Vibrios are often considered animal specialists or generalists. Here, we show that Vibrio breoganii has undergone massive genomic changes to become specialized on algal carbohydrates. Accompanying genomic changes include massive gene import and loss. These vibrios may help us better understand how algal biomass is degraded in the environment and may serve as a blueprint on how to optimize the conversion of algae to biofuels.
KEYWORDS: macroalgal carbohydrates, horizontal gene transfer, metabolic specialization, Vibrionaceae, ecology, adaptation, macroalgae, seaweed, Vibrio, algae, degradation, polysaccharide
ABSTRACT
While most Vibrionaceae are considered generalists that thrive on diverse substrates, including animal-derived material, we show that Vibrio breoganii has specialized for the consumption of marine macroalga-derived substrates. Genomic and physiological comparisons of V. breoganii with other Vibrionaceae isolates revealed the ability to degrade alginate, laminarin, and additional glycans present in algal cell walls. Moreover, the widely conserved ability to hydrolyze animal-derived polymers, including chitin and glycogen, was lost, along with the ability to efficiently grow on a variety of amino acids. Ecological data showing associations with particulate algal material but not zooplankton further support this shift in niche preference, and the loss of motility appears to reflect a sessile macroalga-associated lifestyle. Together, these findings indicate that algal polysaccharides have become a major source of carbon and energy in V. breoganii, and these ecophysiological adaptations may facilitate transient commensal associations with marine invertebrates that feed on algae.
IMPORTANCE Vibrios are often considered animal specialists or generalists. Here, we show that Vibrio breoganii has undergone massive genomic changes to become specialized on algal carbohydrates. Accompanying genomic changes include massive gene import and loss. These vibrios may help us better understand how algal biomass is degraded in the environment and may serve as a blueprint on how to optimize the conversion of algae to biofuels.
INTRODUCTION
Competition for resources is a major driver in the diversification of organisms, affecting metabolic strategies as well as habitat and organismal associations (1–4). While marine bacteria of the family Vibrionaceae are typically considered specialized for animal associations, a remarkable variety of lifestyles have arisen along with the capacity to utilize distinct assortments of diverse substrates during environmental or host interactions (5–10), and these changes yield promising bioengineering insights and applications (11, 12). Accordingly, fine-scale mapping of differences in metabolic capabilities and habitat associations among closely related isolates has enabled the identification of distinct populations with high resolution, offering valuable insights into the selective forces and fitness trade-offs in natural environments (13–16).
Recently, we identified an adaptive radiation among marine vibrios that was mediated by horizontal gene transfers, leading to ecophysiological differentiation and fine-scale resource partitioning of alginate, a brown algal cell wall glycan (17). This differentiation was particularly dramatic in V. breoganii, where diverse carbohydrate-active enzymes (CAZymes) were repeatedly acquired from multiple sources leading to the multicopy presence of each type of enzyme and facilitating the rapid degradation of large alginate polymers (17). This high degree of differentiation led us to hypothesize that, rather than simply obtaining an additional metabolic capability, V. breoganii has evolved into a macroalgal specialist.
This hypothesis initially appeared to be at odds with much of the literature describing V. breoganii, which was first isolated from the guts of clams (18) and subsequently observed in a diverse array of marine invertebrates, including mussels, crabs, and octopi (19, 20). However, Vibrio halioticoli, a close relative of V. breoganii, has a well-characterized association within the abalone gut, where it facilitates host nutrient absorption by degrading macroalgal carbohydrates (21–24). These reports also raise the question of whether the presence of V. breoganii within the guts of diverse invertebrate hosts reflects stable long-term symbioses or merely transient associations due to ingested algal material. Accordingly, we sought to further characterize the metabolic capabilities differentiating V. breoganii from other Vibrio populations and the extent of ecophysiological specialization to a macroalga- or host-associated lifestyle.
Using a combination of substrate-specific growth assays together with an analysis of the CAZyme content in a diverse collection of Vibrio populations, we demonstrate the specialization of V. breoganii to macroalgal carbohydrates, including alginate and laminarin, along with a shift away from animal-derived chitin and glycogen polymers and amino acids. Comparative genomics further reveals changes associated with this metabolic specialization and adaptations to a sessile seaweed-associated lifestyle, including the loss of motility. Together, these findings illustrate how algal polysaccharides became a major source of carbon and energy for V. breoganii, likely driving distinct ecological associations with algal detritus and marine invertebrates.
RESULTS AND DISCUSSION
V. breoganii isolates are macroalgal carbohydrate specialists.
To investigate metabolic specialization within V. breoganii, representative sequenced isolates from diverse Vibrionaceae populations were subjected to growth assays encompassing a wide variety of carbon substrates, including an extensive panel of carbohydrates with environmental relevance. Genomes were also analyzed using the dbCAN database to identify the CAZyme repertoire of each strain, and the Carbohydrate-Active enZymes database was used to identify annotated substrates for predicted enzymes (25, 26). Together, these substrate utilization and CAZyme profiles differentiate V. breoganii from other Vibrio populations and reflect a specialization for macroalgal carbohydrates.
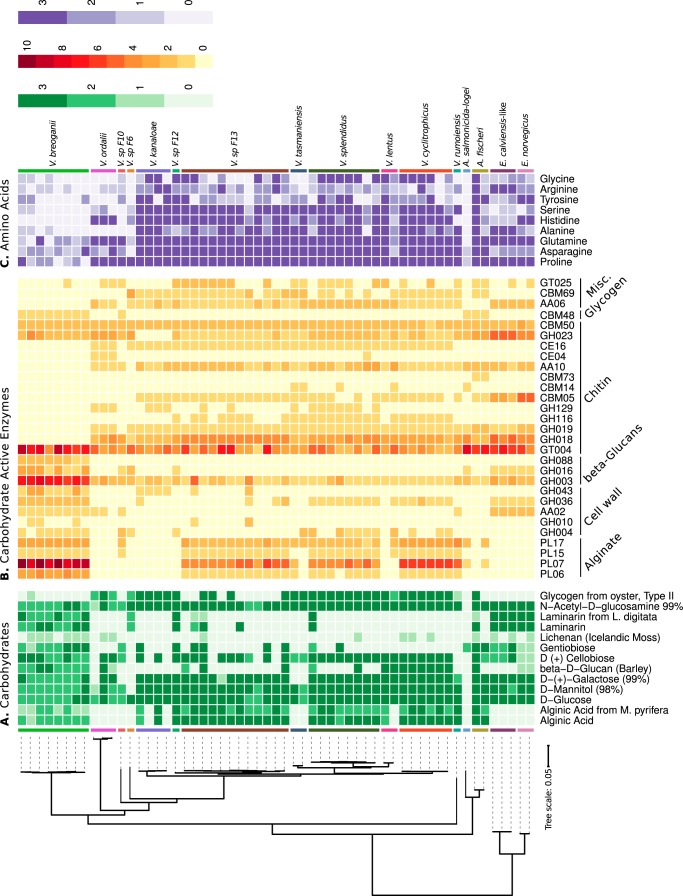
In addition to the previously characterized alginate pathway, V. breoganii and most other Vibrio populations assessed could utilize a wide variety of macroalga-associated substrates as a sole carbon source (Fig. 1A; see also Fig. S1 in the supplemental material). These commonly utilized algal substrates included mannitol, an important sugar alcohol used for carbon storage in brown algae, and galactose, a building block of carrageenans found in red macroalgae.
FIG 1.
Substrate utilization and CAZyme content of diverse Vibrio populations. (A) Differential growth of representative Vibrio populations on diverse carbohydrates. Each square of the heatmap reflects a growth score. Differences between initial and maximum OD600 values for each replicate were scored for growth according to threshold criteria, with average scores shown (<0.05, 0; 0.05 to 0.15, 1; 0.15 to 0.25, 2; >0.25, 3). Rows are ordered according to the phylogenetic species tree of the genomes based on concatenated ribosomal genes. Columns are arranged using hierarchical clustering, such that carbohydrate utilization scores with similar phylogenetic distribution are placed closer together. Only carbohydrates that displayed differential growth among strains are included here. The growth scores for all carbohydrates are shown in Fig. S1. (B) Hierarchical cluster analysis of all CAZymes in representative Vibrio genomes reveals the mosaic presence and absence of GH families as well as cohesive CAZyme repertoires indicating similar glycan catabolism and scavenging strategies within populations. All results are consolidated in a matrix, where rows represent species and columns represent CAZymes. Each cell depicts the absolute abundance of a CAZyme family in a strain. The CAZyme abundance matrix is presented as a heatmap, where rows are ordered according to the phylogenetic species tree of the genomes based on concatenated ribosomal genes. Columns are arranged using hierarchical clustering, such that CAZymes with similar phylogenetic distributions are placed closer together. Thus, absent or present blocks indicate loss or gain of a set of related genes in related species, respectively, e.g., the loss of chitin metabolism (GH18 and GH19) or the acquisition of laminarinases (i.e., GH16) in all Vibrio breoganii strains. The CAZyme contents shown here reflect differences between V. breoganii and other populations. Comprehensive CAZyme content is provided in Fig. S2. (C) Differential growth of representative Vibrio populations on amino acids. Each square of the heatmap reflects a growth score. Differences between initial and maximum OD600 values for each replicate were scored for growth according to threshold criteria, with average scores shown (<0.05, score 0; 0.05 to 0.15, score 1; 0.15 to 0.25, score 2; >0.25, score 3). Rows are ordered according to the phylogenetic species tree of the genomes based on concatenated ribosomal genes. Columns are arranged using hierarchical clustering, such that amino acid utilization scores with similar phylogenetic distribution are placed closer together. The amino acid substrates shown here reflect differences between V. breoganii and other populations. All amino acid substrate results are shown in Fig. S3.
However, V. breoganii isolates differentiated themselves from most Vibrio populations in their ability to utilize a broader variety of beta-glucans, which are an important category of glucose-based polysaccharides composed of beta-glycosidic bonds found primarily in algal cell walls. While most Vibrio populations could grow on the beta-glucan cellobiose, a glucose disaccharide joined with a beta-1,4 linkage, only V. breoganii possessed the capacity to also utilize gentiobiose, the beta-1,6-linked glucose disaccharide. Furthermore, V. breoganii isolates were able to grow on longer beta-d-glucan polysaccharides from barley with a backbone of beta-1,3 linkages and exhibited limited growth on lichenan, a glucose polysaccharide consisting of beta-1,3- and beta-1,4-glucans. Perhaps most importantly, V. breoganii was the only Vibrio population consistently capable of growing on laminarin, the principal storage glycan in brown algae. This abundant glycan is characterized by beta-1,3-glucans with occasional beta-1,6 linkages and branches (27).
Interestingly, the expanded capacity of V. breoganii to degrade beta-glucans appears to be shared with the more distantly related Enterovibrio populations of E. norvegicus and E. calviensis. These Enterovibrio populations could often degrade a similar suite of beta-glucans yet were incapable of degrading alginate polysaccharides. Similarly, although rare, a few non-breoganii Vibrio strains were capable of degrading additional beta-glucans, likely reflecting the independent and rapid horizontal acquisition of novel metabolic capacities.
To investigate the mechanistic basis of the expanded nutritional capacity in V. breoganii, we next analyzed the CAZyme content of each sequenced isolate. We found that V. breoganii isolates encode a CAZyme profile distinct from that of most Vibrio populations (Fig. 1B and S2), primarily defined by the alginate degradation pathway and an expanded repertoire of glycoside hydrolases implicated in the degradation of beta-glucans.
We previously discovered that V. breoganii repeatedly acquired and significantly expanded the alginate degradation pathway (17). Here, we extended our analysis to include additional strains across diverse Vibrio populations and assessed all CAZyme motifs to identify additional evidence of metabolic specialization. The expansion of the alginate degradation pathway was reaffirmed, in that every V. breoganii strain contained between 3 and 5 polysaccharide lyase 6 (PL6) domains, implicated in initiating extracellular polymer cleavage, whereas no other strain representative of other Vibrio populations encoded more than two of these alginate-specific lyases. Similarly, every V. breoganii strain contained more PL7 domains (between 8 and 12), which are important for cleaving mannuronate and guluronate linkages in alginate, than any other assessed Vibrio genome (between 0 and 7), further supporting an expanded capacity for extracellular degradation. The same was true when considering the oligoalginate lyase domains PL15 and PL17, responsible for intracellular degradation of alginate oligosaccharides into monomers, where V. breoganii had more of these alginate-related domains on average than did any other population.
We also found V. breoganii to be enriched in additional glycoside hydrolases (GHs), including a variety with activity relevant to macroalgal carbohydrates (28). These include GH36 (P = 4.6e−5), which have been implicated as galactosidases involved in breaking down carrageenans and carbohydrates found in red macroalgae, as well as others with wide-ranging substrate specificity, like GH4 (P = 0.12) and GH43 (P = 1.1e−8), which are often enriched in cell wall-degrading organisms (29). Additionally, some V. breoganii isolates encoded GH10 domains (P = 0.03) with xylan substrates often found in seaweeds (30), while every representative from this clade contained auxiliary activity 2 (AA2) motifs (P = 0.01), which can confer lignin peroxidase functions that can facilitate the breakdown of cell wall components (31).
Further reinforcing their distinct enzymatic repertoire, V. breoganii isolates encode more hydrolases implicated in beta-glucan (GH88, P = 2.3e−13) and laminarin degradation (GH3, P = 5.0e−15; GH16, P = 1.1e−9) than any other population. This abundance of laminarin-related CAZymes is consistent with V. breoganii growth on laminarin and beta-glucans in substrate utilization assays, and these CAZymes likely provide a mechanism of action for the breakdown of complex glucose-based polysaccharides (32). Furthermore, V. breoganii isolates are enriched in glycosyltransferase 4s (GT4s) (P = 5.6e−4), an enzyme family with activity on diverse substrates employed in the biosynthesis of oligosaccharides, polysaccharides, and glycoconjugates (33). This abundance in GT4 may reflect an increased capacity or specialization for utilizing distinct carbohydrates for anabolism.
V. breoganii cannot degrade chitin or glycogen.
While V. breoganii isolates displayed an expanded capacity to degrade macroalgal carbohydrates, substrate utilization growth assays reveal a diminished capacity to grow on a variety of substrates metabolized by the majority of Vibrionaceae. In particular, V. breoganii isolates were unable to utilize the animal-derived carbohydrates chitin and glycogen.
While most vibrios are widely recognized for their intimate associations with chitin (34–36), V. breoganii isolates are unable to metabolize this abundant marine polysaccharide. Although they have retained the ability to grow on the chitin monomer N-acetyl-d-glucosamine (Fig. 1A), V. breoganii isolates are unable to grow on chitin polymers in liquid culture or degrade them in plate-based chitinase assays (Table S2). Despite the presence of chitin metabolism pathways in more basal Vibrio populations, every V. breoganii strain lacked any GH18, GH19, GH116, or GH129 domain, suggesting a specific loss from this population. While they contain moderate numbers of GH23 domains, to which have been attributed chitinase activities, these enzymes also have annotated activities on lysozyme and peptidoglycan, so these likely reflect general cellular biosynthesis demands.
Carbohydrate binding motifs (CBM) and additional accessory CAZymes also reflect a shift away from chitin utilization. We found that V. breoganii lacked any CBM5, CBM14, or CBM73 motif, each implicated in binding chitin, which were observed in other Vibrio populations. Additional carbohydrate-modifying enzymes working in conjunction with CAZymes also indicate a shift away from chitin utilization. One enzyme with accessory activities implicated in chitin degradation (AA10) was absent in V. breoganii isolates yet widely distributed across most Vibrio populations. Similarly, the carbohydrate esterase 4 (CE4), responsible for the deacetylation of chitin sugars, was absent among V. breoganii, as was the acetylesterase CE16, which has activities on a variety of substrates, yet was widely distributed among other Vibrio populations.
In addition to a loss of chitin degradation ability, V. breoganii isolates were also incapable of using glycogen as a sole carbon source, unlike the majority of other vibrios. Glycogen is the primary energy storage polysaccharide in tissues of bivalves and other marine invertebrates and is composed of linear chains of glucose linked with alpha-1,4-glycosidic bonds with periodic branch points created by alpha-1,6-glycosidic linkages (37). Despite thriving on glucose monomers, V. breoganii isolates failed to degrade glycogen polysaccharides. While V. breoganii does encode a variety of GH13 domains which can include glycogen hydrolases, this family of hydrolases is multifunctional and not specific for glycogen degradation (38). Interestingly, the more basal Enterovibrio populations were also incapable of utilizing glycogen, suggesting that this metabolic capacity might have influenced evolutionary differentiation among the vibrios.
Similarly, most V. breoganii isolates assessed lacked a variety of additional CAZymes otherwise widely distributed throughout most Vibrio populations. V. breoganii isolates lacked any AA6s, which have been characterized as 1,4-benzoquinone reductases and are involved in the biodegradation of aromatic compounds, and they did not encode any CBM69 domains, which have been implicated in binding starch substrates and are also broadly distributed among populations. Most V. breoganii isolates also lacked GT25, which has been implicated as glucosyl- or galactosyltransferases, suggesting a metabolic shift away from substrates commonly utilized by other Vibrio populations.
While lacking the CAZymes required to break down chitin and glycogen, V. breoganii continue to encode binding motifs recognizing these abundant animal-associated marine polysaccharides. Two CBM50 domains with chitin-binding attributes remain in the V. breoganii genomes at an abundance similar to those all other vibrios assessed. Similarly, all V. breoganii genomes encoded CBM48 motifs with glycogen-binding functions, despite their inability to metabolize this carbohydrate. This glycogen-binding motif was less widely distributed and limited to the V. breoganii, Vibrio logei, Vibrio fischeri, and Vibrio sp. strain F-10 populations. The presence of these respective domains might indicate that although V. breoganii isolates have lost the ability to utilize these abundant animal-associated resources, they have maintained the ability to sense and respond to these important environmental cues.
V. breoganii isolates are poor scavengers of amino acids.
In addition to a diminished capacity to degrade and metabolize animal-derived carbohydrates, V. breoganii isolates were also inefficient at utilizing a variety of amino acids (Fig. 1C and S3). While every Vibrio strain grew to high density on media containing diverse amino acids, most V. breoganii strains were comparatively poor at metabolizing many specific amino acids (l-serine, l-tyrosine, l-arginine, l-glycine, l-alanine, l-glutamine, and l-proline) relative to most Vibrio populations. Furthermore, no V. breoganii strain tested was capable of growing on l-histidine as a sole carbon source. Indeed, genomic analysis revealed V. breoganii isolates have lost the ancestral enzymatic machinery required for histidine catabolism (IRP005923, IRP005920, IRP023636, IRP005921, and IRP023637), which is found almost universally among the other Vibrio genomes assessed (Fig. S4).
The diminished capacity of V. breoganii isolates to scavenge amino acids was surprising given how critical nitrogen acquisition can be under nitrogen-limited conditions, but it may be ecologically consistent with specialization for macroalgal carbohydrates. As the protein content in macroalgae biomass is relatively small (39), the impaired ability to scavenge amino acids might reflect their relatively scarce abundance on macroalgal surfaces, which would lead to relaxed selection on scavenging mechanisms (10).
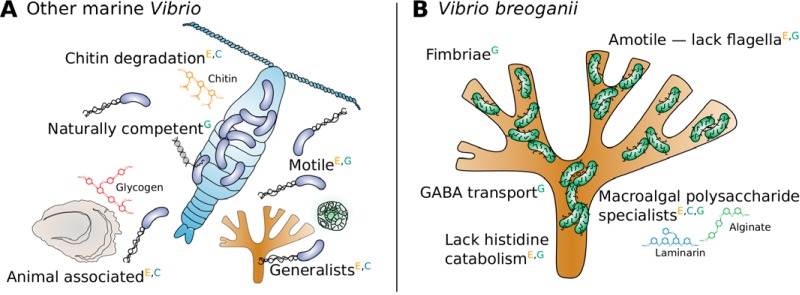
Surprisingly, gene cluster analysis revealed that all V. breoganii isolates encode a transporter for gamma-aminobutyric acid (GABA), a nonprotein amino acid, as well as enzymes for its degradation (Fig. 2 and Table S3). The transporter is absent from all other isolates, and the enzyme was present in 50% of Vibrio kanaloae genomes and 33% of Vibrio sp. strain F12 genomes, yet it was absent from all other populations. GABA is found in the tissue of many macroalgae (40), so the enrichment of these genes could be consistent with macroalgal associations and reflect alternative scavenging mechanisms.
FIG 2.
Features differentiating V. breoganii from other marine Vibrio species. Ecological associations and physiological traits distinguish V. breoganii from most other marine Vibrio species. Here, we illustrate properties common among other marine vibrios but absent in V. breoganii (A), and traits specific to V. breoganii (B). Superscript letters denote the type of supporting evidence for each feature: E, experimental evidence; C, CAZyme database analysis; G, gene cluster analysis.
Ecophysiological adaptations distinguish V. breoganii.
To identify additional distinguishing characteristics of V. breoganii, we expanded our genomic analysis beyond metabolism and assessed physiological traits relevant for habitat associations, summarized in Fig. 2.
Given the well-documented importance of chitin in Vibrio habitat associations and regulatory cues, along with the surprising loss of chitin utilization within V. breoganii, we investigated additional chitin-related functions within the representative genomes. We discovered that nearly all V. breoganii representatives have lost much of the genetic machinery associated with natural competence, transformation efficiency, and motility (Table S3). All V. breoganii isolates investigated lack the natural competence-associated genes comP, pilB, pilM, pilN, and pilP, yet retain some competence components (comEA, comEC, and comF) and pilin genes (pilC, pilD, pilG, pilO, pilQ, pilT, and pilW).
Flagellar proteins have also been lost, and agar stab assays confirmed that all V. breoganii strains were nonmotile, while nearly all representatives from other populations demonstrated motility (Table S4). This loss of motility might be indicative of trade-offs associated with different metabolic and dispersal strategies (15). Extensive environmental sampling has demonstrated that V. breoganii isolates are rarely observed in free-living fractions within the water column, instead favoring a particle-attached lifestyle (17, 19, 41). Metabolic specialization enabling higher growth rates on high-molecular-weight alginate and the capacity to degrade an expanded repertoire of laminarins and beta-glucans likely facilitate this stable association with insoluble macroalgal detritus (17, 19). Accordingly, this shift in nutritional resources enabling a predominantly surface-attached lifestyle might explain the loss of motility, as V. breoganii isolates could rely on passive dispersal via particles and ingestion by detritivore or scavenger hosts.
Conclusions.
While most vibrios display little host preference and a dominance of generalist populations, we demonstrate how V. breoganii strains have become specialized for a macroalga-associated lifestyle, as indicated by a distinct substrate utilization profile, a unique CAZyme repertoire, and ecophysiological adaptations. We also describe a shift away from commonly utilized animal-derived carbohydrates and amino acids, suggesting a transition away from potentially saprophytic associations toward a transient commensal relationship with diverse marine invertebrate hosts.
It has been well documented that most bacteria in marine invertebrate guts are often only transiently associated with their hosts (42–45). However, some vibrios appear to be stable commensal residents that contribute directly to host physiology (46, 47). For example, V. halioticoli strains have a strong association with abalone gut, where they facilitate host nutrient absorption by degrading brown algae (21–24). Although V. breoganii is also highly specialized for macroalgal carbohydrates and falls within the Halioticoli clade, previous observations of V. breoganii within diverse hosts likely reflect merely transient commensalisms.
Unlike V. halioticoli, which has rarely been characterized outside its abalone hosts or aquaculture facilities (23, 48), V. breoganii has been found in the guts of diverse invertebrate hosts (18–20). Furthermore, extensive environmental sampling in Plum Island Estuary (MA) revealed distinct seasonal and habitat associations between V. breoganii and particles within the water column where they appear to be strictly associated with algae and algal detritus (17, 19, 41). Accordingly, invertebrate hosts of V. breoganii likely reflect a transient by-product of algal or detritivore feeding strategies rather than specific host associations.
Here, we demonstrate how V. breoganii isolates have become metabolically specialized for degrading macroalgal carbohydrates, facilitating surface-attached lifestyles on insoluble algal particles. These macroalgal associations likely explain their presence within diverse marine invertebrates following the ingestion of algal detritus. We also show V. breoganii has lost the ability to degrade animal-derived carbohydrates and impaired amino acid scavenging, likely contributing to more benign commensal relationships with diverse hosts, minimizing potentially disruptive impacts of colonization on host physiology. In turn, these diverse hosts may provide stable habitats and useful waste products and facilitate dispersal. Accordingly, the metabolic specialization of V. breoganii to macroalgal carbohydrates appears to drive transient commensal presence within diverse marine invertebrates, illustrating selective pressures and fitness trade-offs driving Vibrio evolution in natural environments.
MATERIALS AND METHODS
Isolates and culture conditions.
The strains tested here originated from previous studies on the ecological population structure of Vibrionaceae. Briefly, isolates were obtained either from size-fractionated water samples, handpicked algal detritus particles and zooplankton, or different body parts of marine invertebrates by plating samples on Vibrio-selective marine thiosulfate-citrate-bile salts-sucrose (TCBS) medium (BD Difco TCBS with 1% NaCl added) (19, 34, 49). Individual colonies were picked and purified by restreaking three times, alternating between 1% tryptic soy broth (TSB) medium (BD Bacto with 2% NaCl added) and marine TCBS medium. Cultured isolates were grown in marine broth 2216 (MB2216, catalog no. 279110; Difco) at room temperature with shaking, unless otherwise specified. Substrate utilization assays were performed in minimal medium adapted from Tibbles-Rawlings medium (TRMM) (50, 51). The strains included in this study had been previously sequenced to enable genomic analyses, and all sequenced V. breoganii isolates were alga associated.
Substrate utilization growth assays.
Frozen glycerol stocks of Vibrio strains were inoculated into 1.5 ml MB2216 in deep-well blocks and grown with shaking for 48 h. Strains were then inoculated with a pin replicator into optically clear 96-well trays containing 200 μl substrate minimal medium solution (1g/liter in TRMM) at an approximately 1:1,000 (vol/vol) final dilution. Substrate minimal medium solutions were made by solubilizing carbohydrates or amino acids in Milli-Q water, filter sterilizing, and adding sterilized Tibbles-Rawlings components. Some substrates were relatively insoluble in water and required additional filters for sterilization, contributing to a lower effective substrate concentration or treatment to improve solubility. A complete list of assessed substrates and their treatment is provided in Table S1 in the supplemental data. Strains were grown in triplicate for each carbohydrate, with plate layouts rearranged in each replicate to control for potential evaporative effects. Culture trays were incubated at room temperature without shaking, and the optical density at 600 nm (OD600) was monitored twice daily for 12 days. Differences between the initial and maximum OD values for each replicate were scored for growth according to threshold criteria (<0.05, score 0; from 0.05 to 0.15, score 1; from 0.15 to 0.25, score 2; >0.25, score 3), and average scores are shown. When scoring growth or no growth among triplicates, any inconsistencies were resolved by removing the anomalous replicate.
CAZyme annotation and analysis.
Hidden Markov models (HMMs) of carbohydrate-active enzyme (CAZyme) domains were obtained from the dbCAN database (25). These were searched against all open reading frames (ORFs) from each genome using hmmscan (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan). The best-scoring domains were then filtered by E value (<10−23) and alignment coverage (>0.8), both of which are more stringent cutoffs than those recommended by dbCAN (http://csbl.bmb.uga.edu/dbCAN/), in an effort to minimize false positives. When multiple domains were observed within an ORF, only the best scores for each category of enzyme are reported. Statistical testing of population-specific CAZyme enrichment was assessed using a predicted binomial distribution model. Annotated CAZyme substrate activities and families are described in the Carbohydrate-Active enZymes database (http://www.cazy.org/) (26).
Chitinase activity.
A plate-based chitin clearing assay was used to assess the ability of representative Vibrio strains to degrade chitin polymers. Dense cultures grown in MB2216 were spotted (10 μl) onto MB2216 agar plates with a top layer supplemented with 2.5% colloidal chitin stained with Remazol brilliant blue R (52). After 5 days of incubation at room temperature, plates were scored for the presence or absence of visible zones of clearing surrounding or below lawns, indicating the degradation of insoluble chitin polymers.
Gene cluster analysis.
All ORFs were clustered at 50% amino acid identity using MMSeqs2. For each genome, the count of each cluster was tabulated, and these counts were compared between V. breoganii genomes and genomes from other populations. Gene clusters that appeared to be either almost exclusive to or almost entirely absent from V. breoganii were considered for further analysis. The ORFs for each genome were annotated with InterProScan version 5.17-56.0 using the iprlookup, goterms, and pathways options. InterProScan is a program from EMBL-EBI that uses the InterPro database for annotations. The InterPro database used contains 15 databases, with TMHMM for predicted transmembrane proteins and SignalP for predicted signal peptide cleavage sites, as well as the 13 default databases which listed at https://github.com/ebi-pf-team/interproscan/wiki/HowToRun#included-analyses.
Motility assay.
A standard agar stab assay was used to assess the potential for motility among isolates. Motility test agar medium was prepared with MB2216, Bacto agar (catalog no. 214010, 0.25% [wt/vol]; BD), and 2,3,5-triphenyltetrazolium chloride solution (catalog no. 17779-10X10ML-F; Sigma-Aldrich) and autoclaved. Medium was aliquoted in autoclaved glass tubes (catalog no. 47729-576; VWR) with plastic closures (catalog no. EW-04500-01, 5 ml of medium per tube; Cole-Parmer) and allowed to cool to room temperature. Using an inoculating needle (catalog no. TL0000; Thomas), a stab inoculation was made from a single colony for each strain into a medium-filled glass tube. The tubes were incubated at room temperature for 7 days before cultures were analyzed. Evidence of motility was assessed visually. If growth occurred only along the stab line, strains were considered nonmotile under these conditions; otherwise, strains were deemed motile. All results were confirmed via microscopy with liquid cultures in MB2216.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the U.S. Department of Energy (grant DE-SC0008743) to M.F.P. and E.J.A. J.-H.H. was partially supported by a fellowship from the Human Frontier Science Program.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00020-18.
REFERENCES
- 1.Futuyma D, Moreno G. 1988. The evolution of ecological specialization. Annu Rev Ecol Syst 19:207–233. doi: 10.1146/annurev.es.19.110188.001231. [DOI] [Google Scholar]
- 2.Shapiro BJ, David LA, Friedman J, Alm EJ. 2009. Looking for Darwin's footprints in the microbial world. Trends Microbiol 17:196–204. doi: 10.1016/j.tim.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Friedman J, Alm EJ, Shapiro BJ. 2013. Sympatric speciation: when is it possible in bacteria? PLoS One 8:e53539. doi: 10.1371/journal.pone.0053539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watkins ER, Maiden MC, Gupta S. 2016. Metabolic competition as a driver of bacterial population structure. Future Microbiol 11:1339–1357. doi: 10.2217/fmb-2016-0079. [DOI] [PubMed] [Google Scholar]
- 5.Thompson F, Iida T, Swings J. 2004. Biodiversity of vibrios. Microbiol Mol Biol Rev 68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson JR, Randa MA, Marcelino LA, Tomita-Mitchell A, Lim E, Polz MF. 2004. Diversity and dynamics of a north Atlantic coastal Vibrio community. Appl Environ Microbiol 70:4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takemura A, Chien D, Polz M. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bongrand C, Koch EJ, Moriano-Gutierrez S, Cordero OX, McFall-Ngai M, Polz MF, Ruby EG. 2016. A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J 10:2907–2917. doi: 10.1038/ismej.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erken M, Lutz C, McDougald D. 2015. Interactions of Vibrio spp. with zooplankton. Microbiol Spectr 3:VE-0003-2014. doi: 10.1128/microbiolspec.VE-0003-2014. [DOI] [PubMed] [Google Scholar]
- 10.Takemura AF, Corzett CH, Hussain F, Arevalo P, Datta M, Yu X, Le Roux F, Polz MF. 2017. Natural resource landscapes of a marine bacterium reveal distinct fitness-determining genes across the genome. Environ Microbiol 19:2422–2433. doi: 10.1111/1462-2920.13765. [DOI] [PubMed] [Google Scholar]
- 11.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. 2012. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335:308–313. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 12.Badur AH, Plutz MJ, Yalamanchili G, Jagtap SS, Schweder T, Unfried F, Markert S, Polz MF, Hehemann JH, Rao CV. 2017. Exploiting fine-scale genetic and physiological variation of closely related microbes to reveal unknown enzyme functions. J Biol Chem 292:13056–13067. doi: 10.1074/jbc.M117.787192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polz M, Hunt D, Preheim S, Weinreich D. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Phil Trans R Soc Lond B Biol Sci 361:2009–2021. doi: 10.1098/rstb.2006.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, Polz MF, Alm EJ. 2012. Population genomics of early events in the ecological differentiation of bacteria. Science 336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yawata Y, Cordero OX, Menolascina F, Hehemann JH, Polz MF, Stocker R. 2014. Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci U S A 111:5622–5627. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferenci T. 2016. Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol 24:209–223. doi: 10.1016/j.tim.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Hehemann JH, Arevalo P, Datta MS, Yu X, Corzett CH, Henschel A, Preheim SP, Timberlake S, Alm EJ, Polz MF. 2016. Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat Commun 7:12860. doi: 10.1038/ncomms12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaz Hidalgo R, Cleenwerck I, Balboa S, Prado S, De Vos P, Romalde JL. 2009. Vibrio breoganii sp. nov., a non-motile, alginolytic, marine bacterium within the Vibrio halioticoli clade. Int J Syst Evol Microbiol 59:1589–1594. doi: 10.1099/ijs.0.003434-0. [DOI] [PubMed] [Google Scholar]
- 19.Preheim SP, Boucher Y, Wildschutte H, David LA, Veneziano D, Alm EJ, Polz MF. 2011. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ Microbiol 13:265–275. doi: 10.1111/j.1462-2920.2010.02328.x. [DOI] [PubMed] [Google Scholar]
- 20.Iehata S, Valenzuela F, Riquelme C. 2015. Analysis of bacterial community and bacterial nutritional enzyme activity associated with the digestive tract of wild Chilean octopus (Octopus mimus Gould, 1852). Aquacult Res 46:861–873. doi: 10.1111/are.12240. [DOI] [Google Scholar]
- 21.Sawabe T, Oda Y, Shiomi Y, Ezura Y. 1995. Alginate degradation by bacteria isolated from the gut of sea-urchins and abalones. Microb Ecol 30:193–202. doi: 10.1007/BF00172574. [DOI] [PubMed] [Google Scholar]
- 22.Sawabe T, Sugimura I, Ohtsuka M, Nakano K, Tajima K, Ezura Y, Christen R. 1998. Vibrio halioticoli sp. nov., a non-motile alginolytic marine bacterium isolated from the gut of the abalone Haliotis discus hannai. Int J Syst Bacteriol 48:573–580. doi: 10.1099/00207713-48-2-573. [DOI] [PubMed] [Google Scholar]
- 23.Sawabe T, Setoguchi N, Inoue S, Tanaka R, Ootsubo M, Yoshimizu M, Ezura Y. 2003. Acetic acid production of Vibrio halioticoli from alginate: a possible role for establishment of abalone-V. halioticoli association. Aquaculture 219:671–679. doi: 10.1016/S0044-8486(02)00618-X. [DOI] [Google Scholar]
- 24.Sawabe T, Thompson F, Austin B, Swings J. 2006. The mutual partnership between Vibrio halioticoli and abalones, p 219–230. In Thompson FL, Austin B, Swings J (ed), The biology of vibrios. ASM Press, Washington, DC. [Google Scholar]
- 25.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombard V, Golaconda Ramulu H, Drula E, Coutinho P, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadam S, Tiwari B, O'Donnell C. 2015. Extraction, structure and biofunctional activities of laminarin from brown algae. Int J Food Sci Technol 50:24–31. doi: 10.1111/ijfs.12692. [DOI] [Google Scholar]
- 28.Mann AJ, Hahnke RL, Huang S, Werner J, Xing P, Barbeyron T, Huettel B, Stüber K, Reinhardt R, Harder J, Glöckner FO, Amann RI, Teeling H. 2013. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl Environ Microbiol 79:6813–6822. doi: 10.1128/AEM.01937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang K, Lin Y, Han Y, Jiao N. 2017. Characterization of potential polysaccharide utilization systems in the marine Bacteroidetes Gramella flava JLT2011 using a multi-omics approach. Front Microbiol 8:220. doi: 10.3389/fmicb.2017.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodd D, Cann IK. 2009. Enzymatic deconstruction of xylan for biofuel production. Glob Change Biol Bioenergy 1:2–17. doi: 10.1111/j.1757-1707.2009.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levasseur A, Drula E, Lombard V, Coutinho P, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabisch A, Otto A, König S, Becher D, Albrecht D, Schüler M, Teeling H, Amann RI, Schweder T. 2014. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J 8:1492–1502. doi: 10.1038/ismej.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen SG, Kim J, Guevarra RB, Lee JH, Kim E, Kim SI, Unno T. 2016. Laminarin favorably modulates gut microbiota in mice fed a high-fat diet. Food Funct 7:4193–4201. doi: 10.1039/C6FO00929H. [DOI] [PubMed] [Google Scholar]
- 34.Hunt D, Gevers D, Vahora N, Polz M. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl Environ Microbiol 74:44–51. doi: 10.1128/AEM.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruzzo C, Vezzulli L, Colwell R. 2008. Global impact of Vibrio cholerae interactions with chitin. Environ Microbiol 10:1400–1410. doi: 10.1111/j.1462-2920.2007.01559.x. [DOI] [PubMed] [Google Scholar]
- 36.Markov E, Kulikalova E, Urbanovich L, Vishnyakov V, Balakhonov S. 2015. Chitin and products of its hydrolysis in Vibrio cholerae ecology. Biochemistry (Mosc) 80:1109–1116. doi: 10.1134/S0006297915090023. [DOI] [PubMed] [Google Scholar]
- 37.Roach P, Depaoli-Roach A, Hurley T, Tagliabracci V. 2012. Glycogen and its metabolism: some new developments and old themes. Biochem J 441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Møller M, Henriksen A, Svensson B. 2016. Structure and function of alpha-glucan debranching enzymes. Cell Mol Life Sci 73:2619–2641. doi: 10.1007/s00018-016-2241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rioux L, Turgeon S, Beaulieu M. 2007. Characterization of polysaccharides extracted from brown seaweeds. Carbohydr Polym 69:530–537. doi: 10.1016/j.carbpol.2007.01.009. [DOI] [Google Scholar]
- 40.Belghit I, Rasinger JD, Heesch S, Biancarosa I, Liland N, Torstensen B, Waagbø R, Lock EJ, Bruckner CG. 2017. In-depth metabolic profiling of marine macroalgae confirms strong biochemical differences between brown, red and green algae. Algal Res 26:240–249. doi: 10.1016/j.algal.2017.08.001. [DOI] [Google Scholar]
- 41.Szabo G, Preheim SP, Kauffman KM, David LA, Shapiro J, Alm EJ, Polz MF. 2013. Reproducibility of Vibrionaceae population structure in coastal bacterioplankton. ISME J 7:509–519. doi: 10.1038/ismej.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris JM. 1993. The presence, nature, and role of gut microflora in aquatic invertebrates–óa synthesis. Microb Ecol 25:195–231. doi: 10.1007/BF00171889. [DOI] [PubMed] [Google Scholar]
- 43.Trabal N, Mazón-Suástegui JM, Vázquez-Juárez R, Asencio-Valle F, Morales-Bojórquez E, Romero J. 2012. Molecular analysis of bacterial microbiota associated with oysters (Crassostrea gigas and Crassostrea corteziensis) in different growth phases at two cultivation sites. Microb Ecol 64:555–569. doi: 10.1007/s00248-012-0039-5. [DOI] [PubMed] [Google Scholar]
- 44.Romalde J, Dieguez A, Lasa A, Balboa S. 2014. New Vibrio species associated to molluscan microbiota: a review. Front Microbiol 4:413. doi: 10.3389/fmicb.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moisander PH, Sexton AD, Daley MC. 2015. Stable associations masked by temporal variability in the marine copepod microbiome. PLoS One 10:e0138967. doi: 10.1371/journal.pone.0138967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruzzo C, Gallo G, Canesi L. 2005. Persistence of vibrios in marine bivalves: the role of interactions with haemolymph components. Environ Microbiol 7:761–772. doi: 10.1111/j.1462-2920.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- 47.Wendling CC, Batista FM, Wegner KM. 2014. Persistence, seasonal dynamics and pathogenic potential of Vibrio communities from Pacific oyster hemolymph. PLoS One 9:e94256. doi: 10.1371/journal.pone.0094256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka R, Sawabe T, Yoshimizu M, Ezura Y. 2002. Distribution of Vibrio halioticoli around an abalone-farming center in Japan. Microbes Environ 17:6–9. doi: 10.1264/jsme2.2002.6. [DOI] [Google Scholar]
- 49.Preheim S, Timberlake S, Polz M. 2011. Merging taxonomy with ecological population prediction in a case study of Vibrionaceae. Appl Environ Microbiol 77:7195–7206. doi: 10.1128/AEM.00665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datta M, Sliwerska E, Gore J, Polz M, Cordero O. 2016. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat Commun 7:11965. doi: 10.1038/ncomms11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tibbles B, Rawlings D. 1994. Characterization of nitrogen-fixing bacteria from a temperate salt-marsh lagoon, including isolates that produce ethane from acetylene. Microb Ecol 27:65–80. doi: 10.1007/BF00170115. [DOI] [PubMed] [Google Scholar]
- 52.Gómez Ramírez M, Rojas Avelizapa LI, Rojas Avelizapa NG, Cruz Camarillo R. 2004. Colloidal chitin stained with Remazol Brilliant Blue R, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J Microbiol Methods 56:213–219. doi: 10.1016/j.mimet.2003.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.