × 10-16 | MPPED2 | GPR50-AS1 | 3.70854 × 10-8 | GPR50 |
| AC010890.1 | 2.07022 × 10-13 | NCKAP5 | RP11-120J1.1 | 3.83071× 10-8 | NFIB |
| AC007966.1 | 5.06794 × 10-13 | FSIP2 | CTC-359M8.1 | 5.22797 × 10-8 | POU4F3 |
| RP11-966I7.1 | 1.70044 × 10-12 | FOXG1 | RP11-379L18.1 | 5.29154 × 10-8 | NOL4 |
| GS1-72M22.1 | 3.62267 × 10-12 | NEFM | CTD-2587H24.5 | 5.48669 × 10-8 | DNAAF3 |
| GRM5-AS1 | 1.10482 × 10-11 | GRM5 | HOXD-AS1 | 5.8567 × 10-8 | HOXD1 |
| CTD-2314G24.2 | 1.20869 × 10-11 | ISL1 | PDZRN3-AS1 | 7.41738 × 10-8 | PDZRN3 |
| PABPC5-AS1 | 1.35322 × 10-11 | PABPC5 | RP6-24A23.3 | 7.45609 × 10-8 | IRS4 |
| RP11-966I7.2 | 1.39785 × 10-11 | LINC01551 | NKX2-1-AS1† | 9.49919 × 10-8 | NKX2–1 |
| FEZF1-AS1† | 1.83805 × 10-11 | FEZF1 | RP11-680F8.1 | 1.55138 × 10-7 | |
| FGF12-AS2 | 1.8678 × 10-11 | FGF12 | AC005754.7 | 1.92123 × 10-7 | PCDHB3 |
| RP11-696P8.2 | 3.89281 × 10-11 | KCNV1 | RP11-91I8.3 | 1.96208 × 10-7 | ARHGAP28 |
| ATXN8OS | 4.06749 × 10-11 | KLHL1 | AC061961.2 | 2.05456 × 10-7 | KCNJ3 |
| RP11-1042B17.3 | 7.09449 × 10-11 | C14orf39 | RP11-62H20.1 | 2.43451 × 10-7 | LRRC9 |
| RP11-22C11.2 | 1.08362 × 10-10 | NEBL-AS1† | 8.57069 × 10-7 | NEBL | |
| RP11-814P5.1 | 1.40192 × 10-10 | ACTC1 | MNX1-AS1† | 1.10435 × 10-6 | MNX1 |
| AC003986.5 | 2.13505 × 10-10 | FERD3L | RP11-7I15.4 | 1.30598 × 10-6 | KCTD14 |
| RP11-92A5.2 | 2.84795 × 10-10 | NPY2R | TMEM5-AS1 | 1.93767 × 10-6 | TMEM5 |
| RP11-797E24.3 | 3.42767 × 10-10 | CELF4 | RP11-445F12.2 | 3.5096 × 10-6 | LHX1 |
| RP11-58K22.5 | 3.46151 × 10-10 | CD82 | ZNF571-AS1 | 4.65623 × 10-6 | ZNF571 |
| RP11-67L3.5 | 6.98329 × 10-10 | DHCR24 | RP11-752G15.3 | 5.04456 × 10-6 | CPEB1 |
| HOXC13-AS | 1.60671 × 10-9 | HOXC13 | AC092669.1† | 6.0995 × 10-6 | MEIS1 |
| AC079154.1 | 1.73361 × 10-9 | CNTNAP5 | MEIS1-AS3† | 6.42939 × 10-6 | MEIS |
| RP11-432I5.2 | 2.12362 × 10-9 | MARVELD3 | SATB2-AS1 | 7.39994 × 10-6 | SATB2† |
| ADAMTS19-AS1 | 2.46417 × 10-9 | ADAMTS19 | AC106786.1 | 7.56599 × 10-6 | PRDM6 |
| GPR158-AS1 | 2.82017 × 10-9 | GPR158 | RP3-326L13.2 | 9.07542 × 10-6 | POU3F4 |
| RP11-62F24.2 | 2.95862 × 10-9 | BNC2 | TBX5-AS1 | 9.84663 × 10-6 | TBX5 |
| RP11-82C23.2 | 4.50715 × 10-9 | LHX5 | RP11-97N19.2 | 1.02924 × 10-5 | SLC24A3 |
| EVX1-AS | 4.82962 × 10-9 | EVX1 | RP11-680F8.3 | 1.16375 × 10-5 | TJP1 |
| DLX6-AS1† | 5.59245 × 10-9 | DLX6 | HOTAIR | 1.35752 × 10-5 | HOXC11 |
| RP11-672L10.3 | 7.01594 × 10-9 | ADCYAP1 | AC010907.5 | 1.39763 × 10-5 | DCDC2C/ALLC |
| DLX6-AS2† | 7.13208 × 10-9 | DLX6 | CTD-2330J20.2 | 1.39942 × 10-5 | ALDH1A2 |
| RP11-137J7.2 | 8.0627 × 10-9 | NMBR | AC005789.9 | 1.448 × 10-5 | GGN |
| RP11-2N1.2 | 9.8052 × 10-9 | CCBE1 | HOXD-AS2 | 1.45228 × 10-5 | HOXD3 |
| RP5-1119D9.4 | 1.20318 × 10-8 | LAMP5 | HAND2-AS1 | 1.90129 × 10-5 | HAND2 |
| AC005281.2 | 1.41782 × 10-8 | SCIN | WWTR1-AS1† | 2.03025 × 10-5 | WWTR1 |
| RP11-964E11.2 | 1.47785 × 10-8 | PAX9 | CTA-363E6.5 | 2.06523 × 10-5 | TMC5 |
| RP11-387A1.5 | 2.38585 × 10-8 | HOXD3 | PGM5-AS1 | 2.3881 × 10-5 | PGM5 |
| CTD-2168K21.2 | 3.29598 × 10-8 | NEFL | MEIS1-AS2† | 2.55497 × 10-5 | MEIS1 |
| RP11-945C19.4 | 3.68047 × 10-8 | TMEM200C | RP11-284H19.1 | 3.00681 × 10-5 | PDE3A |
Bonferroni corrected p-values.
†Genes known to be overexpressed in acute leukemia or other malignancies.
It has been recognized for some time that transcription from an antisense gene may regulate the expression of mRNA from adjacent sense oriented protein coding genes through hybridization with mRNA or its DNA template strand [12]. Therefore, hypermethylation of AS-RNAs may lead to their silencing and the consequent overexpression of the sense protein coding genes under their transcriptional control. It is notable that ten of the 80 (12.5%) most hypermethylated AS-lncRNAs are associated with protein coding genes which have been shown to be overexpressed in acute leukemia or other malignancies. This suggests that methylation of AS-lncRNAs may provide an epigenetic explanation for their expression or suppression and thus lead to the appearance of oncogenic changes within the developmental progression of B-cell lymphoid precursors. Further, the hypermethylation of five (6.25%) AS-lncRNAs (MEIS1-AS2, MEIS1-AS3, AC092669.1, NEBL-AS1 and DLX6-AS1), which are antisense to members of the MLL fusion gene family raises the hypothesis that these proteins may be oncogenic independently of their MLL fusion partner.
Genes potentially regulated by hypermethylated AS-lncRNAs in trans
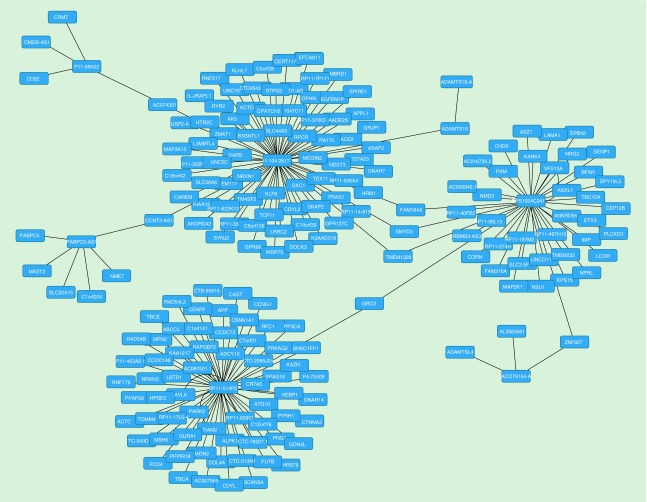
To investigate the potential effect of the most differentially methylated AS-lncRNAs exerted by binding to complementary regions of either DNA or RNA, we blasted the mRNA of the 25 most differentially methylated AS-lncRNAs against the entire genome and identified 368 genes with reverse complementarity indicating potential RNA–DNA binding partners. Reverse complementary alignment sequences were remarkably similar with a median identity of 90.8% over a median span of 122 bp. The number of matches per AS-lncRNA ranged widely from none to 77. The individual clusters were relatively discrete with only two clusters having three genes in common and two clusters with only one common gene. It appears that certain AS-RNA genes have the capability of regulating many others, predominantly protein coding genes (Figure 2).
Figure 2. . Potential interaction network of hypermethylated antisense long ncRNAs as determined by blast confirmation of reverse complementary mRNA–DNA sequences.
The hub of each cluster represents a single antisense long ncRNA.
The major clusters were formed based on genes RP11-1042B17.3 (77 targets), RP11-22C11.2 (73 targets), RP11-814P5.1 (69 targets) and RP5-1024C24.1 (44 targets). The remainder had significantly fewer binding partners ranging from 12 to 0 (two genes). Three of the larger clusters shared genes in common (FAM184A, SMYD3, TMEM132B and GRID2) indicating that the regulation of these genes occurs in coordinated manner under the control of two separate AS-lncRNAs.
Analysis of the function of genes potentially regulated by hypermethylated AS-lncRNAs
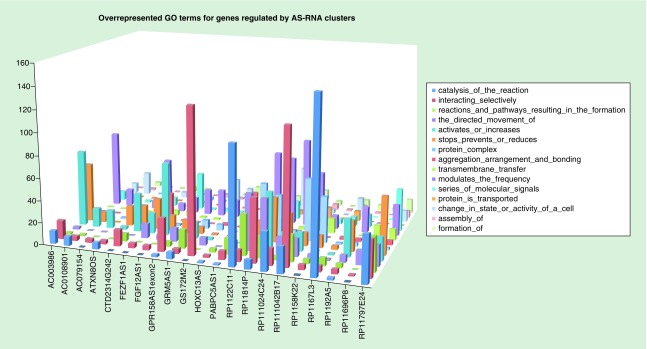
The genes identified as potential binding partners were examined for over-representation of GO terms following the method utilized in Matlab®. The comparison gene population consisted of all protein coding genes listed in the Ensembl biomart database. Over-represented GO terms were then tallied using the text of the GO name of the term, which provided a ready understanding of the gene functions. There was considerable variability in the distribution of these terms; however, overall, they illustrated a predominant regulatory effect again supporting a mechanism for coordinated regulatory modulation of gene expression (Figure 3).
Figure 3. . Representative overexpressed gene ontology terms associated with protein-coding genes potentially regulated in trans by AS-RNA genes due to RNA–DNA binding.
Data are in terms of the number of times a gene ontology term, or its sibling, is associated with protein-coding genes in the antisense-RNA regulatory cluster. These gene ontology terms are a subset of the 15 most common terms of a total of 67.
The total number of over-represented terms for each AS-lncRNA potential interaction cluster ranged from 59 to 922 with a mean of 330. It is evident that any one AS-lncRNA may regulate a large number of coding genes through a range of mechanisms consequent to RNA–DNA binding including splice site modulation and scaffold formation to recruit transcription binding factors, protein activators and suppressors, or chromatin modification genes that can induce histone methylation marks. Such a mechanism for providing centralized control would be advantageous for the cell in order to enable gene expression regulation in a coordinated manner on a large scale.
The over-represented GO terms were then aggregated under the most common ancestor GO term Table 4. This shows a preponderance of terms associated with GO:0050789, (19.15%), ‘regulation of biological process’, which again supports the hypothesis that the AS-RNAs have a significant regulatory impact on protein-coding genes that are themselves capable of regulating other downstream genes.
Table 4. . Most common gene ontology annotation terms for genes potentially regulated by hypermethylated antisense long noncoding RNAs.
| GO_ID | GO_name | Percentage (%) |
|---|---|---|
| GO:0050789 | Regulation of biological process | 19.15 |
| GO:0048856 | Anatomical structure development | 9.88 |
| GO:0003824 | Catalytic activity | 7.18 |
| GO:0005515 | Protein binding | 6.82 |
| GO:0055085 | Transmembrane transport | 5.58 |
| GO:0044238 | Primary metabolic process | 5.06 |
| GO:0043234 | Protein complex | 3.66 |
| GO:0007165 | Signal transduction | 3.34 |
| GO:0051179 | Localization | 3.24 |
| GO:0048731 | System development | 2.87 |
| GO:0050896 | Response to stimulus | 2.33 |
| GO:0016043 | Cellular component organization | 1.34 |
| GO:0065003 | Macromolecular complex assembly | 1.34 |
| GO:0008104 | Protein localization | 1.02 |
Conclusion
In order to avoid the confounding effects of methylation within promoters and other 5′ or 3′ elements, this study focused on evaluating the significance of hypermethylation only within the gene body of AS-lncRNAs in ALL as compared with normal pro-B and pre-BII lymphoblasts and naive B cells. Overall, AS-lncRNAs are significantly less methylated in pro-B cells than in ALL, pre-BII and naive B cells. No difference was observed for methylation of AS-lncRNAs between ALL, pre-BII or naive B cells. The finding of progressive methylation beyond pro-B cells is specific to AS-lncRNAs and is in contrast to an earlier observation of an overall decrease in genome wide methylation of all gene types during the transition from pro-B to pre-BII cells [18]. This finding suggests that hypermethylation may be an important factor in the biological regulation of expression of AS-lncRNAs.
The effect of DNA methylation within gene bodies is poorly understood; however, several potential mechanisms that might cause it to affect gene expression have been proposed including modulation of the elongation phase of transcription [20], controlling alternative RNA splicing [21], selection of alternative promoters [22] or polyadenylation sites [23].
Whatever mechanisms may be involved, analysis of our data indicates that gene body methylation has important consequences for determining the cell fate of early B cells. Unsupervised hierarchical clustering based on average AS-lncRNAs methylation per base pair clearly distinguishes between ALL and normal B-cell lymphoid precursors. Furthermore, it defines three sub clusters of ALL cases, which show evidence of possessing distinct tumor phenotypes based on clinical age, gender and cytogenetic features. Therefore, it appears that classification of subtypes of ALL based on AS-lncRNA methylation profiles reflects the biology of B-cell leukemia and may provide a new means for determining appropriate diagnosis, therapy and prognosis.
Executive summary.
Background
Acute lymphoblastic leukemia (ALL) arises from a developmental abnormality within the progenitor B cells and precursor B-cell lymphoblast that may be the result of epigenetic changes as has been demonstrated in other malignancies.
The recognition of long noncoding RNAs (lncRNAs) as important regulators of gene expression has provided new avenues of research into the etiologic mechanisms of cancer.
The antisense subtype of lncRNAs appears to be an important factor in the regulation of protein coding genes.
The expression of lncRNAs is also subject to DNA methylation.
Materials & methods
Normal immature B-cell lymphoblasts from umbilical cord blood were sorted into four developmental stages by flow cytometry and compared with B-cell ALL samples.
Differentially methylated AS-lncRNAs were identified using MIRA-seq (methylated CpG island recovery assay followed by next generation sequencing).
Protein coding genes potentially regulated in trans by highly methylated AS-lncRNAs were identified by blast searches for reverse complementary nucleotide sequences.
Results & discussion
ALL and normal B lymphoblasts may be readily distinguished by unsupervised hierarchical clustering based on mean AS-lncRNA base pair methylation.
Significant hypermethylation of antisense-lncRNAs (AS-lncRNAs) occurs in ALL, pre-BII and naive B cells as compared with pro-B cells.
Sequence alignments to the 25 most differentially methylated AS-lncRNAs revealed 368 genes, predominantly protein coding, with a median nucleotide identity of 90.8% and span of 122 bp with gene ontology terms related to regulation of biological process (GO:0050789) were highly represented.
Hierarchical clustering of ALL based on hypermethylation of AS-lncRNAS reflects the biology of the disease and may provide new insights into its pathogenesis and therapy.
Footnotes
Financial & competing interests disclosure
This research was supported by grant R00 CA132784 awarded to K Taylor. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Chatterton Z, Burke D, Emslie KR, et al. Validation of DNA methylation biomarkers for diagnosis of acute lymphoblastic leukemia. Clin. Chem. 2014;60(7):995–1003. doi: 10.1373/clinchem.2013.219956. [DOI] [PubMed] [Google Scholar]
- 2.Figueroa ME, Chen SC, Andersson AK, et al. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. J. Clin. Invest. 2013;123(7):3099–3111. doi: 10.1172/JCI66203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev. Cell. 2010;19(5):698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Nordlund J, Backlin CL, Zachariadis V, et al. DNA methylation-based subtype prediction for pediatric acute lymphoblastic leukemia. Clin. Epigenetics. 2015;7(1):11. doi: 10.1186/s13148-014-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstration of the importance of utilizing DNA methylation in the classification of acute lymphoblastic leukemia.
- 5.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Seminal article recognizing the regulatory significance of long noncoding RNAs and their potential ability to function as tumor suppressors and oncogenes.
- 7.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]; • Excellent review of mechanisms of epigenetic regulatory activity by long noncoding RNAs.
- 9.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almamun M, Levinson BT, Van Swaay AC, et al. Integrated methylome and transcriptome analysis reveals novel regulatory elements in pediatric acute lymphoblastic leukemia. Epigenetics. 2015;10(9):882–890. doi: 10.1080/15592294.2015.1078050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013;14(12):880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 12.Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]; •• Early demonstration of the importance of sense-antisense interaction in the regulation of expression of protein coding genes.
- 13.Hong D, Kurzrock R, Kim Y, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015;7(314):314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Discussion of the potential clinical therapeutic applications of antisense oligonucleotides.
- 14.Shearwin KE, Callen BP, Egan JB. Transcriptional interference – a crash course. Trends Genet. 2005;21(6):339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144(3):327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez I, Munita R, Agirre E, et al. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 2015;22(5):370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almamun M, Levinson BT, Gater ST, et al. Genome-wide DNA methylation analysis in precursor B-cells. Epigenetics. 2014;9(12):1588–1595. doi: 10.4161/15592294.2014.983379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung M, Kadam S, Xiong W, Rauch TA, Jin SG, Pfeifer GP. MIRA-seq for DNA methylation analysis of CpG islands. Epigenomics. 2015;7(5):695–706. doi: 10.2217/epi.15.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11(11):1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 21.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23(11):1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collingwood MA, Rose SD, Huang L, et al. Chemical modification patterns compatible with high potency dicer-substrate small interfering RNAs. Oligonucleotides. 2008;18(2):187–200. doi: 10.1089/oli.2008.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]