Formation of membrane vesicles is ubiquitous among bacteria. These vesicles are involved in protein and DNA transfer and offer new approaches for vaccination. Gram-negative bacteria use hydrophobic signaling molecules, among others, for cell-cell communication; however, due to their hydrophobic character, it is unclear how these molecules are disseminated between bacterial cells. Here, we show that the marine pathogen Vibrio harveyi packages one of its QS molecules, the long-chain ketone CAI-1, into outer membrane vesicles (OMVs). Isolated CAI-1-containing vesicles trigger a QS phenotype in CAI-1 nonproducing V. harveyi and also in Vibrio cholerae cells. Packaging of CAI-1 into OMVs not only solubilizes, stabilizes, and concentrates this class of molecules, but facilitate their distribution between bacteria that live in aqueous environments.
KEYWORDS: Vibrio cholerae, autoinducer, cell-cell communication, quorum sensing
ABSTRACT
Many bacteria use extracellular signaling molecules to coordinate group behavior, a process referred to as quorum sensing (QS). However, some QS molecules are hydrophobic in character and are probably unable to diffuse across the bacterial cell envelope. How these molecules are disseminated between bacterial cells within a population is not yet fully understood. Here, we show that the marine pathogen Vibrio harveyi packages the hydrophobic QS molecule CAI-1, a long-chain amino ketone, into outer membrane vesicles. Electron micrographs indicate that outer membrane vesicles of variable size are predominantly produced and released into the surroundings during the stationary phase of V. harveyi, which correlates with the timing of CAI-1-dependent signaling. The large vesicles (diameter, <55 nm) can trigger a QS phenotype in CAI-1-nonproducing V. harveyi and Vibrio cholerae cells. Packaging of CAI-1 into outer membrane vesicles might stabilize the molecule in aqueous environments and facilitate its distribution over distances.
IMPORTANCE Formation of membrane vesicles is ubiquitous among bacteria. These vesicles are involved in protein and DNA transfer and offer new approaches for vaccination. Gram-negative bacteria use hydrophobic signaling molecules, among others, for cell-cell communication; however, due to their hydrophobic character, it is unclear how these molecules are disseminated between bacterial cells. Here, we show that the marine pathogen Vibrio harveyi packages one of its QS molecules, the long-chain ketone CAI-1, into outer membrane vesicles (OMVs). Isolated CAI-1-containing vesicles trigger a QS phenotype in CAI-1 nonproducing V. harveyi and also in Vibrio cholerae cells. Packaging of CAI-1 into OMVs not only solubilizes, stabilizes, and concentrates this class of molecules, but facilitate their distribution between bacteria that live in aqueous environments.
INTRODUCTION
All living organisms employ various types of membrane vesicles to disseminate products into the environment. In particular, it has been known for decades that Gram-negative bacteria naturally shed 20- to 300-nm spherical vesicles from their outer membrane. Formation of outer membrane vesicles (OMVs) is ubiquitous among bacteria, and occurs in liquid and solid cultures, as well as in biofilms (1, 2). Recently, extracellular vesicles (EVs) emanating from organisms with cell walls, e.g., Gram-positive bacteria, mycobacteria, and fungi, have also been described (3). The surface of OMVs is thought to reflect the composition of the bacterial outer membrane and is therefore primarily comprised of phospholipids, outer membrane proteins, and lipopolysaccharides. The lumen of an OMV contains mostly periplasmic components, which are trapped within the vesicle during the release process. Surprisingly, OMVs also include proteins derived from the inner membrane, together with cytoplasmic proteins, chromosomal, plasmid, and viral DNA, RNA, ions, and metabolites (1, 4, 5). The biological function of these vesicles has not been fully elucidated, but it is known that OMVs are involved in protein and DNA transfer, as well as in signaling between bacteria (6). Furthermore, OMVs can also have ecological functions, as they are necessary for survival in particular environments (7). Additionally, in many pathogenic bacteria, OMVs have been shown to act as vehicles for long-distance delivery of toxins and effector proteins to host cells. For example, the main virulence factor in Vibrio cholerae, the cholera toxin, and colonization-promoting factors are associated with OMVs, which deliver them to the intestinal epithelial cells of the host (8, 9). Thereby, OMVs might be promising platforms to develop novel vaccines (10). Thus, OMVs enable the transport of toxins and effectors into host cells without any need for direct contact between pathogenic bacteria and their target host cells. Moreover, OMVs provide a means of stabilizing, concentrating, and protecting bacterial molecules with specific properties until they make contact with their cognate target receptors.
Many bacterial species use low-molecular-weight molecules, such as N-acyl homoserine lactones (AHLs), for cell-cell communication, in a process referred to as quorum sensing (QS) (11). In general, QS molecules are synthesized and released into the environment. They accumulate in a cell-density-dependent manner and are recognized by specific receptors, which may be located either in the inner membrane or the cytoplasm (12). Certain signaling molecules are highly hydrophobic, but the cell envelope of a Gram-negative bacterium acts as an efficient barrier to the diffusion of hydrophobic molecules, due to the presence of the polar lipopolysaccharide layer on the exterior of the cell (13). Hence, it is not clear how they make their way through an aqueous environment to nearby cells and then cross the bacterial outer membrane in order to bind to their cognate receptors in the inner membrane or the cytoplasm. Structurally diverse classes of QS molecules that regulate various phenotypes in Gram-negative bacteria have been identified over the past few decades (14). Several bacterial species use complex QS networks, with different types of signaling molecules which are sensed by specific receptors, for fine-tuning the activation of key phenotypes, such as virulence or biofilm formation (15, 16). Short-chain AHLs are thought to diffuse freely across the bacterial cell envelope, whereas long-chain AHLs are assumed to require transport mechanisms across the cell envelope, owing to their length and inherent hydrophobicity (17, 18). The length and degree of substitution of the N-acyl side chain therefore determine whether an AHL is freely diffusible or requires active export and import mechanisms (17).
Interestingly, until now, few examples of signaling molecules associated with OMVs have been described. The coral-associated bacterium Vibrio shilonii produces OMVs that contain alkaline phosphatase, lipase, and chitinase, as well as AHLs (19). Moreover, the long-chain AHL [C16-N-(hexadecanoyl)-l-homoserine lactone], used for cell-cell communication in the soil bacterium Paracoccus denitrificans PD1222, is mainly released via membrane vesicles (20). Furthermore, the opportunistic human pathogen Pseudomonas aeruginosa packages most of the Pseudomonas quinolone signal (PQS) into OMVs, and a type VI secretion effector recruits PQS-containing OMVs for iron acquisition within the population (4, 21). However, how hydrophobic QS molecules of the CAI-1 type—typically long-chain ketones—cross the bacterial envelope (22) and reach other cells in an aqueous environment is not yet understood.
CAI-1, the predominant QS molecule of the human-pathogenic bacterium V. cholerae, is produced by all Vibrio spp., albeit with different acyl chain lengths and modifications (14). Besides CAI-1 [(Z)-3-aminoundec-2-en-4-one, = Ea-C8-CAI-1], Vibrio harveyi strain BAA-1116 (recently reclassified as Vibrio campbellii [23]) synthesizes two other signaling molecules—HAI-1, the N-3-(hydroxybutyryl)-homoserine lactone, and autoinducer 2 (AI-2), a furanosyl borate diester (22, 24, 25). The use of AI-2 is widespread, and it is thought to be an interspecies signaling molecule (26). In contrast, HAI-1 is specific to V. harveyi and its close relatives (27). In V. harveyi, peak production of these three AIs occurs in different growth phases (28). While AI-2 is synthesized during early exponential growth, HAI-1 and CAI-1 are undetectable prior to the late-exponential growth phase. Furthermore, each QS molecule is perceived by a specific membrane-integrated hybrid sensor kinase in V. harveyi. HAI-1 is sensed by LuxN, AI-2 by LuxQ in combination with the periplasmic binding-protein LuxP, and CAI-1 by CqsS (15, 25, 29, 30). These hybrid sensor kinases channel the information into a shared regulatory pathway. Briefly, at low cell densities and correspondingly low signaling molecule concentrations, the sensors act as kinases and maintain the QS phenotypes in an off state. At high cell densities, upon perception of the cognate QS molecule, the kinase activities are inhibited, and genes whose protein products mediate various phenotypes—such as luminescence, biofilm formation, or proteolysis—are induced (31–33). Additionally, genes for type III secretion and siderophore production are repressed (15, 34).
While detailed knowledge of signal synthesis and integration in the QS cascade of HAI-1, AI-2, and CAI-1 in V. harveyi is now available, most studies have disregarded the potential impact of the physicochemical properties of these signaling molecules on their dissemination in aqueous media. The partition coefficient (P) defines the distribution of a molecule between a lipophilic and an aqueous phase (the logarithm of the ratio is log P) and, based on this criterion, HAI-1 and AI-2 are both hydrophilic (with log P values of −0.94 and −1.25, respectively; for comparison, methanol has a log P of −0.8). CAI-1 has a high lipophilicity (log P of 3.05, which is comparable to that of dichlorobenzene), so it readily partitions into a lipid bilayer; however, once in the bilayer, it might not partition out again, and would thus be unable to cross the polar lipopolysaccharide layer on the outside of the cell (13, 35). Since CAI-1 is known to function as a signaling molecule that influences the phenotypes of remote single cells within the population, it must somehow be conveyed through the aqueous environment. To obtain deeper insight into the trafficking of CAI-1, we investigated the association of CAI-1 with OMVs. Our data indicate that, when grown in a complex, nutrient-rich medium, V. harveyi produces OMVs in the late stationary phase. Moreover, the signaling molecule CAI-1 is packaged into OMVs that are recognized by nonproducing V. harveyi cells and that activate their QS cascade.
RESULTS
V. harveyi naturally releases OMVs in the late stationary growth phase.
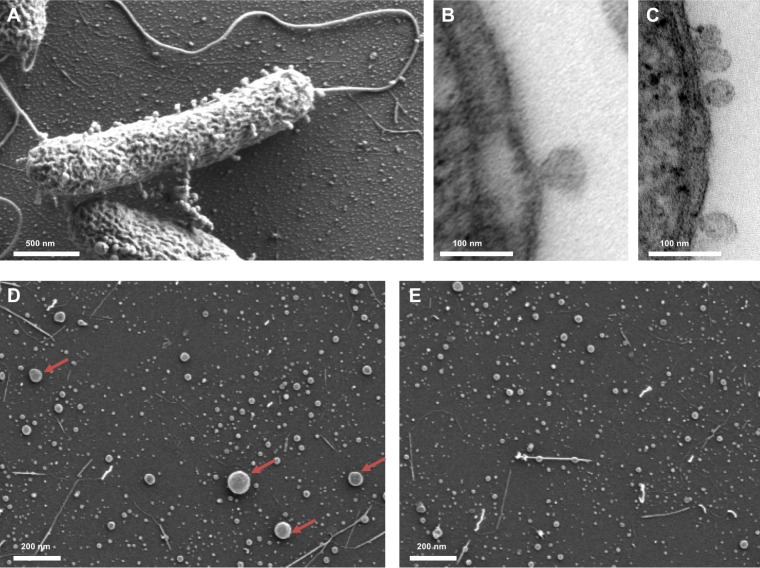
V. harveyi cells cultivated in complex Luria marine (LM) medium naturally produce OMVs, which are visible on the cell surface and in the surrounding medium in stationary phase (Fig. 1A). Furthermore, transmission electron microscopy (TEM) images of ultrathin sections revealed that the OMVs of V. harveyi are indeed derived from the outer membrane of the cells (Fig. 1B and C). The OMVs produced by V. harveyi vary in size from 20 nm to 260 nm. The majority of OMVs that are shed are small, and only 7.6% of them were found to be larger than 55 nm (Fig. 1D and E).
FIG 1.
V. harveyi naturally produces and releases OMVs. (A) Scanning electron micrograph of a whole cell of V. harveyi with attached OMVs. (B, C) Transmission electron micrographs of ultrathin sections of V. harveyi cells showing OMVs formed from the outer membrane. (D) Scanning electron micrograph of the OMV-containing 0.45-μm filtrate obtained from a culture of strain MR17. Isolated OMVs vary widely in size. Arrows indicate examples of OMVs with a diameter of >55 nm. (E) Scanning electron micrograph of the OMV-containing 0.22-μm filtrate obtained from a culture of strain MR17.
CAI-1 is found in V. harveyi OMVs.
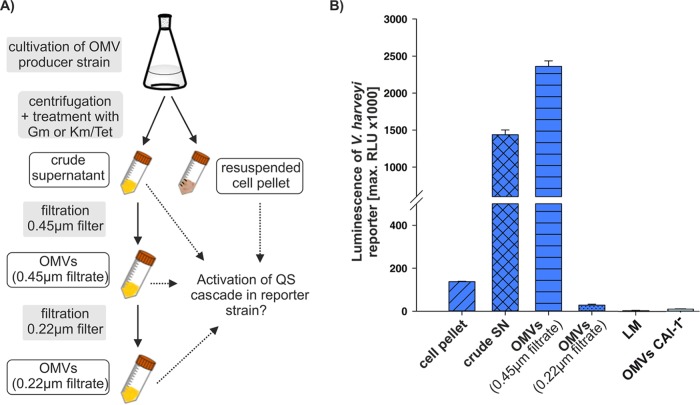
In order to study the impact of OMV release on activation of the QS cascade in V. harveyi, OMVs were harvested in the late stationary growth phase from V. harveyi strain MR17 cells, which synthesize only CAI-1. Cells were pelleted by centrifugation (cell pellet), and the supernatant (SN) was subsequently treated with gentamicin for 3 h at 30°C to inhibit growth of the remaining cells. Afterwards, the supernatant was filtered through a 0.45-μm polyvinylidene difluoride (PVDF) filter to obtain a cell-free fraction that contained OMVs (0.45-μm filtrate). Part of the OMV fraction was then passed through a 0.22-μm PVDF filter, yielding a 0.22-μm filtrate fraction, which mainly contained small OMVs (Fig. 1E). All fractions (OMVs, 0.45-μm and 0.22-μm filtrates) were tested for remaining bacteria by inoculating LM medium, but growth was never observed after 24 h at 30°C. The fractions were then incubated with the gentamicin-resistant V. harveyi reporter strain NL20 (cqsS+ ΔluxN ΔluxPQ), which lacks all three QS synthases (is AI−) and is only able to sense and respond to CAI-1, and luminescence was assessed as the QS readout.
None of the fractions affected the growth of the reporter strain (see Fig. S1 in the supplemental material). Supernatant and OMV-containing 0.45-μm filtrate obtained from V. harveyi strain MR17, which synthesizes only CAI-1, led to QS activation and therefore to luminescence production in the reporter strain NL20, which expresses the CqsS receptor (Fig. 2B). When this OMV-containing suspension was then passed through a 0.22-μm PVDF filter, the resulting filtrate hardly contained enough CAI-1 to induce luminescence production (Fig. 2B). Similarly, OMVs isolated from a CAI-1-nonproducing strain (MR16, cqsA mutant) showed no activation of the QS cascade in the reporter strain NL20.
FIG 2.
OMV-associated CAI-1 activates QS cascade in V. harveyi reporter strain at population level. (A) Schematic depiction of OMV harvesting process by fractionating of the V. harveyi supernatant. (B) Each fraction from MR17 cells (ΔluxM ΔluxS) was tested for QS activation in the reporter strains NL20 (ratio of OMVs to reporter strain culture, 1:6.7) by measuring luminescence as the readout. The reporter strain NL20 senses only CAI-1 and strain MR17 synthesizes only CAI-1. As a control, the OMV-containing 0.45-μm filtrate of the non-CAI-1 producing strain MR16 (ΔcqsA ΔluxM) was tested for QS activation in the reporter strains NL20. Error bars represent the standard deviations of data from three different experiments. RLU, relative light units, expressed in counts per second per milliliter per OD600 unit. SN, culture supernatant of MR17; OMVs (0.45μm filtrate), filtrate of SN of MR17 cells through 0.45-μm filter; OMVs (0.22μm filtrate), filtrate of SN of MR17 cells through an additional 0.22-μm filter; LM, cell-free culture medium as control; OMVs CAI-1−, filtrate of SN of MR16 cells through 0.45-μm filter. Time courses of the growth and luminescence production of the reporter strain NL20 are shown in Fig. S1.
The V. harveyi MR17 cell pellet also contained only a small amount of CAI-1, as indicated by 15-fold lower luminescence production in comparison to that of the OMV fraction of the 0.45-μm filtrate (Fig. 2B). It is important to note that large OMVs (diameter, >55 nm) were only observed in the 0.45-μm filtrate (Fig. 1D). These were hardly found in the 0.22-μm filtrate but were retained on the 0.22-μm PVDF filter (Fig. 1E and Fig. S2 in the supplemental material).
Serial dilutions of OMV suspension (0.45-μm filtrate) of strain MR17 cells show a linear increase of the reporter strain activity at low concentrations. At higher concentrations of the OMV-containing 0.45-μm filtrate (e.g., 15 to 21%), no further increase in QS activation was found, demonstrating the saturation of the reporter strain (see Fig. S3 in the supplemental material).
CAI-1-containing OMVs activate QS in nonproducing V. harveyi at a single-cell level.
We previously demonstrated that the QS cascade of V. harveyi is inherently noisy, which is reflected in a heterogeneous output that becomes homogenous when the available sensors are saturated with their cognate AIs (36). To study QS activation via OMVs at a single-cell level, we therefore used the reporter strain NL20, which contains a chromosomally integrated fusion between the luciferase promoter (PluxC) and mCherry at the attTn7 site (36). Fluorescence of this reporter was induced after exposure of cells to either CAI-1-containing supernatant or OMVs from strain MR17 (csqA+). Furthermore, the luxC promoter was activated homogeneously by the OMV-containing 0.45-μm filtrate of strain MR17 and its supernatant (Fig. 3). These results indicate that OMVs contain sufficient amounts of CAI-1 to generate a homogenous response in this reporter strain.
FIG 3.
Homogeneous OMV-mediated CAI-1 response in V. harveyi at the single-cell level. The V. harveyi reporter strain NL20 (AI− cqsS+ ΔluxN ΔluxPQ PluxC-mCherry) was grown to an optical density at 600 nm (OD600) of 0.5, supernatant or OMVs were added as indicated (also see text), and cells were analyzed for expression of the PluxC-mCherry reporter fusion after further growth for 4.5 h (left panel; PH, phase-contrast images; mCherry, fluorescence images; crude SN, culture supernatant of MR17 cells; OMVs (0.45μm filtrate), filtrate of SN of MR17 cells through a 0.45-μm filter; OMVs (0.22μm filtrate), filtrate of SN of MR17 cells through an additional 0.22-μm filter; OMVs CAI-1−, filtrate of SN of MR16 cells through a 0.45-μm filter.) Average fluorescence of 1,000 cells quantified by ImageJ (arbitrary unit [AU]) is presented in the graphs on the right. Bars, 5 μm.
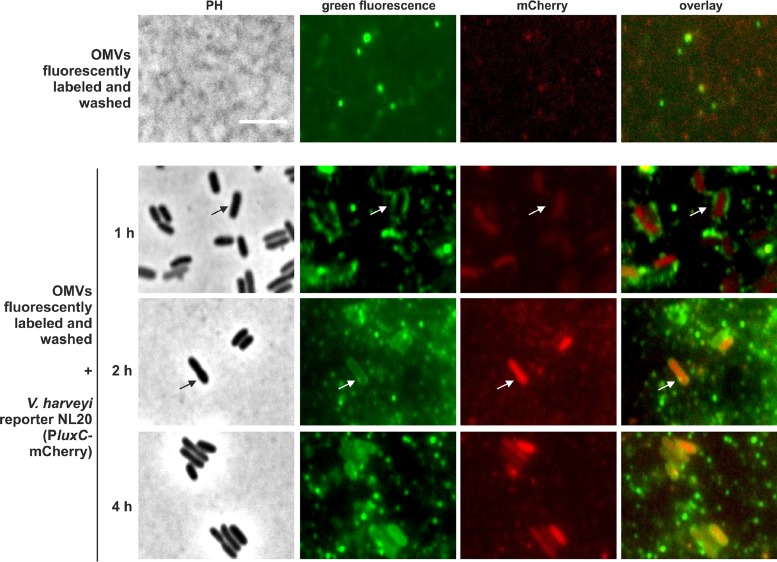
In addition, we fluorescently stained the membrane of the CAI-1-containing OMV fraction (0.45-μm filtrate) of strain MR17 and, after thorough washing, we added these stained OMVs to the reporter strain V. harveyi NL20 PluxC-mCherry, which reports CAI-1-dependent activation of the QS signaling cascade at a single-cell level. By using microscopy, we first saw the attachment of the green fluorescent OMVs (1 h) and subsequent green fluorescence of the membranes of the unlabeled cells (within 2 h). Eventually, the reporter was activated and the cells exhibited mCherry fluorescence and thus activation of the QS cascade (2 to 4 h) (Fig. 4).
FIG 4.
CAI-1 is delivered via OMVs to nonproducing cells and triggers intracellular QS activation. The membrane of CAI-1-containing OMVs (OMV-producing strain MR17 [ΔluxM ΔluxS]) was fluorescently stained, and green fluorescent OMVs were detected by the microscope (top panel). These OMVs were incubated with the nonfluorescent reporter strain V. harveyi NL20 (AI− cqsS+ ΔluxN ΔluxPQ PluxC-mCherry). At the indicated times, cells were analyzed by microscopy for green fluorescence and expression of the PluxC-mCherry reporter fusion (bottom panels). PH, phase-contrast images; green fluorescence, green fluorescence images; mCherry, red fluorescence images. Bar, 5 μm.
Medium composition and growth phase affect the delivery of CAI-1 via OMVs.
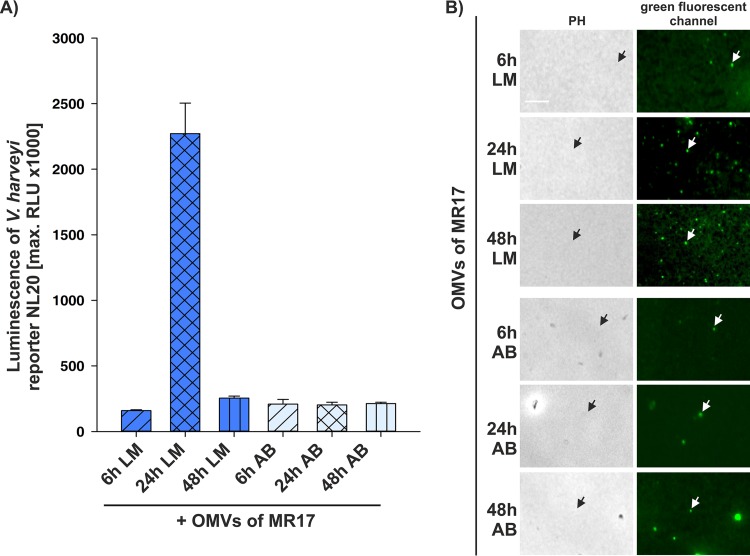
As previous studies have shown that CAI-1 activity peaks in the stationary phase when cells are grown in complex LM medium (15, 28), we tested the influence of different media on the delivery of CAI-1 into OMVs. OMVs were harvested at different time points from V. harveyi MR17 cells grown either in the complex, rich LM medium or in defined, minimal autoinducer bioassay (AB) medium. Maximal activation of CAI-1-dependent QS, as measured by the high luminescence of the reporter strain NL20, was observed with OMVs harvested after 24 h when strain MR17 was cultured in LM medium (Fig. 5A). OMVs harvested at earlier or later time points or after growth in AB medium were far less active (Fig. 5A). According to these results, more CAI-1 is produced by cells grown in rich LM medium than in AB medium, and its level peaks at 24 h. To visualize and quantitatively determine the OMVs produced by V. harveyi MR17, we stained them with a fluorescent lipid-specific dye. Staining of the lipids of OMVs provides a better measure of OMV yield, as the phospholipid content is less influenced by the growth phase than the protein content (37). OMV abundance itself was found to be high in stationary phase, i.e., at 24 h and 48 h, and higher overall after cultivation in LM than in AB medium (Fig. 5B). These results reveal that CAI-1 production coincides with OMV release. In parallel, we did not find a stimulation of OMV production after external addition of CAI-1 (see Fig. S4 in the supplemental material).
FIG 5.
Delivery of CAI-1 into OMVs depends on growth conditions. (A) The NL20 reporter strain, which responds only to CAI-1, was incubated with OMVs harvested from V. harveyi MR17 (0.45-μm filtrate). OMVs were harvested after 6, 24, and 48 h of growth in LM or AB medium. QS activation in the reporter strain NL20 was monitored by measuring luminescence. Error bars represent the standard deviations of data from three different experiments. RLU, relative light units, expressed in counts per second per milliliter per OD600 unit. (B) Images of fluorescently labeled OMVs of MR17 cells harvested after 6, 24, and 48 h of growth in LM or AB medium. Isolated OMVs were stained with the green fluorescent MitoTracker FM dye, which stains lipids, and imaged using a green fluorescent channel. Each white arrow indicates a single fluorescent OMV, and each black arrow shows its respective position in the phase-contrast image. PH, phase-contrast images. Bar, 5 μm.
Interspecies communication.
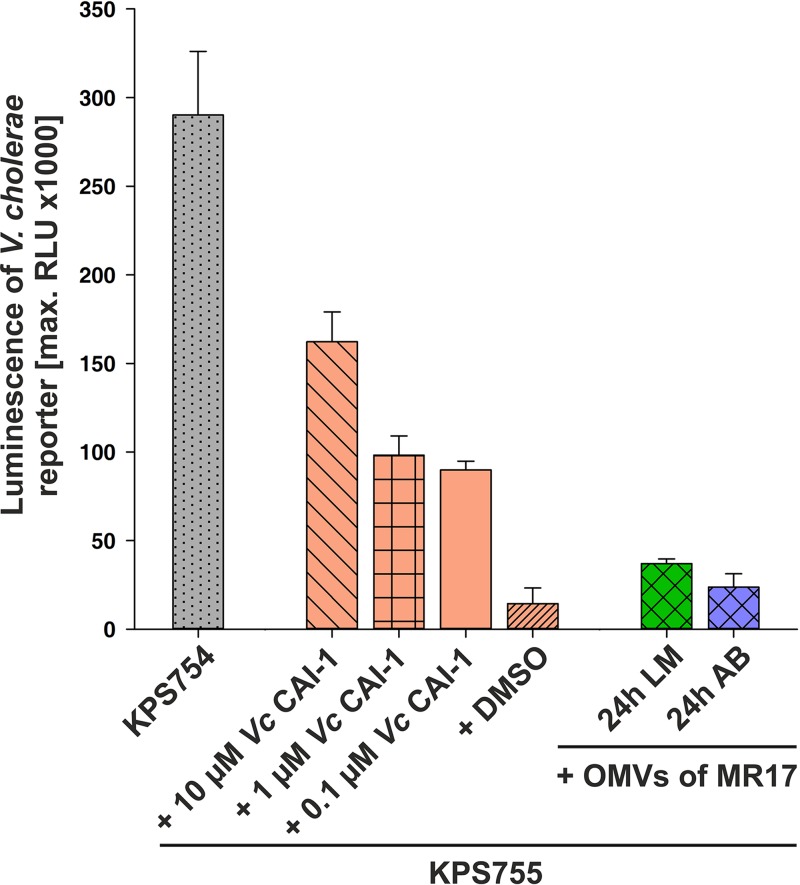
Previously, it was demonstrated that CAI-1 (Ea-C8-CAI-1) produced by V. harveyi is recognized as a QS signal by V. cholerae (15). We therefore tested the ability of OMV-associated CAI-1 from V. harveyi to activate QS in a V. cholerae reporter strain. Although external addition of the V. cholerae-specific CAI-1 [(S)-3-hydroxytridecan-4-one] or its endogenous synthesis induced markedly higher luminescence production in the V. cholerae reporter, OMVs isolated from V. harveyi MR17 grown in LM medium were indeed able to detectably activate QS-dependent luminescence reporter activity in V. cholerae (Fig. 6).
FIG 6.
CAI-1 in OMVs from V. harveyi activates QS cascade in V. cholerae. The V. cholerae reporter strains KPS754 (ΔluxS) and KPS755 (ΔcqsA ΔluxS) carry the pBB1 plasmid, which harbors the V. harveyi luxCDABE operon, enabling V. cholerae to produce luminescence. The V. cholerae reporter strain KPS755 was incubated with different concentrations of V. cholerae-specific CAI-1 (Vc CAI-1) dissolved in DMSO and with different OMV-containing fractions harvested from V. harveyi MR17 cells grown in either LM or AB medium for 24 h. QS activation in these reporter strains was followed by monitoring of luminescence production. Error bars represent the standard deviations of data from three different experiments. RLU, relative light units expressed in counts per second per milliliter per OD600 unit.
DISCUSSION
Production of membrane vesicles is a ubiquitous process among bacterial species, and it occurs in biofilms and in liquid and solid culture (1, 2). Here, we have demonstrated that V. harveyi cells naturally release outer membrane vesicles (OMVs) into their environment during stationary phase (Fig. 1). Furthermore, CAI-1 was found to be associated with OMVs, leading to activation of the QS cascade in a CAI-1-nonproducing strain (Fig. 2B). These OMVs can activate the QS cascade homogeneously in all CAI-1-nonproducing cells (Fig. 3). Thus, V. harveyi OMVs can be used as a vehicle for trafficking of CAI-1 between cells (Fig. 3 and 4). Moreover, the amount of CAI-1 in OMVs was sufficient to saturate the corresponding QS-receptor (see Fig. S3 in the supplemental material), which results in a homogeneous QS response in V. harveyi cells (36).
Growth conditions undoubtedly have an influence on the numbers, size, content, and lipid and protein compositions of OMVs produced. Thereby, the released OMVs can contain different properties that differently influence neighboring cells (2). Likewise, the amount of CAI-1 associated with V. harveyi OMVs was found to be dependent on growth phase and medium composition (Fig. 5). V. harveyi cells released OMVs predominantly in rich medium and at stationary phase (Fig. 5B). Moreover, the CAI-1 content of V. harveyi OMVs varied as well, and it peaked at early stationary phase in rich medium (Fig. 5A). The Pseudomonas quinolone signal (PQS) is required for membrane vesicle formation and seems to dictate OMV biogenesis (38, 39). In contrast, exogenous CAI-1 itself does not stimulate OMV formation in V. harveyi (Fig. S4).
Previously, OMV production was shown to be influenced by antibiotic treatment, preferentially by envelope targeting or by DNA-damaging antibiotics (40). For examples, in P. aeruginosa, OMV formation is increased upon treatment with ciprofloxacin, an antibiotic which leads to DNA damage and which therefore results in the activation of the SOS response (41). Since in our case filtration (0.22-μm filter) removed not only all cells but also the large, CAI-1-containing OMVs, we used gentamicin to kill all remaining cells (Fig. 2A). Although gentamicin inhibits protein synthesis by binding to the bacterial 30S ribosomal subunit (42), it cannot be completely ruled out that this treatment might cause additional release of OMVs of the few remaining cells.
The CAI-1-specific receptor CqsS is located in the inner membrane, and the incoming lipophilic CAI-1 molecule probably interacts with intramembrane segments of the receptor dimer, as extracellular loops for ligand binding are absent (43, 44). Conversely, due to its inherent lipophilic character, CAI-1 is unlikely to diffuse across the polar lipopolysaccharide layer (13) on the outside of the sending cell. It probably remains trapped in the outer and/or inner membrane, which would potentially make it available for incorporation into OMVs. In this form, CAI-1 can be easily transported between cells and can activate the QS cascade in neighboring cells (Fig. 7). In rich medium, external CAI-1 is first detected in the stationary phase (28), which coincides with the onset of OMV formation. OMVs produced by V. harveyi range from 20 nm to 260 nm in diameter (Fig. 1), similar in size to those released by many other bacteria (2). The majority of OMVs that are shed by V. harveyi are small (diameter, <50 nm); however, about 10% of them are large (diameter, 55 to 260 nm), and only those were able to activate CAI-1 dependent QS. It is still unclear whether only the large OMVs are able to fuse with the surface of neighboring cells.
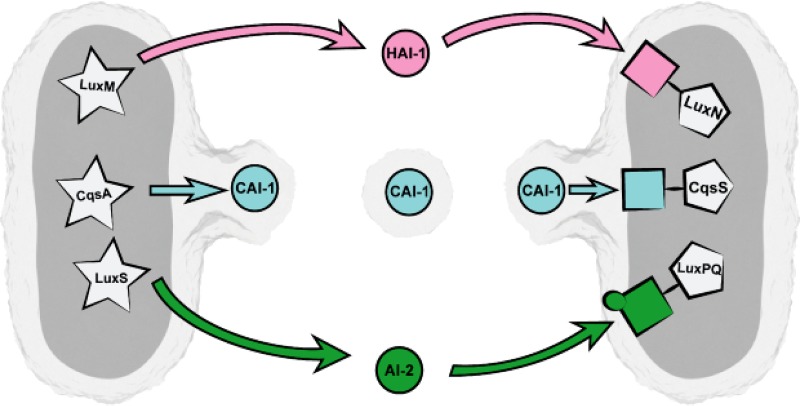
FIG 7.
Model of trafficking of CAI-1 via OMVs between V. harveyi cells. Due to its high lipophilicity, CAI-1 remains trapped in the outer membrane of V. harveyi cells and gets incorporated into OMVs. In this form, CAI-1 can be easily transported between cells and can activate the QS cascade in neighboring cells upon recognition by its specific receptor CqsS. The other QS signaling molecules of V. harveyi, AI-1 and HAI-1, are supposed to be freely diffusible across the cell envelope and are sensed by their specific receptors LuxPQ and LuxN, respectively, which are located in the inner membrane. The AIs HAI-1, CAI-1, and AI-2 are depicted with circles. AI synthases are depicted in stars in the cytoplasm. The AI-specific receptors LuxN, CqsS, and LuxPQ are depicted by rectangles in pink, blue, or green, respectively, connected to pentagons. The image was created by Christoph Hohmann (Nanosystems Initiative Munich).
CAI-1 is a Vibrio-specific, interspecies signaling molecule used by several Vibrio species, although it mediates opposing effects on their QS cascades. Interestingly, V. harveyi CAI-1 can control gene expression in V. cholerae and vice versa (15). Indeed, OMVs released by V. harveyi MR17, which synthesizes only CAI-1, are able to activate QS-dependent luminescence reporter activity in V. cholerae (Fig. 6). Recently, it was shown that OMVs produced by P. denitrificans PD1222 fuse, with various affinities, to different bacterial species (20).
Overall, delivery of CAI-1 via OMVs might provide three major advantages, namely, (i) solubilization and stabilization of CAI-1 in an aqueous environment, (ii) increasing the concentration of CAI-1, and (iii) the unhindered passage of CAI-1 across the polar lipopolysaccharide layer of both the producing and targeted cells.
MATERIALS AND METHODS
Bacterial strains.
The V. harveyi strains MR17 (ΔluxM ΔluxS), MR13 (ΔcqsA), MR16 (ΔcqsA ΔluxM), and NL20 (ΔluxM ΔluxS ΔcqsA ΔluxN ΔluxQ PluxC-mCherry) (36) were cultivated in Luria marine (LM) medium (20 g/liter NaCl, 10 g/liter tryptone, and 5 g/liter yeast extract) or autoinducer bioassay (AB) medium (45) and incubated aerobically on a rotary shaker at 30°C (36). Media used for the strain NL20 were supplemented with 20 μg/ml gentamicin.
The plasmid pBAD-cqsA, used to enable synthesis of CAI-1, and the control plasmid pCMW-1 (46) were introduced into the V. cholerae strain BH-1575 (ΔcqsA ΔluxS) by conjugation. The resulting strains were additionally conjugated with the pBB1 plasmid, which carries the V. harveyi luxCDABE operon (47), in order to use luminescence production as a readout for QS. These V. cholerae reporter strains, named KPS754 (BH-1575/pBAD-cqsA/pBB1) and KPS755 (BH-1575/pCMW-1/pBB1), were grown at 37°C in LB (5 g/liter NaCl, 10 g/liter tryptone, and 5 g/liter yeast extract) supplemented with 50 μg/ml kanamycin and 5 μg/ml tetracycline. For construction of pBAD-cqsA (pKP-420), cqsA was amplified from V. cholerae genomic DNA by PCR, using the primers KPO-0776P (5′-GTCAGCTGGCGTTAAATTTTT-3′) and KPO-0777 (5′-GTTTTTGGTACCCTTTAGGAATAACGTTTAGCAG-3′), and cloned into the KpnI site in pKP-331 (48). Correct insertion was verified by sequence analysis.
Preparation of OMVs.
For OMV preparation, an overnight culture of V. harveyi MR17, MR16, or MR13 was adjusted to an optical density at 600 nm (OD600) of 0.05 in LM or AB medium and incubated at 30°C for 6, 24 or 48 h with shaking at 250 rpm (adapted from reference 49). The V. harveyi strain MR17 produces only CAI-1, whereas the strains MR13 and MR16 do not produce CAI-1. To isolate OMVs from each of these strains, after growing the cells, they were first pelleted at 4°C by centrifugation at 5,000 rpm for 1 h. The supernatant was then treated with 60 μg/ml gentamicin at 30°C for 3 h to inhibit the growth of any remaining cells, which are gentamicin sensitive (in contrast to the gentamicin-resistant reporter strain). This supernatant was then filtered through a 0.45-μm filter (PVDF membrane; Merck Millipore) to obtain a cell-free suspension that contained OMVs (0.45-μm filtrate). This OMV-containing suspension was additionally fractionated via filtration through a 0.22-μm filter (PVDF membrane; Merck Millipore). Equal volumes were maintained for all collected fractions during the preparation of OMVs (see Fig. 2A for an overview).
OMV-containing suspensions prepared for the V. cholerae reporter were treated similarly, except that 150 μg/ml kanamycin and 15 μg/ml tetracycline were added instead of gentamicin.
In order to follow the impact of CAI-1 on OMV formation, V. harveyi strain MR13, which does not synthesize CAI-1, was cultivated in LM at 30°C for 24 h with shaking at 250 rpm and then incubated for 1 and 2 h with externally supplied (10 μM) V. cholerae-specific CAI-1 [(S)-3-hydroxytridecan-4-one] (44). OMVs were then harvested as described above.
Labeling of OMVs.
Isolated OMVs were stained with the MitoTracker green FM dye (Thermo Fisher), which is nonfluorescent in aqueous solution but becomes fluorescent in the presence of phospholipids. Staining with this marker provides a better measure of OMV yield, as the phospholipid content is less influenced by the growth phase (37). A stock solution (1 mM) of the dye was prepared in dimethyl sulfoxide (DMSO), and OMVs were incubated with 0.5 μM of MitoTracker green FM for 40 min at 30°C and washed with phosphate-buffered saline (PBS; pH 7.4). After centrifugation at 35,000 rpm at 4°C for 1 h, the OMV-containing pellet was resuspended in the same volume in 1× PBS and spotted on 1% (wt/vol) agarose pads before microscopy. Images were taken on a Leica DMi8 microscope with a Leica DFC365 FX camera. An excitation wavelength of 485 nm and a 510-nm emission filter with a 75-nm bandwidth were used for GFP fluorescence. In order to exclude the presence of adventitiously fluorescent particles, OMV samples were additionally analyzed for red fluorescence using an excitation wavelength of 546 nm and a 605-nm emission filter with a 75-nm bandwidth (data not shown).
Bioluminescence assay and fluorescence microscopy.
In order to monitor the impact of OMVs on the delivery of CAI-1 produced by V. harveyi, the reporter strain NL20, which expresses the cognate receptor for CAI-1 and therefore can recognize only CAI-1, was used. NL20 additionally caries a chromosomally integrated fusion between the luciferase promoter (PluxC) and mCherry (at the attTn7 site), which allows one to monitor QS activation at the single-cell level in terms of luminescence production (relative light units [RLU]) (36). The V. harveyi reporter strain NL20 was grown to an OD600 of 0.5 in LM medium, and then OMVs and the other fractions were added to a final concentration of 15%. Bioluminescence and growth were determined every 15 min in microtiter plates with a Tecan Infinite F500 system (Tecan) for 0.1 s. In order to quantitatively analyze QS activation of the reporter strain NL20, different volumes of the OMV 0.45-μm filtrate were added (0 to 21%).
The V. cholerae reporter strains KPS755 and KPS754 were cultivated in LB at 37°C, and bioluminescence and growth were determined every 15 min in microtiter plates with a Spark 10M system (Tecan) for 0.1 s. V. cholerae-specific CAI-1, (S)-3-hydroxytridecan-4-one (44), was dissolved in DMSO and used as the positive control. The V. cholerae reporter strains KPS755 and KPS754 were conjugated with the pBB1 plasmid, which carries the V. harveyi luxCDABE operon, so that sensing of CAI-1 could be quantified using luminescence as the readout. Data are reported as relative light units (RLU) in counts per second per milliliter per OD600 unit.
For phase-contrast and fluorescence microscopy, samples were analyzed on 1% (wt/vol) agarose pads, which were placed on microscope slides and covered with a coverslip. Images were taken as described above. An excitation wavelength of 546 nm and a 605-nm emission filter with a 75-nm bandwidth were used for mCherry fluorescence. The V. harveyi reporter strain NL20 was grown to an OD600 of 0.5 in LM medium, and then OMVs and the other fractions were added to a final dilution of 1:15. Fluorescence images of the V. harveyi reporter strain NL20 were taken after 2, 4.5, and 6 h of incubation with the respective OMV fractions. Fluorescence intensities of single cells were quantified using ImageJ (50) and are displayed in arbitrary units (AU) (mean fluorescence/area).
In order to observe the attachment of OMVs to V. harveyi cells, the OMV-containing 0.45-μm filtrate fraction was stained with MitoTracker green FM as described above. The reporter strain NL20 was subsequently incubated with this OMV fraction with a final dilution of 1:15 and incubated for up to 4 h at 30°C. Every hour, samples were taken and analyzed on 1% (wt/vol) agarose pads, which were placed on microscope slides and covered with a coverslip. Images were taken as described above. An excitation wavelength of 546 nm and a 605-nm emission filter with a 75-nm bandwidth were used for mCherry fluorescence in order to follow the behavior of the reporter strain NL20. An excitation wavelength of 485 nm and a 510 nm emission filter with a 75-nm bandwidth were used for green fluorescent protein (GFP) fluorescence in order to visualize fluorescently stained OMVs.
Electron microscopy.
V. harveyi cells were cultivated in LM medium for 24 h. Cells and OMVs were harvested as described above and subsequently fixed with 2.5% (vol/vol) glutardialdehyde in 50 mM sodium cacodylate and 2 mM MgCl2 (pH 7.0) at room temperature for 1 h, then rinsed several times in fixative buffer and postfixed with 1% (wt/vol) osmium tetroxide in fixative buffer at room temperature for 1 h. After two washing steps in distilled water, the cells were stained en bloc with 1% (wt/vol) uranyl acetate in 20% (vol/vol) acetone for 30 min. Dehydration was performed with a graded acetone series. Samples were then infiltrated and embedded in Spurr's low-viscosity resin. Ultrathin sections were cut with a diamond knife and mounted on uncoated copper grids and stained with lead citrate. Micrographs were taken with an EM 912 transmission electron microscope (Zeiss, Oberkochen, Germany) equipped with an integrated Omega energy filter operated in the zero loss mode.
For scanning electron microscopy, drops of the sample were placed on glass slides, covered with a coverslip and rapidly frozen with liquid nitrogen. The coverslip was removed with a razor blade and the slide was immediately fixed with 2.5% (vol/vol) glutaraldehyde in 50 mM cacodylate buffer (pH 7.0), postfixed with osmium tetroxide, dehydrated in a graded series of acetone solutions and critical-point dried from liquid CO2, mounted on stubs, and coated with a 3-nm layer of platinum using a magnetron sputter coater. The specimens were examined with a high-resolution field emission scanning electron microscope (Zeiss Auriga workstation) operated at 1 kV.
Diameters of isolated and attached OMVs were measured from scanning electron micrographs of at least 1,400 vesicles of different replicates using ImageJ (50). OMVs were classified into two groups with diameters of <50 nm or >51 nm. The surface areas of OMVs were calculated by assuming them to be spherical (s = π · d2, where s is surface and d is diameter).
Analysis of the lipophilicity of autoinducers (AIs).
Structures of V. harveyi signaling molecules CAI-1, HAI-1, and AI-2 (22) were analyzed with respect to their lipophilicity (log P) using the online tool ALOGPS 2.1 from the Virtual Computational Chemistry Laboratory (http://www.vcclab.org) (35).
Supplementary Material
ACKNOWLEDGMENTS
We thank Kai Papenfort, LMU Munich, for providing the V. cholerae reporter strains KPS754 and KPS755. We thank Andreas Starick and Silvia Dobler for excellent technical assistance, and we kindly thank Joachim Schultz, University of Tübingen, for providing CAI-1 [(S)-3-hydroxytridecan-4-one].
This work was supported by the Deutsche Forschungsgemeinschaft (Exc114/2, SPP1617 [JU270/13-2] to K.J.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00740-17.
REFERENCES
- 1.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orench-Rivera N, Kuehn MJ. 2016. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol 18:1525–1536. doi: 10.1111/cmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. 2015. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Micro 13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 5.Gaudin M, Krupovic M, Marguet E, Gauliard E, Cvirkaite-Krupovic V, Le Cam E, Oberto J, Forterre P. 2014. Extracellular membrane vesicles harbouring viral genomes. Environ Microbiol 16:1167–1175. doi: 10.1111/1462-2920.12235. [DOI] [PubMed] [Google Scholar]
- 6.Laughlin RC, Alaniz RC. 2016. Outer membrane vesicles in service as protein shuttles, biotic defenders, and immunological doppelgängers. Gut Microbes 7:450–454. doi: 10.1080/19490976.2016.1222345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tashiro Y, Uchiyama H, Nomura N. 2012. Multifunctional membrane vesicles in Pseudomonas aeruginosa. Environ Microbiol 14:1349–1362. doi: 10.1111/j.1462-2920.2011.02632.x. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee D, Chaudhuri K. 2011. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett 585:1357–1362. doi: 10.1016/j.febslet.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Altindis E, Fu Y, Mekalanos JJ. 2014. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc Natl Acad Sci U S A 111:E1548–E1556. doi: 10.1073/pnas.1403683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Lazinski DW, Camilli A. 2017. Immunity provided by an outer membrane vesicle cholera vaccine is due to O-antigen-specific antibodies inhibiting bacterial motility. Infect Immun 85:e00626-. doi: 10.1128/IAI.00626-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan HB, Greenberg EP. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol 163:1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drees B, Reiger M, Jung K, Bischofs IB. 2014. A modular view of the diversity of cell-density-encoding schemes in bacterial quorum-sensing systems. Biophys J 107:266–277. doi: 10.1016/j.bpj.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. 2009. Transmembrane passage of hydrophobic compounds through a protein channel wall. ISME J 458:367–370. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papenfort K, Bassler BL. 2016. Quorum sensing signal–response systems in Gram-negative bacteria. ISME J 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henke JM, Bassler BL. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol 186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Zhang L. 2015. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson JP, van Delden C, Iglewski BH. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 181:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krol E, Becker A. 2014. Rhizobial homologs of the fatty acid transporter FadL facilitate perception of long-chain acyl-homoserine lactone signals. Proc Natl Acad Sci U S A 111:10702–10707. doi: 10.1073/pnas.1404929111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Azam F, Zhang S. 2016. Outer membrane vesicles containing signaling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ Microbiol 18:3850–3866. doi: 10.1111/1462-2920.13344. [DOI] [PubMed] [Google Scholar]
- 20.Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, Inaba H, Schmitt-Kopplin P, Eberl L, Nomura N. 2017. Membrane vesicle-mediated bacterial communication. ISME J 11:1504–1509. doi: 10.1038/ismej.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian P-Y, Luo Z-Q, Shen X. 2017. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun 8:14888. doi: 10.1038/s41467-016-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng W-L, Perez LJ, Wei Y, Kraml C, Semmelhack MF, Bassler BL. 2011. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol Microbiol 79:1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin B, Wang Z, Malanoski AP, O'Grady EA, Wimpee CF, Vuddhakul V, Alves N Jr, Thompson FL, Gomez-Gil B, Vora GJ. 2009. Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ Microbiol Rep 2:81–89. doi: 10.1111/j.1758-2229.2009.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao J-G, Meighen EA. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem 264:21670–21676. [PubMed] [Google Scholar]
- 25.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 26.Pereira CS, Thompson JA, Xavier KB. 2013. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev 37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 27.Bassler BL, Greenberg EP, Stevens AM. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol 179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anetzberger C, Reiger M, Fekete A, Schell U, Stambrau N, Plener L, Kopka J, Schmitt-Kopplin P, Hilbi H, Jung K. 2012. Autoinducers act as biological timers in Vibrio harveyi. PLoS One 7:e48310. doi: 10.1371/journal.pone.0048310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freeman JA, Lilley BN, Bassler BL. 2000. A genetic analysis of the functions of LuxN: a two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi. Mol Microbiol 35:139–149. doi: 10.1046/j.1365-2958.2000.01684.x. [DOI] [PubMed] [Google Scholar]
- 30.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. 2005. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell 18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 31.Bassler BL, Wright M, Silverman MR. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol 13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 32.Anetzberger C, Pirch T, Jung K. 2009. Heterogeneity in quorum sensing-regulated bioluminescence of Vibrio harveyi. Mol Microbiol 73:267–277. doi: 10.1111/j.1365-2958.2009.06768.x. [DOI] [PubMed] [Google Scholar]
- 33.Mok KC, Wingreen NS, Bassler BL. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J 22:870–881. doi: 10.1093/emboj/cdg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lilley BN, Bassler BL. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and Sigma-54. Mol Microbiol 36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 35.Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, Palyulin VA, Radchenko EV, Zefirov NS, Makarenko AS, Tanchuk VY, Prokopenko VV. 2005. Virtual computational chemistry laboratory – design and description. J Comput Aided Mol Des 19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- 36.Plener L, Lorenz N, Reiger M, Ramalho T, Gerland U, Jung K. 2015. The phosphorylation flow of the Vibrio harveyi quorum-sensing cascade determines levels of phenotypic heterogeneity in the population. J Bacteriol 197:1747–1756. doi: 10.1128/JB.02544-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klimentová J, Stulík J. 2015. Methods of isolation and purification of outer membrane vesicles from gram-negative bacteria. Microbiol Res 170:1–9. doi: 10.1016/j.micres.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Mashburn-Warren L, Howe JR, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M. 2008. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol 69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Florez C, Raab JE, Cooke AC, Schertzer JW. 2017. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio 8:e01034-17. doi: 10.1128/mBio.01034-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. ISME J 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maredia R, Devineni N, Lentz P, Dallo SF, Yu J, Guentzel N, Chambers J, Arulanandam B, Haskins WE, Weitao T. 2012. Vesiculation from Pseudomonas aeruginosa under SOS. ScientificWorldJournal 2012:402919. doi: 10.1100/2012/402919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshizawa S, Fourmy D, Puglisi JD. 1998. Structural origins of gentamicin antibiotic action. EMBO J 17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng W-L, Perez L, Cong J, Semmelhack MF, Bassler BL. 2012. Broad spectrum pro-quorum-sensing molecules as inhibitors of virulence in Vibrios. PLoS Pathog 8:e1002767-14. doi: 10.1371/journal.ppat.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beltz S, Bassler J, Schultz JE. 2016. Regulation by the quorum sensor from Vibrio indicates a receptor function for the membrane anchors of adenylate cyclases. Elife 5:e13098-17. doi: 10.7554/eLife.13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenberg EP, Hastings JW, Ulitzur S. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol 120:87–91. doi: 10.1007/BF00409093. [DOI] [Google Scholar]
- 46.Waters CM, Bassler BL. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303–314. doi: 10.1016/S0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 48.Papenfort K, Förstner KU, Cong J-P, Sharma CM, Bassler BL. 2015. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci U S A 112:E766–E775. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadurugamuwa JL, Beveridge TJ. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol 177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Meth 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.