Abstract
True physiological imaging of subcellular dynamics requires studying cells within their parent organisms, where all the environmental cues that drive gene expression, and hence the phenotypes that we actually observe, are present. A complete understanding also requires volumetric imaging of the cell and its surroundings at high spatiotemporal resolution, without inducing undue stress on either. We combined lattice light-sheet microscopy with adaptive optics to achieve, across large multicellular volumes, noninvasive aberration-free imaging of subcellular processes, including endocytosis, organelle remodeling during mitosis, and the migration of axons, immune cells, and metastatic cancer cells in vivo. The technology reveals the phenotypic diversity within cells across different organisms and developmental stages and may offer insights into how cells harness their intrinsic variability to adapt to different physiological environments.
A common tenet, oft repeated in the field of bioimaging, is “seeing is believing.” But when can we believe what we see? The question becomes particularly relevant when imaging subcellular dynamics by fluorescence microscopy. Traditional imaging tools such as confocal microscopy are often too slow to study fast three-dimensional (3D) processes across cellular volumes, create out-of-focus photoinduced damage (1, 2) and fluorescence photobleaching, and subject the cell at the point of measurement to peak intensities far beyond those under which life evolved. In addition, much of what fluorescence microscopy has taught us about subcellular processes has come from observing isolated adherent cells on glass. True physiological imaging requires studying cells within the organism in which they evolved, where all the environmental cues that regulate cell physiology are present (3). Although intravital imaging achieves this goal (4, 5) and has contributed pivotally to our understanding of cellular and developmental biology, the resolution needed to study minute subcellular processes in 3D detail is compromised by the optically challenging multicellular environment.
Two imaging tools have recently been developed to address these problems: Lattice light-sheet microscopy (LLSM) (6) provides a noninvasive alternative for volumetric imaging of whole living cells at high spatiotemporal resolution, often over hundreds of time points, and adaptive optics (AO) (7) corrects for sample-induced aberrations caused by the inhomogeneous refractive index of multicellular specimens and recovers resolution and signal-to-background ratios comparable to those attained for isolated cultured cells. The remaining challenge is to combine these technologies in a way that retains their benefits and thereby enables the in vivo study of cell biology at high resolution in conditions as close as possible to the native physiological state. Here we describe a technique based on an adaptive optical lattice light-sheet microscope designed for this purpose (AO-LLSM) and demonstrate its utility through high-speed, high-resolution, 3D in vivo imaging of a variety of dynamic subcellular processes.
Lattice light-sheet microscope with two-channel adaptive optics
Although several AO methods have been demonstrated in biological systems (7), including in the excitation (8) or detection (9) light paths of a light-sheet microscope, we chose an approach where the sample-induced aberrations affecting the image of a localized reference “guide star” created through two-photon excited fluorescence (TPEF) within the specimen are measured and then corrected with a phase modulation element (10). By scanning the guide star over the region to be imaged (11), an average correction is measured that is often more accurate than single-point correction—which is essential, because a poor AO correction is often worse than none at all. Scanning also greatly reduces the photon load demanded from any single point. Coupled with correction times as short as 70 ms (11), this AO method is compatible with the speed and noninvasiveness of LLSM.
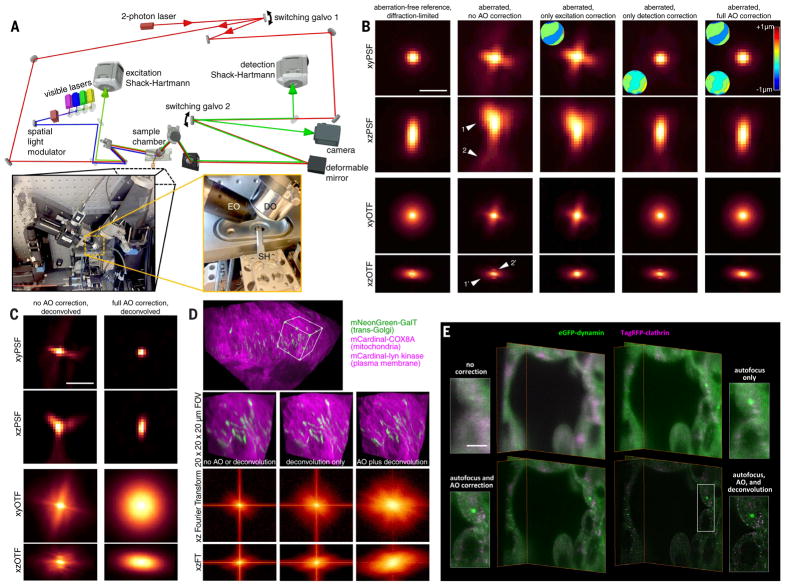
In LLSM, light traverses different regions of the specimen for excitation and detection and therefore is subject to different aberrations. Hence, independent AO systems are needed for each. This led us to design a system (Fig. 1A, supplementary note 1, and fig. S1) where light (red) from a Ti:Sapphire ultrafast laser is ported to either the excitation or detection arm of a LLS microscope (left inset, Fig. 1A) by switching galvanometer 1. In the detection case, TPEF (green) generated within a specimen by scanning the guide star across the focal plane of the detection objective (DO) is descanned (11) and sent to a Shack-Hartmann wavefront sensor (DSH) via switching galvanometer 2 (SG2). We then apply the inverse of the measured aberration to a deformable mirror (DM) placed conjugate to both the DSH and the rear pupil plane of the DO (supplementary note 2). Because the signal (also green) generated by the LLS when in imaging mode travels the same path through the specimen as the guide star, and reflects from the same DM, the corrective pattern that we apply to the DM produces an AO-corrected image of the current excitation plane within the specimen on the camera when ported there by the SG2.
Fig. 1. Adaptive optical lattice light-sheet microscopy (AO-LLSM).
(A) Simplified microscope schematic (fig. S1 shows a detailed version). EO, excitation objective; DO, detection objective; SH, sample holder. (B) xy and xz maximum intensity projections (MIPs) of the point spread function (PSF; top two rows) and corresponding optical transfer function (OTF; bottom two rows) of the microscope under five different degrees of AO correction (columns), as measured from a 200-nm fluorescent bead in an aberrating agarose gel. Insets show the corrective wavefronts applied. Arrowheads indicate lateral and axial aberrations. Scale bar, 1 μm. (C) MIPs and corresponding OTFs of the uncorrected (left column) and fully corrected (right column) bead images from (B), after deconvolution using the aberration-free reference PSF. Scale bar, 1 μm. (D) Cellular trans-Golgi, mitochondria, and plasma membranes in the spine of a live zebrafish embryo 24 hpf. Shown are unprocessed data without AO correction (left column), deconvolved data without AO correction (center), and deconvolved data after AO correction (top and right) (movie S2). MIP views (bottom two rows) of the Fourier transform (FT) of the data in all three cases indicate their respective degrees of information recovery. (E) Four different levels of correction, shown for orthoslices of a live human stem cell–derived organoid grown in Matrigel and gene-edited to express endogenous levels of clathrin and dynamin in coated endocytic pits (Movie 1). Scale bar, 5 μm.
Similarly, for excitation correction, we send descanned TPEF generated and collected through the excitation objective (EO) to a second Shack-Hartmann sensor. However, because LLS excitation is confined to a thin annulus at the rear pupil of the DO (6), a DM placed conjugate to this pupil would be ineffective for AO correction over most of its surface. Instead, we apply wavefront correction at the same sample-conjugate spatial light modulator (SLM) that creates the light sheet itself, thereby enlisting thousands of independently corrective pixels. To do so, we subtract the measured phase aberration from the phase of the Fourier transform (FT) of the ideal, aberration-free SLM pattern, then inverse-transform the result back to the sample-conjugate SLM plane (supplementary note 3 and fig. S2).
Lastly, optimal resolution requires the LLS to be coincident with the focal plane of the DO to less than ~0.46 μm over the entire field of view (FOV), whereas refractive index differences between the specimen and the surrounding media lead to tip-tilt alignment or axial displacement errors for the light sheet that might exceed this (12). Fortunately, we find that only displacement is a concern over FOVs typical of LLSM (supplementary note 4 and fig. S3) and that it can be robustly corrected by imaging, edge-on through the EO, the offset of the plane of TPEF that we generate when measuring the detection aberration relative to the plane of fluorescence generated by the LLS (supplementary note 5 and fig. S4).
As an example (Fig. 1B), after correction for aberrations in the microscope itself, the point spread function (PSF) and corresponding optical transfer function (OTF), measured from a 200-nm bead, indicate nearly diffraction-limited performance (first column). However, when a similar bead is placed in 2% low-melt agarose, we observe substantial aberration (second column), both laterally (arrowheads 1 and 1') and axially (arrowheads 2 and 2'). Correcting only the excitation aberration improves the axial resolution (third column) by returning the light sheet to its original width (fig. S2). Conversely, correcting only the detection aberration improves primarily the lateral performance (fourth column). However, when we combine the excitation and detection corrections, the image of the bead is returned to its diffraction-limited form (fifth column), with an eightfold recovery to its original peak intensity. Furthermore, the same correction proves valid over a 30-μm field of beads in agarose (movie S1).
One of the key advantages of complete AO correction is that it enables accurate deconvo-lution (Fig. 1C), giving the most truthful representation of the specimen possible within the limits of diffraction (second column). In contrast, applying deconvolution to an aberrated image gives a distorted result, because the OTF can then fall below the noise floor asymmetrically and at spatial frequencies well below the diffraction limit (first column), after which higher spatial frequencies cannot be recovered by de-convolution. These same trends can be seen in densely labeled specimens as well, such as across mitochondria, Golgi, and plasma membranes (PMs) of cells near the spinal midline in a living zebrafish embryo 24 hours postfertilization (hpf) (Fig. 1D, bottom three rows, and movie S2), where the greatest information content as seen in the FT of image volume is obtained by combined AO and deconvolution.
Next, we imaged organoids differentiated from human stem cells, gene-edited to express endogenous levels of red fluorescent protein (tagRFP)–clathrin and enhanced green fluorescent protein (EGFP)–dynamin in endocytic pits (clathrin-coated pits; CCPs). Such organoids permit the study of human cellular differentiation and tissue morphogenesis in vitro at subcellular resolution, with an experimental accessibility that is difficult to achieve in vivo. However, the extracellular matrix in which they are grown introduces considerable aberrations, and the fast dynamics and limited number of fluorophores in each CCP demand high-speed imaging with low photobleaching. The system is therefore well suited to the capabilities of AO-LLSM, with the CCPs doubling as distributed puncta of subdiffractive size to evaluate imaging performance.
Without AO or focus correction (upper left of Fig. 1E and Movie 1), no CCPs are visible, and the cell boundaries are poorly defined. Autofocus alone (upper right) reveals larger patches of clathrin and dynamin but must be combined with complete AO correction (excitation and detection, lower left) to identify individual CCPs. At that point, the imaging is diffraction-limited, so the data can be deconvolved using the system-corrected PSF to compensate for the spatial filtering properties of the microscope. Both dynamin and clathrin puncta then stand out clearly above the cytosolic background (lower right), allowing us to quantitatively measure their lifetimes and diffusion tracks (Movie 1). The increasing recovery of information as we progress through these four cases can also be seen quantitatively in their corresponding OTFs (fig. S5).
Movie 1. Endocytosis in a human stem cell–derived organoid.
Gene-edited clathrin (magenta) and dynamin (green) before and after adaptive optical correction and deconvolution, showing comparative xy and xz orthoslices, volume renderings, and postcorrection tracking of the motion and lifetimes of individual CCPs and CCVs over 120 time points at 1.86-s intervals (Fig. 1E and fig. S5).
Clathrin dynamics in zebrafish
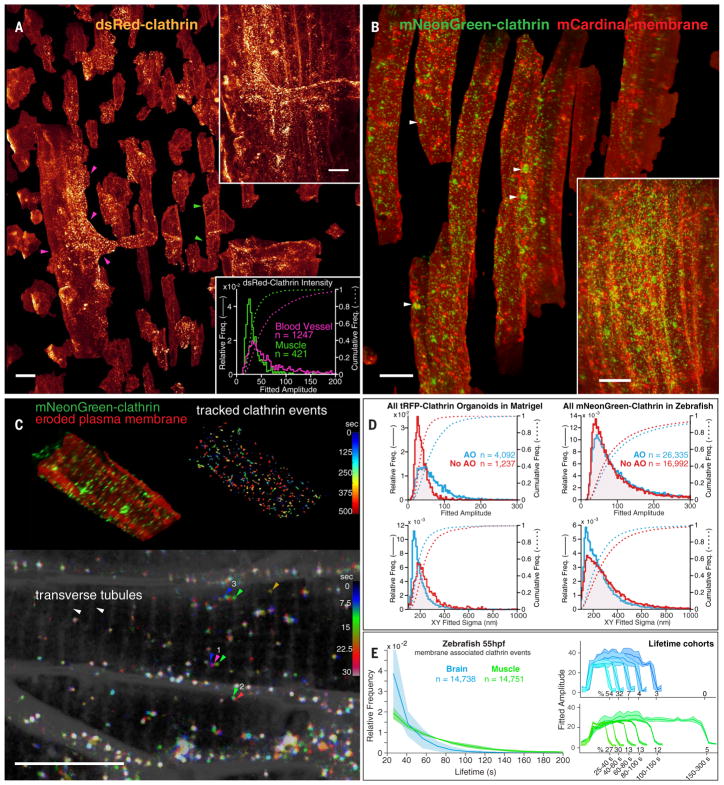
For transparent model organisms, we can apply AO-LLSM in vivo, where the complete physiological environment is preserved. The zebrafish has become the most widely used nonmammalian vertebrate model organism. We took advantage of its small size and transparency to visualize the formation of endocytic CCPs and clathrin-coated vesicles (CCVs) in the context of the developing organism. We first imaged a volume in the dorsal tail region of a fish larva 80 hpf stably expressing dsRed–clathrin light chain A (CLTA) (13) (Fig. 2A and Movie 2). We observed a high density of diffraction-limited clathrin spots that, after computational separation of all cells, were found to be mostly colocalized with the PM. These spots appeared and disappeared with the formation of new CCPs and their eventual uncoating. We determined that the areal density of CCPs was similar in muscle fibers (e.g., green arrowheads, Fig. 2A) and endothelial cells (magenta arrowheads) lining blood vessels, but the distribution of their intensities was broader in the latter (lower inset, Fig. 2A), which had a subpopulation of pits that were up to sixfold as bright as the median CCP intensity in the former. Because CCP size is proportional to intensity (14), these results indicate the presence of larger structures, possibly clusters of CCPs (15), in the vasculature endothelium.
Fig. 2. Clathrin dynamics in zebrafish.
(A) Computationally separated muscle fibers (e.g., green arrowheads) and vascular endothelial cells (e.g., magenta arrowheads), both expressing DsRed-CLTA to highlight CCPs and CCVs, from muscle tissue in a 75-μm by 99-μm by 41-μm region (upper inset) of the tail of a developing zebrafish larva 80 hpf (Movie 2). Brighter clathrin puncta were observed in the endothelial cells (lower inset). Scale bars, 10 μm. (B) Computationally separated muscle fibers from a region (lower inset) in the tail of a zebrafish embryo 50 hpf coexpressing an mCardinal-PM marker (red) and mNeonGreen-CLTA (green). Individual CCPs and CCVs and larger clathrin-rich vesicles (arrowheads) are visible (Movie 3). Scale bars, 10 μm. (C) Spatial distribution and dynamics of CCPs and CCVs tracked for 12 min at 7.5-s intervals in one muscle fiber from (B), showing CCPs localized at t-tubules (top left) and diffusion and lifetime characteristics for CCPs and CCVs across the cell (top right). A MIP through a 2-μm-thick slab at three consecutive time points (bottom) shows examples of a pinned CCV (arrowheads 2) and slowly diffusing (arrowheads 1) or rapidly shuttling CCVs (arrowheads 3) (movie S3). Scale bar, 10 μm. (D) Effect of AO on the measured quantity, intensity, and localization precision of CCPs and CCVs in the organoid in Fig. 1E and the zebrafish in (B). (E) Comparative distribution of CCP and CCV lifetimes (left) and intensity cohorts grouped by their lifetimes (right) in the brain and muscle of a developing zebrafish embryo 55 hpf.
Movie 2. Clathrin-mediated endocytosis in vivo.
Dynamics of CCPs and CCVs over 15 min at 10-s intervals in the dorsal tail region of a zebrafish embryo 80 hpf. Segmented cells reveal brighter clathrin puncta at the vascular endothelium than at muscle fibers (Fig. 2A).
To track CCPs for longer times, we turned to zebrafish embryos mRNA-injected to express the brighter and more photostable fluorescent fusion protein mNeonGreen-CLTA. These embryos also expressed mCardinal targeted to the PM, facilitating the computational separation of cells. Embryos imaged in the tail (Fig. 2B, fig. S6A, and Movie 3) and in the hindbrain (fig. S6B) displayed a variety of morphologies, trafficking behaviors (Fig. 2C), and lifetime distributions (Fig. 2D). Large, micron-scale intracellular spots of limited mobility (arrowheads, Fig. 2B) probably represent clathrin-rich vesicles clustered at the trans-Golgi network, whereas mobile diffraction-limited spots (Fig. 2C, arrowhead group 1) likely correspond to endosomal carriers similar to those seen in cultured mammalian cells (16). Diffraction-limited spots at the PM (Fig. 2C, arrowhead group 2) likely represent individual CCPs and CCVs. We also found CCPs at the t-tubules spanning muscle fibers (Fig. 2C, top left), in contrast to the diffuse clathrin signal observed using immunofluorescence in fixed rat muscle fibers (17). Most of these were pinned, but occasionally they would break free from a t-tubule and move rapidly along the fiber axis (Fig. 2C, arrowhead group 3, and movie S3), possibly by active transport along myofibrils or within the sarcoplasmic reticulum.
Movie 3. Clathrin localization in muscle fibers.
PM (red) and clathrin (green) in the tail of a zebrafish embryo 50 to 55 hpf, showing xy and xz orthoslices before and after AO correction and deconvolution, dynamics of individual CCPs and CCVs at and between t-tubules, large clathrin clusters and small clathrin puncta in volume-rendered and segmented cells, and tracked CCPs and CCVs in a segmented cell (Fig. 2, B to E; fig. S8; and movie S3).
AO allowed us to detect more CCPs (fig. S7) and track all CCPs with higher precision (Fig. 2C, top right; fig. S8, A to C; and Movie 3). Comparing CCPs in muscle fibers and the brain, we found that, although their initiation frequencies and fluorescence intensities were similar, brain CCPs tended to internalize faster (Fig. 2E). Assuming that clathrin puncta in muscle and brain cells lasting >21 s corresponded to successful coated vesicles, each with an assumed membrane diameter of 60 nm, ~0.1% of the PM was internalized through clathrin-mediated endocytosis every minute (fig. S8D). This is similar to the values derived from measurements in cultured SUM-159 (18) or htertRPE-1 (19) mammalian cells at 37°C.
In vivo imaging of organelle morphology and dynamics during embryogenesis
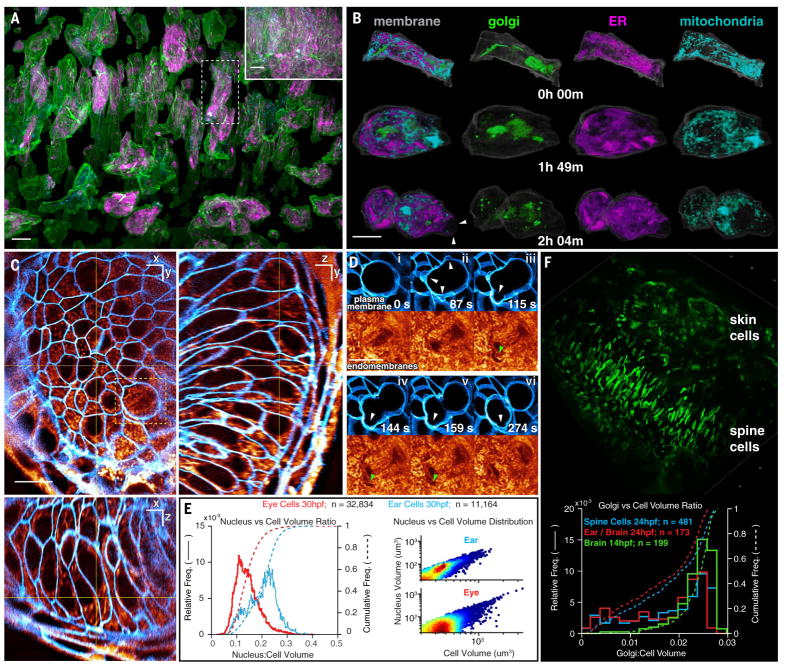
A major focus in cell biology is the study of the structure and function of organelles within the living cell. To study the dynamics of multiple organelles simultaneously throughout the cell cycle across a population of cells in vivo, we imaged brain progenitor cells with markers for the trans-Golgi, endoplasmic reticulum (ER), mitochondria, and PM for 200 image volumes at 44-s intervals (Fig. 3, A and B, and Movie 4). In interphase, we observed multiple trans-Golgi segments in most cells, often appearing as long filaments preferentially aligned along the axis of cell polarization (Fig. 3A) that fragmented during mitosis (Fig. 3B). The ER and mitochondria largely recapitulated the forms that they commonly take in cultured cells: The ER established a reticular network in interphase and sheetlike cisternae during mitosis (20), whereas mitochondria formed punctate structures near the surface and longer tubules in the subset of more deeply buried interphase cells that were well labeled. Analyzing one such cell, we found that all three organelles were distributed uniformly between the PM and the nucleus in interphase (fig. S9A), but mitochondria were preferentially located nearer the PM during mitosis (fig. S9A, 109 min).
Fig. 3. Organelle morphologies and dynamics in zebrafish.
(A) Computationally separated neural progenitor cells from a 70-μm by 35-μm by 35-μm region (inset) in the brain of a developing zebrafish embryo expressing GalT-mNeonGreen, tagRFP-Sec61β, and Citrine as markers of the trans-Golgi, ER, and PM, respectively, with additional labeling by MitoTracker Deep Red dye (Movie 4). Scale bars, 10 μm. (B) Changing morphologies of the organelles in the specific cell outlined in (A) at three time points through mitosis. Arrowheads indicate mitotic blebs. Scale bar, 10 μm. (C) MIP views from 1-μm-thick orthogonal slabs within the eye of a zebrafish embryo 30 hpf, showing PM (blue) and endomembranes (orange) (Movie 5). Scale bars, 10 μm. (D) Six time points from Movie 5, showing PM blebs (white arrowheads) during mitosis and the exclusion of endomembranes in early blebs (green arrowheads). (E) Correlation between nuclear volume and total cell volume in the eye and ear (Pearson’s coefficient, 0.9 and 0.8, respectively). (F) Different morphologies of trans-Golgi (top) near the spine of a zebrafish embryo 24 hpf and distribution of trans-Golgi volume in different cell types and at different developmental stages (bottom).
Movie 4. Subcellular imaging of organelle dynamics in the early zebrafish brain.
Dynamics of PM (green or gray) and trans-Golgi (green), ER (magenta or red), and mitochondria (cyan) within neural progenitor cells over 200 time points at 44-s intervals from 14.0 hpf, showing complexity within the tissue, cross-sectional slab views through cells, sequential division of adjacent cells, segmentation and separation of all cells, and morphological changes to organelles during mitosis in one such cell (Fig. 3, A and B, and figs. S9 and S10).
The early synchrony of cell division is lost in zebrafish at the midblastula transition (3 hpf). Nevertheless, at 14 hpf, we observed instances of cascading cell division, where adjacent cells underwent mitosis one after another (fig. S10 and Movie 4). Mitotic cells, as seen previously in cell cultures (18, 21), decreased their surface area (fig. S9E) as they assumed a spheroidal shape and produced transient blebs before cytokinesis, but then recovered their initial area after division. Total cellular volume remained constant throughout mitosis (fig. S9E). We also identified instances of asymmetric cytokinesis (Fig. 3B and Movie 4), where the two daughter cells differed in surface area by ~50% (fig. S9F). When we quantified organelle intensity in one such instance, we also observed asymmetric fractional partitioning of the Golgi and mitochondria between the daughter cells during cytokinesis (fig. S9G).
Mitotic cells in the developing eye 30 hpf (white arrowheads in Fig. 3D and Movie 5) also produced transient blebs before cytokinesis. We discovered upon labeling with Bodipy–tetramethylrhodamine (TMR) that these blebs created voids (green arrowheads in Fig. 3D) that only slowly filled with endomembranes.
Movie 5. Membrane dynamics in the zebrafish eye.
PM (blue) and the endomembrane system (orange) 30 hpf viewed as xy orthoslices, cell divisions in a 1-μm-thick slab, and volume-rendered PM dynamics across the eye at 43.8-s intervals for 200 time points (Fig. 3, C to E).
Lastly, we observed considerable variability in the size and morphology of specific subcellular features across different organs and developmental stages. Bodipy-TMR negative staining (e.g., Fig. 3, C and D) revealed that the nuclei of ear cells 30 hpf were nearly twice as large as those of eye cells when normalized by the total cellular volume (Fig. 3E, left) and that nuclear volume and cellular volume were positively correlated (Fig. 3E, right). Likewise, we found that the median trans-Golgi volume as a percentage of total cellular volume in brain progenitor cells 14 hpf was larger [2.46%; median absolute deviation (MAD) = 0.26%] than in ear, brain, or spine cells 24 hpf (2.0%; MAD = 0.66%) (Fig. 3F). Golgi took many forms, from the aforementioned narrow polarized filaments in the early brain (Fig. 3A) to shorter segments clustered near the midline in the spine and nuclear-wrapping filaments in skin cells (Fig. 3F).
Tiled acquisition for aberrations varying in space and time
Because the refractive index profile can vary across a specimen and can also vary as the specimen develops, the AO corrections required can vary in both space and time. Unfortunately, it is difficult to estimate a priori the size or temporal stability of the isoplanatic patch (the FOV over which a single AO correction is valid). Empirically, we found in zebrafish embryos less than 72 hpf that a single excitation-detection correction pair obtained by scanning and descanning over the FOV is usually valid across 30 to 60 μm in each direction for at least 1 hour, provided that the light does not intersect the yolk. Fortuitously, these dimensions are comparable to those over which a LLS of submicron thickness does not deviate substantially in width. The examples shown above largely fall within these limits and hence, for them, a single AO correction pair at a single time point sufficed.
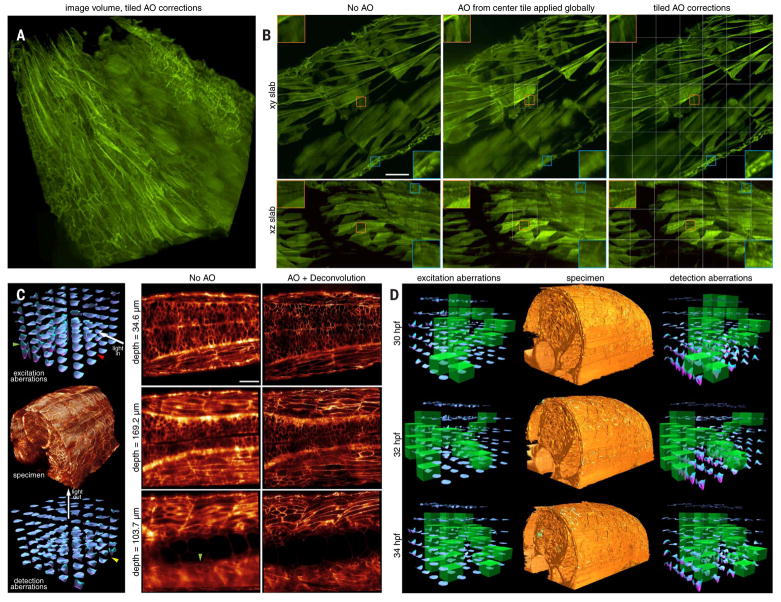
In other cases, however, we may wish to cover much larger FOVs. To do so, we must stitch together data from multiple image subvolume tiles, each with its own independent AO correction. To demonstrate the necessity of this, we imaged a 213-μm by 213-μm by 113-μm volume (Fig. 4A and movie S4) comprising 7 by 7 by 3 tiles in the tail region of a zebrafish embryo 96 hpf expressing membrane-EGFP, using three different protocols: no AO correction (Fig. 4B, left column), AO correction from the center tile applied to all tiles (middle column), and independent AO correction in each tile (right column). When viewed across 3-μm-thick slabs perpendicular (xy; Fig. 4B, top row) or parallel (xz; bottom row) to the detection axis, a small volume within the center tile (small orange boxes) showed substantial improvement by either center-tile or all-tiles AO correction, in both the xy lateral (upper-left orange boxes, top row) and xz axial (upper-left orange boxes, bottom row) planes. This is to be expected, because the site of AO correction coincided with the viewing area in these two cases. However, in a small volume at the edge of the fish (small blue boxes), only the data taken using individual AO corrections in each tile recovered optimal resolution in all directions (lower-right blue boxes, right column), because this volume was outside the isoplanatic patch over which the center-tile AO correction is valid. Applying the center correction across the larger stitched volume often results in greater wavefront errors (fig. S11) and poorer resolution (lower-right blue boxes, middle column) than applying no correction at all (lower-right blue boxes, left column), highlighting the importance of accurate and robust correction, if AO is to be applied.
Fig. 4. AO-LLSM over large volumes.
(A) Aberration-corrected volume rendering over 213 μm by 213 μm by 113 μm in the tail region of a live zebrafish embryo 96 hpf expressing PM-targeted EGFP, assembled from independently corrected subvolumes of 7 by 7 by 3 tiles (movie S4). (B) Increasing effectiveness of correction, as seen in orthogonal MIPs from 3-μm-thick slabs, under different scenarios: no AO (left column), AO correction from the center tile applied globally (middle column), and independent AO correction in each tile (right column) (fig. S11). Insets compare, at higher magnification, the quality of correction at the center tile (orange boxes) versus at the tiles at the periphery of the tail (blue boxes). Tile boundaries are shown in white. Scale bar, 30 μm. (C) A 5 by 4 by 7 set of measured excitation (left column, top) and detection (left column, bottom) aberrations which, after AO correction, yields diffraction-limited imaging over a 170-μm by 185-μm by 135-μm volume (left column, center) in the spine of a zebrafish embryo 30 hpf (Movie 6). Red and green arrowheads indicate excitation aberrations in specific tiles before and after passage through the notocord, respectively. The yellow arrowhead indicates a tile with a large detection aberration deep within the specimen. Orthoslices before (middle column) and after (right column) AO correction show increased aberration but continued recovery of high resolution at progressively greater depth. Scale bar, 30 μm. (D) Aberration-corrected volume renderings over 156 μm by 220 μm by 162 μm in the spine of a zebrafish embryo, at three points from a time series at 30-min intervals (movie S5), flanked by excitation and detection path aberrations at those points. Those tiles whose corrections were updated at a given time point are marked in green.
Empirically, the largest aberrations that we observed in zebrafish embryos occur at regions of high curvature between the embryo and the surrounding media or at regions of rapid refractive index change, such as near the notochord (Fig. 4C and Movie 6). For example, when the LLS penetrates the embryo near-perpendicularly to its surface, the excitation aberration is initially small (Fig. 4C, red arrowhead, left column, top). However, after the light sheet passes through the notochord, it encounters substantial aberration, as seen in both the measured wavefront (green arrowhead, left column, top) and the uncorrected image (green arrowhead, middle column, bottom). On the detection side, aberrations increase with increasing depth in the embryo (left column, bottom, and middle column, top to bottom). In addition, substantial aberrations occur when the edge of the embryo is imaged tangentially (yellow arrowhead, left column, bottom), so that part of the detection light cone intersects the embryo and part does not.
Movie 6. Tiled AO correction for imaging large volumes.
A 170-μm by 185-μm by 135-μm volume from the dorsal surface to the notochord in a PM-labeled zebrafish embryo, showing increasing aberration but continued full correction at increasing depth; corrective excitation and detection wavefronts in each of the tiled isoplanatic subvolumes of 5 by 4 by 7 tiles; and four views of PM dynamics within the complete volume from 30 to 39.5 hpf, imaged at 7.5 min intervals (Fig. 4C).
Provided that the specimen does not shift by more than a fraction of the isoplanatic patch size during imaging, a given set of tiled AO corrections can remain valid for hours (Movie 6). However, growth during development can cause an embryo to change its shape, position, or refractive index profile so that new corrections are occasionally needed. Fortunately, these changes often occur on a time scale that is slow compared with that needed to image even a large FOV by LLSM. In such cases, it is sufficient to update the correction at only a subset of different tiles at each time point, as long as all subsets together encompass all tiles in the FOV before the previous round of corrections becomes inaccurate (Fig. 4D and movie S5). Usually, we chose subsets that broadly covered the FOV to monitor where the aberrations change the fastest.
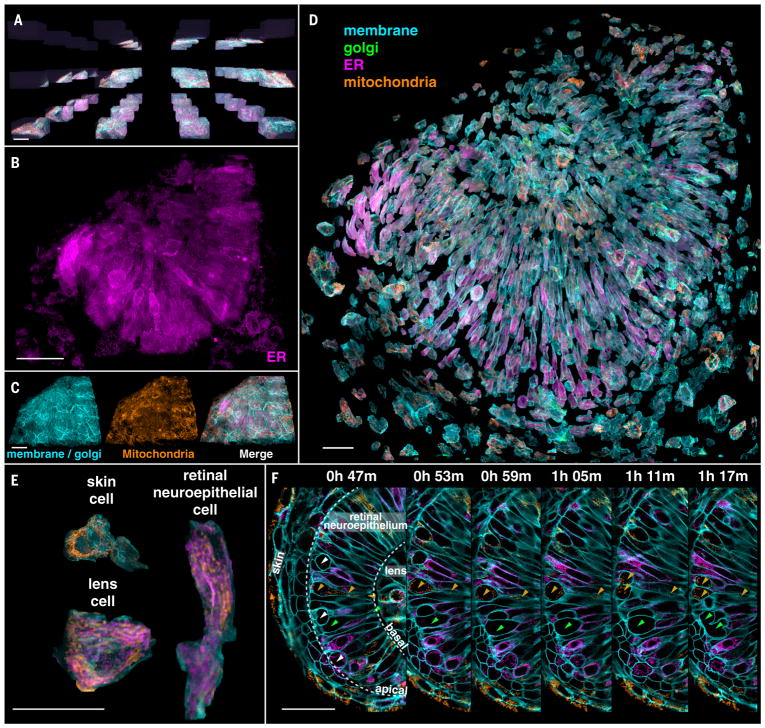
One region that involves substantial specimen curvature, large spatial variation of the refractive index, and gradual aberration change is the eye of the developing zebrafish. Although in vivo optic cup development has been studied at sub-cellular 3D resolution by conventional (22, 23) and multiview (24) light-sheet microscopy, tiled AO-LLSM permits a more detailed look at cellular morphology and organelle distributions during this process. We imaged across 4 by 4 by 3 tiles (Fig. 5A and Movie 7) spanning most of the eye of an embryo 24 to 27 hpf at 6-min intervals to study differences in the intracellular organization of various organelles (Fig. 5, B and C). Transgenic labeling of the PM allowed us to segment, isolate (Fig. 5D), and characterize each cell by type (Fig. 5E). Skin cells exhibited mitochondria clustered in the perinuclear region, like mitochondria seen in flat and thin cultured cells, to which these skin cells are morphologically similar. In contrast, mitochondria in retinal neuroepithelial (RNE) cells were generally longer, distributed across the length of the cell, and polarized along the same axis as the cell itself. The ER in RNE cells, although broadly distributed, was usually densest around the nucleus and least dense near the polarized ends. In mitotic cells, however, we again observed (20) that the ER remodeled into sheetlike structures concentrated near the PM (Movie 7).
Fig. 5. Organelle diversity across the zebrafish eye.
(A) Tiled array used to provide AO correction across the eye of a developing zebrafish embryo 24 hpf (Movie 7). Scale bar, 30 μm. (B and C) Distribution of three different types of organelles across the volume assembled from the tiles in (A). Scale bars, 30 μm. (D) Computationally separated cells across the eye, with the organelles colored as indicated. Scale bar, 30 μm. (E) Organelle morphologies in cells of three different types within the eye. Scale bar, 30 μm. (F) Orthoslices at six different time points highlighting cell divisions (white and green arrowheads, left panel) at the apical surface of the retinal neuro-epithelium and mitochondria (orange arrowheads) present from the apical to the basal surface in one dividing cell. Scale bar, 30 μm.
Movie 7. Organelle dynamics across the zebrafish eye.
PM (cyan), trans-Golgi (green), ER (magenta), and mitochondria (brown) across a 128-μm by 150-μm by 75-μm volume assembled from subvolumes of 4 by 4 by 3 tiles, showing orthoslices in a single tile, volume-rendered tiles before assembly into the combined volume, organelles in the combined volume, dynamics over 30 time points from 24.0 to 26.8 hpf in a 1-μm-thick slab through the combined volume, dynamics in perpendicular orthoslices, and organelle morphologies in different cell types in the computationally expanded volume (Fig. 5).
By imaging over time (Fig. 5F), we could follow the stages of RNE cell division (green arrowheads). As reported elsewhere (25), we found that, before mitosis, the nucleus retracts to the apical side of the retina (leftmost orange arrowheads), while the cell maintains a thin connection to the basal side (rightmost orange arrowheads). Despite its narrowness, mitochondria remain in this region. RNE cell divisions then occur at the apical surface (white arrowheads). This process has been found to be necessary to maintain the integrity of retinal tissue (26).
3D cell migration in vivo
In vivo 3D migration of a cell in the densely crowded environment of living tissue involves forces, constraints, elasticity and adhesion heterogeneity, and chemical cues not found in the simple 2D environment on a cover glass. Furthermore, cell migration involves intricate and rapid remodeling of membranes, organelles, and the cytoskeleton that requires high spatiotemporal resolution to observe. It is therefore a problem well suited to AO-LLSM.
An example of this problem involves the wiring of neuronal circuits during development. To help them establish precise connections, axons are tipped with a highly complex and motile structure, the growth cone. This structure functions as both a sensor and a motor, driving the growth of neurites on the basis of environmental cues (27). Although its dynamics have been shown to be important for its proper function (28–30), its 3D dynamics in an intact animal have been difficult to study because of the lack of techniques for imaging the structure with sufficient resolution in vivo.
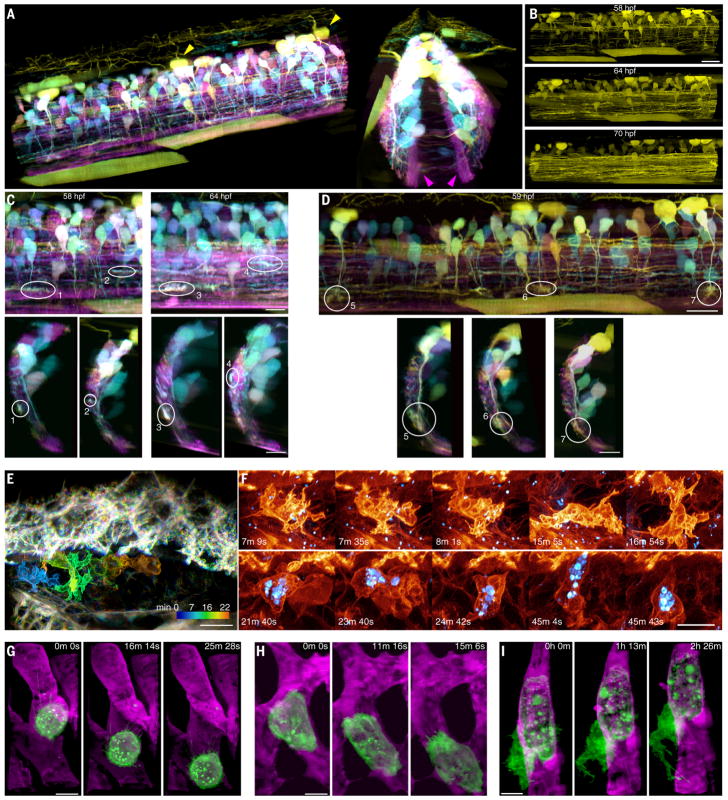
To address this, we used AO-LLSM to image growth cones in the spinal cord of a zebrafish embryo in which a subset of newly differentiated neurons expressed stochastic combinations of three different fluorophores via Autobow (31) (Fig. 6A and fig. S12), so that they could be spectrally distinguished from earlier differentiated neurons expressing only mCherry [e.g., those within the medial neuropil of the reticulospinal tract (magenta arrowheads, Fig. 6A)]. The Autobow-labeled neurons included Rohon-Beard sensory neurons in the dorsal spinal cord (e.g., yellow arrowheads, Fig. 6A) and interneurons with commissural axons. By imaging over more than two spinal segments at 10.4-min intervals from 58 to 70 hpf (Fig. 6B and Movie 8), we observed that the growth cones of axons migrating in the rostrocaudal direction primarily probed in the direction of their motion (Fig. 6C, top, and movie S6), whereas the growth cones of dorsoventrally aligned axons probed across a broader 2D fan (Fig. 6D, top). Transverse views (Fig. 6, C and D, bottom) revealed that most, if not all, growth cones of both types were located close to the surface of the spinal cord, with their filopodia preferentially aligned parallel to the surface, even though the dorsoventrally projecting axons had to pass through the spinal cord to reach its surface. This is consistent with the previous observation that the neurites of late-born V2a ipsilateral projecting interneurons are located lateral to the preexisting ones, forming a layer-like organization based on the age of neurons (32), and extends this notion to other classes of spinal neurons. This also raises an interesting possibility that the shape of the growth cone is actively controlled in vivo to keep its exploration within a layer of its own age group.
Fig. 6. 3D cell migration in vivo.
(A) Two views of newly differentiated neurons highlighted by Autobow labeling in a 60-μm by 224-μm by 180-μm section of the spinal cord of a zebrafish embryo 58 hpf (Movie 8). Magenta and yellow arrowheads show neurons differentiated before and after Autobow expression, respectively. (B) Increase in the density of rostro-caudally projecting axons over time. Scale bar, 20 μm. (C) Sagittal (top) and transverse (bottom) views of the growth cones of four rostrocaudally projecting axons. Scale bars, 10 μm. (D) Sagittal (top) and transverse (bottom) views of the growth cones of three dorsoventrally projecting axons. Scale bars, 10 μm. (E) Time-coded color overlay of an immune cell migrating within the perilymphatic space next to the inner ear of a live transgenic zebrafish embryo 70 hpf expressing PM-targeted Citrine (Movie 9 and fig. S12). Texas Red dextran particles are shown in blue. Scale bar, 10 μm. (F) Changing morphologies of two different immune cells (top and bottom rows), one showing internalized dextran particles (blue) (fig. S13). Scale bar, 5 μm. (G) MDA-MB-231 human breast cancer cell (green) rolling in a blood vessel (magenta) in a zebrafish embryo 48 hpf. (H) Another MDA-MB-231 cell crawling through a blood vessel. (I) A partially extravasated MDA-MB-231 cell, showing an increasingly complex morphology over time (Movie 10 and fig. S1). Scale bars, 10 μm in (G) to (I).
Movie 8. In vivo imaging of spinal cord neural circuit development.
Autobow-labeled, newly differentiated neurons expressing stochastic combinations of three fluorophores in a zebrafish embryo, showing corrective excitation and detection wavefronts in subvolumes of 5 by 2 by 1 tiles, with scrolling updates at one tile (green box) per time point; AO-corrected orthoslices and volume-rendered views in each color channel 58 hpf; and axon pathfinding in each color channel from 58 to 70 hpf (Fig. 6, A to D; fig. S12; and movie S6).
Cell migration is also a key aspect of the innate immune system. Neutrophils, for example, migrate from the vasculature through the endothelium to reach and engulf infectious targets (33). To study this process in vivo, we imaged endogenously produced immune cells moving through the perilymphatic space of the ear in a transgenic zebrafish larva ~80 hpf expressing the fluorescent protein Citrine in the PM of all cells (Fig. 6E and Movie 9). Acquiring 438 image volumes at 13-s intervals allowed us to accurately measure the 3D position and speed (fig. S13, A and B) of the cellular center of mass. Across five trials involving different embryos at 22°C, immune cells adopted a halting search pattern involving frequent changes in direction and speed (fig. S13, C to G), from nearly motionless to 10 μm/min. In contrast, neutrophilic mammalian HL-60 cells imaged in a collagen matrix at 37°C (6) exhibited peak speeds of ~25 μm/min (fig. S13H).
Immune cells next to the ear were remarkable for their rapidly changing and complex 3D morphologies (Fig. 6F). Their surface areas changed as much as 25% in 2 min (fig. S13I) as they remodeled themselves to exhibit a variety of protrusive features, including lamellar sheets, blebs, and short filopodia. Frequently, they also trailed a long filopodium behind them as they migrated to new regions (fig. S14). After injecting fluorescent Texas Red dextran into the heart, we also observed several immune cells containing granules of dextran up to several microns in size (light blue, Fig. 6F, and Movie 9, part 1), presumably ingested earlier by phagocytosis. However, these did not noticeably affect the motility of the cells or their ability to navigate through tight interstitial spaces.
Apart from immune cells, 3D AO-LLSM movies (e.g., Movie 9, part 2) of the developing ear region revealed a wealth of cellular morphologies and behaviors, including filopodial oscillations at the dorsal surface of skin cells, gradual inflation of the perilymphatic volume, rapid passage of cells through blood vessels, long and active filopodia on endothelial cells, and cellular motion in the hindbrain. With AO, the spatiotemporal resolution and noninvasiveness that we achieved was comparable to what we obtained when imaging cultured cells with our original LLSM approach (6), allowing us, for example, to follow the detailed morphological changes in a single endothelial cell lining the hindbrain over the entire course of its division (fig. S15 and Movie 9, part 2).
As a final example of 3D migration, during cancer metastasis, circulating tumor cells (CTCs) extravasate and seed new tumor formation at sites distant from the primary tumor (34). Extravasation has been studied extensively in vitro, but little is known about the process in vivo, owing to the highly dynamic nature of cells in circulation and the low density of CTCs in the vasculature. A long-standing hypothesis (34) based on in vitro studies is that CTCs co-opt a three-step process used by leukocytes to extravasate at sites of inflammation (34): Circulating leukocytes initially form tethers to weakly adhere to the vascular endothelium, causing them to roll slowly downstream (35); eventually, they attach and crawl along the endothelial wall; and finally, they penetrate the endothelium and migrate into the tissues beyond.
To determine whether CTCs follow this same pattern in vivo, we used a xenograft model (36), where we injected PM-labeled human breast cancer cells (MDA-MB-231) into the vasculature of 2-day-old zebrafish embryos that were trans-genic for an endothelial reporter (kdrl:gfp). As hypothesized, we observed all three leukocyte migration behaviors in the cancer cells. First, we recorded MDA-MB-231 cells rolling through the blood vessels (Fig. 6G), trailing microvilli that adhered to the endothelium and stretched several microns before detaching as the cell continued to move downstream (Fig. 6G and Movie 10, part 1). Second, we visualized MDA-MB-231 cells crawling along the endothelium (Fig. 6H and Movie 10, part 2). Last, we observed an MDA-MB-231 cell actively engaged in transendothelial migration, with the portion of the cell outside the blood vessel projecting actin-rich extensions into the surrounding tissue (Fig. 6I and Movie 10, part 3) as the area of the cell increased by ~50% over 2 hours (fig. S16).
Discussion
AO-LLSM enables minimally invasive high-speed 3D imaging of subcellular dynamics within optically challenging living specimens while maintaining diffraction-limited resolution, even over large FOVs. It corrects not only sample-induced aberrations, but also those introduced by mounting and immersion media (e.g., Fig. 1E) or imperfections in the optical path through the microscope. It therefore can provide practical 3D resolution exceeding that of nominally higher numerical aperture (NA) confocal or spinning disk microscopes, even in the comparatively benign optical environment encountered when imaging isolated adherent cells on cover slips.
This performance does not come without caveats, however. Because the fluorescence induced by the light sheet is captured with wide-field optics, only weakly scattering specimens can be imaged. In addition, extremely sparse and/or weakly emitting fluorescent targets may require colabeling with a second, brighter color channel to provide a sufficient guide star signal for accurate wavefront measurement. Highly absorbing structures such as large blood vessels or melanin bodies within the detection light cone can block guide star light from reaching enough cells of the sensor for accurate wavefront measurement, although wavefront reconstruction algorithms might be made more robust against such missing information. Wavefront aberration can vary considerably across the specimen, and at present, this variation can only be determined empirically for each specimen type and developmental stage to determine how to subdivide the desired image volume into tiled isoplanatic subvolumes of relatively uniform aberration. Fortunately, such tile maps tend to be consistent between specimens of the same type and age, given similar mounting geometries. Lastly, specimens imaged after muscle development must be anesthetized and immobilized, or else a new correction must be measured and applied whenever sample motion exceeds the size of a given isoplanatic patch.
Another caveat is that all but one of the above described involved imaging subcellular dynamics within zebrafish embryos. Although we have shown that we can achieve substantial gains in imaging performance in both Caenorhabditis elegans larvae (fig. S17 and movie S7) and Arabidopsis thaliana leaves (fig. S18 and movie S8), Danio rerio represents an ideal model system in which to study cell and developmental biology in vivo, because it is a rapidly developing transparent vertebrate that is amenable to genetic manipulation. Furthermore, zebrafish embryos are small enough that most regions are optically accessible far into development, yet large enough to exhibit smoothly varying refractive index profiles that result in isoplanatic patch sizes that are comparable to imaging fields typical of LLSM. In contrast, C. elegans larvae and adults exhibit larger and more rapid spatial variations in refractive index, particularly near the gut, that can require a denser mesh of AO corrections, despite this organism’s reputation as an optically tractable model.
Conventional light-sheet microscopy using weakly focused Gaussian beams is also susceptible to aberrations and would therefore also benefit from AO correction (8, 9). However, conventional systems typically cover much wider FOVs and often operate at greater depth in larger organisms, such as in applications involving functional imaging of whole neural circuits (37) or in toto cellular tracking during development (38). They therefore usually image over regions much larger than a single isoplanatic patch, making it difficult to retain even cellular-level resolution at all locations and compromising the accuracy and resolution of approaches based on multiview fusion (39–41). A single AO correction would provide at best only partial correction, and a tiled AO approach, such as we use with LLSM, would negate the high-speed, large-field advantages of the conventional light-sheet microscopy. On the other hand, simultaneous full-field AO correction would likely require multiconjugate adaptive optics (42), substantially increasing cost and complexity.
Perhaps the greatest challenge of AO-LLSM involves mining the immense and complex data that it produces to extract as much biological insight as possible. In Fig. 6, for example, panels A to D represent a minute fraction of a 0.62-terabyte raw data set which first had to be deconvolved, creating a second copy, and then imported into 3D visualization software, generating a third. We deconvolve and store data in real time, but importation and visualization can take many hours, preventing meaningful real-time feedback on whether the biological structure and dynamic process of interest are being optimally recorded. If history is any guide, problems of petabyte-scale data storage and visualization at reasonable cost will yield to continued advancements in commercial hardware, but problems of image analysis and meaningful quantification of data at this scale may prove far less tractable. Although we have demonstrated quantification on a smaller scale through single-particle (Fig. 2, D and E, and fig. S8) and single-cell (fig. S13) tracking, segmentation (Figs. 2B, 3A, and 5D), and measurement of area and volume (Fig. 3, E and F, and figs. S9 and S16), the diversity of questions that can be asked when modern genetic and pharmacological tools are combined with high-resolution 5D in vivo data spanning hundreds of cells over many hours will demand bioinformatics expertise, machine learning, and custom algorithm development on an unprecedented level. Nevertheless, such efforts promise to offer insights into how cells harness their intrinsic variability to adapt to different physiological environments and have the potential to reveal the phenotypic diversity of organelle morphologies, intracellular dynamics, extracellular communication, and collective cell behavior across different cell types, organisms, and developmental stages.
Materials and methods
Lattice light-sheet subsystem
The lattice light-sheet excitation path of the AO-LLSM was designed as described previously (6). Noted here are the changes introduced in the AO-LLSM. The collinear laser beams from the combiner were first expanded using a pair of cylindrical lenses and aligned such that up to three different wavelengths illuminated three vertically separated thin stripes on spatial light modulator SLM, (Holoeye, PLUTO-Vis-014 1920 × 1080 pixels; fig. S1). As a grayscale phase modulation device, SLM was introduced to not only create the light sheet but to correct sample-induced aberrations as well. The diffraction orders reflected from SLM were then filtered using annular mask MSK (Photo Sciences) as before and conjugated to galvanometer scanning mirrors G3 and G4 (3 mm mirror, Cambridge Technology, 8315H) to scan the light sheet along the x and the z axis. During imaging, different offset voltages were applied to the z galvo to sequentially realign the light sheet from each laser to the same plane within the specimen. Sample plane conjugate resonant galvanometer RG (Electro-Optical Products Corp. 7 × 8 mm, SC-30) was also added prior to the excitation objective (Special Optics, 0.65 NA, 3.74 mm WD) to wobble the light sheet in the xy plane and thereby minimize stripe artifacts due to localized absorbing or scattering objects in the specimen. The fluorescence generated in the excitation plane was collected with detection objective DO (Nikon, CFI Apo LWD 25XW, 1.1 NA, 2 mm WD) and reflected off deformable mirror DM (ALPAO 97-15) conjugate to the rear focal plane of DO before being imaged at sCMOS camera CAM 1 (Hamamatsu Orca Flash 4.0 v2). Complete details of the optical design are given in supplementary note 1.
Adaptive optics subsystems
In principle, independent AO corrective systems are needed for excitation and detection in LLSM, since light traverses different regions of the specimen in each case and hence is subject to different aberrations. However, given that: (i) aberrations decrease quickly with decreasing numerical aperture (NA) (7); (ii) we use at most 0.6 NA for excitation, versus 1.1 NA for detection in LLSM; and (iii) only aberrations within a narrow annulus at the rear pupil of the excitation objective will affect a lattice light sheet, it is not obvious that AO correction of the light sheet itself is necessary. To check, we simulated the effect of aberrations consisting of random combinations of the 55 lowest-order Zernike modes up to a root mean square (RMS) amplitude of two wavelengths (λ). We found (fig. S19) that aberrations at this level could expand a 0.7-μm-thick lattice light sheet to as much as 20 μm, and displace it perpendicular to its plane by up to ±8 μm, indicating that correction of excitation as well as detection is essential.
Hence, during aberration measurement, light from Ti:Sapphire ultrafast pulsed laser 2PL (Coherent Cameleon Ultra II) was ported to either the excitation or detection arm by switching galvanometer SG1 (fig. S1). In either case, TPEF generated within the specimen by scanning the guide star focused by EO or DO was collected by the same objective, descanned (11) and sent to homebuilt Shack-Hartmann wavefront sensor ESH or DSH, each consisting of a square microlens array (Edmund Optics) focused onto an EMCCD camera (Andor iXon). Corrective wavefronts were then applied to SLM or DM as described in supplementary notes 3 or 2, respectively. Further hardware details are given in supplementary note 1.
Autofocus measurement was achieved by viewing, side-on through EO, both the light-sheet fluorescence and the plane of fluorescence generated by guide star TPE excitation through DO on camera CAM4, and correcting for any displacement between them as outlined in supplementary note 5.
Zebrafish immobilization, mounting, and imaging conditions
Zebrafish embryos were paralyzed with ~1 ng of α-bungarotoxin protein injected prior to imaging (43) or anesthetized using tricaine (0.16 mg/ml) for 15 min. 12-mm-diameter glass coverslips were precleaned as follows: ~20 coverslips were placed in a 50 ml Falcon tube containing 0.1M NaOH and the tube placed in a sonicator for 15 min, followed by at least 5 consecutives washes with Mili-Q water and then immobilized in the sample holder using superglue. An agarose holder containing narrow groves for mounting the embryos was created by solidifying a few drops of 0.5 to 2% (wt/wt) low-melting agarose between the coverslip and a mold containing ridges. For the immune cell experiments, larvae were placed in 3D-printed volcano-shaped mounts (https://www.shapeways.com/shops/megason-lab). A homemade hair-loop was used to position the embryo in the mold, which was then stabilized with a thin layer of agarose made by applying on top of the immobilized embryo ~10 to 20 μl 1% low-melting agarose at 37° to 40°C and then wicking the excess. After solidification, the sample holder was bolted onto a three-axis set of sample stages (Attocube, ECS3030 for x and y, ECS3050 for z) and submerged in a sample bath containing ~8 ml of 1x Danieau buffer. This assembly was then raised by a motorized actuator (Newport, LTA-HS Actuator, integrated with CONEX-CC Controller, CONEX-LTA-HS) until EO and DO were immersed in the media. The sample stages then positioned the desired FOV to the mutual focal point of the objectives. Detailed imaging conditions for each experiment discussed in the paper, including excitation power, imaging time, image, tile and voxel sizes, fluorophores, and proteins, are in table S1. Additional preparation conditions are discussed in supplementary note 6.
Supplementary Material
Acknowledgments
We thank the Shared Resource teams at the Janelia Research Campus for their skill and dedication in specimen handling and preparation, K. Schaefer for the preparation of cancer cells for the xenograph experiments, and the Instrument Design and Fabrication team for their manufacturing expertise. We also gratefully acknowledge the support of the Janelia Visitor Program. S.U. thanks H. Elliott, D. Richmond, and R. Gao for discussions and acknowledges the MATLAB code repository received from the Computational Image Analysis Workshop supported by NIH grant GM103792.
Funding: T.-L.L., D.E.M., V.S., J.S., M.K., E.M.M., and E.B. are funded by the Howard Hughes Medical Institute (HHMI). T.K. and S.U. are funded by grants from Biogen, Ionis Pharmaceuticals, and NIH grant R01GM075252 (to T.K.). S.U. is a Fellow at the Image and Data Analysis core at Harvard Medical School. S.S. is funded by a Living Systems Institute start-up grant, University of Exeter. K.R.M., I.A.S., Z.M.C., T.W.H., and S.G.M. were supported by NIH grant R01DC015478. D.Q.M. is funded by the NIH (5R00CA154870-05 and 1R01GM121597-01). D.Q.M. and B.L.M. are funded by the Carol M. Baldwin Foundation and are Damon Runyon-Rachleff Innovators supported (in part) by the Damon Runyon Cancer Research Foundation (DRR-47-17). B.L.M. is also funded by the NSF (IOS1452928). D.H. is a Pew-Stewart Scholar for Cancer Research supported by the Pew Charitable Trusts and NIH grant R01CA196884. D.D. was supported by a Human Frontier Science Program Fellowship. D.G.D. was supported by NIH grant R35GM118149.
Footnotes
Author contributions: E.B. supervised the project and wrote the manuscript with input from all coauthors. T.-L.L. built the microscope with input from E.B., D.E.M., and K.W. and performed all microscope characterization experiments. D.E.M. created the instrument control software. T.-L.L., V.S., and S.U. acquired all biological data with coauthors. D.D., D.G.D., R.F., and D.H. provided organoids and led related experiments. T.K., S.U., B.C., and S.S. provided transgenic zebrafish and AP2 cells and led related clathrin experiments. Z.M.C., T.W.H., and S.G.M. created the mCardinal-PM transgenic zebrafish. T.K., S.G.M., S.U., and I.A.S. provided zebrafish and led related organelle dynamics and in vivo immune cell experiments. J.S. and M.K. created the Autobow zebrafish, and M.K. led related experiments. A.Q.K., T.N.M., and D.Q.M. provided C. elegans specimens and led related experiments. B.L.M. provided MDA-MB-231 cells and vasculature-labeled zebrafish and led related experiments with D.Q.M. Y.R., H.Y., and E.M.M. provided Arabidopsis specimens and led related experiments. S.U., K.R.M., T.-L.L., and V.S. processed all image data. S.U. performed quantitative analysis of all image data. S.U., T.-L.L., and E.B. produced all figures and movies
Competing interests: Portions of the technology described herein are covered by U.S. Patent 7,894,136 issued to E.B., assigned to Lattice Light of Ashburn, VA, and licensed to Carl Zeiss Microscopy; U.S. Patents 8,711,211 and 9,477,074 issued to E.B., assigned to HHMI, and licensed to Carl Zeiss Microscopy; U.S. Patent application 13/844,405 filed by E.B. and K.W. and assigned to HHMI; and U.S. Patent 9,500,846 issued to E.B. and K.W. and assigned to HHMI.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the supplementary materials. Documentation for construction of a lattice light-sheet microscope can be obtained by execution of a research license agreement with HHMI.
REFERENCES AND NOTES
- 1.Icha J, Weber M, Waters JC, Norden C. Phototoxicity in live fluorescence microscopy, and how to avoid it. BioEssays. 2017;39:1700003. doi: 10.1002/bies.201700003. [DOI] [PubMed] [Google Scholar]
- 2.Laissue PP, Alghamdi RA, Tomancak P, Reynaud EG, Shroff H. Assessing phototoxicity in live fluorescence imaging. Nat Methods. 2017;14:657–661. doi: 10.1038/nmeth.4344. [DOI] [PubMed] [Google Scholar]
- 3.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 4.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigert R, Sramkova M, Parente L, Amornphimoltham P, Masedunskas A. Intravital microscopy: A novel tool to study cell biology in living animals. Histochem Cell Biol. 2010;133:481–491. doi: 10.1007/s00418-010-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen BC, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji N. Adaptive optical fluorescence microscopy. Nat Methods. 2017;14:374–380. doi: 10.1038/nmeth.4218. [DOI] [PubMed] [Google Scholar]
- 8.Wilding D, Pozzi P, Soloviev O, Vdovin G, Verhaegen M. Adaptive illumination based on direct wavefront sensing in a light-sheet fluorescence microscope. Opt Express. 2016;24:24896–24906. doi: 10.1364/OE.24.024896. [DOI] [PubMed] [Google Scholar]
- 9.Bourgenot C, Saunter CD, Taylor JM, Girkin JM, Love GD. 3D adaptive optics in a light sheet microscope. Opt Express. 2012;20:13252–13261. doi: 10.1364/OE.20.013252. [DOI] [PubMed] [Google Scholar]
- 10.Aviles-Espinosa R, et al. Measurement and correction of in vivo sample aberrations employing a nonlinear guide-star in two-photon excited fluorescence microscopy. Biomed Opt Express. 2011;2:3135–3149. doi: 10.1364/BOE.2.003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, et al. Rapid adaptive optical recovery of optimal resolution over large volumes. Nat Methods. 2014;11:625–628. doi: 10.1038/nmeth.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Royer LA, et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat Biotechnol. 2016;34:1267–1278. doi: 10.1038/nbt.3708. [DOI] [PubMed] [Google Scholar]
- 13.Arbizzani F, Mayrhofer M, Mione M. Novel transgenic lines to fluorescently label clathrin and caveolin endosomes in live zebrafish. Zebrafish. 2015;12:202–203. doi: 10.1089/zeb.2015.1501. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich M, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Li D, et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:aab3500. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kural C, et al. Dynamics of intracellular clathrin/AP1- and clathrin/AP3-containing carriers. Cell Rep. 2012;2:1111–1119. doi: 10.1016/j.celrep.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vassilopoulos S, et al. Actin scaffolding by clathrin heavy chain is required for skeletal muscle sarcomere organization. J Cell Biol. 2014;205:377–393. doi: 10.1083/jcb.201309096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguet F, et al. Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Mol Biol Cell. 2016;27:3418–3435. doi: 10.1091/mbc.E16-03-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadlecova Z, et al. Regulation of clathrin-mediated endocytosis by hierarchical allosteric activation of AP2. J Cell Biol. 2017;216:167–179. doi: 10.1083/jcb.201608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Ladinsky MS, Kirchhausen T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol Biol Cell. 2009;20:3471–3480. doi: 10.1091/mbc.E09-04-0327;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci USA. 2007;104:7939–7944. doi: 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Icha J, Kunath C, Rocha-Martins M, Norden C. Independent modes of ganglion cell translocation ensure correct lamination of the zebrafish retina. J Cell Biol. 2016;215:259–275. doi: 10.1083/jcb.201604095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sidhaye J, Norden C. Concerted action of neuroepithelial basal shrinkage and active epithelial migration ensures efficient optic cup morphogenesis. eLife. 2017;6:e22689. doi: 10.7554/eLife.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Icha J, et al. Using light sheet fluorescence microscopy to image zebrafish eye development. J Vis Exp. 2016;110:e53966. doi: 10.3791/53966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer FC. Mitosis in the neural tube. J Comp Neurol. 1935;62:377–405. doi: 10.1002/cne.900620207. [DOI] [Google Scholar]
- 26.Strzyz PJ, et al. Interkinetic nuclear migration is centrosome independent and ensures apical cell division to maintain tissue integrity. Dev Cell. 2015;32:203–219. doi: 10.1016/j.devcel.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Lowery LA, Van Vactor D. The trip of the tip: Understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentley D, Toroian-Raymond A. Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia by cytochalasin treatment. Nature. 1986;323:712–715. doi: 10.1038/323712a0. [DOI] [PubMed] [Google Scholar]
- 29.Chien CB, Rosenthal DE, Harris WA, Holt CE. Navigational errors made by growth cones without filopodia in the embryonic Xenopus brain. Neuron. 1993;11:237–251. doi: 10.1016/0896-6273(93)90181-P. [DOI] [PubMed] [Google Scholar]
- 30.Zheng JQ, Wan JJ, Poo MM. Essential role of filopodia in chemotropic turning of nerve growth cone induced by a glutamate gradient. J Neurosci. 1996;16:1140–1149. doi: 10.1523/JNEUROSCI.16-03-01140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10:540–547. doi: 10.1038/nmeth.2450. [DOI] [PubMed] [Google Scholar]
- 32.Kinkhabwala A, et al. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc Natl Acad Sci USA. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Oliveira S, Rosowski EE, Huttenlocher A. Neutrophil migration in infection and wound repair: Going forward in reverse. Nat Rev Immunol. 2016;16:378–391. doi: 10.1038/nri.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reymond N, d–Água BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- 35.Marki A, Buscher K, Mikulski Z, Pries A, Ley K. Rolling neutrophils form tethers and slings under physiologic conditions in vivo. J Leukoc Biol. 2017;103:67–70. doi: 10.1189/jlb.1AB0617-230R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoletov K, et al. Visualizing extravasation dynamics of metastatic tumor cells. J Cell Sci. 2010;123:2332–2341. doi: 10.1242/jcs.069443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 38.Tomer R, Khairy K, Amat F, Keller PJ. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat Methods. 2012;9:755–763. doi: 10.1038/nmeth.2062. [DOI] [PubMed] [Google Scholar]
- 39.Chhetri RK, et al. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat Methods. 2015;12:1171–1178. doi: 10.1038/nmeth.3632. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A, et al. Dual-view plane illumination microscopy for rapid and spatially isotropic imaging. Nat Protoc. 2014;9:2555–2573. doi: 10.1038/nprot.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swoger J, Verveer P, Greger K, Huisken J, Stelzer EH. Multi-view image fusion improves resolution in three-dimensional microscopy. Opt Express. 2007;15:8029–8042. doi: 10.1364/OE.15.008029. [DOI] [PubMed] [Google Scholar]
- 42.Simmonds RD, Booth MJ. Modelling of multi-conjugate adaptive optics for spatially variant aberrations in microscopy. J Opt. 2013;15:094010. doi: 10.1088/2040-8978/15/9/094010. [DOI] [Google Scholar]
- 43.Swinburne IA, Mosaliganti KR, Green AA, Megason SG. Improved long-term imaging of embryos with genetically encoded α-bungarotoxin. PLOS ONE. 2015;10:e0134005. doi: 10.1371/journal.pone.0134005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.