Abstract
Light activated compounds are powerful tools and potential agents for medical applications, as biological effects can be controlled in space and time. Ruthenium polypyridyl complexes can induce cytotoxic effects through multiple mechanisms, including acting as photosensitizers for singlet oxygen (1O2) production, the generation of other reactive oxygen species (ROS), the release of biologically active ligands, or through the creation of reactive intermediates that form covalent bonds to biological molecules. A structure activity relationship (SAR) study was performed on a series of Ru(II) complexes containing isomeric tetramethyl-substituted bipyridyl-type ligands. Three of the ligand systems studied contained strain-inducing methyl groups and created photolabile metal complexes, which can form covalent bonds to biomolecules upon light activation, while the fourth was unstrained and resulted in photostable complexes, which can generate 1O2. The compounds studied included both bis-heteroleptic complexes containing two bipyridine ligands and a third, substituted ligand, and tris-homoleptic complexes containing only the substituted ligand. The photophysics, electrochemistry, photochemistry, and photobiology were assessed. Strained heteroleptic complexes were found to be more photo-active and cytotoxic then tris-homoleptic complexes, and bipyridine ligands were superior to bipyrimidine. However, the homoleptic complexes exhibited an enhanced ability to inhibit protein production in live cells. Specific methylation patterns were associated with improved activation with red light, and photolabile complexes were generally more potent cytotoxic agents than the photostable 1O2 generating compounds.
Introduction
Research in the area of medicinal inorganic chemistry generally involves the generation and testing of metal complexes that function as prodrugs, undergoing chemical reactions as a result of thermal ligand exchange, electron transfer reactions, or photophysical or photochemical reactions. There is an added benefit of potential selectivity for tumor tissues when working with compounds which are triggered by electrons, photons, or other species that can be controlled externally.1 Alternatively, environmental features, such as hypoxia, which are associated with the tumor microenvironment, can also provide for selectivity.2, 3 In the context of controlling reactivity through photons, ruthenium polypyridyl complexes have been the subject of extensive study, as the complexes absorb visible light and have multiple excited state relaxation pathways that can be harnessed to induce cytotoxicity. This has been a very productive area of research, with contributions from many groups applying a variety of photochemical, photophysical, and biological approaches and mechanisms; a limited selection of references closely related to this current work is provided.4-26
Three main excited state reaction pathways are currently utilized. The first is Type II photoreactivity, where the triplet metal-to-ligand charge transfer (3MLCT) excited state sensitizes tissue oxygen, producing singlet oxygen (1O2).7 The second is electron transfer reactions, which can be oxidative or reductive, as Ru(II) polypyridyl complexes are both strong oxidizing and reducing agents in the excited state.27 The third is a photodissociative pathway, where the 3MLCT can relax to a formally antibonding metal centered (3MC) state, which results in ligand loss. While this is a possible relaxation pathway in all Ru(II) polypyridyl complexes, the efficiency can be radically enhanced and controlled by the incorporation of sterically hindered ligands, as elegantly demonstrated by Jean-Pierre Sauvage.28, 29 We have previously utilized this photochemical reactivity to develop potent cytotoxic agents.30-33
In the present study, we systematically investigated the effect of methylation patterns on bidentate ligands with regard to electrochemistry, photophysics, photochemistry, and photobiological activity of the corresponding Ru(II) complexes. In addition to the bipyridine ligand, bipyrimidine was also explored. Five of the complexes contained strain-inducing ligands and were photolabile. Subsequently the coordinatively unsaturated complex can form covalent bonds to biomolecules after light activation. Two of the studied complexes were unstrained and can generate 1O2 under irradiation. Surprising trends emerged that resulted in the identification of specific methylation patterns and symmetry features that correlated with reactivity and specific biological effects.
Results
Synthesis and Characterization
The Ru(II) complexes discussed in this paper are depicted in Chart 1. The symmetrical tetramethyl derivatives of 2,2′-bipyridine (bpy) used in complexes 1 – 6 have been reported (4,4′, 6,6′-tetramethyl-2,2′-bipyridine,34-37 5,5′, 6,6′-tetramethyl-2,2′-bipyridine,38 and 4,4′, 5,5′-tetramethyl-2,2′-bipyridine39) and were prepared by the oxidative homo-coupling of the corresponding dimethylpyridine under the influence of Pd/C at elevated temperature. The 4,4′,6,6′-tetramethyl-2,2′-bipyrimidine ligand used in complex 7 was prepared by reported procedures.40 All complexes were synthesized as racemic mixtures of the Δ and Λ enantiomers. The synthesis, isolation, and characterization of the complexes was performed under low light conditions. The heteroleptic complexes 1 – 3 and 7 were generated in good yields by treating [Ru(bpy)2Cl2] ·2H2O with one equivalent of the appropriate ligand and precipitation of the complex with NH4PF6. The preparation of the homoleptic Ru-complexes proved to be more challenging. Complexes 4 – 6 were prepared by treating RuCl33H2O with 3.1 equivalents of the appropriate ligand under microwave irradiation followed by precipitation of the desired complex with NH4PF6. The reactions involving tetramethylbpy (tmbpy) ligands carrying methyl groups ortho to the nitrogens proceeded in low yields; 17% for 4 and 3% for 5, probably due to steric hindrance by the methyl groups. Indeed, structural studies have revealed significant distortions in homoleptic complexes containing related strain-inducing ligands.41
Chart 1.
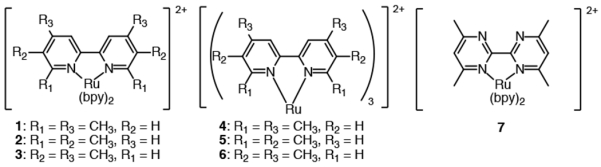
Structure of compounds included in this study.
The Ru complexes were characterized by their 1H NMR spectra that showed very clear first order behavior (see SI). The electrochemical redox potentials were measured in acetonitrile and all complexes exhibited one reversible oxidation wave and, with the exception of complex 6, two reversible and one quasi-reversible reduction waves (Table 1). The UV-visible absorption spectra of the complexes were measured for 10−5 M acetonitrile solutions at 20 °C, and the data is recorded in Table 1. All the complexes showed a characteristic long wavelength absorption band typically associated with a metal-to-ligand charge transfer (MLCT) involving the promotion of an electron from a metal-based HOMO to a ligand-based LUMO. For all the complexes, this absorption occurs over a narrow range of 444–455 nm with very comparable molar extinction constants.
Table 1.
Electronic absorption, emission, and ground state electrochemical potential data for all Ru complexes.
| Compound | λmax (log ε)a | λem (20 °C)b | E1/2ox (ΔE)c | E1/2red (ΔE)c |
|---|---|---|---|---|
| 1 | 244 (4.36), 288 (4.86), 451 (4.05) | non-emissive | 1.23 (81) | −1.37 (80), −1.59 (97), −1.87 (108) |
| 2 | 245 (4.09), 289 (4.57), 452 (3.80) | non-emissive | 1.25 (80) | −1.37 (74), −1.59 (98), −1.95 (124) |
| 3 | 254 (4.41), 288 (4.94), 451 (4.12) | 625 | 1.15 (78) | −1.41 (64), −1.61 (84), −1.97 (112) |
| 4 | 257 (4.25), 297 (4.79), 455 (4.02) | non-emissive | 1.18 (76) | −1.48 (69), −1.70 (74), −1.96 (99) |
| 5 | 270 (4.41), 307 (4.67), 445 (3.89) | non-emissive | 1.26 (83) | −1.57ir |
| 6 | 266 (4.48), 291 (4.90), 450 (4.13) | 599 | 0.99 (73) | −1.65 (83), −1.83 (89), −2.07 (107) |
| 7 | 246 (4.49), 286 (4.82), 444 (4.12) | non-emissive | 1.33 (76) | −1.20 (71), −1.51 (76), −1.75 (80) |
“Measured in CH3CN (1.0 × 10−5 M) at 20 °C; λ in nm and log ε in L·mol−1·cm−1.
Measured in CH3CN at 20 °C; excitation at absorption maxima; λ in nm.
Measured with a glassy-carbon electrode at 100 mV/s in CH3CN containing 0.1 M NBu4PF6 and E1/2 reported in volts relative to SCE; E1/2 = (Epa + Epc)/2 in volts, and ΔE = (Epa − Epc) in mV; ir = irreversible.
The complexes containing methyl groups ortho to the coordinating nitrogen were anticipated to be susceptible to ligand loss upon irradiation, as these methyl groups introduce steric interference near the metal center. This interference activates a photodissociative pathway from a triplet metal centered (3MC) excited state.42, 43 The 3MC is populated from the 3MLCT state, and provides an additional, irreversible non-radiative decay pathway that overrides the radiative relaxation pathway. As a result, none of the complexes containing methyl groups at the 6,6′ showed any emission. In contrast, the unstrained heteroleptic and homoleptic complexes of 4,4′:5,5′-tetramethylbpy (3 and 6) were emissive; they also did not show any significant photodissociation even after 6 h of irradiation.
Photochemical Characterization
All the complexes containing strain-inducing ligands were photoactive, and underwent light-induced ligand exchange. The behavior was similar to that of [Ru(bpy)2dmbpy]2+ (dmbpy = 6,6′-dimethyl-2,2′-bipyridine), which was used as a control for all studies; we have previously demonstrated strain-induced photochemical ligand loss with this complex.30 The different complexes exhibited half-life (t1/2) values that were dependent on the wavelength of light and power used for the photochemical reaction. Several different light sources and wavelengths were evaluated, as shown in Table 2. The photochemical ligand release was always fastest with the Indigo LED (Loctite), which gave the highest power (485 mW/cm2). Next was the blue long pass filter (80 mW/cm2), followed by green (68 mW/cm2) and then red (79 mW/cm2). While only small changes in the t1/2 values were observed moving from the blue to the green cut-off filter, the red filter resulted in significant reductions in the rates of the photochemical reactions. This is likely related to the very low absorptivity of the complexes at wavelengths > 600 nm.
Table 2.
Photoejection half-life analysis using different color light sources.a
| Compound | Indigo | Blue | Green | Red |
|---|---|---|---|---|
| 1 | < 2 s | 14.4 s | 35 s | 37 m |
| 2 | < 2 s | 10.5 s | 26 s | 49 m |
| 3 | - | - | - | - |
| 4 | 10.3 s | 4.5 m | 8.7 m | 250 m |
| 5 | 4.4 s | 2.9 m | 7.8 m | 200 m |
| 6 | - | - | - | - |
| 7 | < 2 s | 32 s | 78 s | 44 m |
| Ru(bpy)2dmbpy | < 2 s | 9.1 s | 29 s | 72.3 m |
Measured using the Loctite Indigo LED or a slide projector equipped with different long pass filters; see Experimental Section.
The quantum yield of photolysis was determined for the compounds using the Indigo LED, assumed to be 450 nm monochromatic light.44 The measured values were 0.08. 0.09, 0.004, 0.01, and 0.05 for compounds 1, 2, 4, 5, and 7, respectively. As a control, [Ru(bpy)2dmbpy]2+ was analyzed, and gave a value of 0.10. This is somewhat dissimilar to a previously reported value of 0.19 for this compound.45 The discrepancy may be due to inaccuracies in the measurement stemming from the rapid photoejection with the Indigo LED.
Cell Cytotoxicity
Given the strain-mediated photochemical reactivity of the family of compounds, light-induced cytotoxicity was investigated using the HL60 leukemia cell line. The complex [Ru(bpy)2dmbpy]2+ was used as a control, as this compound is a potent light-activated cytotoxic agent that has minimal toxicity in the dark.30 All compounds were incubated for 1 h in the dark with the cells before irradiation. Cells were then exposed to light, or alternatively, kept in the dark. Cell survival was quantified 72 h later. As shown in Table 3, cell death was observed following light irradiation, with six of the seven compounds exhibiting IC50 values below 10 μM, though compound 6 only exhibited this activity upon irradiation with the Indigo LED. Only one compound exhibited significant cytotoxicity in the dark, with IC50 values below 100 μM. Notably, compound 7, which contained the bipyrimidine ligand, was one of the least potent of the light-activated complexes.
Table 3.
Cellular cytotoxicity values in HL60 cells using different light sources.a
| Compound | Dark IC50 (μM) | Indigo IC50 (μM) (% Photoejected)a | Blue IC50 (μM) (% Photoejected) a | Green IC50 (μM) (% Photoejected) a | Red IC50 (μM) (% Photoejected) a |
|---|---|---|---|---|---|
| 1 | >100 | 3.0 ± 0.2 (100) | 3.9 ± 0.2 (100) | 3.8 ± 0.4 (98) | 6.8 ± 0.7 (9.3) |
| 2 | >100 | 1.2 ± 0.2 (100) | 1.8 ± 0.1 (100) | 2.9 ± 0.1 (99.5) | 3.9 ± 0.9 (7.4) |
| 3 | >100 | 0.5 ± 0.2 (0) | 3.0 ± 0.5 (0) | 7.7 ± 1.3 (0) | >100 (0) |
| 4 | ~100 | 5.8 ± 1.2 (100) | 7.4 ± 1.2 (55) | 14.0 ± 2 (38) | 60 ± 5b (2.8) |
| 5 | 42.2 ± 9.3 | 3.1 ± 1.2 (100) | 4.7 ± 1.1 (72) | 6.0 ± 1.3 (40) | 20.4 ± 2.3 (2.5) |
| 6 | >100 | 2.9 ± 0.9 (0) | 32 ± 3.5 (0) | ~ 36 ± 3.9 (0) | ~ 43 ± 4.2 (0) |
| 7 | >100 | 4.9 ± 0.8 (100) | ND (100) | ND (100) | >100 (7.3) |
| Ru(bpy)2dmbpy | >100 | 3.4 ± 1.1 (100) | 3.6 ± 1.1 (100) | 3.7 ± 1.1 (99.5) | 11.7 ± 1.4 (4) |
| Cisplatin | 3.1 ± 0.3 | ndc | nd | nd | nd |
| ALA | >100 | nd | 16.2 ± 3.2 | nd | nd |
Measured using the Loctite Indigo LED or a slide projector equipped with different long pass filters; see Experimental Section.
Only 60% cell death achieved at highest dose point, so the IC50 represents 30% cell death.
nd=not determined.
Given the need to use longer wavelengths of light to activate compounds in deeper tissues, the ability of these complexes to be activated by red light was investigated. Following an established procedure, the cells were treated with compounds and then exposed to red light (using a >600 nm cutoff filter).31 Despite the fact that none of the compounds have strong absorption features in this region of the spectrum, compounds 1 and 2 maintained the ability to kill cells using this low energy light. The efficacy is particularly notable as less that 10% of the compound was activated with this light dose, but the IC50 only increased by 2–3 fold.
Recent reports have demonstrated significant cytotoxicity for bipyridine-type ligands containing methy substituents at the positions ortho to the nitrogen.46, 47 The cytotoxicity of the strained, ejecting ligands for compounds 1 and 4, 2 and 5, and 7 were tested, and no cytotoxicity was observed at concentrations up to 30 μM. Accordingly, the activity of the complexes was attributed to the biological interactions of the Ru(II) fragment.48
Singlet Oxygen Production
While compounds that photoeject ligands are likely to induce cell cytotoxicity through formation of covalent adducts, compounds 3 and 6 do not photoeject and thus are anticipated to work through an alternative mechanism. As unstrained Ru(II) complexes possess long-lived 3MLCT excited states, they are able to generate reactive oxygen species. Accordingly, Singlet Oxygen Sensor Green, a fluorescent reporter for the production of singlet oxygen (1O2), was used to determine the ability of the different compounds to create this reactive oxygen species. Tris(bathophenanthroline)Ru(II) ([Ru(dpp)3]2+) was used as a positive control, as this compound has a quantum yield for 1O2 production (ΔO) of 0.42.49 In addition, [Ru(bpy)2dmbpy]2+ was investigated as a negative control, and to determine if photoejection of a ligand had any impact on 1O2 levels.
As shown in Figure 1, all unstrained compounds are able to produce 1O2 upon illumination. Somewhat surprisingly, even excitation with wavelengths > 600 nm produced significant levels of this reactive oxygen species with compound 6, though the Indigo LED was more efficient. [Ru(dpp)3]2+, compound 3, and compound 6 all produced 1O2 in a concentration dependent manner. Both compounds 3 and 6 were more potent than [Ru(dpp)3]2+ with the Indigo LED, but only compound 6 was able to produce an appreciable amount of 1O2 at a concentration of 10 μM using red light.
Figure 1.
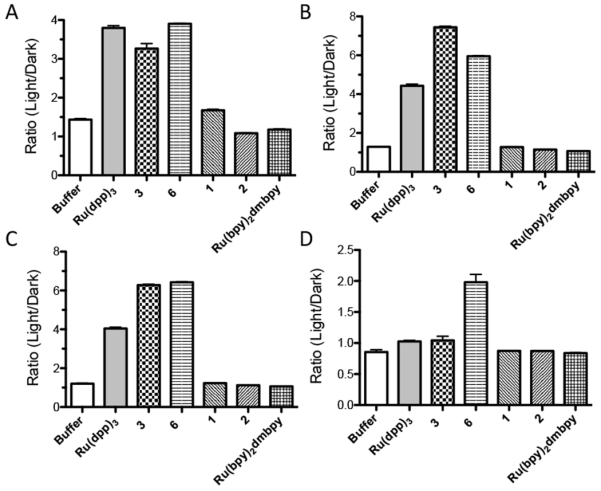
Irradiation-induced 1O2 production as determined by the Singlet Oxygen Sensor Green assay as a function of light source (compound concentrations = 20 μM). A) Indigo LED; B) Blue cut off filter; C) Green cut off filter; D) Red cut off filter.
Unexpectedly, both the photoejecting control compound [Ru(bpy)2dmbpy]2+ and compound 2 slightly quenched the 1O2 in the buffer when irradiated with the Indigo LED. As these compounds photoeject rapidly, it appears that the quenching may be related to the presence of the photoejected product in solution.
Inhibition of Transcription and Translation
Ru(II) complexes that become ligand deficient upon irradiation have been demonstrated to directly damage DNA30, 31, 50 and RNA.51 Rather than probing damage to nucleic acids through detection of structural changes in the biomolecules induced by metal complexes, functional assays can provide a more sensitive and meaningful report for types of biological interactions that inhibit functions essential for cell health and survival. DNA replication52 and transcription53 and translation assays have been used, along with in-cell transcription and translation assays.54 The in vitro transcription and translation (IVTT) assay assesses nucleic acid damage via a readout of production of green fluorescent protein (GFP), providing both ease of compound analysis and high sensitivity.51 All compounds were incubated with the plasmid DNA containing the gene for GFP (pCFE-GFP) for 12 hrs at 22 °C, either in the dark or after activation with light using the Indigo LED. After this, the plasmid DNA solution was added to the IVTT reaction solution and GFP emission was quantified after a 2 hr incubation at 30 °C to allow sufficient time for transcription and translation of GFP.
As shown in Figure 2A, neither compounds 2 nor 6 inhibited protein production when incubated with the DNA in the dark. In marked contrast, all protein synthesis was inhibited for all compounds when exposed to light. Thus, at concentrations of 5 μM the compounds are able to cause the complete cessation of protein synthesis. Interestingly, the only compound that affected protein synthesis in the dark, compound 3, inhibited ca. 40% of the control level of GFP production.
Figure 2.
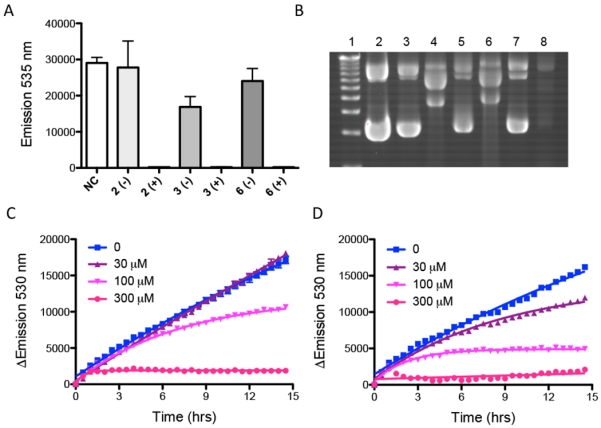
Irradiation-induced DNA damage and inhibition of transcription and translation. A) Inhibition of GFP synthesis following treatment of the pCFE-GFP plasmid with various compounds at 5 μM concentration. B) Agarose gels showing the DNA damage with 40 μg mL−1 pCFE-GPF plasmid used in the IVTT reactions. Lane 1: DNA Ladder; Lane 2: pCFE-GFP control; Lane 3: compound 3, no light; Lane 4: compound 3, light; Lane 5: compound 6, no light; Lane 6: compound 6, light; Lane 7: compound 2, no light; Lane 8: compound 2, light. C) Time dependent inhibition of production of Dendra2 by compound 1 following irradiation in HEK293 T-REx cells. D) Time dependent inhibition of production of Dendra2 by compound 4 following irradiation in HEK293 T-REx cells. The compounds were dosed at the indicated concentrations. The Indigo LED was used for all sample irradiation for both in vitro and in cell experiments.
When the plasmids used in the IVTT experiment were analyzed for structural damage, the different mechanisms could be visualized by differences in DNA migration and EtBr staining. Both compounds 3 and 6 induced strand breaks in the DNA upon irradiation, as evidenced by the loss of the primary, supercoiled band for the pCFE-GFP plasmid and the appearance of new bands. In marked contrast, compound 2 induced covalent adducts, as indicated by the smearing and reduced mobility on the gel, and the loss of the EtBr signal, which has been previously demonstrated is not due to a loss of the DNA, but rather that the presence of the Ru(II) adduct either inhibits EtBr binding or quenches its emission.31, 53
The ability of the compounds to inhibit protein production was also assessed in live cells using an assay that reports on the production of a photoconvertable protein, Dendra2.55 Dendra2 is a fluorescent protein that is expressed with a chromophore that can be excited at 485 nm to induce emission in the green region of the spectrum at 530 nm, similar to GFP. However, the chromophore can be subjected to a photochemical reaction by exposure to 405 nm light, resulting in emission at 595 nm. This creates a stable pool of protein with emission in the red region of the spectrum, while newly synthesized protein will emit at 530 nm. The analysis of the ratio of the two emission features allows for correction for cell number and cell health over the time course of the experiment.55
As shown in Figure 2C and 2D, photoejecting compounds were able to completely abrogate new protein synthesis at the highest dose point of 300 μM. Inhibition was observed for compound 4 at concentrations down to 30 μM, in contrast to compound 1, where inhibition was only observed at 100 μM and higher doses. Compound 4 also inhibited protein production slightly at 300 μM in the dark (see Figure S30). Similar behavior was observed for compound 5. Compound 2 was similar to compound 1, though less potent (Figure S31). Unfortunately, compounds 3 and 6 gave inconclusive data due to the fact that the compounds are emissive.
Discussion
The series of compounds studied involved bis-heteroleptic complexes incorporating two bpy ligands and a third, tetramethyl-substituted ligand, and homoleptic complexes incorporating only the tetramethyl-substituted bidentate ligand. The two compounds with the smallest differences in activity under irradiation vs. in the dark were compounds 4 and 5. These are both homoleptic complexes containing strained 4,4′, 6,6′-tetramethyl-2,2′-bipyridyl and 5,5′, 6,6′-tetramethyl-2,2′-bipyridyl ligands. Interestingly, while these ligands are able to induce intramolecular strain, the homoleptic complexes eject far less efficiently than the corresponding bis-heteroleptic complexes 1 and 2, which was associated with a ca. 20-fold difference in the t1/2 values. This is qualitatively consistent with a report from Hauser, who compared compound 4 to a tris complex containing the asymmetric ligand 6-methyl-2,2′-bipyridyl.56 The 3MC state was stabilized for compound 4 in comparison to the complex containing fewer strain-inducing methyl groups, and there was a concomitant decrease in the lifetime of the dissociative excited state, which correlated with a decrease in the photodissociative quantum yield. Presumably, the same effect would occur in complexes that contain one or more unstrained ligands.
The heteroleptic complexes 1 and 2 provided better PI values than the associated tris complexes, with greater than 33- and 83-fold differences in toxicity in the dark and with the indigo light. In addition, both 1 and 2 could be activated with red light, with only a 2–3-fold reduction in potency from the values obtained with the indigo light. From an analysis of these four compounds, it appears that heteroleptic compounds containing the 5,5′, 6,6′-tetramethyl-2,2′-bipyridine ligand provide improved biological properties for compounds with larger therapeutic windows.
Of the compounds that do not undergo light-induced ligand dissociation, the bis-heteroleptic complex (compound 3) and the tris-homoleptic complex (compound 6) had IC50 values that increased ca. 10-fold upon irradiation with indigo light, which is similar to the values observed for compounds like [Ru(bpy)3]2+, which have a moderate ability to produce singlet oxygen. Compound 6 maintained the ability to generate 1O2 even under red light irradiation, in contrast to the control compound [Ru(dpp)3]2+, though this did not translate to cytotoxicity under irradiation with the same light. Finally, compound 7 is an outlier, as it exhibited essentially poor toxicity upon irradiation with red light, despite the fact that the compound photoejects to give the same amount of the activated species as for compound 2. We are not able to explain this finding, but it is possible that photoejection is reduced in the biological environment of the cell.
A specific mechanism of DNA damage is not required to inhibit the processes of transcription and translation using an in vitro assay. Both the covalent adducts formed by compound 2, visualized by the loss of signal of the EtBr stained DNA, and ROS mediated DNA damage produced by compounds 3 and 6, which led to strand scission, were equally effective in eliminating the production of GFP (Figure 2A). However, the nature of the IVTT assay can bias the results towards the observation of DNA damage, as the compounds are incubated with the nucleic acid with no other competing biomolecules in solution.
An in-cell assay we recently developed to monitor effects on protein production55 gave intriguing results for the different compounds as a function of compound structure. The bis-heteroleptic compound 1, containing the 4,4′, 6,6′-tetramethyl-2,2′-bipyridine ligand was able to completely abrogate protein production at high concentrations when irradiated, but its tris-homoleptic analogue 4 was even more potent, with effects observed at the 30 μM concentration point (Figure 2C and D). This compound, however, also has some inhibitory activity at high concentrations in the dark. While the strain-inducing ligand is the same in the two complexes, compound 4 transforms into the “active” diaqua species that still contains two 4,4′, 6,6′-tetramethyl-2,2′-bipyridyl ligands, in contrast to compound 1, which becomes the less sterically encumbered [Ru(bpy)2(H2O)2]2+ upon light activation. Clearly, the presence of the 4,4′, 6,6′-tetramethyl-2,2′-bipyridine ligands increases the potency of the aqua species for inhibition of protein production, and the presence of three of these ligands imparts some activity (or affinity for a biological target) before irradiation.
Also notable is the fact that compounds 1 and 4 have a rapid impact on protein production, with effects observed as soon as 2 hr after treatment. This observation may not be fully consistent with a mechanism where the compounds only inhibit transcription, as the mRNA for the Dendra2 reporter protein is present in the cells for hours after its production, and thus, transcriptional inhibition is unlikely be observed until several hours have passed. It is possible this effect is indicative of either damage to the mRNA encoding the Dendra2 protein, or direct inhibition of translation.
It also is apparent that the cytotoxic potency of compounds 1 and 2 (IC50 values of 3 and 1.2 μM) is not matched by the potency for inhibition of protein synthesis, as no effects are observed for either compound at concentrations below 30 μM. The values for compound 4 are in slightly better agreement, which may suggest that inhibition of protein production is an important component of the mechanism of action of this compound. However, the different time scales for the two experiments (15 hrs for the protein production assay, and 72 hrs for cytotoxicity) may result in an amplification of what appears to be a small effect over a short time period for the translation assay.
In conclusion, comparison of various complexes containing methylated bipyridine and bipyrimidine ligands revealed unanticipated structure-activity relationships. In all cases, bis-heteroleptic complexes were more potent in cytotoxicity assays, regardless of the nature of the reactive species produced (1O2 or ligand deficient metal centers). The heteroleptic complexes ejected more rapidly and generally exhibited lower dark toxicities. Complexes containing bipyridine ligands were preferable to the bipyrimidine ligand containing the same methylation pattern (compound 1 vs. compound 7). Analysis of effects on transcription and translation potentially suggest that specific methylation patterns on the ligands increase potency, but the complexes may also have undesired activity in the dark, which increases with the number of methylated ligands. Finally, several complexes could be activated with red light, which was not anticipated based on the absorption profile. This phenomena has been described before, notably by the groups of Harry Morrison,57, 58 Peter Sadler,59 and Sherri McFarland,60-62 with the explanation of direct excitation to the triplet excited states despite the low oscillator strength. In such cases, the “action spectrum” does not recapitulate the absorbance spectrum. This appears to be the case for several of the methylated Ru(II) complexes in this report. The 5,5’, 6,6’-tetramethyl-2,2’-bipyridine ligand is a lead compound for incorporation into heteroleptic complexes to provide good PI values and activation with red light, and the 4,4’, 6,6’-tetramethyl-2,2’-bipyridine ligand results in the most efficacious inhibition of protein production in live cells. Complexes containing these ligands or their derivatives will be the subject of further research.
Experimental section
The symmetrical tetramethyl derivatives of 2,2’-bipyridine (bpy), 4,4’, 6,6’-tetramethyl-2,2’-bipyridine,34-37 5,5’, 6,6’-tetramethyl-2,2’-bipyridine,38 and 4,4’, 5,5’-tetramethyl-2,2’-bipyridine39 have been reported and were prepared by the oxidative homo-coupling of the corresponding dimethylpyridine under the influence of Pd/C at elevated temperature. The 4,4’,6,6’-tetramethyl-2,2’-bipyrimidine was prepared by reported procedures.40
The 1H NMR spectra were recorded on a JEOL ECX-400 or an ECA-500 spectrometer operating at 400 or 500 MHz, respectively. The chemical shifts were reported in parts per million (ppm) and were referenced to the solvent residue peaks, which were referenced to tetramethylsilane. CV measurements were carried out on a Bioanalysis BAS Epsilon Electroanalytical System. The CV experiments were performed in a one-compartment cell equipped with a glassy carbon working electrode, a SCE, and a platinum wire auxiliary electrode. For compound synthesis, a household microwave oven (Samsung, Model MW 2000 U) was modified according to a previously published description.63
Absorption spectra were obtained on an Agilent 8453 diode array spectrometer. Samples were read at a concentration of 20 μM in a quartz cuvette at room temperature; the absorption was corrected for the background absorption of the solvent. For Indigo light, samples were irradiated with a LOCTITE LED Flood System Indigo Array Light. For the red, green and blue light samples, the compounds were irradiated in a 410 Watt 3M 955 overhead projector with the stage glass removed. Edmund Optics filters (item numbers NT43-941, NF49-935, NT43-947, and NT43-954) were used to cut off the appropriate sections of the UV and visible spectrum for the respective colors. Prism software was used to analyze data and the kinetic half-lives were measured from fitting the data points to a one-phase association curve.
Quantum yields of photolysis were determined as described in the literature.13 Briefly, ferrioxalate was used as an actinomiter to determine the photon flux of the Indigo LED, and as Φλ (the quantum yield of the actinometer at 450 nm, the wavelength of the LED lamp) has not been reported, Φ436 was used with a reported value of 1.11.64 The decrease in absorption of each complex was determined as a function of irradiation time, taking data every second. The value at each time point for moles of compound was then plotted against the moles of photons, and the initial linear region was used to determine the slope of the line, which provides the quantum yield. Significant uncertainty is associated with compounds with t1/2 values of less than 10 s.
All synthesized compounds were isolated in > 94 % purity, as determined by analytical HPLC. For HPLC analysis, the ruthenium complexes were injected into an Agilent 1100 series HPLC equipped with a model G1311 quaternary pump, G1315B UV diode array detector, and ChemStation software version B.01.03. Chromatographic conditions were optimized on a Phenomenex C18(2), 100 A (250 mm × 4.6 mm inner diameter, 5 μM) fitted with a Phenomenex C18 (4 mm × 3 mm) guard column. Injection volumes of 20 μL of 100 μM solutions of the complex were used. The detection wavelength was 280 nm. Mobile phases were: mobile phase A, formic acid (0.1 %) in distilled water (dH2O); mobile phase B, formic acid (0.1 %) in HPLC grade acetonitrile. The mobile phase flow rate was 1.0 mL min–1. The following mobile phase gradient was used: 98–95% A (containing 2–5% B) from 0 to 5 min; 95–70% A (5–30% B) from 5 to 15 min; 70–40 % A (30–60 % B) from 15 to 20 min; 40–5% A (60–95% B) from 20 to 30 min; 5–98% A (95–2% B) from 30 to 35 min; re-equilibration at 98 % A (2 % B) from 35 to 40 min.
Before performing photochemical and biological experiments, all compounds were converted to Cl− salts by dissolving 5–20 mg of product in 1–2 mL methanol. The dissolved product was loaded onto an Amberlite IRA-410 chloride ion exchange column, eluted with methanol, and the solvent was removed in vacuo.
Compound 1 – [Ru(bpy)2(4,4’, 6,6’-tetramethyl-2,2’-bipyridine)](PF6)2
[Ru(bpy)2Cl2]·2H2O (258 mg, 0.496 mmol) and 4,4’,6,6’-tetramethyl-2,2’-bipyridine (119 mg, 0.559 mmol) were suspended in ethylene glycol (10 mL) and the reaction mixture was irradiated with microwaves for 30 min (2 × 5 min, 2 × 10 min). The reaction mixture was allowed to cool to room temperature and NH4PF6 (400 mg) dissolved in water (20 mL) was added. The precipitate was filtered, washed with water and diethyl ether, and dried. It was purified by chromatography on alumina, with CH2Cl2/acetone as eluent, followed by recrystallization from acetone/diethyl ether to provide the desired complex as an orange solid (300 mg, 66%): 1H NMR (acetone-d6): δ 8.83 (2H, d, J = 7.8 Hz), 8.76 (2H, d, J = 7.8 Hz), 8.51 (2H, s), 8.29-8.23 (4H, m), 8.13 (2H, dt, J = 7.8, 1.4 Hz), 8.00 (2H, d, J = 5.5 Hz), 7.65 (2H, dt, J = 6.6, 1.4 Hz), 7.43 (2H, dt, J = 6.6, 1.4 Hz), 7.33 (2H, s), 2.50 (6H, s), 1.80 (6H, s). ESI MS calcd for C34H32N6Ru [M]2+ 313.1; found 313.0 [M]2+. Purity by HPLC: 95.1% by area.
Compound 2 – [Ru(bpy)2(5,5’, 6,6’-tetramethyl-2,2’-bipyridine)](PF6)2
[Ru(bpy)2Cl2]·2H2O (126.6 mg, 0.236 mmol) and 5,5’, 6,6’-tetramethyl-2,2’-bipyridine (50.0 mg, 0.236 mmol) were treated as described above for 1 to provide a crude product that was filtered, washed with water and diethyl ether, and dried. The product was isolated as an orange solid (198 mg, 92%): 1H NMR (CD3CN): δ 8.47 (2H, d, J = 8.2 Hz), 8.39 (2H, d, J = 8.2 Hz), 8.12-8.06 (4H, m), 7.96-7.91 (4H, m), 7.75 (2H, d, J = 8.2 Hz), 7.63 (2H, d, J = 5.0 Hz), 7.46 (2H, dt, J = 6.7, 1.4 Hz), 7.21 (2H, dt, J = 5.7, 1.4 Hz), 2.19 (6H, s), 1.59 (6H, s). ESI MS calcd for C34H32N6Ru [M]2+ 313.1; found 313.1 [M]2+. Purity by HPLC: 98.6% by area.
Compound 3 – [Ru(bpy)2(4,4’, 5,5’-tetramethyl-2,2’-bipyridine)](PF6)2
[Ru(bpy)2Cl2]·2H2O (122.6 mg, 0.236 mmol) and 4,4’, 5,5’-tetramethyl-2,2’-bipyridine (50.0 mg, 0.236 mmol) were treated as described above for 1 to provide a crude product that was filtered, washed with water and diethyl ether, and dried. The product was isolated as an orange solid (189 mg, 88%): 1H NMR (acetone-d6): δ 8.81 (2H, d, J = 8.2 Hz), 8.80 (2H, d, J = 8.2 Hz), 8.59 (2H, s), 8.23-8.17 (4H, m), 8.05 (2H, d, J = 5.5 Hz), 8.03 (2H, d, J = 6.0 Hz), 7.68 (2H, s), 7.59-7.53 (4H, m), 2.49 (6H, s), 2.12 (6H, s). ESI MS calcd for C34H32N6Ru [M]2+ 313.1; found 313.1 [M]2+. Purity by HPLC: 97.0% by area.
Compound 4 – [Ru(4,4’, 6,6’-tetramethyl-2,2’-bipyridine)3](PF6)2
RuCl3·3H2O (37.3 mg, 0.143 mmol) and 4,4’, 6,6’-tetramethyl-2,2’-bipyridine (100.0 mg, 0.471 mmol) were suspended in ethylene glycol (10 mL) and the reaction mixture was irradiated with microwaves for 20 min (4 × 5 min). The reaction mixture was allowed to cool to room temperature and NH4PF6 (400 mg) dissolved in water (20 mL) was added. The precipitate was filtered, washed with water and diethyl ether, and dried. Chromatography on alumina, eluting first with CH2Cl2/acetone (4:1) and then CH2Cl2/acetone (1:1) followed by recrystallization from acetone/diethyl ether afforded the product as an orange solid (25 mg, 17%): 1H NMR (acetone-d6): δ 8.26 (6H, s), 7.30 (6H, s), 2.49 (18H, s), 1.91 (18H, s). ESI MS calcd for C42H48N6Ru [M]2+ 369.1; found 369.2 [M]2+. Purity by HPLC: 94.6% by area.
Compound 5 – [Ru(5,5’, 6,6’-tetramethyl-2,2’-bipyridine)3](PF6)2
RuCl3·3H2O (37.3 mg, 0.143 mmol) and 5,5’, 6,6’-tetramethyl-2,2’-bipyridine (100.0 mg, 0.471 mmol) were treated as described above for 1 to provide the product as an orange solid (5 mg, 3%): 1H NMR (acetone-d6): δ 8.26 (6H, d, J = 8.2 Hz), 7.88 (6H, d, J = 8.2 Hz), 2.22 (18H, s), 1.95 (18H, s). ESI MS calcd for C42H48N6Ru [M]2+ 369.1; found 369.0 [M]2+. Purity by HPLC: 92.5% by area.
Compound 6 – [Ru(4,4’, 5,5’-tetramethyl-2,2’-bipyridine)3](PF6)2
RuCl3·3H2O (37.3 mg, 0.143 mmol) and 4,4’, 5,5’-tetramethyl-2,2’-bipyridine (100.0 mg, 0.471 mmol) were treated as described above for 1 to provide the product as an orange solid: (140 mg, 95%): 1H NMR (acetone-d6): δ 8.55 (6H, s), 8.63 (6H, s), 2.48 (18H, s), 2.10 (18H, s). ESI MS calcd for C42H48N6Ru [M]2+ 369.1; found 369.1 [M]2+. Purity by HPLC: 97.4% by area.
Compound 7 – Ru(bpy)2(4,4’, 6,6’-tetramethyl-2,2’-bipyrimidine)](PF6)2
[Ru(bpy)2Cl2]·2H2O (122.6 mg, 0.236 mmol) and 4,4’, 6,6’-tetramethyl-2,2’-bipyrimidine (50 mg, 0.236 mmol) were treated as described above for 1 to provide the product as an orange solid (134 mg, 65%): 1H NMR (acetone-d6): δ 8.83 (2H, d, J = 8.2 Hz), 8.76 (2H, d, J = 8.2 Hz), 8.36 (2H, d, J = 6.0 Hz), 8.27 (2H, t, J = 7.8 Hz), 8.15 (2H, t, J = 7.8 Hz), 8.02 (2H, d, J = 5.5 Hz), 7.64 (2H, t, J = 6.8 Hz), 7.51 (2H, s), 7.45 (2H, t, J = 6.9 Hz), 2.65 (6H, s), 1.83 (6H, s). ESI MS calcd for C32H30N8Ru [M]2+ 314.1; found 314.0 [M]2+. Purity by HPLC: 97.1% by area.
Cell Culture
HL60 human leukemic cells were obtained from ATCC and maintained in Iscove’s media and supplemented with 10% FBS and 1× Penicillin-Streptomycin. HEK293 T-Rex cells were maintained in DMEM media with 10% FBS. Cells were incubated at 37 °C and 5% CO2. An extracellular solution was used for cell cytotoxicity studies in place of opi-MEM to prevent cellular damage from light irradiation. Extracellular solution was made with 10 mM HEPES, 10 mM Glucose, 1.2 mM CoCl2, 1.2 mM MgCl2, 3.3 mM KH2PO4, 0.83 mM K2HPO4 and 145 mM NaCl in water.
Cell Cytotoxicity Assay
Cells were plated at a density of 30,000 cells per well in 96-well plates in extracellular solution. Compounds were added to cells and incubated for one hour before irradiating with light. The time used for irradiation was varied based on the wavelength and intensity of the light source. Cells were irradiated under the indigo light for one minute (29.1 J/cm2), blue light for five minutes (24 J/cm2), green light for five minutes (20.4 J/cm2) and red light for six minutes (28.4 J/cm2). Cells were irradiated under projector light at thirty second intervals with light exposure never exceeding six minutes to ensure cell viability. Once irradiated with light, opti-MEM containing 2% FBS was added to the cells and incubated at 37 °C for 72 hours. Cell viability was determined using resazurin where the dye was added at a final concentration of 70 μM. Cell viability was quantified after three hours using a SpectraFlour Plus (Tecan) Plate Reader with an excitation wavelength of 530 nm and emission measured at 590 nm. Background fluorescence was measured in wells that did not contain cells and subtracted from the rest of the values. Prism software was used to analyze data and the IC50 was measured from fitting the data points to a sigmoidal dose response curve.
Singlet Oxygen Assay
The Singlet Oxygen Sensor Green kit was obtained from Molecular Probes. The singlet oxygen sensor green (SOSG) was diluted in 33 μL ethanol to create a 5 mM stock solution. Compounds were diluted in extracellular solution to mirror the cell cytotoxicity assay and added to a 96 well plate followed by the addition of SOSG. The SOSG in the assay was at a concentration of 5 μM. To measure background fluorescence before subjecting the samples to light irradiation, the plate was read in a SpectraFluor plus plate reader with an excitation of 485 nm and an emission of 530 nm. The plate was then irradiated and read on the plate reader a second time. Compounds were irradiated under the indigo light for one minute (29.1 J/cm2), blue light for five minutes (24 J/cm2), green light for five minutes (20.4 J/cm2) and red light for six minutes (28.4 J/cm2). The singlet oxygen level was determined by calculating the ratio from the signal observed before and after light irradiation for each sample.
In-Vitro Transcription/Translation Assay
1-Step Human Coupled IVT Kit – DNA (Thermo Scientific) was used to carry out experiments. 0.5 μg of pCFE-GFP plasmid was used for all reactions and compounds were used at 5 μM. Compounds were irradiated with one minute indigo light (29.1 J/cm2) then incubated with plasmid overnight. The reaction mix was made according to the procedure described by Thermo Scientific and all reactions were scaled down to 12.5 μL. Reactions were incubated in a water bath for two hours at 30°C then read in a Greiner-Bio One 384-well small volume plate. All samples were read on a SpectraFluor Plus (Tecan) Plate Reader with a 485 nm excitation and 530 nm emission.
DNA Damage Assay
A 1% agarose gel was made with 1x Tris Acetate buffer. An aliquot of the plasmid and compound solution prepared for IVTT was used for the assay after being diluted with SDS-BME dye. A 12 μL sample was loaded on the agarose gel and the samples resolved by running the gel at 100 V for an hour. The gel was stained with 150 mL Tris-Acetate buffer with 0.5 μg/mL ethidium bromide for 45 minutes. The gel was then de-stained in Tris-Acetate buffer for an hour. The gel was imaged on a Chemi-Doc (Bio-Rad) using the UV Trans illumination light and 605/50nm filter.
In Cell Transcription and Translation Assay
A 96-well plate was coated with matrigel, followed by the addition of HEK293 T-Rex-dendra2 cells at a density of 30,000 cells/well and incubated with 1 μg/mL tetracycline for 16 hours. Media was aspirated and 50 μL of L-15 media with 1 μg/mL tetracycline was added to the cells. The plate was then irradiated with one minute of 405 nm light to photoconvert the Dendra2 in the cells. After this, 50 μL of compound was added to each well and incubated with the cells for two hours. The plate was the irradiated with one minute of indigo light from the LED to activate compounds. The plate was set in the SpectraFluor Plus (Tecan) plate reader and the green channel was read with an excitation of 485 nm and emission of 530 nm while the red channel was read with an excitation of 530 nm and emission of 595 nm. A scan was taken every 30 minutes for fifteen hours. Prism software was used to analyze the data.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health (5R01GM107586). L.K., P.V., and R.P.T. also thank the Robert A. Welch foundation (grant E-621) and the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences (award number DE-FG02-07ER15888) for support.
Footnotes
ASSOCIATED CONTENT
Supporting Information. NMR spectra, dose response curves, and inhibition of production of Dendra2. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1. Knoll JD Turro C Control and utilization of ruthenium and rhodium metal complex excited states for photoactivated cancer therapy Coord. Chem. Rev 2015. 282-283 110–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yadav A Janaratne T Krishnan A Singhal SS Yadav S Dayoub AS Hawkins DL Awasthi S MacDonnell FM Regression of lung cancer by hypoxia-sensitizing ruthenium polypyridyl complexes Mol. Cancer. Ther 2013. 12 5 643–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Griffith C Dayoub AS Jaranatne T Alatrash N Mohamedi A Abayan K Breitbach ZS Armstrong DW MacDonnell FM Cellular and cell-free studies of catalytic DNA cleavage by ruthenium polypyridyl complexes containing redox-active intercalating ligands Chem. Sci 2017. 8 5 3726–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith NA, Sadler PJ. Photoactivatable metal complexes: from theory to applications in biotechnology and medicine. Philos. Trans. A Math. Phys. Eng. Sci. 2013;371(1995):20120519. doi: 10.1098/rsta.2012.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mari C Pierroz V Ferrari S Gasser G Combination of Ru(ii) complexes and light: new frontiers in cancer therapy Chem. Sci 2015. 6 5 2660–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glazer EC Light-Activated Metal Complexes that Covalently Modify DNA Isr. J. Chem 2013. 53 6-7 391–400 [Google Scholar]
- 7. Shi GM,S Hennigar R Colpitts J Fong J Kasimova K Yin H DeCoste R Spencer C Chamberlain L Mandel A Lilge L McFarland SA Ru(II) dyads derived from α-oligothiophenes: A new class of potent and versatile photosensitizers for PDT Coord. Chem. Rev 2015. 282-3 127–138 [Google Scholar]
- 8. Singh TN Turro C Photoinitiated DNA binding by cis-[Ru(bpy)(2)(NH3)(2)](2+) Inorg. Chem 2004. 43 23 7260–7262 [DOI] [PubMed] [Google Scholar]
- 9. Garner RN Gallucci JC Dunbar KR Turro C [Ru(bpy)2(5-cyanouracil)2]2+ as a potential light-activated dual-action therapeutic agent Inorg. Chem 2011. 50 19 9213–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higgins SL Tucker AJ Winkel BS Brewer KJ Metal to ligand charge transfer induced DNA photobinding in a Ru(II)-Pt(H) supramolecule using red light in the therapeutic window: a new mechanism for DNA modification Chem. Commun 2011. 48 1 67–9 [DOI] [PubMed] [Google Scholar]
- 11. Zhu J Rodriguez-Corrales JA Prussin R Zhao Z Dominijanni A Hopkins SL Winkel BS Robertson JL Brewer KJ Exploring the activity of a polyazine bridged Ru(ii)-Pt(ii) supramolecule in F98 rat malignant glioma cells Chem. Commun 2016. 53 1 145–148 [DOI] [PubMed] [Google Scholar]
- 12. Karaoun N Renfrew AK A luminescent ruthenium(II) complex for light-triggered drug release and live cell imaging Chem. Commun 2015. 51 74 14038–41 [DOI] [PubMed] [Google Scholar]
- 13. Chan H Ghrayche JB Wei J Renfrew AK Photolabile Ruthenium(II)–Purine Complexes: Phototoxicity, DNA Binding, and Light-Triggered Drug Release Eur. J. Inorg. Chem 2017. 2017 12 1679–1686 [Google Scholar]
- 14. Hufziger KT Thowfeik FS Charboneau DJ Nieto I Dougherty WG Kassel WS Dudley TJ Merino EJ Papish ET Paul JJ Ruthenium dihydroxybipyridine complexes are tumor activated prodrugs due to low pH and blue light induced ligand release J. Inorg. Biochem 2014. 130 103–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qu F Park S Martinez K Gray JL Thowfeik FS Lundeen JA Kuhn AE Charboneau DJ Gerlach DL Lockart MM Law JA Jernigan KL Chambers N Zeller M Piro NA Kassel WS Schmehl RH Paul JJ Merino EJ Kim Y Papish ET Ruthenium Complexes are pH-Activated Metallo Prodrugs (pHAMPs) with Light-Triggered Selective Toxicity Toward Cancer Cells Inorg. Chem 2017. 56 13 7519–7532 [DOI] [PubMed] [Google Scholar]
- 16. Respondek T Garner RN Herroon MK Podgorski I Turro C Kodanko JJ Light activation of a cysteine protease inhibitor: caging of a peptidomimetic nitrile with Ru(II)(bpy)2 J Am Chem Soc 2011. 133 43 17164–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herroon MK Sharma R Rajagurubandara E Turro C Kodanko JJ Podgorski I Photoactivated inhibition of cathepsin K in a 3D tumor model Biol. Chem 2016. 397 6 571–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monro S Scott J Chouai A Lincoln R Zong R Thummel RP McFarland SA Photobiological activity of Ru(II) dyads based on (pyren-1-yl)ethynyl derivatives of 1,10-phenanthroline Inorg. Chem 2010. 49 6 2889–900 [DOI] [PubMed] [Google Scholar]
- 19. Lincoln R Kohler L Monro S Yin H Stephenson M Zong R Chouai A Dorsey C Hennigar R Thummel RP McFarland SA Exploitation of long-lived 3IL excited states for metal-organic photodynamic therapy: verification in a metastatic melanoma model J. Am. Chem. Soc 2013. 135 45 17161–75 [DOI] [PubMed] [Google Scholar]
- 20. Poteet SA Majewski MB Breitbach ZS Griffith CA Singh S Armstrong DW Wolf MO MacDonnell FM Cleavage of DNA by proton-coupled electron transfer to a photoexcited, hydrated Ru(II) 1,10-phenanthroline-5,6-dione complex J. Am. Chem. Soc 2013. 135 7 2419–22 [DOI] [PubMed] [Google Scholar]
- 21. Hess J Huang H Kaiser A Pierroz V Blacque O Chao H Gasser G Evaluation of the Medicinal Potential of Two Ruthenium(II) Polypyridine Complexes as One- and Two-Photon Photodynamic Therapy Photosensitizers Chemistry 2017. 23 41 9888–9896 [DOI] [PubMed] [Google Scholar]
- 22. Bonnet S Limburg B Meeldijk JD Gebbink RJ Killian JA Ruthenium-decorated lipid vesicles: light-induced release of [Ru(terpy)(bpy)(OH2)]2+ and thermal back coordination J. Am. Chem. Soc 2011. 133 2 252–61 [DOI] [PubMed] [Google Scholar]
- 23.Lameijer LN, Ernst D, Hopkins SL, Meijer MS, Askes SHC, Le Devedec SE, Bonnet S. A Red-Light-Activated Ruthenium-Caged NAMPT Inhibitor Remains Phototoxic in Hypoxic Cancer Cells. Angew. Chem. Int. Ed. Engl. 2017 doi: 10.1002/anie.201703890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Betanzos-Lara S Salassa L Habtemariam A Sadler PJ Photocontrolled nucleobase binding to an organometallic Ru(II) arene complex Chem. Commun 2009. 43 6622–4 [DOI] [PubMed] [Google Scholar]
- 25. Barragan F Lopez-Senin P Salassa L Betanzos-Lara S Habtemariam A Moreno V Sadler PJ Marchan V Photocontrolled DNA binding of a receptor-targeted organometallic ruthenium(II) complex J. Am. Chem. Soc 2011. 133 35 14098–108 [DOI] [PubMed] [Google Scholar]
- 26. Smith NA Zhang P Greenough SE Horbury MD Clarkson GJ McFeely D Habtemariam A Salassa L Stavros VG Dowson CG Sadler PJ Combatting AMR: photoactivatable ruthenium(II)-isoniazid complex exhibits rapid selective antimycobacterial activity Chem. Sci 2017. 8 1 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Juris A Balzani V Barigelletti F Campagna S Belser P Vonzelewsky A Ru(II) Polypyridine Complexes - Photophysics, Photochemistry, Electrochemistry, and Chemi-Luminescence Coord. Chem. Rev 1988. 84 85–277 [Google Scholar]
- 28. Baranoff E Collin JP Furusho J Furusho Y Laemmel AC Sauvage JP Photochemical or thermal chelate exchange in the ruthenium coordination sphere of complexes of the Ru(phen)(2)L family (L = diimine or dinitrile ligands) Inorg. Chem 2002. 41 5 1215–1222 [DOI] [PubMed] [Google Scholar]
- 29. Mobian P Kern JM Sauvage JP Light-driven machine prototypes based on dissociative excited states: Photoinduced decoordination and thermal recoordination of a ring in a ruthenium(II)-containing [2]catenane Angew. Chem. Int. Ed 2004. 43 18 2392–2395 [DOI] [PubMed] [Google Scholar]
- 30. Howerton BS Heidary DK Glazer EC Strained ruthenium complexes are potent light-activated anticancer agents J. Am. Chem. Soc 2012. 134 20 8324–7 [DOI] [PubMed] [Google Scholar]
- 31. Wachter E Heidary DK Howerton BS Parkin S Glazer EC Light-activated ruthenium complexes photobind DNA and are cytotoxic in the photodynamic therapy window Chem. Commun 2012. 48 77 9649–51 [DOI] [PubMed] [Google Scholar]
- 32. Hidayatullah AN Wachter E Heidary DK Parkin S Glazer EC Photoactive Ru(II) complexes with dioxinophenanthroline ligands are potent cytotoxic agents Inorg. Chem 2014. 53 19 10030–2 [DOI] [PubMed] [Google Scholar]
- 33. Havrylyuk D Heidary DK Nease L Parkin S Glazer EC Photochemical Properties and Structure–Activity Relationships of RuII Complexes with Pyridylbenzazole Ligands as Promising Anticancer Agents Eur. J. Inorg. Chem 2017. 2017 12 1687–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaw AP Ghosh MK Tornroos KW Wragg DS Tilset M Swang O Heyn RH Jakobsen S Rock ‘n’ Roll With Gold: Synthesis, Structure, and Dynamics of a (bipyridine)AuCl3 Complex Organometallics 2012. 31 20 7093–7100 [Google Scholar]
- 35. Sasse WHF Whittle CP Synthetical applications of activated metal catalysts. Part XII. The preparation of symmetrically substituted 2,2′-bipyridyls J. Chem. Soc 1961. 1347–1350 [Google Scholar]
- 36. Linnell R Notes - 4,4′,6,6′-Tetramethyl-2,2′-bipyridine J. Org. Chem 1957. 22 12 1691–1692 [Google Scholar]
- 37. Rapoport H Iwamoto R Tretter JR The Synthesis of 2,2′-Biquinolyls and Related Compounds by Catalytic Dehydrogenation J. Org. Chem 1960. 25 3 372–373 [Google Scholar]
- 38. Lehn J-M Raymond Z Efficient Synthesis of 1,2-Bis(2,2’-bipyridinyl)ethane and 1,2-Bis(1,10-phenanthroIinyl)ethane Ligands by Oxidative Coupling of the Corresponding Monomeric Methylene Carbanions Helv. Chim. Acta 1988. 71 1511–1516 [Google Scholar]
- 39. Patterson BT Keene FR Isolation of Geometric Isomers within Diastereoisomers of Dinuclear Ligand-Bridged Complexes of Ruthenium(II) Inorg. Chem 1998. 37 4 645–650 [Google Scholar]
- 40. Inagaki A Yatsuda S Edure S Suzuki A Takahashi T Akita M Synthesis of Pd Complexes Combined with Photosensitizing of a Ruthenium(II) Polypyridyl Moiety through a Series of Substituted Bipyrimidine Bridges. Substituent Effect of the Bridging Ligand on the Photocatalytic Dimerization of α-Methylstyrene Inorg. Chem 2007. 46 7 2432–2445 [DOI] [PubMed] [Google Scholar]
- 41. Onggo D Scudder ML Craig DC Goodwin HA The influence of ortho-substitution within the ligand on the geometry of the tris(2,2′-bipyridine)ruthenium(II) and tris(1,10-phenanthroline)ruthenium(II) ions J. Mol. Struct 2005. 738 1–3 129–136 [Google Scholar]
- 42. Durham B Caspar JV Nagle JK Meyer TJ Photochemistry of Ru(Bpy)32+ J. Am. Chem. Soc 1982. 104 18 4803–4810 [Google Scholar]
- 43. Ford PC The Ligand-Field Photosubstitution Reactions of D6 Hexacoordinate Metal-Complexes Coord. Chem. Rev 1982. 44 1 61–82 [Google Scholar]
- 44.All other light sources utilized long pass filters, making quantum yield determination inaccurate. In addition, the values for the Indigo LED are also subject to imprecision due to the rapid photochemical reaction.
- 45. Word TA Whittington CL Karolak A Kemp MT Woodcock HL van der Vaart A Larsen RW Photoacoustic calorimetry study of ligand photorelease from the Ru(II)bis(2,2′-bipyridine)(6,6′-dimethyl-2,2′-bipyridine) complex in aqueous solution Chem. Phys. Lett 2015. 619 214–218 [Google Scholar]
- 46. Cuello-Garibo JA Meijer MS Bonnet S To cage or to be caged? The cytotoxic species in ruthenium-based photoactivated chemotherapy is not always the metal Chem. Commun 2017. 53 50 6768–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azar DF, Audi H, Farhat S, El-Sibai M, Abi-Habib RJ, Khnayzer RS. Phototoxicity of strained Ru(II) complexes: is it the metal complex or the dissociating ligand? Dalton Trans. 2017 doi: 10.1039/c7dt02255g. [DOI] [PubMed] [Google Scholar]
- 48.Additional details and a mechanistic study of the seperate cytotoxicities of specific bipyridyl ligands and the metal fragment will be the subject of a seperate report.
- 49. Garcia-Fresnadillo D Georgiadou Y Orellana G Braun AM Oliveros E Singlet-oxygen ((1)Delta(g)) production by ruthenium(II) complexes containing polyazaheterocyclic ligands in methanol and in water Helv. Chim. Acta 1996. 79 4 1222–1238 [Google Scholar]
- 50. Wachter E Howerton BS Hall EC Parkin S Glazer EC A new type of DNA “light switch”: a dual photochemical sensor and metalating agent for duplex and G-quadruplex DNA Chem. Commun 2014. 50 3 311–13 [DOI] [PubMed] [Google Scholar]
- 51. Heidary DK Glazer EC A light-activated metal complex targets both DNA and RNA in a fluorescent in vitro transcription and translation assay Chembiochem 2014. 15 4 507–11 [DOI] [PubMed] [Google Scholar]
- 52. Wang J Newman J Jr. Higgins SL Brewer KM Winkel BS Brewer KJ Red-light-induced inhibition of DNA replication and amplification by PCR with an Os/Rh supramolecule Angew. Chem. Int. Ed. Engl 2013. 52 1262–1265 [DOI] [PubMed] [Google Scholar]
- 53. Betanzos-Lara S Salassa L Habtemariam A Novakova O Pizarro AM Clarkson GJ Liskova B Brabec V Sadler PJ Photoactivatable Organometallic Pyridyl Ruthenium(II) Arene Complexes Organometallics 2012. 31 9 3466–3479 [Google Scholar]
- 54. Sandman KE Marla SS Zlokarnik G Lippard SJ Rapid fluorescence-based reporter-gene assays to evaluate the cytotoxicity and antitumor drug potential of platinum complexes Chem. Biol 1999. 6 8 541–51 [DOI] [PubMed] [Google Scholar]
- 55. Heidary DK Fox A Richards CI Glazer EC A High-Throughput Screening Assay Using a Photoconvertable Protein for Identifying Inhibitors of Transcription, Translation, or Proteasomal Degradation SLAS Discov 2017. 22 4 399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun Q Mosquera-Vazquez S Daku LM Guenee L Goodwin HA Vauthey E Hauser A Experimental evidence of ultrafast quenching of the 3MLCT luminescence in ruthenium(II) tris-bipyridyl complexes via a 3dd state J. Am. Chem. Soc 2013. 135 37 13660–3 [DOI] [PubMed] [Google Scholar]
- 57. Loganathan D Morrison H ‘Photocisplatin’ reagents Curr. Opin. Drug. Discov. Devel 2005. 8 4 478–86 [PubMed] [Google Scholar]
- 58. Loganathan D Morrison H Effect of ring methylation on the photophysical, photochemical and photobiological properties of cis-dichlorobis(1,10-phenanthroline)rhodium(III)chloride Photochem. Photobiol 2006. 82 1 237–247 [DOI] [PubMed] [Google Scholar]
- 59. Mackay FS Woods JA Heringova P Kasparkova J Pizarro AM Moggach SA Parsons S Brabec V Sadler PJ A potent cytotoxic photoactivated platinum complex Proc. Natl. Acad. Sci. U.S.A 2007. 104 52 20743–20748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yin H Stephenson M Gibson J Sampson E Shi G Sainuddin T Monro S McFarland SA In vitro multiwavelength PDT with 3IL states: teaching old molecules new tricks Inorg. Chem 2014. 53 9 4548–59 [DOI] [PubMed] [Google Scholar]
- 61. Reichardt C Sainuddin T Wachtler M Monro S Kupfer S Guthmuller J Grafe S McFarland S Dietzek B Influence of Protonation State on the Excited State Dynamics of a Photobiologically Active Ru(II) Dyad J. Phys. Chem. A 2016. 120 32 6379–88 [DOI] [PubMed] [Google Scholar]
- 62. Smithen DA Yin H Beh MH Hetu M Cameron TS McFarland SA Thompson A Synthesis and Photobiological Activity of Ru(II) Dyads Derived from Pyrrole-2-carboxylate Thionoesters Inorg. Chem 2017. 56 7 4121–4132 [DOI] [PubMed] [Google Scholar]
- 63.Arai TM,T, Oka T. Kagaku To Kyouiku. Chemical Education, Japanese. 1993;41:278. [Google Scholar]
- 64.Montalti M, Credi A, Prodi L, Gandolfi MT. Handbook of Photochemistry. Third ed. CRC Press; Florida: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


