Phages have been increasingly recognized for the significance of their interactions with bacterial cells in multiple environments. Bacteria use myriad strategies to defend against phage infection, including restriction modification, abortive infection, phase variation of cell surface receptors, phage-inducible chromosomal islands, and clustered regularly interspaced short palindromic repeat(s) (CRISPR)-Cas systems. The data presented here suggest that the apparently passive process of OMV release can also contribute to phage defense. By considering the effect of OMVs on V. cholerae infection by three unique virulent phages, ICP1, ICP2, and ICP3, we show that, in vitro, a reproducible reduction in bacterial killing is both dose and phage receptor dependent. This work supports a role for OMVs as natural decoys to defend bacteria from phage predation.
KEYWORDS: Vibrio cholerae, bacteriophages, outer membrane proteins, outer membrane vesicle, phage defense
ABSTRACT
Novel preventatives could help in efforts to limit Vibrio cholerae infection and the spread of cholera. Bacteriophage (phage) treatment has been proposed as an alternative intervention, given the rapid replication of virulent phages, prey specificity, and relative ease of finding new virulent phages. Phage tropism is dictated in part by the presence of phage receptors on the bacterial surface. While many phages that can kill V. cholerae have been isolated, whether this pathogen is able to defend itself by neutralizing phage binding is unknown. Here, we show that secreted outer membrane vesicles (OMVs) act as a defense mechanism that confers protection to V. cholerae against phage predation and that this OMV-mediated inhibition is phage receptor dependent. Our results suggest that phage therapy or prophylaxis should take into consideration the production of OMVs as a bacterial decoy mechanism that could influence the outcome of phage treatment.
IMPORTANCE Phages have been increasingly recognized for the significance of their interactions with bacterial cells in multiple environments. Bacteria use myriad strategies to defend against phage infection, including restriction modification, abortive infection, phase variation of cell surface receptors, phage-inducible chromosomal islands, and clustered regularly interspaced short palindromic repeat(s) (CRISPR)-Cas systems. The data presented here suggest that the apparently passive process of OMV release can also contribute to phage defense. By considering the effect of OMVs on V. cholerae infection by three unique virulent phages, ICP1, ICP2, and ICP3, we show that, in vitro, a reproducible reduction in bacterial killing is both dose and phage receptor dependent. This work supports a role for OMVs as natural decoys to defend bacteria from phage predation.
INTRODUCTION
Cholera, which is caused by toxigenic strains of V. cholerae, is a diarrheal disease endemic to many countries with poor sanitation infrastructure and limited access to clean water and food (1). It is estimated that 1.3 to 4 million cases of cholera occur annually, with 23,000 to 143,000 deaths (2). Cholera symptoms can be ameliorated with the standard care of oral rehydration therapy or intravenously administered fluids for severe cases; yet, if left untreated, it can result in death within several hours (3).
Given the explosive rapid onset of disease and risk of mortality, control efforts have focused on generating and administering vaccines against cholera. Whole-cell killed and live attenuated cholera oral vaccines are available (reviewed in reference 4). Additional alternative approaches are being pursued to prevent V. cholerae infection, including phage prophylaxis (5, 6) and an outer membrane vesicle (OMV)-based vaccine (7–10). Recently, oral administration of a cocktail of three virulent phages (named ICP1, ICP2, and ICP3) which were isolated from rice-water stool samples of cholera patients in Bangladesh (11), was shown to protect against V. cholerae infection in infant mice and infant rabbit models of colonization and disease (6). Potentially, phages could also be used to reduce the numbers of V. cholerae or other waterborne bacterial pathogens in aquatic reservoirs.
OMVs are nonreplicating, spherical nanostructures produced during bacterial growth. These outer membrane-enclosed structures are composed of outer membrane lipids, outer membrane proteins, and periplasmic components. It has been shown that OMVs play several roles in bacterial biology, including assisting interactions between bacterial cells and delivery of cargo proteins, toxins, and nucleic acids to neighboring bacteria and host cells (12–15). V. cholerae OMVs are immunogenic, and it has been shown that mucosal immunization of adult female mice with OMVs provides maternal protection from V. cholerae colonization to their suckling pups (7–10). Similar immunization strategies and outcomes have been observed with OMVs against other pathogenic bacteria (16, 17).
Given that OMVs are comprised of bacterial membrane components, their potential to act as decoys—defense mechanisms that bacteria employ to protect themselves against external insults, including antibiotics, antimicrobial peptides, and bacteriophage infection—has been explored by others (14, 18). In this work, we tested whether V. cholerae OMVs can act as a defense mechanism against predation by the virulent phages ICP1, ICP2, and ICP3. We show that OMVs produced by two representative modern cholera pandemic strains can partially inhibit (neutralize) all three phages, and we show this neutralization is receptor dependent for two of the phages whose receptors have been identified.
RESULTS
V. cholerae OMVs inhibit phage infection.
To evaluate whether V. cholerae OMVs inhibit phage predation, OMVs isolated from spent culture supernatants of wild-type V. cholerae O1 El Tor strain E7946 (19) were incubated with the V. cholerae O1-specific virulent phage ICP1 (11, 20). Bacterial killing was measured using plaque formation on the V. cholerae E7946 strain as a proxy (Fig. 1). We observed a significant and dose-dependent inhibition of plaque formation by OMVs. It has been shown that the O1 antigen of the V. cholerae lipopolysaccharide (LPS) is the receptor for ICP1 (11, 21). To test whether the observed phage neutralization is receptor dependent, we performed plaque assays using OMVs from an acapsular derivative of the V. cholerae O139 serogroup strain MO10, as well as OMVs from an E7946 ΔwbeL rough mutant that fails to synthesize O1 antigen (21). Addition of OMVs from either the O139 strain or the E7946 rough mutant did not reduce ICP1 infection, indicating that the presence of O1 antigen in OMVs is required to neutralize this phage (Fig. 1).
FIG 1.
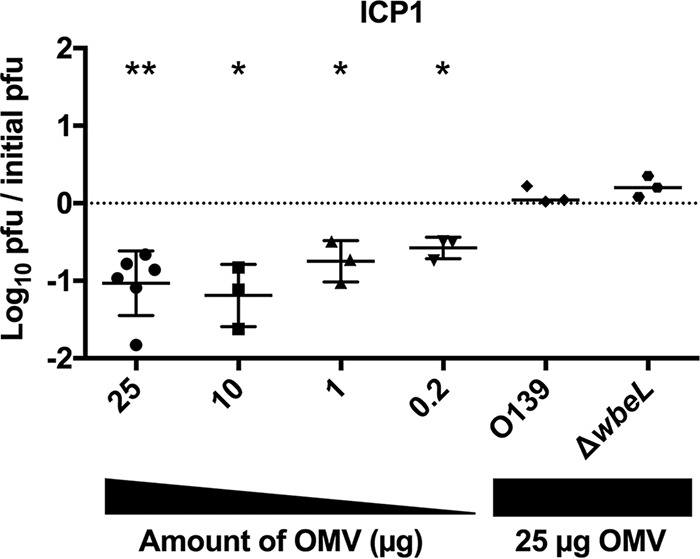
LPS O1-antigen in OMVs is required to neutralize ICP1. Titration of V. cholerae E7946 OMVs or of OMVs from V. cholerae O139 or E7946 ΔwbeL on a fixed concentration of prey (E7946) and phage ICP1 (∼10,000 PFU). n = 3 for all, except for 25 μg of OMVs, where n = 6. The horizontal solid lines indicate means ± standard deviations. **, P = 0.0018 for 25 μg; *, P = 0.036 for 10 μg, P = 0.0398 for 1 μg, and P = 0.019 for 0.2 μg.
To investigate whether the neutralizing effect observed for V. cholerae O1 OMVs on ICP1 was a general phenomenon, we performed similar neutralization assays using two additional virulent phages, ICP2 and ICP3. We observed that while there was a significant, dose-dependent inhibition of ICP2 (reduced by 0.5 log), the magnitude of inhibition was approximately half of that observed for ICP1 and only at the two highest OMV doses tested (25 and 10 μg) (Fig. 2). The receptor for ICP2 was previously shown to be the outer membrane porin OmpU (22). We hypothesize that the lower level of neutralization of ICP2 by OMVs may be due to the known low level of expression of OmpU during growth in vitro in Luria-Bertani Miller (LB) broth (23). Despite the low level of expression of OmpU under the conditions tested, we observed that OMVs purified from E7946 OmpU G325D, which is an ICP2-resistant point mutant (22), failed to neutralize ICP2 at the highest OMV dose tested (Fig. 2). These results indicate that OMV-mediated neutralization of ICP2 is dependent on the presence of the ICP2 receptor OmpU.
FIG 2.
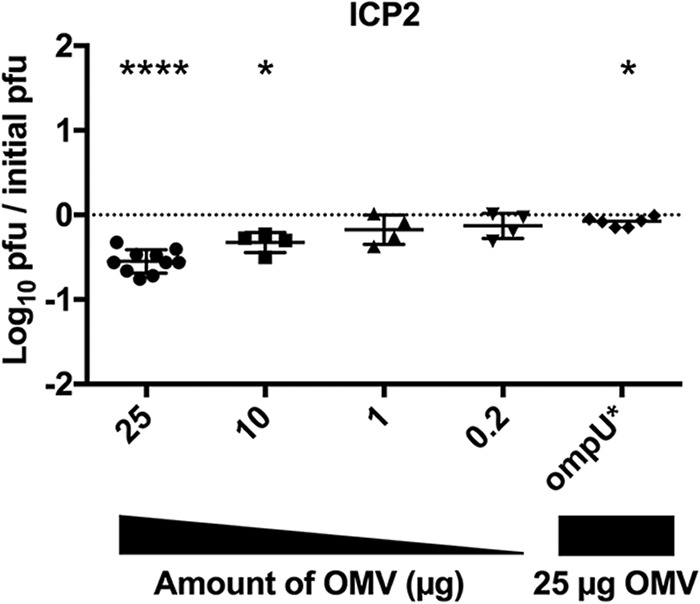
OmpU in OMVs is required to neutralize ICP2. Titration of V. cholerae E7946 OMVs, or OMVs from E7946 OmpU G235D mutant (OmpU*), on a fixed concentration of prey (E7946) and phage ICP2 (∼10,000 PFU). n = 10 for 25 μg; n = 4 for 10, 1, and 0.2 μg of OMVs; and n = 6 for OmpU G325D. The horizontal solid lines indicate means ± standard deviations. ****, P = <0.0001 for 25 μg; *, P = 0.0121 for 10 μg and P = 0.0135 for OmpU G325D.
As observed for ICP1 and ICP2 neutralization, addition of OMVs showed a dose-dependent reduction in the ability of ICP3 to infect (Fig. 3), albeit again to a lower magnitude than ICP1 inhibition (Fig. 1). The receptor for ICP3 is currently unknown.
FIG 3.
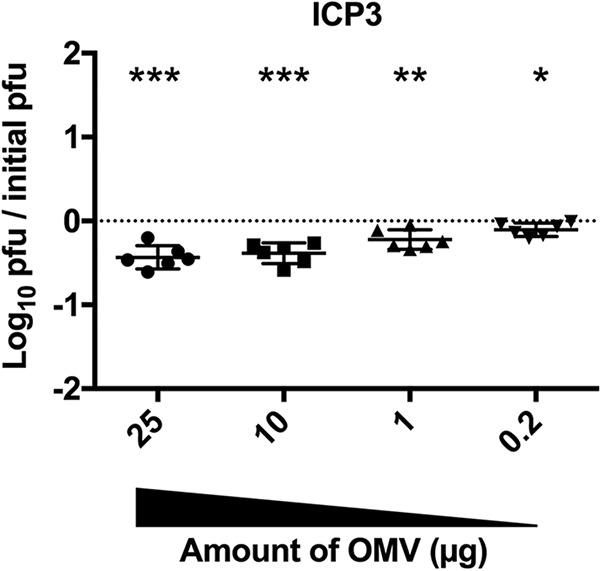
OMVs neutralize ICP3. Titration of V. cholerae E7946 OMVs on a fixed concentration of prey (E7946) and ICP3 (∼10,000 PFU). n = 6 for all. The horizontal solid lines indicate means ± standard deviations. ***, P = 0.0006 for 25 and 10 μg; **, P = 0.0053 for 1 μg; and *, P = 0.0224 for 0.2 μg.
Modern (7th pandemic) cholera epidemics have been attributed to a succession of three types of V. cholerae O1 El Tor strains, called waves 1, 2, and 3, with V. cholerae E7946 belonging to wave 1 (24). Wave 2 strains began to replace wave 1 strains in the late 1970s, and wave 3 strains subsequently began to replace wave 2 strains in the late 1980s. Wave 3 strains, which now dominate as the cause of cholera epidemics in South Asia, Africa, and Haiti, have recently been shown to be hypervirulent (25). To evaluate whether ICP1, ICP2, and ICP3 can also be inhibited by OMVs from a wave 3 strain, plaque assays were performed using a V. cholerae clinical isolate, HC1037, from the ongoing epidemic in Haiti. We observed an order of magnitude reduction in plaque formation for all three phages (Fig. 4), indicating that OMVs from distinct V. cholerae strains, including the most current pandemic strain, can neutralize virulent phages.
FIG 4.
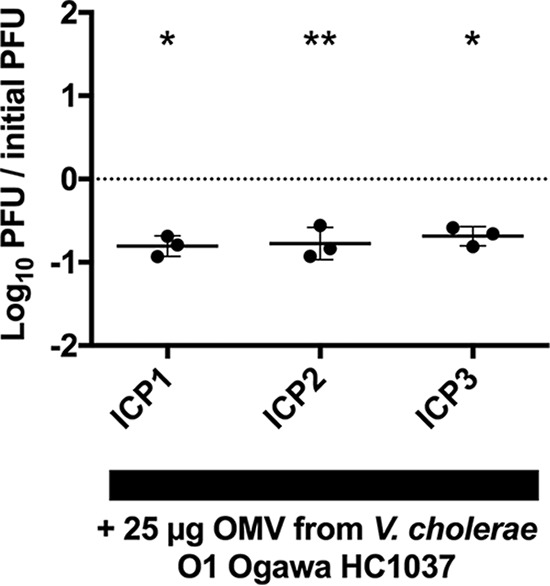
OMVs from a V. cholerae wave 3 pandemic strain neutralizes all three ICP phages. Titration of V. cholerae HC1037 OMVs on a fixed concentration of prey (HC1037) and ICP1, ICP2, and ICP3 (∼10,000 PFU). n = 3 for all. The horizontal solid lines indicate means ± standard deviations. *, P = 0.0076 for ICP1 and P = 0.0093 for ICP3; **, P = 0.02 for ICP2.
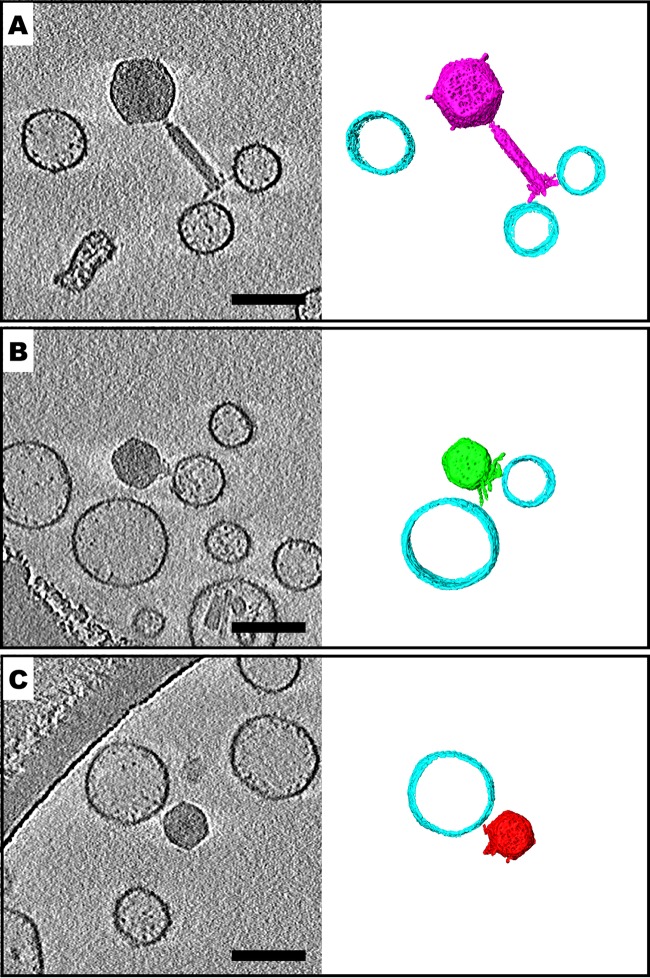
Our results thus far suggest that V. cholerae OMVs are able to neutralize phages by acting as decoys, whereby presumably the phage tail fibers bind to receptors on the OMV surface, therefore precluding the tail fibers from interacting with bacteria. We sought to directly visualize and confirm these interactions using cryo-electron tomography (cryo-ET). OMVs purified from the wave 3 strain HC1037 were used to neutralize phages ICP1, ICP2, and ICP3, and samples were applied to copper transmission electron microscopy (TEM) grids, immediately flash frozen in liquid ethane, and stored in liquid nitrogen. Grids were then imaged by cryo-electron microscopy (cryo-EM) to observe phages and OMVs. For a subset of individual phages bound to OMVs, tilt series were acquired and reconstructed via cryo-ET. Three representative tomograms of phages bound to OMVs are shown in Fig. 5. A slice through the three-dimensional volume is displayed, and so the OMVs appear as circles. The sizes and morphologies of all three phages were consistent with previous work using traditional transmission electron microscopy (11). Contact between a subset of the phage tail fibers and individual OMVs can be seen in each example, with the bound OMV being offset from the central axis of the phage tail for ICP1 and ICP3. We resolved ICP2 phages bound to OMVs along the central axis of the phage tail, as well as those offset from the central axis of the phage tail. The majority of phages that were bound to OMVs retained DNA in their capsids. Consistent with this, we did not observe contraction of the ICP1 tail in phages attached to OMVs. ICP2 and ICP3, which are members of the short-tailed family Podoviridae, do not have contractile tails.
FIG 5.
Tail fibers of ICP phages interact with V. cholerae outer membrane vesicles (OMVs). Cryo-electron tomography slices and three-dimensional (3D) segmentations of V. cholerae HC1037 OMVs free or bound to tail fibers of ICP1 (A), ICP2 (B), and ICP3 (C) are shown. Not all OMVs are shown in the 3D segmentation panels. Bars, 100 nm.
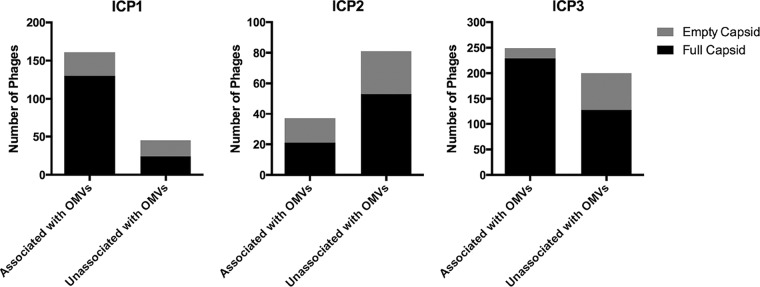
Quantification of OMV-phage interactions was performed, wherein phages with full or empty capsids, either associated or unassociated with OMVs were evaluated based on cryo-EM montage imaging. The results are shown graphically in Fig. 6, and the percentages for each group are presented in Table 1. Under the conditions of this assay, between 31 and 78% of phages were bound to OMVs, with ICP2 exhibiting the lowest percentage of binding, consistent with low expression of its OmpU receptor. Of the phages bound to OMVs, between 57 and 92% retained DNA in their capsids. This is consistent with the failure of most phages to engage all of their tail fibers productively with receptors on the OMVs. DNA ejection by phages T4 and T7 has been shown to require a proton motive force across the inner membrane of E. coli (36–39). Since ICP1 is related to T4 and ICP3 is related to T7, it is additionally possible that one or more of our phages has a similar requirement that cannot be met by an OMV. Phages that were not associated with OMVs also had between 35 and 47% empty capsids, which is consistent with our phage stocks containing a large fraction of phages with empty capsids. However, we cannot rule out transient on-off interactions of the phages with OMVs, which in some way leads to instability of some of the phages and subsequent loss of DNA.
FIG 6.
Quantification of ICP1, ICP2, and ICP3 interactions with OMVs, visualized by cryo-EM.
TABLE 1.
Presence of DNA within capsids of phages bound or unbound to OMVs
| Phage | No. (%) of capsids of phages: |
Total no. of capsids | |||||
|---|---|---|---|---|---|---|---|
| Associated with OMVs |
Unassociated with OMVs |
||||||
| Full | Empty | Subtotal | Full | Empty | Subtotal | ||
| ICP1 | 130 (81) | 31 (19) | 161 (78) | 24 (53) | 21 (47) | 45 (22) | 206 |
| ICP2 | 21 (57) | 16 (43) | 37 (31) | 53 (65) | 28 (35) | 81 (69) | 118 |
| ICP3 | 229 (92) | 20 (8) | 249 (56) | 128 (64) | 72 (36) | 200 (454) | 449 |
Altogether, our data indicate that OMVs can neutralize virulent phages by partial engagement of phage tail fibers with phage receptors on the OMV surface. This engagement presumably prevents or interferes with productive phage adsorption to the V. cholerae surface.
DISCUSSION
OMVs are naturally secreted by Gram-negative bacteria and have been implicated in a diversity of biological roles, including cell-to-cell communication and biofilm integrity (reviewed in references 12–15). Because OMVs have essentially the same surface protein, lipid, and carbohydrate structures as the bacterial cells that produce them, they may serve as natural decoys to defend the bacteria that secrete them against harmful products. Indeed, OMVs have been described as a virulence mechanism, where their secretion dampens the effects of host antimicrobial peptides and administered antibiotics, which target the OMVs instead of the bacterial cells (reviewed in reference 14).
Similarly, OMVs would seem to be natural decoys to protect bacteria from virulent phages. However, there have been few reports demonstrating this. Escherichia coli OMVs were previously shown to neutralize bacteriophage T4 (18, 26), while the cyanophage PHM-2 can directly interact with Prochlorococcus, resulting in contraction of the phage tail, and ultimately in DNA release (27). Here, we provide further support for this biological function of OMVs by showing that OMVs produced by pathogenic strains of V. cholerae can neutralize three different virulent phages. The three virulent phages we tested, ICP1, ICP2, and ICP3, are naturally found in association with pathogenic V. cholerae in patient stool samples from Bangladesh, and thus the frequency of interaction between them is likely high. In this work, we show that OMVs reduced phage predation in a dose- and receptor-dependent manner. That we see these effects with three unique phages from two distinct phage families (Myoviridae for ICP1, and Podoviridae for ICP2 and ICP3) that likely use different infection mechanisms suggests that OMV phage neutralization is a general defense strategy.
Because the concentrations of OMVs that naturally accumulate in the vicinity of V. cholerae in aquatic reservoirs, in biofilms, and in the human intestinal tract are unknown, it is not possible at present to positively state that V. cholerae OMVs neutralize phages in nature. Additionally, mutants of V. cholerae that fail to make OMVs have not been isolated as yet, and thus it is not possible to quantify the role of OMVs in protection from phage predation during infection or in aquatic environment microcosms. However, because V. cholerae does produce OMVs, it is reasonable to assume that OMVs protect V. cholerae from phage predation to an extent commensurate with the rate of OMV production and accumulated OMV concentration.
Multiple phage defense strategies are used by bacteria, each targeting different stages of phage infection. Phase variation or mutation of cell surface receptors, restriction-modification systems, abortive infection, clustered regularly interspaced short palindromic repeat(s) (CRISPR)-Cas, phage-inducible chromosomal islands, and bacteriophage exclusion systems have all been shown to contribute to the protection of bacterial cells (28, 40). How these processes are regulated and integrated to ensure bacterial cell survival remains to be fully understood. We suggest that the data presented here further support the idea that OMVs can also contribute to phage defense by acting as decoys in a receptor-dependent manner.
Our results add to the increasing recognition of OMVs as important bacterial secreted products. OMVs have potentially important differences from other canonical secretion systems, including continuous OMV production, exacerbated OMV production under conditions stressful to the bacterium, and encasing of virulence factors and signaling molecules by a membrane lipid bilayer that is protective against degradative enzymes (29, 30). Our findings add to a growing body of data showing that OMVs act as a first line of defense against external insults.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For all experiments, V. cholerae and isogenic mutants were routinely grown in Luria-Bertani Miller (LB) broth or on LB agar (Thermo Fisher Scientific) at 37°C with aeration. V. cholerae strains and phages used in this study are listed in Table 2. V. cholerae HC1037 was isolated in January of 2014 at the Cholera Treatment Center, Jacmel, Haiti, from a cholera patient rice-water stool sample.
TABLE 2.
Bacterial strains and bacteriophages used in this study
| Strain | Genotype and description | Reference(s) or source |
|---|---|---|
| AC53 | V. cholerae O1 El Tor Ogawa E7946 | 19 |
| AC5423 | V. cholerae O1 El Tor Ogawa HC1037 | This study |
| AC4653 | E7946 ΔwbeL | 21 |
| AC3276 | V. cholerae O139 MO10 ontA::pGP | 35 |
| AC4682 | E7946 OmpU G325D | 22 |
| ICP1 | Bacteriophage ICP1_2011_A; Myoviridae family | 11, 20 |
| ICP2 | Bacteriophage ICP2_2004; Podoviridae family | 11 |
| ICP3 | Bacteriophage ICP3_2007; Podoviridae family | 11 |
Bacteriophage stock preparation.
To obtain high-titer phage stocks, 40-ml cultures of V. cholerae E7946 were grown to mid-exponential phase (absorbance measured by optical density at 600 nm [OD600], 0.2 to 0.3). A single-plaque stock was inoculated into the culture and supplemented with calcium chloride at a final concentration of 5 mM to aid phage adsorption. The infected culture was incubated at 37°C with aeration for 1.5 h and then treated with sodium citrate at a final concentration of 50 mM to block further phage adsorption. The infected culture was centrifuged for 15 min at 8,000 × g, and the supernatant was filter-sterilized using a 0.22-μm filter (Millipore). A volume of 5× phage precipitation buffer (20% polyethylene glycol 8000, 2.5 M NaCl) was added and mixed by inversion. The mixture was incubated for 24 to 48 h at 4°C to precipitate the phage, after which the phage was pelleted by centrifugation for 15 min at 10,000 × g at 4°C using a Beckman Coulter floor centrifuge with a F13-14x50Cy rotor. The supernatant was removed by decanting, and the tube was centrifuged again for 1 min at 7,000 × g to allow subsequent removal of the last traces of supernatant. The phage pellet was resuspended in sodium chloride Tris-EDTA (STE) buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA) supplemented with 10 mM MgSO4 to help stabilize the phage and stored at 4°C until use.
Outer membrane vesicle isolation.
OMV isolation was adapted from Schild et al. (7). Briefly, V. cholerae cells were struck out on an LB agar plate and incubated overnight at 37°C. The following day, an individual colony was inoculated into 6 ml of LB broth and grown for ∼8 h at 37°C with aeration in a roller drum. This culture was then subcultured in a 1:100 ratio in 600 ml of LB broth and incubated overnight at 37°C with aeration. Cultures were chilled in an ice-water bath for ∼10 min and then centrifuged for 10 min at 10,000 × g to pellet bacterial cells. The supernatants, which contained the OMVs, were filtered through a SteriCup 0.22-μm filter system (Millipore) to remove any remaining bacterial cells, and an EDTA-free protease inhibitor cocktail (Roche) was added. OMVs were pelleted from culture filtrates by centrifugation at 28,000 rpm for 3 h and 15 min, using a Beckman Coulter Optima L-90K ultracentrifuge with an SW 32 Ti rotor. After removal of supernatants by decanting, OMV pellets originating from one 600-ml culture were resuspended in a total of 0.5 ml of phosphate-buffered saline (Boston BioProducts) and stored in aliquots at −80°C. OMV protein concentration was measured using the modified Lowry protein assay kit (Thermo Fisher Scientific).
Phage neutralization assays.
For phage neutralization assays, 10,000 PFU of phage were added to OMVs at the specified amount (25, 10, 1, or 0.2 μg) in a final volume of 200 μl of LB broth and vortexed to mix. Mixed samples were incubated for 1 h at room temperature. Mid-exponential-growth-phase cultures of wild-type V. cholerae E7946 or HC1037 were used as host bacteria for plaque assays. Cultures of the host bacteria were centrifuged for 5 min at 4,000 × g, washed once in LB broth, and adjusted to an absorbance (OD600) of 0.5. Phage alone or a phage-OMV mixture in a total volume of 90 μl was added to 100 μl of host V. cholerae and was allowed to adsorb for 10 min at room temperature. The mixtures were then transferred to 6-well, clear, untreated tissue culture plates (Corning). To each well, 3 ml of molten 47°C 0.3% agarose in LB broth was added, mixed by gentle swirling, and allowed to cool and solidify. The plates were incubated for ∼4 h at 37°C to allow plaque formation. After visually counting plaques, the titer of PFU per ml of phage added was calculated. Results are shown as log change in PFU · ml−1 compared to the phage-only control (no OMVs added).
Bacteriophage-OMV imaging using cryo-EM and cryo-ET.
The interactions between OMVs and ICP phages were characterized using cryo-electron tomography (cryo-ET). One microliter of V. cholerae HC1037 OMVs at a concentration of 8.4 mg · ml−1 (protein content) was added to 100 μl of phage at a titer of ∼1010 PFU · ml−1 in phosphate-buffered saline and incubated for 1 h at room temperature. After incubation, 10 nm of bovine serum albumin (BSA)-treated colloidal gold fiducials (Electron Microscopy Sciences, Hatfield, PA) was added as alignment markers. The sample (4 μl) was applied to glow-discharged, 200-mesh copper Quantifoil R 2/1 TEM grids (Quantifoil Micro Tools Gmbh, Jena, Germany) before plunge freezing with a Vitrobot Mark III system (FEI, Hillsboro, OR). Images were acquired on a JEM-2200FS field emission TEM (JEOL Ltd., Japan) operating at 200 kV and equipped with an in-column Omega energy filter (slit width of 20 eV) and a Direct Electron DE-20 (Direct Electron, LP, San Diego, CA). Bidirectional tilt series were collected semiautomatically at 10,000× nominal magnification (6.14 Å/pixel) using the SerialEM software package (31). Data sets were acquired with a tilt range of −62° to +62° at 2° increments, a defocus of −6 μm, and a total electron dose of ∼120 e−/Å2 (32, 33). Images were recorded at 12 frames per second and motion corrected using scripts provided by Direct Electron. IMOD software (34) was used for contrast transfer function (CTF) correction of the images by phase inversion and for three-dimensional (3D) reconstruction of the tomograms by radial-weighted back-projection. 3D segmentations were done manually using the Amira package (FEI Visualization Sciences Group, Hillsboro, OR). For quantification of OMV-phage interactions, 10,000× nominal magnification (11.8Å/pixel) cryo-electron microscopy (cryo-EM) polygon montages were acquired on a US4000 4k × 4k charge-coupled device (CCD) camera (Gatan, Inc., Pleasanton, CA) using SerialEM software (31). Montages were stitched using the IMOD sloppyblend.com script prior to quantification (34).
Statistical analyses.
Graphs were plotted using Prism 7 software (GraphPad), and statistical analyses were done using Student's two-tailed t test.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant R01 GM104540, NSF grant 0923395, grants from Emory University, Children's Healthcare of Atlanta, the Georgia Research Alliance, the Center for AIDS Research at Emory University (grant P30 AI050409), and the James B. Pendleton Charitable Trust to E.R.W; NIH grant T32 AI007329 to T.R.-R.; a Pew Latin American Fellows Program in the Biomedical Sciences and a CONICYT Becas Chile postdoctoral fellowship to C.A.S.-V.; NIH grant R01 AI12875 to A.A.; and NIH grant R01 AI055058 to A.C. A.C. is a Howard Hughes Medical Institute Investigator.
We thank the Robert P. Apkarian Integrated Electron Microscopy Core facility of Emory University for microscopy services and support.
REFERENCES
- 1.Clemens JD, Nair GB, Ahmed T, Qadri F, Holmgren J. 2017. Cholera. Lancet 390:1539–1549. doi: 10.1016/S0140-6736(17)30559-7. [DOI] [PubMed] [Google Scholar]
- 2.Ali M, Nelson AR, Lopez AL, Sack DA. 2015. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis 6:e0003832. doi: 10.1371/journal.pntd.0003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. 2012. Cholera. Lancet 379:2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop AL, Camilli A. 2011. Vibrio cholerae: lessons for mucosal vaccine design. Expert Rev Vaccines 10:79–94. doi: 10.1586/erv.10.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal A, Koley H, Ghosh A, Palit A, Sarkar B. 2013. Efficacy of cocktail phage therapy in treating Vibrio cholerae infection in rabbit model. Microbes Infect 15:152–156. doi: 10.1016/j.micinf.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Yen M, Cairns LS, Camilli A. 2017. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat Commun 8:14187. doi: 10.1038/ncomms14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schild S, Nelson EJ, Camilli A. 2008. Immunization with Vibrio cholerae outer membrane vesicles induces protective immunity in mice. Infect Immun 76:4554–4563. doi: 10.1128/IAI.00532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop AL, Schild S, Patimalla B, Klein B, Camilli A. 2010. Mucosal immunization with Vibrio cholerae outer membrane vesicles provides maternal protection mediated by antilipopolysaccharide antibodies that inhibit bacterial motility. Infect Immun 78:4402–4420. doi: 10.1128/IAI.00398-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Lazinski DW, Camilli A. 2017. Immunity provided by an outer membrane vesicle cholera vaccine is due to O-antigen-specific antibodies inhibiting bacterial motility. Infect Immun 85:e00626-. doi: 10.1016/S0140-6736(17)30559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop AL, Tarique AA, Patimalla B, Calderwood SB, Qadri F, Camilli A. 2012. Immunization of mice with Vibrio cholerae outer-membrane vesicles protects against hyperinfectious challenge and blocks transmission. J Infect Dis 205:412–421. doi: 10.1093/infdis/jir756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seed KD, Bodi KL, Kropinski AM, Ackermann H-W, Calderwood SB, Qadri F, Camilli A. 2011. Evidence of a dominant lineage of Vibrio cholerae-specific lytic bacteriophages shed by cholera patients over a 10-year period in Dhaka, Bangladesh. mBio 2:e00334-10. doi: 10.1128/mBio.00334-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jan AT. 2017. Outer membrane vesicles (OMVs) of Gram-negative bacteria: a perspective update. Front Microbiol 8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, Wang S, Yao Y, Xia Y, Yang X, Li K, Sun P, Liu C, Sun W, Bai H, Chu X, Li Y, Ma Y. 2016. Employing Escherichia coli-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Sci Rep 6:37242. doi: 10.1038/srep37242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roier S, Leitner DR, Iwashkiw J, Schild-Prüfert K, Feldman MF, Krohne G, Reidl J, Schild S. 2012. Intranasal immunization with nontypeable Haemophilus influenzae outer membrane vesicles induces cross-protective immunity in mice. PLoS One 7:e42664. doi: 10.1371/journal.pone.0042664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine MM, Black RE, Clements ML, Cisneros L, Saah A, Nalin DR, Gill DM, Craig JP, Young CR, Ristaino P. 1982. The pathogenicity of nonenterotoxigenic Vibrio cholerae serogroup O1 biotype El Tor isolated from sewage water in Brazil. J Infect Dis 145:296–299. doi: 10.1093/infdis/145.3.296. [DOI] [PubMed] [Google Scholar]
- 20.Seed KD, Lazinski DW, Calderwood SB, Camilli A. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seed KD, Faruque SM, Mekalanos JJ, Calderwood SB, Qadri F, Camilli A. 2012. Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog 8:e1002917. doi: 10.1371/journal.ppat.1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seed KD, Yen M, Shapiro BJ, Hilaire IJ, Charles RC, Teng JE, Ivers LC, Boncy J, Harris JB, Camilli A. 2014. Evolutionary consequences of intra-patient phage predation on microbial populations. Elife 3:e03497. doi: 10.7554/eLife.03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bact 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JLN, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satchell KJF, Jones CJ, Wong J, Queen J, Agarwal S, Yildiz FH. 2016. Phenotypic analysis reveals that the 2010 Haiti cholera epidemic is linked to a hypervirulent strain. Infect Immun 84:2473–2481. doi: 10.1128/IAI.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loeb MR. 1974. Bacteriophage T4-mediated release of envelope components from Escherichia coli. J Virol 13:631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. 2014. Bacterial vesicles in marine ecosystems. Science 343:183–186. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- 28.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnington KE, Kuehn MJ. 2014. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta 1843:1612–1619. doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerrero-Mandujano A, Hernández-Cortez C, Ibarra JA, Castro-Escarpulli G. 2017. The outer membrane vesicles: secretion system type zero. Traffic 18:425–432. doi: 10.1111/tra.12488. [DOI] [PubMed] [Google Scholar]
- 31.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Hampton CM, Guerrero-Ferreira RC, Storms RE, Taylor JV, Yi H, Gulig PA, Wright ER. 2017. The opportunistic pathogen Vibrio vulnificus produces outer membrane vesicles in a spatially distinct manner related to capsular polysaccharide. Front Microbiol 8:2177. doi: 10.3389/fmicb.2017.02177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB, Brun YV. 2017. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358:535–538. doi: 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 35.Nesper J, Kraiss A, Schild S, Blass J, Klose KE, Bockemühl J, Reidl J. 2002. Comparative and genetic analyses of the putative Vibrio cholerae lipopolysaccharide core oligosaccharide biosynthesis (wav) gene cluster. Infect Immun 70:2419–2433. doi: 10.1128/IAI.70.5.2419-2433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labedan B, Goldberg EB. 1979. Requirement for membrane potential in injection of phage T4 DNA. Proc Natl Acad Sci U S A 76:4669–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimkus AZ, Zavriev SK, Grinius LL. 1986. The role of ATP and membrane potential in the penetration of phage T17 DNA into the cell during infection. Mol Biol (Mosk) 20:185–191. (In Russian.) [PubMed] [Google Scholar]
- 38.Kemp P, Gupta M, Molineux IJ. 2004. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol 53:1251–1265. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- 39.Hu B, Margolin W, Molineux IJ, Liu J. 2015. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci U S A 112:E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seed KD. 2015. Battling Phages: How Bacteria Defend against Viral Attack. PLoS Pathog 11:e1004847. doi: 10.1371/journal.ppat.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]