Abstract
Introduction
The long non-coding RNAs (lncRNAs) urothelial cancer associated 1 (UCA1) and metastasis associated lung adenocarcinoma transcript 1 (MALAT1) are known to impact cancer cell regulation. The aim of the present study was to determine the relationship between the expression of these lncRNAs in esophageal squamous cell carcinoma (ESCC) tissues and disease prognosis.
Material and methods
The expression of UCA1 and MALAT1 lncRNAs was assessed in ESCC and adjacent carcinoma tissues (5 cm away from the tumor) and evaluated in relation to overall survival (OS) and disease-free survival (DFS) of patients. This prospective study included 100 ESCC patients who were admitted to the First Hospital of Yulin City between January 2007 and January 2014.
Results
The expression levels of UCA1 and MALAT1 lncRNAs in ESCC tissues were significantly higher than those in adjacent carcinoma tissues, and there were statistically significant differences in TNM staging between the patients with high lncRNA expression and low lncRNA expression. The OS and DFS of patients with high UCA1 and MALAT1 lncRNA expression levels were significantly shorter than those with low expression levels. Furthermore, the OS and DFS of ESCC patients appeared to be correlated with TNM staging.
Conclusions
These results imply that the up-regulation of UCA1 and MALAT1 lncRNAs in ESCC tissues can impact the degree of tumor progression and is predictive of postoperative survival. Therefore, the expression levels of these lncRNAs can be used as measurement indexes to determine the prognosis of ESCC patients.
Keywords: long non-coding RNA, urothelial cancer associated 1, metastasis associated lung adenocarcinoma transcript 1, esophageal squamous cell carcinoma, prognosis, survival analysis
Introduction
Esophageal carcinoma (EC) is one of the most serious malignant tumors of the digestive system. The morbidity rate of EC ranks eighth and the mortality rate ranks sixth among malignant tumors worldwide [1, 2]. Two histologic types are typical among EC patients: esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). Patients in Western countries predominantly develop EAC, while Chinese patients typically develop ESCC. About 50% of ESCCs are formed in the middle esophagus [3]. Surgical removal is currently the most common treatment for ESCC, but the prognosis after a radical operation is less than ideal. According to recent studies, surgical treatment only gives ESCC patients at stages IIA–III a 5-year survival rate of 20–34% and fails to effectively eliminate distal metastasis and local tumor recurrence [4]. Currently, the TNM classification of malignant tumors serves as the primary basis for determining ESCC prognosis. The lack of objective quantitative indicators to accurately predict prognosis lends itself to opportunities for improvement [5].
Long noncoding RNAs (lncRNA) have emerged as influential factors in the occurrence and development of tumors. LncRNAs are more than 200 nucleotides in length and in most cases are produced by RNA polymerase II and undergo post-transcriptional modifications, such as capping and intron splicing. LncRNA genes usually contain few introns and have low levels of transcription, low conservation among different species, and in most cases have spatiotemporal expression specificity [6, 7]. Although this type of RNA does not have protein-encoding function, it can regulate gene expression at the chromosome, chromatin, transcriptional, and post-transcriptional levels and exerts significant regulatory effects through, inter alia, genomic imprinting, cell differentiation, immune reactions, tumor occurrence, and embryonic stem cell multi-potency. Recent studies have shown that multiple lncRNAs can produce small molecular peptides through transcription to become the driving force for the generation of new proteins [8–10]. In multiple large-scale screening studies, the expression of over 100 lncRNAs in ESCC tissues has been found to be altered. The abnormal expression of lncRNAs is mostly present during the evolution of highly differentiated intraepithelial neoplasia into invasive carcinoma, reflecting the important role of lncRNA in the development of ESCC [11–13].
Two genes known to encode lncRNAs are urothelial cancer associated 1 (UCA1) and metastasis associated lung adenocarcinoma transcript 1 (MALAT1). Li et al. reported that the relative level of UCA1 was significantly higher in ESCC tissues compared to the adjacent non-tumor tissue, and remarkably higher expression of UCA1 was found in esophageal cancer cell lines compared with the immortalized esophageal epithelial cell line NE1 [14], and it was found to be deregulated in cisplatin-resistant cells compared with their parent cells [15]. MALAT1 was over-expressed in 46.3% of ESCC tissues. The enhanced MALAT1 expression levels were positively correlated with clinical stages, primary tumor size, and lymph node metastasis, and inhibition of MALAT1 suppressed tumor proliferation in vitro and in vivo, as well as the migratory and invasive capacity [16]. Furthermore, silencing of MALAT1 could significantly suppress the proliferation of ESCC cells through the arrest of the G2/M cell cycle, which may be due to MALAT1-mediated up-regulation of p21 and p27 expression and the inhibition of B-MYB expression [17]. Recently, animal experiments showed that knockdown of MALAT1 decreased tumor formation and improved survival [18]. Here, we describe the relationship between the expression of UCA1 and MALAT1 lncRNAs in ESCC tissues and the associated prognosis.
Material and methods
Tissues samples
The tumor tissues and para-carcinoma tissues were taken from 100 ESCC patients who were admitted to the First Hospital of Yulin City between January 2007 and January 2014 and treated as participants of this case-control prospective study.
The adjacent carcinoma tissues had been obtained at least 5 cm away from the tumor. Patients were diagnosed with ESCC based on a postoperative pathological examination. Clinical characteristics of participants are shown in Table I. Patients were monitored after surgery by telephone follow-up or visits to the patient’s home, and the total survival (OS) and disease free survival (DFS) status was observed, with OS representing the time from surgery to death or the last follow-up visit and DFS representing the time from the surgery to the local recurrence, distal metastasis, or the last follow-up visit. This study was approved by the Ethics Committee of the First Hospital of Yulin City, and informed consent was given by all participants.
Table I.
Clinical characteristics of study participants
| Characteristics | Number of cases | Percentage |
|---|---|---|
| Gender: | ||
| Male | 75 | 75.0 |
| Female | 25 | 25.0 |
| Age [years]: | ||
| ≤ 60 | 68 | 68.0 |
| > 60 | 32 | 32.0 |
| Histological grade: | ||
| 1–2 | 41 | 41.0 |
| 3–4 | 59 | 59.0 |
| TNM stage: | ||
| I–II | 73 | 73.0 |
| III | 27 | 27.0 |
| Pathological type: | ||
| Ulcerative carcinoma | 65 | 65.0 |
| Others | 35 | 35.0 |
| Adjuvant chemotherapy or radiotherapy: | ||
| Yes | 59 | 59.0 |
| No | 41 | 41.0 |
Detection of lncRNAs
Expression levels of UCA1 and MALAT1 lncRNAs were detected in the tumor tissues and para-carcinoma tissues as follows: total RNA was extracted by RNAiso Plus (code no. 9108Q; TaKaRa Biotechnology Limited Company, Dalian, China), the PrimeScript RT reagent kit (code no. RR037A, TaKaRa Bio.) was used to transcribe and synthesize cDNA, and real-time quantitative PCR (q-RT PCR) was used to quantify target lncRNA with 18S as the internal reference. The primer sequences are shown in Table II. The total reaction volume was 10 μl, including of 5 μl of SYBR Green I Master Mix, 1 μl of RNAiso Plus, 0.6 μl of forward primers (10 μmol/l), 0.6 μl of reverse primers (10 μmol/l), 1 μl of cDNA, and 1.8 μl of dH2O deionized water. The reaction conditions were: pre-degeneration at 95ºC for 10 s, 95ºC for 5 s, 60ºC for 5 s, and 72ºC for 31 s, and amplification for 40 cycles. The differences between the target lncRNA cycle threshold values (Ct values) and 18S Ct value were determined (ΔCt) and the 2–ΔΔCt method was used to determine the relative expression of target lncRNA. Compared to the mean expression levels in ESCC tissues, the patients were categorized as having high UCA1 expression, low UCA1 expression, high MALAT1 expression, and low MALAT1 expression.
Table II.
Primer sequences of target lncRNA
| LncRNAs | Primer | Sequence |
|---|---|---|
| UCA1 | Forward | 5′-CTCTCCATTGGGTTCACCATTC-3′ |
| Reverse | 5′-GCGGCAGGTCTTAAGAGATGAG-3′ | |
| MALAT1 | Forward | 5′-CAGTGGGGAACTCTGACTCG-3′ |
| Reverse | 5′-GTGCCTGGTGCTCTCTTAC C-3′ | |
| 18 S | Forward | 5′-CAGCCACCCGAGATTGAGCA-3′ |
| Reverse | 5′-TAGTAGCGACGGGCGGTGTG-3′ |
Statistical analysis
Data were analyzed using SPSS 19.0 statistical software (IBM, Armonk, NY). Measurement data were expressed as mean ± standard deviation and an independent sample t test was used to compare groups. Kaplan-Meier survival analysis was used to compare OS and DFS and the log-rank test was used to determine statistical significance. The Cox risk model was used to analyze factors influencing OS and DFS. A p level < 0.05 indicated statistical significance.
Results
UCA1 and MALAT1 lncRNAs are higher in ESCC tissues than in para-carcinoma tissues
The relative expression levels of UCA1 were 0.485 ±0.248 in ESCC tissues and 0.199 ±0.122 in adjacent carcinoma tissues, with a significant difference (t = 10.348, p < 0.05). Similarly, the expression levels of MALAT1 were 3.842 ±0.415 in ESCC tissues and 1.451 ±0.136 in adjacent carcinoma tissues, with a significant difference (t = 54.750, p < 0.05).
UCA1 and MALAT1 lncRNA expression is correlated with TNM staging in ESCC tissues
Patients were categorized as having high UCA1 expression (49 cases), low UCA1 expression (51 cases), high MALAT1 expression (50 cases), and low MALAT1 expression (50 cases) compared to the mean expression levels in ESCC tissues. Clinical characteristics were compared between groups and the difference in TNM staging between the high UCA1 expression group and the low UCA1 expression group as well as between the high MALAT1 expression group and the low MALAT1 expression group was statistically significant (χ2 = 12.257, 26.839, p < 0.05). The difference in other clinical characteristics was not found to be statistically significant (Tables III and IV).
Table III.
UCA1 lncRNA expression in ESCC tissues and clinical characteristics
| Characteristics | Number of cases | UCA1 high expression(N = 49) | UCA1 low expression(N = 51) | χ2 | P-value |
|---|---|---|---|---|---|
| Gender: | |||||
| Male | 75 | 35 | 40 | 0.654 | > 0.05 |
| Female | 25 | 14 | 11 | ||
| Age [years]: | |||||
| > 60 | 68 | 31 | 37 | 0.990 | > 0.05 |
| < 60 | 32 | 18 | 14 | ||
| Histological grade: | |||||
| 1–2 | 41 | 22 | 19 | 0.603 | > 0.05 |
| 3–4 | 59 | 27 | 32 | ||
| TNM stage: | |||||
| I–II | 73 | 28 | 45 | 12.257 | < 0.05 |
| III | 27 | 21 | 6 | ||
| Pathological type: | |||||
| Ulcerative carcinoma | 65 | 33 | 32 | 0.233 | > 0.05 |
| Others | 35 | 16 | 19 |
Table IV.
MALAT1 lncRNA expression in ESCC tissues and clinical characteristics
| Characteristics | Number of cases | MALAT1 high expression(N = 50) | MALAT1 low expression(N = 50) | χ2 | P-value |
|---|---|---|---|---|---|
| Gender: | |||||
| Male | 75 | 39 | 36 | 0.480 | > 0.05 |
| Female | 25 | 11 | 14 | ||
| Age [years]: | |||||
| > 60 | 68 | 36 | 32 | 0.735 | > 0.05 |
| < 60 | 32 | 14 | 18 | ||
| Histological grade: | |||||
| 1–2 | 41 | 19 | 22 | 0.372 | > 0.05 |
| 3–4 | 59 | 31 | 28 | ||
| TNM stage: | |||||
| I–II | 73 | 25 | 48 | 26.839 | < 0.05 |
| III | 27 | 25 | 2 | ||
| Pathological type: | |||||
| Ulcerative carcinoma | 65 | 31 | 34 | 0.396 | > 0.05 |
| Others | 35 | 19 | 16 |
UCA1 and MALAT1 lncRNA expression in ESCC tissues is predictive of patient survival
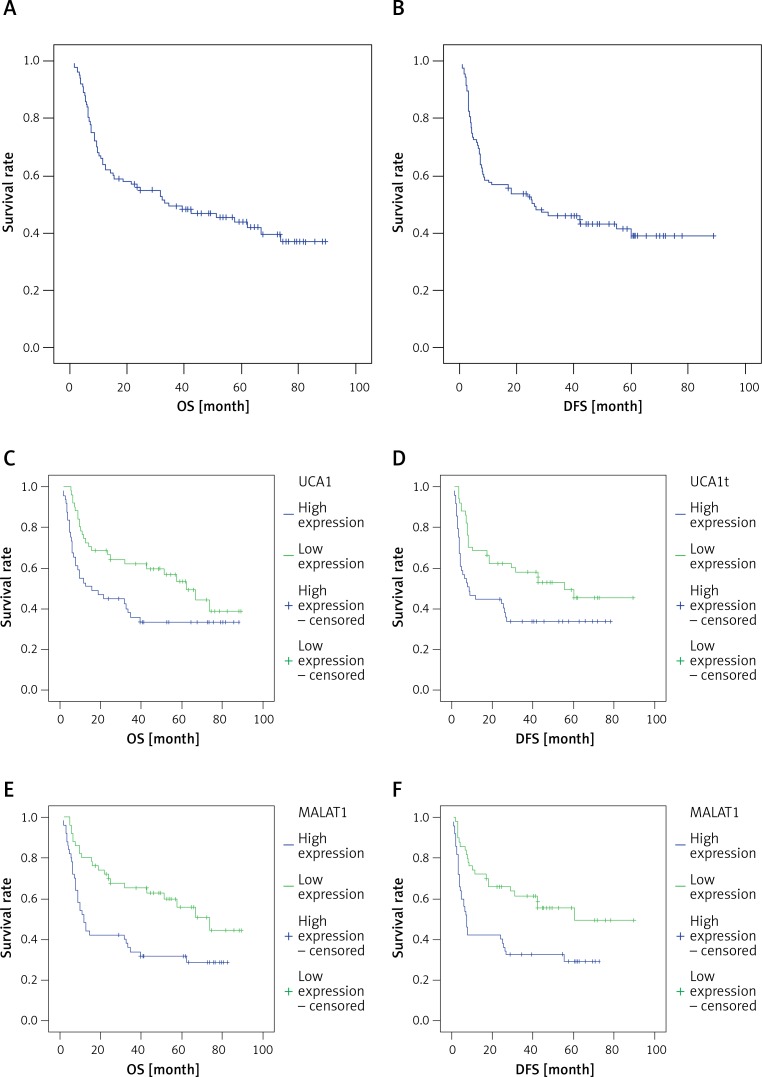
The OS and DFS of patients with high UCA1 and MALAT1 lncRNA expression in ESCC tissues were significantly lower than those of patients with low expression levels (χ2 = 4.880, 5.894, 8.635, 8.906, p < 0.05). OS, DFS, and 95% confidence intervals (CI) are shown in Tables V, VI and VII, and the Kaplan-Meier statistical significance analytic curves are shown in Figure 1.
Table V.
OS and DFS of ESCC patients
| Survival | Estimate | Std. error | 95% CI | |
|---|---|---|---|---|
| Lower bound | Upper bound | |||
| OS [month] | 46.069 | 3.815 | 38.591 | 53.547 |
| DFS [month] | 43.457 | 3.996 | 35.625 | 51.288 |
Table VI.
OS and DFS of patients with different UCA1 lncRNA expression levels
| Groups | Survival | Estimate | Std. Error | 95% CI | |
|---|---|---|---|---|---|
| Lower bound | Upper bound | ||||
| UCA1 high expression | OS | 53.754 | 5.096 | 43.766 | 63.741 |
| DFS | 51.636 | 5.403 | 41.046 | 62.225 | |
| UCA1 low expression | OS | 37.538 | 5.365 | 27.023 | 48.052 |
| DFS | 31.855 | 4.891 | 22.269 | 41.442 | |
Table VII.
OS and DFS of patients with different MALAT1 lncRNA expression levels
| Groups | Survival | Estimate | Std. error | 95% CI | |
|---|---|---|---|---|---|
| Lower bound | Upper bound | ||||
| MALAT1 high expression | OS | 57.089 | 5.136 | 47.022 | 67.156 |
| DFS | 54.440 | 5.540 | 43.582 | 65.299 | |
| MALAT1 low expression | OS | 32.999 | 4.760 | 23.670 | 42.329 |
| DFS | 27.819 | 4.289 | 19.413 | 36.225 | |
Figure 1.
A – Kaplan-Meier survival curve of OS of patients, B – Kaplan-Meier survival curve of DFS of patients, C – Kaplan-Meier survival curve of OS of patients with different UCA1 lncRNA expression levels, D – Kaplan-Meier survival curve of DFS of patients with different UCA1 lncRNA expression levels, E – Kaplan-Meier survival curve of OS of patients with different MALAT1 lncRNA expression levels, F – Kaplan-Meier survival curve of DFS of patients with different MALAT1 lncRNA expression levels
Multiple factors influence the survival of ESCC patients
Cox survival models revealed that OS and TNM staging (HR = 2.516) were significantly correlated (p < 0.05) with UCA1 lncRNA (HR = 1.638) and MALAT1 LncRNA (HR = 1.553) in ESCC patients. In addition, DFS and TNM staging (HR = 2.881) were also correlated with UCA1 lncRNA (HR = 1.662) and MALAT1 lncRNA (HR = 1.713), as shown in Tables VIII and IX.
Table VIII.
Multiple factors influence OS in ESCC patients
| Item | β | χ2 | HR | p-value |
|---|---|---|---|---|
| TNM stage (stage III) | 0.794 | 5.116 | 2.516 | < 0.05 |
| UCA1 high expression | 0.436 | 6.025 | 1.638 | < 0.05 |
| MALAT1 high expression | 0.237 | 4.915 | 1.553 | < 0.05 |
Table IX.
Multiple factors influence DFS in ESCC patients
| Item | β | χ2 | HR | p-value |
|---|---|---|---|---|
| TNM stage (stage III) | 0.662 | 4.628 | 2.881 | < 0.05 |
| UCA1 high expression | 0.168 | 5.604 | 1.662 | < 0.05 |
| MALAT1 high expression | 0.339 | 6.113 | 1.713 | < 0.05 |
Discussion
In this study, UCA1 and MALAT1 lncRNA in ESCC tissues were found to have increased expression levels compared to adjacent carcinoma tissues and were significantly correlated with TNM staging. The results of this study indicate that the expression levels of these two lncRNAs are up-regulated in ESCC tissues and may play important roles in ESCC pathogenicity.
The OS and DFS of patients with high expression of UCA1 and MALAT1 lncRNAs in ESCC tissues were found to be significantly lower than those of patients with low expression levels, suggesting that the high expression of these two types of lncRNA played a role in determining OS and DFS. The degree to which UCA1 and MALAT1 lncRNAs changed in ESCC tissues was associated with the progression of the tumors and the patient’s postoperative survival, suggesting that the expression of these lncRNAs might be used as auxiliary indicators to evaluate tumor progression and prognosis.
Relevant studies have revealed that UCA1 can promote the expression of WNT6 and inhibit the expression of the p27 gene, and it can influence the stage of tumor cells through the PI3K/Akt signaling pathway to exert cancer-promoting effects on proliferation and apoptosis in a variety of cancer types [19, 20]. UCA1 can also regulate the expression of tumor drug resistance genes [21, 22], giving it the ability to enhance the proliferation, infiltration, and migration of ESCC cells and lower the sensitivity to anti-tumor drugs. Our results support previous studies suggesting that measuring UCA1 expression could be a target for the treatment of ESCC and the evaluation of patient condition [23].
MALAT1 can react with multiple proteins in the cell nucleus to influence gene regulation and cancer cell movement capability [24]. Previous studies have found that the inhibition of MALAT1 can up-regulate the expression of caspase-8, caspase-3 and Bax, and down-regulate the expression of Bcl-2 and Bcl-xL, all of which were related to the development ESCC [25, 26]. The expression of MALAT1 in ESCC tissues at the middle-advanced stage has been found to be higher than that at the early stage, and the amplitude of the up-regulation is associated with the progression of tumor TNM staging and lymph node metastasis, suggesting that the inhibition of MALAT1 could stagnate the proliferation of ESCC tumor cells [27]. Therefore, abnormal expression of MALAT1 in malignant tumor tissues and its involvement with cell cycle stage make it a potential target for the treatment of ESCC [28].
In conclusion, we found that the expression of UCA1 and MALAT1 lncRNAs is up-regulated in ESCC tissues and that their expression can impact the degree of tumor progression. This in turn impacts patients’ postoperative survival time and can serve as an auxiliary index for predicting the prognosis of ESCC patients. Both lncRNAs are potential targets for ESCC treatments, and future studies will investigate the feasibility of interfering with their expression in a clinical setting.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diakowska D, Markocka-Maczka K, Nienartowicz M, et al. Increased level of serum prostaglandin-2 in early stage of esophageal squamous cell carcinoma. Arch Med Sci. 2014;10:956–61. doi: 10.5114/aoms.2013.34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Jiang Y, Wu C, et al. Comparison of clinicopathologic features and survival between eastern and western population with esophageal squamous cell carcinoma. J Thorac Dis. 2015;7:1780–6. doi: 10.3978/j.issn.2072-1439.2015.10.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takada A, Nakamura T, Takayama K, et al. Preliminary treatment results of proton beam therapy with chemoradiotherapy for stage I-III esophageal cancer. Cancer Med. 2016;5:506–15. doi: 10.1002/cam4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han D, Yuan Y, Song X, et al. What is the appropriate clinical target volume for esophageal squamous cell carcinoma? Debate and consensus based on pathological and clinical outcomes. J Cancer. 2016;7:200–6. doi: 10.7150/jca.13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng C, Zhou Y, Li H, et al. Whole-genome sequencing reveals diverse models of structural variations in esophageal squamous cell carcinoma. Am J Hum Genet. 2016;98:256–74. doi: 10.1016/j.ajhg.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen R, Ghosal S, Das S, et al. Competing endogenous RNA: the key to posttranscriptional regulation. Sci World J. 2014;2014:896206. doi: 10.1155/2014/896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan F, Yao J, Chen Y, et al. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:2838–49. [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNA and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitra SA, Mitra AP, Tfiche TJ. A central role for long non-coding RNA in cancer. Genetics. 2012;3:17. doi: 10.3389/fgene.2012.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo JC, Li CQ, Wang QY, et al. Protein-coding genes combined with long non-coding RNAs predict prognosis in esophageal squamous cell carcinoma patients as a novel clinical multi-dimensional signature. Mol Biosyst. 2016;12:3467–77. doi: 10.1039/c6mb00585c. [DOI] [PubMed] [Google Scholar]
- 13.Yao J, Huang JX, Lin M, et al. Microarray expression profile analysis of aberrant long non-coding RNAs in esphageal squamous cell carcinoma. Int J Oncol. 2016;48:2543–57. doi: 10.3892/ijo.2016.3457. [DOI] [PubMed] [Google Scholar]
- 14.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:7938–44. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou XL, Wang WW, Zhu WG, et al. High expression of long-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol Carcinog. 2016;55:2095–105. doi: 10.1002/mc.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu L, Wu Y, Tan D, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Li M, Wang Z, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–35. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Zhu Y, Li S, et al. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting b-catenin via Ezh2. Oncotarget. 2016;7:25668–82. doi: 10.18632/oncotarget.8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Zhou N, Watabe K, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Z, Wu L, Wang L, Yang Y, Meng Y, Yang H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: a possible correlation with cancer metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117:89–95. doi: 10.1016/j.oooo.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Han Y, Yang YN, Yuan HH, et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Li X, Wu S, Xue M, Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the Mtor-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951–5. doi: 10.1111/cas.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:7938–44. [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding RNA MALAT1 indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–92. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 25.Gutschner T, Diederichs S. The hallmarks of cancer: a long noncoding RNA point of view. RNA Biol. 2012;9:703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren S, Liu Y, Xu M, et al. Long non-coding RNA MALAT1 is a new potential therapeutic target for castration resistant prostate cell. J Urol. 2013;190:2278–87. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Gutsehner T, Hammerle M, Eissmanm M, et al. The non-coding RNA MALAT1 is acritical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao X, Zhao R, Chen Q, et al. MALAT1 might be a predictive marker of poor prognosis in patients who underwent radical resection of middle thoracic esophageal squamous cell carcinoma. Cancer Biomark. 2015;15:717–23. doi: 10.3233/CBM-150513. [DOI] [PubMed] [Google Scholar]