Summary
MicroRNAs (miRNAs) are small non-coding RNAs, of typically 20–24 nt, that regulate gene expression post-transcriptionally through sequence complementarity. Since the identification of the first miRNA, lin-4, in the nematode Caenorhabditis elegans in 1993, thousands of miRNAs have been discovered in animals and plants, and their regulatory roles in numerous biological processes have been uncovered. In plants, research efforts have established the major molecular framework of miRNA biogenesis and modes of action, and are beginning to elucidate the mechanisms of miRNA degradation. Studies have implicated restricted and surprising subcellular locations in which miRNA biogenesis or activity takes place. In this article, we summarize the current knowledge on how plant miRNAs are made and degraded, and how they repress target gene expression. We discuss not only the players involved in these processes, but also the subcellular sites in which these processes are known or implicated to take place. We hope to raise awareness that the cell biology of miRNAs holds the key to a full understanding of these enigmatic molecules.
Keywords: ARGONAUTE1, DICER-LIKE1, dicing body, endoplasmic reticulum (ER), HYPONASTIC LEAVES 1, membrane-bound polysome, microRNA, phased small interfering RNA
I. MicroRNA biogenesis in plants
MicroRNA (MIR) genes encoding microRNAs (miRNAs) are transcribed by RNA polymerase II (Pol II) into primary miRNAs (pri-miRNAs). The stem loop-containing pri-miRNAs are processed by the RNase III family enzyme DICER-LIKE1 (DCL1) into miRNA/miRNA* duplexes. These duplexes are 2′-O-methylated at the 3′ ends by the methyltransferase HUA ENHANCER1 (HEN1). One strand from the duplex is incorporated into ARGONAUTE1 (AGO1) to form an active RNA-induced silencing complex (RISC) (reviewed in Rogers & Chen, 2013) (Fig. 1). The following section focuses on complexity and regulation in miRNA biogenesis unveiled by recent studies.
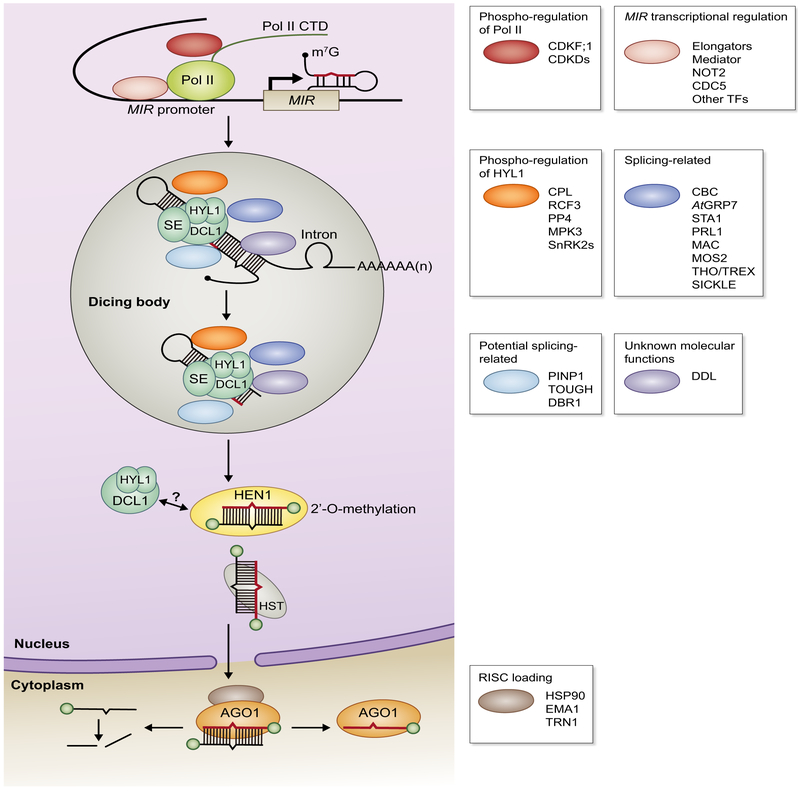
Fig. 1.
Illustrations of major steps in microRNA (miRNA) biogenesis. RNA polymerase II (Pol II)-mediated miRNA gene (MIR) transcription is regulated by multiple transcription factors (TFs). Pol II activity itself is also subjected to phospho-regulation at its C-terminal domain (CTD). miRNA precursors are processed at the dicing bodies by the dicing complex, which is mainly composed of DICER-LIKE 1 (DCL1), HYPONASTIC LEAVES 1 (HYL1) and SERRATE (SE). Many other protein factors contribute to miRNA precursor processing through phospho-regulation, RNA splicing and other unknown molecular mechanisms. It remains unclear whether the dicing complex interacts with HUA ENHANCER 1 (HEN1) (question mark) and contributes to miRNA/miRNA* duplex export and RNA-induced silencing complex (RISC) assembly. During RISC loading, one strand of the small RNA duplex is selected as the guide strand (red) and incorporated into ARGONAUTE 1 (AGO1) to form a functional RISC, whereas the other strand (the passenger strand) is removed and degraded. Proteins are color-coded according to their known molecular functions in phospho-regulation of Pol II (red), MIR transcription (pink), phospho-regulation of HYL1 (orange), splicing/RNA-binding(dark blue)and potentially splicing/RNA-binding (light blue), and RISC assembly (brown). The core dicing complex components are colored green and protein with unknown molecular functions is colored purple. m7G,7-methylguanylate cap at the 5′ end of primary miRNAs; CDKF;1, CYCLIN-DEPENDENT KINASE F;1; CDKDs, CYCLIN-DEPENDENT KINASE D; NOT2, NEGATIVE ON TATA LESS 2; CDC5, CELL DIVISION CYCLE 5; CPL, C-TERMINAL DOMAIN PHOSPHATASE-LIKE; RCF3, REGULATOR OF CBF GENE EXPRESSION 3; PP4, Protein Phosphatase 4 complex; MPK3, MITOGEN-ACTIVATED PROTEIN KINASE 3; SnRK2s,SNF1-related protein kinase subfamily 2; CBC, Cap Binding Complex; AtGRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7; STA1, STABILIZED 1; PRL1, PROTEIN PLEIOTROPIC REGULATORY LOCUS 1; MAC, MOS4-associated Complex; MOS2, MODIFIER OF SNC1, 2; THO/TREX, suppressor of the Transcription defects of Hpr1 mutants by Overexpression/TRanscription-EXport complex; PINP1, PSR1-INTERACTING PROTEIN 1; DBR1, LARIAT DEBRANCHING ENZYME 1; DDL, DAWDLE; HST, HASTY; HSP90, HEAT SHOCK PROTEIN 90; EMA1, ENHANCED MIRNA ACTIVITY 1; TRN1, TRANSPORTIN 1.
1. MIR transcription and transcriptional regulation
Similar to protein coding genes, most MIR genes contain the TATA-box motif and transcription factor binding motifs, such as those of Auxin Response Factors (ARFs) and MYC2, in their promoters, indicating that MIR transcription is regulated by general and specific transcription factors (Xie et al., 2005a; Megraw et al., 2006).
Mediator, a general transcriptional coactivator, helps recruit Pol II to MIR loci (Kim et al., 2011). Other factors promoting general MIR transcription include NEGATIVE ON TATA LESS2 (NOT2), the putative MYB domain-containing DNA-binding protein CELL DIVISION CYCLE 5 (CDC5) and the Elongator complex, which is thought to assist transcriptional elongation (Wang et al., 2013; Zhang et al., 2013; Fang et al., 2015). NOT2, CDC5 and Elongator all interact with Pol II and the dicing complex (the plant miRNA precursor processing complex), implying their functions in bridging Pol II transcription and pri-miRNA processing (Wang et al., 2013; Zhang et al., 2013; Fang et al., 2015). Pol II activity in MIR transcription is probably subject to phospho-regulation. miRNA levels are significantly reduced in mutants of CDKF;1 (CYCLIN-DEPENDENT KINASE F;1) and CDKD (CYCLIN-DEPENDENT KINASE D) genes. These mutants also have reduced phosphorylation marks at the Pol II C-terminal domain (CTD) (Hajheidari et al., 2012; reviewed in Hajheidari et al., 2013).
Factors specifically controlling the transcription of certain miRNAs within an miRNA family have also been characterized. For instance, POWERDRESS promotes the transcription of MIR172a, b and c by enhancing Pol II occupancy at their promoters, without affecting MIR172d or e. Under phosphate starvation, the MYB2 transcription factor binds to the promoter of MIR399fto promote its transcription (Baek et al., 2013; reviewed in Rogers & Chen, 2013; Yumul et al., 2013).
2. miRNA precursor processing
The dicing complex Nascent pri-miRNAs are capped at the 5′ end and polyadenylated at the 3′ end, and intron-containing pri-miRNAs are spliced or alternatively spliced (Xie et al., 2005a; Szarzynska et al., 2009; Zielezinski et al., 2015; reviewed in Stepien et al., 2016). pri-miRNAs are processed by the dicing complex, which contains DICER-LIKE1 (DCL1), HYPONASTIC LEAVES1 (HYL1) and SERRATE (SE) as core components, to yield mature miRNA/miRNA* duplexes (Park et al., 2002; Reinhart et al., 2002; Kurihara & Watanabe, 2004; reviewed in Fukudome & Fukuhara, 2017) (Fig. 1).
Of the four DCL RNase III family endonucleases in Arabidopsis, DCL1 is the predominant miRNA precursor processing enzyme (Park et al., 2002; Reinhart et al., 2002). DCL2, DCL3 and DCL4 produce various types of small interfering RNAs (siRNAs), including endogenous siRNAs, as well as viral and transgene siRNAs (Gasciolli et al., 2005; Xie et al., 2005b; Bouche et al., 2006; Mlotshwa et al., 2008; reviewed in Fukudome & Fukuhara, 2017). A notable exception is that several young miRNAs, such as miR822 and miR839, are generated by DCL4 instead of DCL1 (Rajagopalan et al., 2006). DCL proteins appear to function as molecular rulers that measure and cleave small RNA duplexes at a specific length (Macrae et al., 2006). DCL1 mainly processes pri-miRNAs in a base-to-loop manner in two steps. The first cut is 15–17 nt away from the base of the stem or a bulge or unstructured region within the loop-distal stem. The resulting precursor-miRNA (pre-miRNA) is further cleaved by DCL1 to produce a 21-nt miRNA/miRNA* duplex (Song et al., 2010; Liu et al., 2012; Zhu et al., 2013). Alternative processing modes include loop-to-base processing (Bologna et al., 2009).
In a five-member family of DOUBLE-STRANDED RNA-BINDING PROTEINS (DRBs), HYL1/DRB1 is a major miRNA biogenesis factor and DRB2 affects the accumulation of a few miRNAs (Hiraguri et al., 2005; Curtin et al., 2008; Eamens et al., 2012). HYL1 interacts with DCL1 to facilitate efficient and precise miRNA precursor processing (Kurihara et al., 2006; Dong et al., 2008; Manavella et al., 2012a; Yang et al., 2014). Homodimerization of HYL1 is essential for its functions in miRNA precursor processing (Yang et al., 2010,2014). HYL1 also affects the splicing of some pri-miRNAs and strand selection from miRNA/miRNA* duplexes in AGO1 loading (Szarzynska et al., 2009; Manavella et al., 2012a; Ben Chaabane et al., 2013).
Recent research has uncovered regulatory mechanisms impacting the activity, stability and nuclear localization of HYL1 in miRNA precursor processing (Manavella et al., 2012a; Cho et al., 2014; Karlsson et al., 2015; Raghuram et al., 2015; Zhang et al., 2017a). C-TERMINAL DOMAIN PHOSPHATASE-LIKE (CPL) proteins dephosphorylate HYL1 to facilitate accurate miRNA precursor processing and strand selection during AGO loading (Manavella et al., 2012a). The K homology (KH) domain protein REGULATOR OF CBF GENE EXPRESSION 3 (RCF3) promotes HYL1 dephosphorylation through interaction with CPL proteins (Karlsson et al., 2015). In addition, a PP4 (Protein Phosphatase 4) complex targets HYL1 for dephosphorylation and stabilizes HYTL1 (Su et al., 2017). This dephosphorylation is antagonized by the protein kinases MITOGEN-ACTIVATED PROTEIN KINASE 3 (MPK3) and SNF1-related protein kinase subfamily 2 (SnRK2) (Raghuram et al., 2015; Yan et al., 2017). Phospho-regulation affects not only HYL1 activity, but also its protein stability, for example an snrk2 mutation leads to reduced levels of HYL1 (Yan et al., 2017). HYL1 protein stability is regulated by the RING-finger E3 ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) through light signaling. Specifically, under light conditions, COP1 shifts to the cytoplasm and suppresses HYL1 cleavage by an unidentified protease, whereas, in darkness, COP1 enters the nucleus thereby releasing the protease that cleaves HYL1 (Cho et al., 2014). KETCH1 (KARYOPHERIN ENABLING THE TRANSPORT OF THE CYTOPLASMIC HYL1), a well-conserved importin-beta protein, transports HYL1 from the cytoplasm to the nucleus to form the dicing complex. KETCH1 knockdown mutants show similar phenotypes to miRNA biogenesis mutants, including reduced miRNA levels and compromised pri-miRNA processing, indicating the importance of HYL1’s nuclear localization in miRNA biogenesis (Zhang et al., 2017a).
In addition to DCL1 and HYL1, SE is also considered as a core member of the miRNA processing complex in Arabidopsis. A mutation in SE results in reduced levels of mature miRNAs, increased levels of pri-miRNAs and defects in pri-miRNA splicing (Grigg et al., 2005; Lobbes et al., 2006; L. Yang et al., 2006; Laubinger et al., 2008). As a zinc-finger protein with RNA-binding activity, SE also functions outside of miRNA biogenesis. For example, SE interacts with U1 small nuclear ribonucleoprotein (snRNP) components (Knop et al., 2016). Different from DCL1 or HYL1, SE is distributed in a heterogeneous subnuclear pattern, reminiscent of nuclear speckles in which serine/arginine (SR) splicing factors are enriched (Fang & Spector, 2007). SE affects the alternative splicing of some Arabidopsis mRNAs (Laubinger et al., 2008; Raczynska et al., 2014).
Other proteins that influence miRNA precursor processing and/or MIR transcription In the past decade, many proteins that influence miRNA precursor processing or MIR gene transcription have been identified (Fig. 1). On the one hand, these proteins promote miRNA biogenesis in general, as most miRNAs accumulate to lower levels in mutants of these genes. On the other hand, the functions of these genes are not specific to miRNAs; indeed, many have functions in precursor mRNA (pre-mRNA) splicing. In terms of miRNA biogenesis, these proteins appear to act before or during pri-miRNA processing, as mutants in these genes have either higher or lower levels of pri-miRNAs. Below, we categorize these proteins into two groups based on their effects on pri-miRNA accumulation and discuss their potential roles in miRNA biogenesis.
A large group of proteins appears to promote pri-miRNA processing, as loss/reduction-of-function mutants in the corresponding genes have reduced levels of mature miRNAs and increased levels of pri-miRNAs. Proteins belonging to this group include CAP-BINDING PROTEIN 80 (CBP80) and CAP-BINDING PROTEIN 20 (CBP20) (Laubinger et al., 2008; reviewed in Gonatopoulos-Pournatzis & Cowling, 2014), STABILIZED1 (STA1) (Ben Chaabane et al., 2013), THO1/HPR1/EMU and THO2 (Furumizu et al., 2010; Francisco-Mangilet et al., 2015), SICKLE (Zhan et al., 2012), TOUGH (Ren et al., 2012b), PSR1-INTERACTING PROTEIN 1 (PINP1) (Qiao et al., 2015) and MODIFIER OF SNC1, 2 (MOS2) (X. Wu et al., 2013). GLYCINE-RICH RNA-BINDING PROTEIN 7 (At GRP7) may repress miRNA biogenesis, as its overexpression causes reduced levels of mature miRNAs and increased accumulation of pri-miRNAs (Koster et al., 2014). All of these proteins have demonstrated or predicted ability to associate with or act on RNAs. CBP80 and CBP20 are subunits of the nuclear Cap Binding Complex (CBC), which interacts with SE (Laubinger et al., 2008). The human and yeast homolog of STA1 is the U5 snRNP-associated protein Pre-mRNA Processing Factor 6 (PRPF6), a confirmed splicing factor (reviewed in Will & Luhrmann, 2011; Ben Chaabane et al., 2013). THO1/HPR1/EMU and THO2 are subunits of the THO/TREX (suppressor of the Transcription defects of Hpr1 mutants by Overexpression/TRanscription-EXport) complex, a conserved multi-subunit complex involved in pre-mRNA co-transcriptional processing and mRNA export in yeast and animals (reviewed in Heath et al., 2016). SICKLE is a plant-specific protein that interacts with many RNA processing proteins (Zhan et al., 2012; Marshall et al., 2016). TOUGH and MOS2 are RNA-binding proteins (Ren et al., 2012b; X. Wu et al., 2013). PINP1 is a putative RNA helicase (Qiao et al., 2015). At GRP7 is a heterogeneous nuclear ribonucleoprotein (hnRNP)-like glycine-rich RNA-binding protein (Streitner et al., 2012; Koster et al., 2014). The molecular functions of these proteins in pri-miRNA processing are currently unknown.
A second group of proteins may act differently from the previously discussed group in miRNA biogenesis. Loss/reduction-of-function mutants in genes in this second group have reduced accumulation of both mature miRNAs and pri-miRNAs. Proteins in this group include CDC5, NOT2, Elongator, PRL1 (PROTEIN PLEIOTROPIC REGULATORY LOCUS 1) and DDL (DAWDLE) (Yu et al., 2008; Wang et al., 2013; Zhang et al., 2013, 2014; Fang et al., 2015). Although the reduced pri-miRNA accumulation in the mutants suggests a role of the proteins in MIR transcription or pri-miRNA stability, these proteins also seem to affect miRNA precursor processing. For example, mutants in NOT2 show an increase in the number of dicing bodies, whereas mutants in MOS2 and Elongator have a reduced number of dicing bodies. CDC5, NOT2, Elongator, PRL1 and DDL were all found to interact with DCL1 and may help to recruit DCL1 to pri-miRNAs (Yu et al., 2008; Wang et al., 2013; Zhang et al., 2013, 2014; Fang et al., 2015). Perhaps the most parsimonious hypothesis is that these proteins promote miRNA biogenesis by recruiting the dicing complex to nascent pri-miRNAs during transcription.
A prominent feature of these two groups of proteins, regardless of their effects on pri-miRNAs, is their demonstrated or predicted roles in splicing. Splicing defects in both pri-miRNAs and pre-mRNAs were detected in abh1/cbp80, cbp20, sic-1 and sta1-1 mutants (Laubinger et al., 2008; reviewed in Gonatopoulos-Pournatzis & Cowling, 2014). AtGRP7 overexpression results in changes in pri-miRNA splicing (Streitner et al., 2012; Koster et al., 2014) MOS2 is required for appropriate splicing of SNC1 (SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1), which encodes a Toll Interleukin 1 Receptor Nucleotide Binding Leucine-Rich Repeat (TIR-NB-LRR) class of protein involved in plant defense responses (Zhang et al., 2005; Copeland et al., 2013). Alternatively spliced SR gene transcripts were detected in tho1 and tho2 mutants (Furumizu et al., 2010; Francisco-Mangilet et al., 2015). PRL1 belongs to the NineTeen Complex (NTC) or MOS4-associated complex (MAC), a protein complex with 19 conserved members in yeast, human and plants. MAC is involved in spliceosome assembly and pre-mRNA splicing in all eukaryotic model organisms (Monaghan et al., 2009; reviewed in Johnson et al., 2011 and Koncz et al., 2012). Arabidopsis TOUGH colocalizes or interacts with splicing factor SR proteins, indicating a potential role in general pre-mRNA splicing (Calderon-Villalobos et al., 2005; Ren et al., 2012b). Although there is no direct evidence demonstrating PINP1’s involvement in pre-mRNA processing in Arabidopsis, its yeast homolog, Prp16, is a confirmed splicing factor (Wang et al., 1998).
The large number of proteins acting in both RNA splicing and miRNA biogenesis begs the question of whether or how these two nuclear RNA processing events are related. Some pri-miRNAs harbor introns (Szarzynska et al., 2009), and thus splicing may be an essential step in miRNA biogenesis. However, pri-miRNAs without introns are also affected in mutants in some of the aforementioned genes. For example, the levels of pri-miR159a, which contains no introns, were altered in the mutants of CBC, STA1, PRL1, MOS2, THO2 and SICKLE (Laubinger et al., 2008; Szarzynska et al., 2009; Zhan et al., 2012; Ben Chaabane et al., 2013; Copeland et al., 2013; X. Wu et al., 2013; Zhang et al., 2014; Francisco-Mangilet et al., 2015). At least for pri-miRNAs without introns, the aforementioned proteins cannot act in miRNA biogenesis through their functions in RNA splicing. Another formal possibility is that these proteins only act in splicing; in loss/reduction-of-function mutants of these genes, the accumulation of unspliced introns sequesters the dicing complex and thus inhibits miRNA biogenesis. It was found that a mutation in the intron lariat debranching gene DBR1 (LARIAT DEBRANCHING ENZYME 1) results in the over-accumulation of intronic RNAs, which compete with pri-miRNAs for the dicing complex (Z. Li et al., 2016). However, many of the aforementioned proteins interact with the dicing complex or pri-miRNAs, which implies a direct role in miRNA biogenesis. Perhaps many of the proteins act broadly in nuclear RNA metabolism, with RNA splicing and miRNA biogenesis being two independent processes in which they participate.
3. miRNA stabilization and RISC formation
The miRNA/miRNA* duplex is stabilized through 3′-terminal 2′-O-methylation by HEN1 (Fig. 1). HEN1 was first discovered in Arabidopsis as a methyltransferase that specifically methylates small RNAs (Yu et al., 2005; Yang Z et al., 2006). HEN1 homologs with similar functions were later discovered in other plants, animals and fungi (Kirino & Mourelatos, 2007; Saito et al., 2007; reviewed in Huang, 2012). The crystal structure of an Arabidopsis HEN1-small RNA complex suggests that the small RNA duplex is bound by the HEN1 double-strand RNA-binding domains (dsRBD), with one terminus being in the methyltransferase (MTase) active site and methylated in an Mg2+-dependent manner (Huang et al., 2009). A recent study has suggested that HEN1 might interact with DCL1 and HYL1 based on yeast two-hybrid and in vitro pull-down assays (Baranauske et al., 2015). However, further in vivo analysis is needed to confirm this interaction.
During AGO loading, one strand of the small RNA duplex is selected as the guide strand, whereas the passenger strand is removed (Fig. 1). The current model for Arabidopsis RISC loading is as follows. (1) AGO1 and a dimer of HEAT SHOCK PROTEIN 90 (HSP90) form a complex. (2) The binding of adenosine triphosphate (ATP) to HSP90 causes a conformational change of AGO1 that allows the small RNA duplex to be incorporated into the AGO1–HSP90 protein complex. (3) ATP hydrolysis induces AGO1 dissociation from HSP90. (4) The AGO1 conformational change caused by HSP90 dissociation removes the passenger strand and results in a mature RISC (Iki et al., 2010).
Two AGO1-interacting importin-beta family proteins, ENHANCED MIRNA ACTIVITY1 (EMA1) and TRANSPORTIN1 (TRN1), negatively and positively regulate miRNA loading into AGO1, respectively. As importin-beta family proteins, the most intuitive expectation would be that they mediate AGO1’s or miRNA’s nuclear-cytoplasmic shuttling. However, the nuclear-cytoplasmic partitioning of AGO1 or miRNAs is unchanged in these mutants (Wang et al., 2011; Cui et al., 2016). Nevertheless, the fact that importin-beta family proteins affect the loading of miRNAs into AGO1 suggests that RISC formation occurs at specific subcellular locations.
The selection of miRNA guide strands is not random. In Arabidopsis, guide strand selection is known to be affected by miRNA precursor processing factors, the nature of the 5′ end nucleotide and the structure of the small RNA duplex. HYL1 and the HYL1 phosphatase CPL1 facilitate guide strand selection, with hyl1 and cpl1 mutants exhibiting accumulated miRNA* strands (Eamens et al., 2009; Manavella et al., 2012a). The nature of the 5′ nucleotides directs AGO loading. Most miRNA guide strands start with a 5′-terminal uridine and are incorporated into AGO1. By contrast, few miRNA star strands have 5′-terminal uridine. miRNA star strands with 5′-terminal adenosine are largely associated with AGO2, whereas those with 5′-terminal cytosine are associated with AGO5 (Mi et al., 2008). The loading of miRNAs into AGO proteins is also affected by the bulges in miRNA/miRNA* duplex structures. AGO2 favors miRNA duplexes without central mismatches, whereas AGO1 prefers duplexes with central mismatches, and the preference of AGO10 for miR165/6 relies on the internal base mismatches of the miRNA166 precursor (Zhu et al., 2011; Ren et al., 2014).
4. The cell biology of miRNA biogenesis: dicing bodies, nuclear export and RISC loading
The nucleus is the site of pri-miRNA processing in Arabidopsis (Papp et al., 2003). Live-cell imaging revealed that DCL1 and HYL1 colocalize in round and membrane-less nuclear bodies, namely dicing bodies, which range in number from zero to four in each cell (Han et al., 2004; Fang & Spector, 2007; Song et al., 2007). DCL1 and HYL1 also exhibit diffuse patterns in the nucleoplasm, but are excluded from nucleoli (Fang & Spector, 2007; Z. Li et al., 2016). In vivo tracking of a pri-miRNA showed its colocalization with dicing bodies, indicating the role of dicing bodies in pri-miRNA processing (Fang & Spector, 2007). Dicing bodies resemble Cajal bodies in shape, size and number. However, colocalization analysis demonstrated that dicing bodies and Cajal bodies are distinct structures (Fang & Spector, 2007; Song et al., 2007) (Fig. 1).
Different mechanisms have been proposed for the formation of membrane-less nuclear bodies (e.g. dicing bodies), including stochastic assembly, ordered assembly and seeded assembly (reviewed in Mao et al., 2011). Low-complexity sequences, which are enriched in many RNA- and DNA-binding proteins, contribute to the formation of higher order RNA- and protein-containing structures (Han et al., 2012; Kato et al., 2012). Arabidopsis DCL1 contains two dsRBDs. The second dsRBD is truncated in the dcl1-9 mutant, which exhibits severe miRNA biogenesis defects (Park et al., 2002; reviewed in Schauer et al., 2002); moreover, the truncated DCL1-9 protein fails to localize to dicing bodies (Fang & Spector, 2007). Similarly, the N-terminal dsRBDs of HYL1 are essential for HYL1’s localization to dicing bodies (Wu et al., 2007).
Many other miRNA biogenesis factors, such as SE, RCF3 and THO2, largely form splicing speckles and partially colocalize with dicing bodies (Fang & Spector, 2007; Francisco-Mangilet et al., 2015; Karlsson et al., 2015). NOT2, MOS2 and PINP1 show diffuse nucleoplasmic patterns and also partially colocalize with dicing bodies (Wang et al., 2013; X. Wu et al., 2013; Qiao et al.,2015). The subcellular localization patterns of the above factors implicate their roles in both miRNA and mRNA biogenesis. Dicing body formation is affected by several miRNA biogenesis factors. Mutants of MOS2 and Elongator subunits have a reduced number of dicing bodies, whereas those of PINP1, NOT2 and DBR1 have more dicing bodies than the wild-type; thus, opposite effects are observed, although all of the above mutants have compromised miRNA levels (Wang et al., 2013; X. Wu et al., 2013; Fang et al., 2015; Qiao et al., 2015; Z. Li et al., 2016).
The export of miRNAs from the nucleus to the cytoplasm is fundamental for miRNA activity (Lund et al., 2004; Park et al., 2005; reviewed in Kohler & Hurt, 2007 and Rogers & Chen, 2013). Exportin 5, a RanGTP-dependent dsRNA-binding protein, mediates the nuclear export of pre-miRNAs in animals (Yi et al., 2003; Bohnsack et al., 2004; Lund et al., 2004). In Arabidopsis, miRNA/miRNA* duplexes are probably excised from pre-miRNAs in the nucleus (as DCL1 acts in the nucleus) and are thought to be transported to the cytoplasm by HASTY (Papp et al., 2003; Park et al., 2005) (Fig. 1). In the hasty mutant, the nuclear-cytoplasmic partitioning of miRNAs is not altered (Park et al., 2005). Therefore, the functions of HASTY in miRNA nuclear export in Arabidopsis still require further investigation. THO/TREX complex components are required for miRNA biogenesis (Furumizu et al., 2010; Francisco-Mangilet et al., 2015). As THO/TREX mediates transcription-coupled mRNA export through interactions with the nuclear pore complex, it is also possible that THO/TREX plays a role in miRNA export (reviewed in Kohler & Hurt, 2007).
It is still unclear whether miRNAs are exported to the cytoplasm before RISC loading or whether RISC loading precedes export to the cytoplasm. However, the involvement of cytoplasmic HSP90 in RISC loading is one line of evidence in favor of RISC loading in the cytoplasm (reviewed in Krishna & Gloor, 2001; Mi et al., 2008).
II. Modes of action of miRNAs
Plant miRNAs regulate target genes at the post-transcriptional level via two major mechanisms: transcript cleavage and translation repression (reviewed in Chen, 2005, Chen, 2009, Voinnet, 2009 and Rogers & Chen, 2013) (Fig. 2). For small RNAs in general, the degree of sequence complementarity between small RNAs and their targets influences the particular mode of action in which the small RNAs can engage, with transcript cleavage requiring a high degree of sequence complementarity (Hutvagner & Zamore, 2002). In plants, miRNAs and their target mRNAs have nearly perfect complementarity, and, because of this, transcript cleavage was thought to be the predominant mode of action of plant miRNAs (reviewed in Chen, 2005, Jones-Rhoades et al., 2006, Chen, 2009 and Voinnet, 2009). However, this is a mis-conception. Although a high degree of sequence complementarity is conducive to RNA cleavage, it is not necessarily refractory to translational repression. Indeed, targets that have been experimentally validated to undergo miRNA-mediated translation inhibition pair with miRNAs with a high degree of sequence complementarity (Brodersen et al., 2008; Yang et al., 2012; Li et al., 2013). Examples are APETALA2 (AP2), SCARECROW-LIKE PROTEIN 4 (SCL4), COPPER/ZINC SUPEROXIDE DISMUTASE 2 (CSD2) and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (SPL3), targeted by miR172, miR171, miR398 and miR156, respectively (Aukerman & Sakai, 2003; Chen, 2004; Gandikota et al., 2007; Brodersen et al., 2008). Indeed, the same mRNAs also undergo cleavage caused by the same miRNAs (Li et al., 2013; Hou et al., 2016; Yu et al., 2016). Thus, sequence complementarity is not the factor that dictates the mode of action in which plant miRNAs engage. Emerging findings of miRNA target transcripts bound by ribosomes or ribosomes on the endoplasmic reticulum (ER) (Hou et al., 2016; S. Li et al., 2016; Yu et al., 2016) imply that translation inhibition may occur at a larger number of miRNA targets than expected.
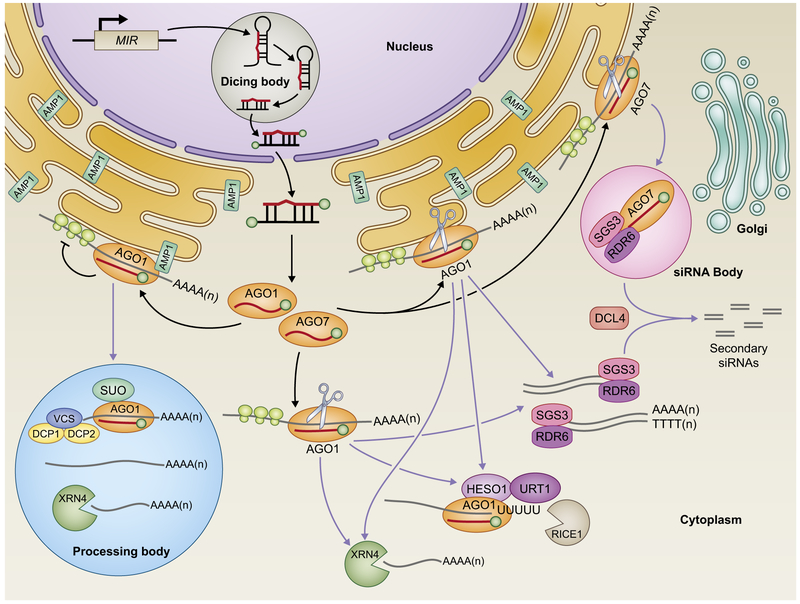
Fig. 2.
Overview of microRNA (miRNA) modes of action in plants. Mature miRNAs are incorporated into ARGONAUTE proteins to direct post-transcriptional gene silencing (PTGS) via transcript cleavage and translation repression or trigger the biogenesis of secondary small interfering RNAs(siRNAs). ARGONAUTE 1 (AGO1) mediates miRNA target cleavage followed by degradation of the cleavage fragments. The cytoplasmic location of this event is unclear, but the uridylation and turnover of 5′ cleavage fragments occur on AGO1. Translation repression takes place on membrane-bound polysomes (MBPs), and requires endoplasmic reticulum (ER)-localized ALTERED MERISTEM PROGRAM 1 (AMP1). Components of Processing body (P-body) are also involved in this process, although the function of these factors and their connection to ER remain mysterious. ARGONAUTE 7 (AGO7) cleaves miR390 targets that are associated with MBPs, and forms siRNA bodies together with SUPPRESSOR OF GENE SILENCING 3 (SGS3) and RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) that are adjacent to the cis-Golgi. Other TAS transcripts that generate phased secondary siRNAs (phasiRNAs) in response to AGO1-mediated cleavage also associate with MBPs. Events are classified with colored lines according to miRNA-mediated actions (dark lines) and subsequent processing (light purple lines) of their targets. DCL4 DICER-LIKE4; HESO1, HEN1 SUPPRESSOR 1; URT1, UTP:RNA URIDYLYLTRANSFERASE 1; XRN4, EXORIBONUCLEASE4; SUO, a GW-repeat protein; VCS, VARICOSE; DCP1, DECAPPING 1; DCP2, DECAPPING 2; RICE1, RISC-INTERACTING CLEARING 3′–5′ EXORIBONUCLEASE 1.
1. Transcript cleavage
miRNA-guided RNA cleavage, also known as slicing, occurs at a precise position in the target mRNA (Llave et al., 2002). Genome-wide identification of RNAs with a 5′ monophosphate (the 3′ cleavage fragments have a 5′ monophosphate) found that most plant miRNA targets undergo transcript cleavage (German et al., 2008). Cleavage is accomplished by the PIWI domain of AGO proteins, which forms an RNase H-like fold and exhibits endonuclease activity; this activity has been demonstrated for Arabidopsis AGO1, the major miRNA effector, together with AGO2, AGO4, AGO7 and AGO 10 (Mi et al., 2008;Montgomery et al., 2008;Takeda et al., 2008; Ji et al., 2011; Maunoury & Vaucheret, 2011; Zhu et al., 2011).
On slicing, the 5′ and 3′ cleavage fragments are subsequently degraded by exonucleases (Fig. 2). In Arabidopsis, EXORIBONUCLEASE 4 (XRN4), a 5′-to-3′ exonuclease, is responsible for degrading the 3′ fragments (Souret et al., 2004). Unlike the 3′ fragments, which are usually detectable in wild-type plants, the 5′ fragments are barely detected, probably as a result of rapid degradation. In Chlamydomonas reinhardtii, the 5′ fragments are polyadenylated by the nucleotidyl transferase MUT68, followed by degradation by the cytoplasmic exosome (Ibrahim et al., 2006). HEN1 SUPPRESSOR 1 (HESO1), an Arabidopsis homolog of MUT68, and its paralog UTP:RNA URIDYLYLTRANSFERASE 1 (URT1) polyuridylate the 5′ fragments in vivo and in vitro (Ren et al., 2014; Wang et al., 2015). RISC-INTERACTING CLEARING 3′– 5′ EXORIBONUCLEASE 1 (RICE1) is responsible for the degradation of uridylated 5′ fragments in Arabidopsis, because these uridylated fragments are over-accumulated in plants ectopically expressing a catalytically inactive RICE1 (Zhang et al., 2017b). The cytoplasmic exosome may also play a role, as its cofactor’s subunits, SUPERKILLER2 (SKI2), SKI3 and SKI8, are required for the degradation of RISC-generated 5′ fragments (Branscheid et al., 2015).
2. Translation inhibition
miRNA-mediated translation repression was initially proposed to account for the disproportionate effects of miRNAs on target gene repression at the protein vs mRNA level (Aukerman & Sakai, 2003; Chen, 2004; Gandikota et al., 2007). In plants, translation repression is less frequently observed than transcript cleavage, possibly owing to the universal presence of miRNA-guided cleavage, coupled with difficulty in determining protein levels because of the absence of high-quality antibodies.
Early examples of miRNA-mediated translation inhibition in plants were AP2 and SPL3 regulated by miR172 and miR156/7, respectively (Aukerman & Sakai, 2003; Chen, 2004; Gandikota et al., 2007). When miR172 and miR156/7 accumulated abnormally, AP2 and SPL3 transcript levels were comparable with those of the wild-type, but their protein levels were altered (Chen, 2004; Gandikota et al., 2007). Subsequently, similar observations were made for other miRNAs, including miR159 (Alonso-Peral et al., 2010), miR164, miR165/6 (Li et al., 2013), miR171, miR395, miR398 andmiR834 (Brodersen et al., 2008). Moreover, the study by Li et al. (2013) went beyond observations of effects of miRNAs on target gene expression at the transcript vs protein level by showing that miR398 and miR165/6 inhibit protein synthesis from their target genes CSD2 and PHB, respectively.
Known factors required for miRNA-mediated translation inhibition include the microtubule-severing enzyme KATANIN 1 (KTN1) (Brodersen et al., 2008), the processing body (P body) component VARICOSE (VCS) (Brodersen et al., 2008), the GW-repeat protein SUO (Yang et al., 2012) and the ER membrane protein ALTERED MERISTEM PROGRAM 1 (AMP1) (Li et al., 2013) (Fig. 2). Mutations in these genes selectively interfere with miRNA-guided repression at the protein level, suggesting that transcript cleavage and translation repression are two independent modes of action. Based on the finding that the recruitment of miRNA target transcripts throughout the polysome fractions is enhanced in the amp1 mutant compared with the wild-type (Li et al., 2013), plant miRNAs may repress translation initiation, but other possibilities exist. Genome-wide analyses of RNA degradation through the profiling of RNAs with 5′ monophosphate in Arabidopsis show that co-translational mRNA degradation occurs for most genes, including a large number of miRNA targets (Hou et al., 2016; Yu et al., 2016). The cleavage of presumably ribosome-bound AP2 and SPL3 transcripts at the corresponding miRNA binding sites was observed (Yu et al., 2016). An important lesson is that, even for the ‘RNA cleavage’ mode of action of miRNAs, translating mRNAs are the targets. This is consistent with findings that AGO1 and miRNAs associate with polysomes (Lanet et al., 2009; S. Li et al., 2016). Although the molecular mechanisms underlying miRNA-mediated translation repression are far from clear, an in vitro analysis indicated that plant miRNAs could inhibit translation initiation or hinder the movement of ribosomes (Iwakawa & Tomari, 2013). Proposed mechanisms for miRNA-mediated translation repression in animals include the dissociation of translation initiation complexes, recruitment of translational repressors or displacement of polyA binding proteins from mRNAs (reviewed in Iwakawa & Tomari, 2015). The activities of animal miRNAs require a scaffold protein GW182 (reviewed in Iwakawa & Tomari, 2015), which is lacking in plants.
3. Biogenesis of secondary siRNAs
In addition to mRNA cleavage and translation repression, some miRNAs also trigger the production of phased secondary siRNAs (phasiRNAs) from their target transcripts (Fig. 2), and this is a widespread and conserved phenomenon in plants (reviewed in Chen, 2005, Chen, 2009 and Rogers & Chen, 2013). In Arabidopsis, after AGO-mediated slicing, either the 5′ or 3′ fragment is stabilized by SUPPRESSOR OF GENE SILENCING 3 (SGS3), which associates with RISC by recognizing specific features of the 22-nt miRNA/target duplex to protect the cleavage fragment from degradation (Yoshikawa et al., 2005, 2013). RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) is recruited to convert the cleavage fragment into dsRNA which is later diced into phasiRNAs at a 21-nt interval (Yoshikawa et al., 2005). This phasing requires AGO1-mediated cleavage: in an ago1 mutant with defective slicing activity, secondary siRNAs are generated, but the phasing is disrupted (Arribas-Hernandez et al., 2016).
The phasiRNAs generated from four families of non-coding TAS genes (TAS1 to TAS4) in Arabidopsis were termed tasiRNAs at the time of discovery because of their in-trans mode of action similar to miRNAs (Allen et al., 2005; Yoshikawa et al., 2005; Montgomery et al., 2008; Chen et al., 2010; Cuperus et al., 2010). Two mechanisms of tasiRNA production are based on the number of miRNA binding sites within the target transcripts. The predominant mechanism, known as the ‘one-hit model’, entails one miRNA binding site in the target transcript and a 22-nt miRNA (Allen et al., 2005; Yoshikawa et al., 2005; Chen et al., 2010). The ‘two-hit model’ requires two miRNA binding sites within the target transcript (Axtell et al., 2006). This is observed for TAS3 transcripts, which contain two miR390 binding sites. AGO7, instead of AGO1, mediates the cleavage at the 3′ site, but not at the 5′ site (Axtell et al., 2006) (Fig. 2).
In addition to the length of the miRNAs triggering phasiRNA biogenesis, other factors may also be influential. The asymmetric bulge structure within miRNA/miRNA* and the degree of complementarity in miRNA–target pairing affect tasiRNA production (Manavella et al., 2012b; Yoshikawa et al., 2013). The position of the miR173 binding site relative to the short open reading frame in TAS2 or a transgene containing TAS1c sequence was found to be important, as tasiRNA abundance decreased when premature stop codons were introduced further upstream of the miR173 binding site (Zhang et al., 2012; Yoshikawa et al., 2016), suggesting a relationship between translation and tasiRNA biogenesis.
phasiRNAs are not generated from most miRNA target transcripts. Most miRNAs are 21 nt in length and do not trigger phasiRNA biogenesis from their targets. Genome-wide small RNA sequencing and bioinformatic analysis identified a small number of protein coding genes, including immune receptor NUCLEOTIDE-BINDING LEUCINE-RICH REPEAT (NBS-LRR) and PENTATRICOPEPTIDE REPEAT (PPR) genes, as targets of 22-nt miRNAs for phasiRNA production in Arabidopsis (reviewed in Fei et al., 2013). Monouridylation of miR171 catalyzed by URT1 in the hen1 mutant leads to a 22-nt miR171 that is able to trigger the production of phasiRNAs (Zhai et al., 2013). A larger number of phasiRNAs, as well as the loci that generate them (PHAS loci), have been identified in many non-Brassicaceae plants (reviewed in Fei et al., 2013). The phasiRNAs are derived from transcripts of protein coding genes, such as NBS-LRR and PPR genes, or long non-coding RNAs (Fei et al., 2013; Zhai et al., 2015; Fan et al., 2016). Although the targets of many phasiRNAs are still unclear, miRNA-triggered production of phasiRNAs is nevertheless hypothesized to act in beneficial microbial interactions or plant defense, or have other long-term evolutionary benefits (reviewed in Fei et al., 2013).
4. Subcellular locations of miRNA activities
Several studies in Arabidopsis link the sites of miRNA activity to polysomes (Lanet et al., 2009), the ER membrane (Li et al., 2013) and membrane-bound polysomes (S. Li et al., 2016; Yu et al., 2016) (Fig. 2).
Because AGO1 is the major miRNA effector, the subcellular localization of AGO1 is an important clue for uncovering the sites of miRNA activity. Previous studies have shown that AGO1 is detectable in the cytosol, but excluded from the nucleus, by fluorescence microscopy analysis, and AGO1 is enriched around the nuclear envelope in some cells (Derrien et al., 2012). AGO1 is a peripheral membrane protein, based on fractionation experiments after high-salt or high-pH treatments (Brodersen et al., 2012; Li et al., 2013). The link between AGO1’s membrane localization and the rough endoplasmic reticulum (rER) is based on fluorescence microscopy analysis showing that AGO1 accumulates in cytoplasmic granules that colocalize with an ER marker (Li et al., 2013). The association of AGO1 with the rER is further supported by its interaction with AMP1, an integral membrane protein localized to the rER (Li et al., 2013).
Subcellular fractionation detected the association of miRNAs and AGO1 with polysomes (Lanet et al., 2009). Further fractionation revealed the association of AGO1 with membrane-bound polysomes (MBPs) and the preferential association of miRNAs with MBPs rather than polysomes in general (Li et al., 2013; S. Li et al., 2016). AMP1 and its paralog LIKE AMP1 (LAMP1) are both required for miRNA-guided translation repression, but not transcript cleavage (Li et al., 2013). In the amp1 lamp1 double mutant, miRNA target transcripts are associated with total polysomes as in wild-type plants (Li et al., 2013). However, these transcripts are more enriched on MBPs in amp1 lamp1 than in the wild-type (Li et al., 2013). Thus, miRNA-mediated translation repression probably occurs on the rER.
How AGO1 associates with the endomembrane is unknown, but it may be independent of AMP1 (Li et al., 2013) or target mRNAs (S. Li et al., 2016). Several ago1 mutants harboring various point mutations display compromised membrane association, and this association is further reduced by knocking down HYDROXY METHYLGLUTARYL COA REDUCTASE 1 (HMG1), which encodes an isoprenoid biosynthesis enzyme (Brodersen et al., 2012). Thus, aside from AGO1 itself, isoprenoid may influence the membrane association of AGO1. Loss of function in HMG1 also leads to defective miRNA activity (Brodersen et al., 2012), further suggesting that the membrane association of AGO1 is essential for its role in miRNA-directed activities.
AGO1 also associates with P bodies (reviewed in Xu & Chua, 2011). An Arabidopsis P body-localized protein, VCS, was found to play a role in miRNA-guided translation inhibition (Brodersen et al., 2008). VCS is a component of the decapping complex, which is required for 5′-to-3′ exonucleolytic degradation of mRNA. Loss of VCS results in elevated protein levels of several miRNA targets with subtle or no increases in their corresponding mRNA levels (Brodersen et al., 2008). Similar effects were observed for loss of function in KTN1, which encodes the P60 subunit of a microtubule-severing enzyme (Brodersen et al., 2008). However, the mechanisms by which VCS and KTN1 influence miRNA-mediated translation repression and the connection of P bodies or microtubules with this process are still unknown.
Unlike translation repression, few reports have directly addressed the site of miRNA-guided transcript cleavage. However, the reduced cleavage efficiency observed in the hmg1 mutant (Brodersen et al., 2012) and the ER association of AGO1 (Li et al., 2013) suggest that polysomes and the rER are potential sites. In addition, genome-wide analyses of RNA degradation products suggest that miRNA targets undergo cleavage when bound by translating ribosomes (Hou et al., 2016; Yu et al., 2016). Furthermore, 3′ cleavage fragments from a few miRNA targets were detectable in the MBP fraction (S. Li et al., 2016). Therefore, transcripts targeted by miRNAs may undergo co-translational degradation, and at least a fraction of miRNA-guided cleavage may take place on the rER.
The biogenesis of phasiRNAs probably occurs on membrane structures. SGS3 and RDR6, two essential proteins required for phasiRNA biogenesis, form cytoplasmic siRNA bodies that also contain AGO7 (Kumakura et al., 2009; Jouannet et al., 2012). Moreover, both SGS3 and AGO7 are in the microsomal fraction, and AGO7 tends to be adjacent to vesicles decorated by a cis-Golgi marker (Jouannet et al., 2012) (Fig. 2). All miRNAs, including 22-nt miRNAs, are enriched on MBPs, and reduced membrane association of 22-nt miRNAs correlates with decreased levels of phasiRNAs (S. Li et al., 2016). TAS transcripts are bound by ribosomes (Hou et al., 2016) and MBPs (S. Li et al., 2016). These findings suggest that the initial miRNA-guided cleavage step of phasiRNA biogenesis occurs on MBPs and the subsequent steps occur on certain membrane structures.
III. Turnover of miRNAs
The levels of miRNAs must be precisely and dynamically regulated in vivo and miRNA turnover is a mechanism to regulate miRNA levels. Studies of the hen1 mutant revealed two major mechanisms underlying miRNA degradation in Arabidopsis: 3′-to-5′ truncation and 3′ uridylation (Li et al., 2005; Yu et al., 2005; Yang Z et al., 2006) (Fig. 3). A few genes responsible for miRNA degradation via these two mechanisms have been identified (Ramachandran & Chen, 2008; Zhao et al., 2012b; Tu et al., 2015; Wang et al., 2015), but the full picture remains elusive.
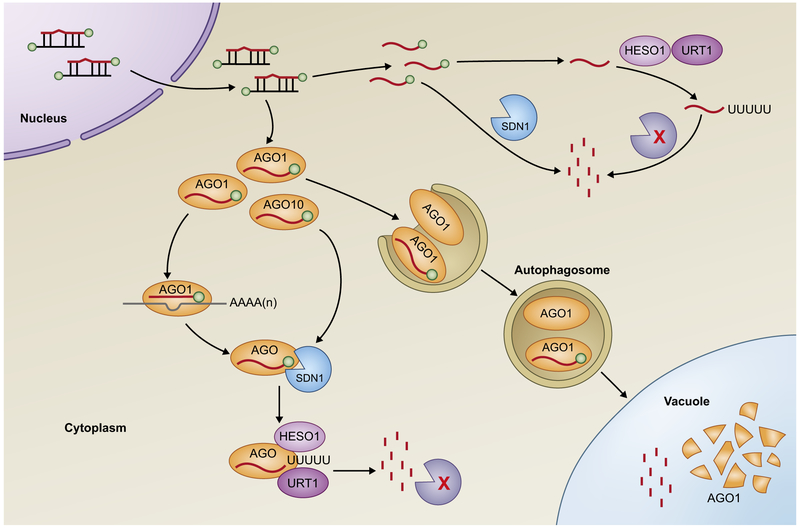
Fig. 3.
Mechanisms of plant microRNA (miRNA) turnover. miRNA degradation starts with the removal of the methyl group at the 3′ end by SMALL RNA DEGRADING NUCLEASE 1 (SDN1), which is followed by 3′ uridylation through HEN1 SUPPRESSOR 1 (HESO1) and/or UTP:RNA URIDYLYLTRANSFERASE 1 (URT1). The tailed miRNAs are subsequently degraded by an unknown exonuclease. SDN1 and nucleotidyl transferases (HESO1 and URT1) can act on both ARGONAUTE (AGO)-bound miRNAs and free miRNAs in the cytoplasm. Free miRNAs are also degraded by SDN1 directly. The degradation of AGO1 via autophagy may also contribute to miRNA turnover.
1. miRNA stabilization by 3′ methylation
Mature miRNAs are protected by 3′ end methylation catalyzed by HEN1. Loss of function in HEN1 results in reduced abundance of almost all miRNAs, which are also heterogeneous in size as a result of 3′ truncation and 3′ tailing (predominantly uridylation) (Li et al., 2005; Yu et al., 2005; Yang Z et al., 2006). Similarly, loss of function in HEN1 homologs in other eukaryotes, including rice (Abe et al., 2010), Drosophila (Horwich et al., 2007; Saito et al., 2007), C. elegans (Billi et al., 2012), zebra fish (Kamminga et al., 2010) and mouse (Kirino & Mourelatos, 2007), also leads to miRNA or piRNA (piwi-interacting RNA) 3′ truncation and 3′ uridylation. Therefore, HEN1-mediated 2′-O-methylation plays a general role in protecting the 3′ ends of small RNAs.
2. Exonucleases in miRNA degradation
The SMALL RNA DEGRADING NUCLEASE (SDN) family encodes four 3′-to-5′ exonucleases that function redundantly in degrading both miRNAs and siRNAs (Ramachandran & Chen, 2008) (Fig. 3). Single and double sdn mutants resemble wild-type plants, but knockdown of three SDN family members leads to severe pleiotropic developmental defects and elevated miRNA accumulation (Ramachandran & Chen, 2008). In vitro enzymatic assays show that SDN1 specifically acts on short single-stranded RNAs, and the exonuclease activity is partially inhibited by the methyl group at the 3′ end of miRNAs (Ramachandran & Chen, 2008).
SDNs are responsible for the 3′ truncation of miRNAs in both hen1 and wild-type plants (Yu et al., 2017). Comparing miRNA profiles of hen1 and hen1 sdn1 sdn2 plants by small RNA-seq showed that the 3′ truncation of some miRNAs is reduced when SDN1 and SDN2 are both absent (Yu et al., 2017). Similar results were observed when comparing the miRNA profiles of wild-type and sdn1 sdn2 plants, although 3′ truncated miRNAs have very low abundance in the wild-type (Yu et al., 2017). The fact that only a small number of miRNAs are affected by the absence of SDN1 and SDN2 could be a result of the redundant function of other SDN members or of non-SDN exonucleases.
SDN1 is unable to degrade U-tailed miRNAs in vitro (Ramachandran & Chen, 2008), and so it does not appear to be responsible for the degradation of uridylated miRNA species. Although it has not been reported in Arabidopsis, several exonucleases in other eukaryotes prefer uridylated RNAs as substrates. The 3′-to-5′ exonuclease DIS3-like 2 (DIS3L2) degrades uridylated RNAs in mammals and yeast, including uridylated pre-let-7 in mammals (Chang et al., 2013; Ustianenko et al., 2013). In Chlamydomonas, depletion of the exosome subunit Ribosomal RNA-Processing Protein 6 (RRP6) results in elevated accumulation of small RNAs in vivo, and RRP6 degrades 3′ uridylated miRNAs rather than non-uridylated miRNAs in vitro (Ibrahim et al., 2010). The Arabidopsis orthologs of DIS3L2 and RRP6 are SUPPRESSOR OF VARICOSE (SOV) and three RRP6-LIKE (PPR6L) genes, respectively, and are therefore the prime candidates for the degradation of uridylated miRNAs.
3. Non-templated tailing of miRNAs
3′ Non-templated tailing is a widespread phenomenon and a common post-transcriptional modification that regulates miRNA biogenesis, stability or activity in diverse model organisms (Wyman et al., 2011). Adenylation and uridylation are the two major types of 3′ tailing and are catalyzed by nucleotidyl transferases including non-canonical PolyA polymerases (PAPs) and terminal uridylyl transferases (TUTases), respectively (reviewed in Martin & Keller, 2007).
In Chlamydomonas, uridylation of miRNAs and siRNAs is catalyzed by the nucleotidyl transferase MUT68 (Ibrahim et al., 2010). MUT68 promotes the in vitro degradation of uridylated miRNAs through the exosome subunit RRP6 (Ibrahim et al., 2010). MUT68 and RRP6 appear to act only on unmethylated miRNAs, as 2′-O-methylated miR912 oligonucleotides failed to be uridylated and degraded in vitro (Ibrahim et al., 2010).
3′ Uridylation of miRNAs in Arabidopsis, rice and maize is widely observed in hen1 mutants in which miRNA methylation is abolished (Li et al., 2005; Yu et al., 2005; Yang Z et al., 2006; Abe et al., 2010; Zhai et al., 2013). (Fig. 3). In Arabidopsis, HESO1 and URT1 uridylate unmethylated miRNAs in the hen1 mutant, leading to miRNA degradation (Ren et al., 2012a; Zhao et al., 2012b; Tu et al., 2015; Wang et al., 2015). Loss of function in both HESO1 and URT1 rescues the developmental defects of the hen1 mutant, accompanied by elevated miRNA accumulation and reduced 3′ uridylation (Ren et al., 2012a; Zhao et al., 2012b; Tu et al., 2015; Wang et al., 2015). In vitro, both HESO1 and URT1 exhibit nucleotidyl transferase activities on unmethylated RNA oligonucleotides, but not 3′ methylated RNAs (Ren et al., 2012a; Zhao et al., 2012b; Tu et al., 2015; Wang et al., 2015). Although HESO1 and URT1 both prefer U over the other three nucleotides, they have different substrate specificities and cooperatively tail different forms of the same miRNAs in vivo. Although HESO1 prefers U-ending miRNAs as substrates, URT1 favors A-ending miRNAs. Given the observation of substantial monouridylated miRNAs in the hen1 heso1 double mutant (Ren et al., 2012a; Zhao et al., 2012b; Tu et al., 2015; Wang et al., 2015), one possibility is that URT1 first uridylates unmethylated miRNAs to generate monoU-tailed forms, the preferred substrates for HESO1, to produce longer U tails.
3′ Uridylation may also affect miRNA activity. When AGO1-bound miR165/6 was uridylated by URT1 in vitro, the slicer activity was reduced (Tu et al., 2015). The monouridylation of miR171a by URT1 in hen1 makes it capable of triggering the biogenesis of secondary phasiRNAs (Zhai et al., 2013).
In Populus trichocarpa (black cottonwood), a few miRNAs undergo 3′ adenylation, although the corresponding enzymes remain unknown (Lu et al., 2009). Synthesized miRNA oligonucleotides with 3′ adenylation were degraded at a slower rate in plant extracts than were those without it (Lu et al., 2009), indicating that adenylation contributes to miRNA stabilization.
4. AGO proteins in miRNA stability
In addition to its key role in miRNA-mediated activities, AGO1 shelters its associated miRNAs from degradation, based on the reduced abundance of many miRNAs in ago1 null mutants (Vaucheret et al., 2004). It is therefore counterintuitive that the weak allele ago1-11 suppresses the 3′ truncation and 3′ uridylation of miRNAs in the hen1 mutant (Zhai et al., 2013). In addition, both truncated and tailed miRNA species associate with AGO1 in vivo (Zhao et al., 2012a; Zhai et al., 2013). This implies that, during miRNA degradation, SDN1 and HESO1/URT1 act on AGO1-bound miRNAs. Indeed, both HESO1 and URT1 are able to tail AGO1-bound miRNAs in vitro, and the tailed miRNAs remain associated with AGO1 (Ren et al., 2014; Tu et al., 2015; Wang et al., 2015). The interactions between HESO1/URT1 and AGO1 are evidenced by reciprocal co-immunoprecipitation (Ren et al., 2014; Wang et al., 2015). Although SDN1–AGO1 interaction has not been reported, SDN1 acts on AGO1-bound miRNAs in vitro to generate truncated miRNAs of heterogeneous sizes that remain bound to AGO1 (Yu et al., 2017). Given that 2′-O-methylation of miRNAs completely inhibits the activity of HESO1 and URT1, but not SDN1 (Ramachandran & Chen, 2008; Zhao et al., 2012b; Tu et al., 2015), one possibility is that SDN1 and HESO1/URT1 cooperate in degrading AGO1-bound miRNAs that are methylated: SDN1 removes the methyl group from these miRNAs, and HESO1/URT1 cause subsequent uridylation. This would lead to miRNA degradation by an unknown exonuclease that prefers U-tailed RNAs (Fig. 3). This hypothesis is supported by the following observations: 3′ truncated-only and 3′ truncated-and-tailed miRNA species are reduced in the hen1 sdn1 sdn2 triple mutant compared with hen1, whereas 3′ tailed species are reduced in hen1 heso1 with the concomitant increase in 3′ truncated-only forms (Yu et al., 2017). Free miRNAs, on the other hand, can be degraded solely by SDN1 or sequentially by SDN1 and HESO1/URT1.
As the closest paralog of AGO1 amongst the 10 Arabidopsis AGO proteins, AGO10 is only expressed in certain cells and acts in stem cell maintenance in the shoot apical meristem (SAM) and in leaf polarity specification (Moussian et al., 1998; Lynn et al., 1999; Mallory et al., 2004). Such functions were found to be achieved through repression of the activity of miR165/166 (Liu et al., 2009; Zhu et al., 2011). AGO10 has a higher binding affinity than AGO1 to miR165/6 and, rather than protecting this miRNA, AGO10 promotes its degradation (Zhu et al., 2011; Zhou et al., 2015). In ago10 mutants, miR165/6 accumulation is sufficiently increased that it can be detected by in situ hybridization in AGO10-expressing cells, which is not the case in the wild-type (Liu et al., 2009). AGO10 overexpression results in the degradation of miR165/6 by SDN1 and SDN2 (Yu et al., 2017). An in vitro assay further suggested that AGO10-bound miR165/6 is more susceptible than AGO1-bound miR165/6 to SDN1-mediated truncation (Yu et al., 2017). Promotion of miR165/6 degradation probably contributes to AGO10-mediated maintenance of stem cells and the specification of leaf polarity.
5. Effect of target transcripts on miRNA stability
Although the enzymes for miRNA 3′ truncation or 3′ uridylation act on many miRNAs, specificity in miRNA degradation may be achieved through target RNAs or non-coding RNAs. In Arabidopsis, miR399 is regulated by a native transcript with a miR399 binding site from the IPS1 (INDUCED BY PHOSPHATE STARVATION 1) locus (Franco-Zorrilla et al., 2007). A3-nt bulge at the cleavage site within the IPS1 transcript abolishes miR399-mediated cleavage, thereby rendering the IPS1 transcript a target mimic (TM) that sequesters miR399 from its other targets and reduces its activity (Franco-Zorrilla et al., 2007). Genome-wide bioinformatic analyses indicate that many transcripts, from either non-coding genomic regions or annotated genes, can serve as potential endogenous TMs to regulate miRNA activity (Ivashuta et al., 2011; H.J. Wu et al., 2013). Intriguingly, in transgenic lines with artificial TMs, the levels of the corresponding miRNAs are reduced (Ivashuta et al., 2011; H.J. Wu et al., 2013). Similar results were observed in transgenic lines expressing short tandem target mimic (STTM) RNAs, which contain two tandem miRNA binding sites with mismatches at the cleavage positions (Tang et al., 2012; Yan et al., 2012). STTM-triggered miRNA degradation requires the activity of SDN1 and SDN2 in vivo (Yan et al., 2012).
Target-induced miRNA turnover is conserved across flies and mammals. In animals, miRNAs recognize their targets through pairing at the seed region (miRNA nucleotides 2–7) (reviewed in Bartel, 2009). Extensive pairing between miRNAs and artificial target transcripts leads to 3′ trimming and tailing of miRNAs in Drosophila and humans (Ameres et al., 2010; Cazalla et al., 2010; Marcinowski et al., 2012). Based on crystal structure analysis in Thermus thermophilus (Sheng et al., 2014), the conformation of AGO is altered after a highly complementary target is in RISC such that the 3′ end of the guide is released from the binding pocket in AGO. Thus, it is deduced that the 3′ end of an AGO1-bound miRNA would be exposed to SDNs, HESO1 or URT1 on recognition of highly complementary targets in Arabidopsis.
6. Subcellular sites of miRNA turnover
The subcellular localization of SDNs, HESO1 and URT1 may offer clues for the subcellular sites of miRNA turnover. In addition, because AGO1-bound miRNAs can be truncated and uridylated, AGO1 localization is another important indicator for the sites of miRNA turnover. Although the localization patterns of SDNs are unknown, HESO1 and URT1 colocalize in cytoplasmic foci, where AGO 1 is also localized (Wang et al., 2015). In addition, both enzymes interact with AGO1, and uridylated miRNAs remain bound by AGO1 (Ren et al., 2014; Wang et al., 2015). Based on these findings, the cytoplasmic foci are potential sites of miRNA degradation. In addition to uridylating unmethylated miRNAs, HESO1 and URT1 also catalyze the uridylation of the 5′ cleavage fragments from miRNA target transcripts, leading to their degradation (Ren et al., 2014). Given that a fraction of AGO1-mediated cleavage takes place on MBPs (S. Li et al., 2016), the undefined cytoplasmic foci may contain MBPs.
The post-translational regulation of AGO1 protein may also provide a clue about the sites of miRNA turnover (Fig. 3). In pathogenic and viral contexts, AGO1 is ubiquitinylated by the polerovirus-encoded F-box protein P0 and subsequently degraded through autophagy, a process in which cytosolic proteins are delivered to lysosomes for degradation (Derrien et al., 2012). AGO1 is also regulated by another F-box protein, F-box and WD-repeat domain-containing protein 2 (FBW2), which also leads to AGO1 degradation via autophagy (Earley et al., 2010). The colocalization of AGO1 and AUTOPHAGY 8 (ATG8), an autophagosomal membrane protein (Derrien et al., 2012), further indicates that AGO1 is associated with autophagosomes. As AGO1 degradation would indisputably impair the stability of its associated miRNAs, miRNA degradation may occur concomitantly with AGO1 autophagy.
IV. Concluding remarks
Although many players involved in miRNA biogenesis, degradation and activity have been discovered, much is unknown regarding the subcellular locations in which these processes take place. For example, it is unknown how D-bodies containing the dicing complex are formed, how AGO1, a presumably soluble protein, associates with ER and membrane-bound polysomes, and how membrane-bound polysomes affect miRNA-guided phasiRNA biogenesis. As AGO1 associates not only with miRNAs, but also with siRNAs from endogenous sequences, such as transposons and phasiRNA loci, as well as from exogenous sequences, such as viruses and transgenes, the subcellular partitioning of AGO1 between the cytosol and endomembranes and between the nucleus and the cytoplasm probably influences the activities of various types of small RNAs. The limited knowledge of the subcellular locations of miRNA biogenesis, degradation and activity precludes a full understanding of miRNAs, as well as the crosstalk between miRNAs and siRNAs.
Acknowledgements
Research in the Chen laboratory is supported by a grant from the National Institutes of Health (GM061146).
References
- Abe M, Yoshikawa T, Nosaka M, Sakakibara H, Sato Y, Nagato Y, Itoh J. 2010. WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining microRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiology 154: 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Alonso-Peral MM, Li J, Li Y,Allen RS, Schnippenkoetter W, Ohms S,White RG, Millar AA. 2010. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiology 154: 757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD.(2010).Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Hernandez L, Marchais A, Poulsen C, Haase B, Hauptmann J, Benes V, Meister G, Brodersen P. 2016. The slicer activity ofARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28: 1563–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Baek D, Kim MC, Chun HJ, Kang S, Park HC, Shin G, Park J, Shen M, Hong H, Kim WY et al. 2013. Regulation of miR399ftranscription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiology 161: 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranauske S, Mickute M, Plotnikova A, Finke A, Venclovas C, Klimasauskas S, Vilkaitis G.2015. Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Research 43: 2802–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Chaabane S, Liu R, Chinnusamy V, Kwon Y, Park JH, Kim SY, Zhu JK, Yang SW, Lee BH. 2013. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Research 41: 1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK. 2012. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genetics 8: e1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Mateos JL, Bresso EG, Palatnik JF. 2009. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO Journal 28: 3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO Journal 25: 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscheid A, Marchais A, Schott G, Lange H, Gagliardi D, Andersen SU, Voinnet O,Brodersen P. 2015. SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Research 43: 10975–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. 2008. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Schaller H, Khafif M, Schott G, Bendahmane A, Voinnet O. 2012. Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109: 1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Villalobos LI, Kuhnle C, Dohmann EM, Li H, Bevan M, Schwechheimer C. 2005. The evolutionarily conserved TOUGH protein is required for proper development of Arabidopsis thaliana. Plant Cell 17: 2473–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328: 1563–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. 2013. A role for the Perlman syndrome exonuclease Dis3 l2 in the Lin28-let-7 pathway. Nature 497: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 2010. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proceedings of the National Academy of Sciences, USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X 2005. MicroRNA biogenesis and function in plants. FEBS Letters 579: 5923–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X 2009. Small RNAs and their roles in plant development. Annual Review of Cell and Developmental Biology 25: 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ben Chaabane S, Shah P, Poulsen CP, Yang SW. 2014. COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nature Communications 5: 5867. [DOI] [PubMed] [Google Scholar]
- Copeland C, Xu S, Qi Y, Li X. 2013. MOS2 has redundant function with its homolog MOS2H and is required for proper splicing of SNC1. Plant Signaling & Behavior 8: e25372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Fang X, Qi Y. 2016. TRANSPORTIN1 promotes the association of microRNA with ARGONAUTE1 in Arabidopsis. Plant Cell 28: 2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. 2010. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature Structural & Molecular Biology 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM. 2008. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Letters 582: 2753–2760. [DOI] [PubMed] [Google Scholar]
- Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, Genschik P. 2012. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proceedings of the National Academy of Sciences, USA 109: 15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N. 2008. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proceedings of the National Academy of Sciences, USA 105: 9970–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens AL, Kim KW, Curtin SJ, Waterhouse PM. 2012. DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS ONE 7: e35933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. 2009. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Smith M, Weber R, Gregory B, Poethig R. 2010. An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Yang J, Mathioni SM, Yu J, Shen J, Yang X, Wang L, Zhang Q, Cai Z, Xu C et al. 2016. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proceedings of the National Academy of Sciences, USA 113: 15144–15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Cui Y, Li Y, Qi Y. 2015. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nature Plants 1: 15075. [DOI] [PubMed] [Google Scholar]
- Fang Y, Spector DL. 2007. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Current Biology 17: 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. 2013. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco-Mangilet AG, Karlsson P, Kim MH, Eo HJ, Oh SA, Kim JH, Kulcheski FR, Park SK, Manavella PA. 2015. THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant Journal 82: 1018–1029. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 39:1033–1037. [DOI] [PubMed] [Google Scholar]
- Fukudome A, Fukuhara T. 2017. Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis. Journal of Plant Research 130: 33–44. [DOI] [PubMed] [Google Scholar]
- Furumizu C, Tsukaya H, Komeda Y. 2010. Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA 16: 1809–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. 2007. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant Journal 49: 683–693. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Current Biology 15: 1494–1500. [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R et al. 2008. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nature Biotechnology 26: 941–946. [DOI] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, Cowling VH. 2014. Cap-binding complex (CBC). Biochemical Journal 457: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M. 2005. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026. [DOI] [PubMed] [Google Scholar]
- Hajheidari M, Farrona S, Huettel B, Koncz Z, Koncz C. 2012. CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell 24: 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M, Koncz C, Eick D. 2013. Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends in Plant Science 18: 633–643. [DOI] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N. 2004. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proceedings of the National Academy of Sciences, USA 101: 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S,Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G et al. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149: 768–779. [DOI] [PubMed] [Google Scholar]
- Heath CG, Viphakone N, Wilson SA. 2016. The role of TREX in gene expression and disease. Biochemical Journal 473: 2911–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. 2005. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Molecular Biology 57: 173–188. [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline pi RNAs and single-stranded siRNAs in RISC. Current Biology 17: 1265–1272. [DOI] [PubMed] [Google Scholar]
- Hou CY, Lee WC,Chou HC,Chen AP,Chou SJ,Chen HM. 2016. Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28:2398–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RH. 2012. Unique 2′-O-methylation byHen1 in eukaryotic RNA interference and bacterial RNA repair. Biochemistry 51: 4087–4095. [DOI] [PubMed] [Google Scholar]
- Huang Y, Ji LJ, Huang QC, Vassylyev D, Chen XM, Ma JB. 2009. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461: 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Ibrahim F,Rohr J,Jeong WJ, Hesson J, Cerutti H. 2006. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 314: 1893. [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. 2010. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proceedings of the National Academy of Sciences, USA 107: 3906–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. 2010. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Molecular Cell 39: 282–291. [DOI] [PubMed] [Google Scholar]
- Ivashuta S, Banks IR, Wiggins BE, Zhang Y, Ziegler TE, Roberts JK, Heck GR. 2011. Regulation of gene expression in plants through miRNA in activation. PLoS ONE 6: e21330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. 2013. Molecular insights into microRNA-mediated translational repression in plants. Molecular Cell 52: 591–601. [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. 2015. The functions of microRNAs: mRNA decay and translational repression. Trends in Cell Biology 25: 651–665. [DOI] [PubMed] [Google Scholar]
- Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng B et al. 2011. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genetics 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KCM, Dong OX, Li X. 2011. The evolutionarily conserved MOS4-associated complex. Central European Journal of Biology 6: 776–784. [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. 2006. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology 57: 19–53. [DOI] [PubMed] [Google Scholar]
- Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A. 2012. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO Journal 31: 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF. 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO Journal 29: 3688–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson P, Christie MD, Seymour DK, Wang H, Wang X, Hagmann J, Kulcheski F, Manavella PA. 2015. KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proceedings of the National Academy of Sciences, USA 112: 14096–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X. 2011. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO Journal 30: 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. 2007. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13: 1397–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop K, Stepien A, Barciszewska-Pacak M, Taube M, Bielewicz D, Michalak M, Borst JW, Jarmolowski A, Szweykowska-Kulinska Z. 2016. Active 5′ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Research 45: 2757–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nature Reviews Molecular Cell Biology 8: 761–773. [DOI] [PubMed] [Google Scholar]
- Koncz C, Dejong F, Villacorta N, Szakonyi D, Koncz Z. 2012. The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Frontiers in Plant Science 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster T, Meyer K, Weinholdt C, Smith LM, Lummer M, Speth C, Grosse I, Weigel D, Staiger D. 2014. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Research 42: 9925–9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P, Gloor G. 2001. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress and Chaperones 6: 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. 2009. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Letters 583: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y. 2006. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. 2004. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proceedings of the National Academy of Sciences, USA 101: 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crete P, Voinnet O, Robaglia C. 2009. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, Weigel D. 2008. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 105: 8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Current Biology 15: 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Le B, Ma X, Li S, You C, Yu Y, Zhang B, Liu L, Gao L, Shi T et al. 2016. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 5: e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B et al. 2013. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang S, Cheng J, Su C, Zhong S, Liu Q, Fang Y, Yu Y, Lv H, Zheng Y et al. 2016. Intron lariat RNA inhibits microRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLoS Genetics 12: e1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Axtell MJ, Fedoroff NV. 2012. The helicase and RNaseIIIa domains of Arabidopsis Dicer-Like1 modulate catalytic parameters during microRNA biogenesis. Plant Physiology 159: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H. 2009. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant Journal 58: 27–40. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. 2006. SERRATE: a new player on the plant microRNA scene. EMBO Reports 7: 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL. 2009. Adenylation of plant miRNAs. Nucleic Acids Research 37: 1878–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. 2004. Nuclear export of microRNA precursors. Science 303: 95–98. [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. 1999. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. 2006. Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. 2004. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO Journal 23: 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D. 2012a. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151: 859–870. [DOI] [PubMed] [Google Scholar]
- Manavella PA, Koenig D, Weigel D. 2012b. Plant secondary siRNA production determined by microRNA-duplex structure. Proceedings of the National Academy of Sciences, USA 109: 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. 2011. Biogenesis and function of nuclear bodies. Trends in Genetics 27: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinowski L, Tanguy M, Krmpotic A, Radle B, Lisnic VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C et al. 2012. Degradation of cellular miR-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathogens 8: e1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CM, Tartaglio V, Duarte M, Harmon FG. 2016. The Arabidopsis sickle mutant exhibits altered circadian clock responses to cool temperatures and temperature-dependent alternative splicing. Plant Cell 28: 2560–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W. 2007. RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunoury N,Vaucheret H. 2011. AGO1 and AGO2 act redundantly in miR408-mediated Plantacyanin regulation. PLoS ONE 6: e28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG. 2006. MicroRNA promoter element discovery in Arabidopsis. RNA 12: 1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C et al. 2008Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Peragine A, Endres MW, Li J, Chen X, Poethig RS, Bowman LH, Vance V. 2008. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS ONE 3: e1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Xu F, Gao M, Zhao Q, Palma K, Long C, Chen S, Zhang Y, Li X. 2009. Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathogens 5: e1000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. 2008. Specificity of ARGONAUTE7–miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141. [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. 1998. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO Journal 17: 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I, Mette MF,Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJ. 2003. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiology 132: 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. 2005. Nuclear processing and export of microRNAs in Arabidopsis. Proceedings of the National Academy of Sciences, USA 102: 3691–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Current Biology 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Shi J, Zhai Y, Hou Y, Ma W. 2015. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proceedings of the National Academy of Sciences, USA 112: 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska KD, Stepien A, Kierzkowski D, Kalak M, Bajczyk M, McNicol J, Simpson CG, Szweykowska-Kulinska Z, Brown JW, Jarmolowski A. 2014. The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Research 42: 1224–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram B, Sheikh AH, Rustagi Y, Sinha AK. 2015. MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS Journal 282: 521–536. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Development 20:3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. 2008. Degradation of micro RNAs by a family of exoribonucleases in Arabidopsis. Science 321: 1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. 2002. MicroRNAs in plants. Genes & Development 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Chen X, Yu B. 2012a. Uridylation of miRNAs by HEN1 SUPPRESSOR1 in Arabidopsis. Current Biology 22: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B. 2012b. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109: 12817–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Zhang S, Vinovskis C, Chen X, Yu B. 2014. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proceedings of the National Academy of Sciences, USA 111: 6365–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. 2013. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. 2007. Pimet, the Drosophila homolog of HEN1, mediates 2’-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes & Development 21: 1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. 2002. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends in Plant Science 7:487–491. [DOI] [PubMed] [Google Scholar]
- Sheng G, Zhao H, Wang J, Rao Y, Tian W, Swarts DC, van der Oost J, Patel DJ, Wang Y. 2014. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proceedings of the National Academy of Sciences, USA 111: 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Axtell MJ, Fedoroff NV. 2010. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Current Biology 20: 37–41. [DOI] [PubMed] [Google Scholar]
- Song L, Han MH, Lesicka J, Fedoroff N. 2007. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proceedings of the National Academy of Sciences, USA 104: 5437–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ. 2004. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Molecular Cell 15: 173–183. [DOI] [PubMed] [Google Scholar]
- Stepien A, Knop K, Dolata J, Taube M, Bajczyk M, Barciszewska-Pacak M, Pacak A, Jarmolowski A, Szweykowska-Kulinska Z. 2016. Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdisciplinary Reviews: RNA 8: e1403. [DOI] [PubMed] [Google Scholar]
- Streitner C, Koster T, Simpson CG, Shaw P, Danisman S, Brown JW, Staiger D. 2012. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Research 40: 11240–11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Li Z, Cheng J, Li L, Zhong S, Liu L, Zheng Y, Zheng B. 2017. The Protein Phosphatase 4 and SMEK1 complex dephosphorylates HYL1 to promote miRNA biogenesis by antagonizing the MAPK cascade in Arabidopsis. Developmental Cell 41: e525. [DOI] [PubMed] [Google Scholar]
- Szarzynska B, Sobkowiak L, Pant BD, Balazadeh S, Scheible WR, Mueller-Roeber B, Jarmolowski A, Szweykowska-Kulinska Z. 2009. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Research 37: 3083–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S,Watanabe T, Utsumi M,Watanabe Y. 2008. The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant & Cell Physiology 49: 493–500. [DOI] [PubMed] [Google Scholar]
- Tang G, Yan J, Gu Y, Qiao M, Fan R, Mao Y, Tang X. 2012. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58: 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B, Liu L, Xu C, Zhai J, Li S, Lopez MA, Zhao Y, Yu Y, Ramachandran V, Ren G et al. 2015. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genetics 11: e1005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. 2013. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 mi RNAs. RNA 19: 1632–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. 2004. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Development 18: 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- Wang L, Song X, Gu L, Li X, Cao S, Chu C, Cui X, Chen X, Cao X. 2013. NOT2 proteins promote polymerase II-dependent transcription and interact with multiple microRNA biogenesis factors in Arabidopsis. Plant Cell 25: 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ye R, Xin Y, Fang X, Li C, Shi H, Zhou X, Qi Y. 2011. An importin β protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell 23: 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang S, Dou Y, Zhang C, Chen X, Yu B, Ren G. 2015. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ tailing of small RNAs in Arabidopsis. PLoS Genetics 11: e1005091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wagner JD, Guthrie C. 1998. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Current Biology 8: 441–451. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harbor Perspectives in Biology 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Yu L, Cao W, Mao Y, Liu Z, He Y. 2007. The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19: 914–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Wang ZM, Wang M, Wang XJ. 2013. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiology 161: 1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shi Y, Li J, Xu L, Fang Y, Li X, Qi Y. 2013. A role for the RNA-binding protein MOS2 in micro RNA maturation in Arabidopsis. Cell Research 23: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. 2011. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Research 21: 1450–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. 2005a. Expression of Arabidopsis MIRNA genes. Plant Physiology 138: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. 2005b. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH. 2011. Processing bodies and plant development. Current Opinion in Plant Biology 14: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G. 2012. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24:415–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang P, Wang B, Hsu CC, Tang K, Zhang H, Hou YJ, Zhao Y, Wang Q, Zhao C et al. 2017. The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genetics 13: e1006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H. 2006. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant Journal 47: 841–850. [DOI] [PubMed] [Google Scholar]
- Yang L, Wu G, Poethig RS. 2012. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109: 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA. 2010. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ren W, Zhao Q, Zhang P, Wu F, He Y. 2014. Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Research 42: 12224–12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X. 2006. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Research 34: 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Development 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Iki T, Numa H, Miyashita K, Meshi T, Ishikawa M. 2016. A short open reading frame encompassing the microRNA173 target site plays a role in trans-acting small interfering RNA biogenesis. Plant Physiology 171: 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Iki T, Tsutsui Y, Miyashita K, Poethig RS, Habu Y, Ishikawa M. 20133′ fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proceedings of the National Academy of Sciences, USA 110: 4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. 2005. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & Development 19: 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC et al. 2008. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proceedings of the National Academy of Sciences, USA 105: 10073–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. 2005. Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Willmann MR., Anderson SJ, Gregory BD. 2016. Genome-wide mapping of uncapped and cleaved transcripts reveals a role for the nuclear mRNA cap-binding complex in cotranslational RNA decay in Arabidopsis. Plant Cell 28: 2385–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ji L, Le BH, Zhai J, Chen J, Luscher E, Gao L, Liu C, Cao X, Mo B et al. 2017. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biology 15: e2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumul RE, Kim YJ, Liu X, Wang R, Ding J, Xiao L, Chen X. 2013. POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PLoS Genetics 9: e1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan G-L, Walbot V, Meyers BC. 2015. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proceedings of the National Academy of Sciences, USA 112:3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Zhao Y, Simon SA, Huang S, Petsch K, Arikit S, Pillay M, Ji L, Xie M, Cao X et al. 2013. Plant microRNAs display differential 3’ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25: 2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Wang B, Li H, Liu R, Kalia RK, Zhu JK, Chinnusamy V. 2012. Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proceedings of the National Academy of Sciences, USA 109: 18198–18203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Ng DWK, Lu J, Chen ZJ. 2012. Roles of target site location and sequence complementarity in trans-acting siRNA formation in Arabidopsis. Plant Journal 69: 217–226. [DOI] [PubMed] [Google Scholar]
- Zhang S, Liu Y, Yu B. 2014. PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genetics 10: e1004841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xie M, Ren G, Yu B. 2013. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proceedings of the National Academy of Sciences, USA 110: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng YT, Bi D, Palma K, Li X 2005. MOS2, a protein containing G-patch and KOW motifs, is essential for innate immunity in Arabidopsis thaliana. Current Biology 15: 1936–1942. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Guo X, Ge C, Ma Z, Jiang M, Li T, Koiwa H, Yang SW, Zhang X. 2017a. KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 114: 4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hu F, Sung MW, Shu C, Castillo-González C, Koiwa H, Tang G, Dickman M, Li P, Zhang X. 2017b. RISC-interacting clearing 3′-5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. eLife 6: e24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Mo B, Chen X 2012a. Mechanisms that impact microRNA stability in plants. RNA Biology 9: 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X 2012b. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Current Biology 22: 689–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Honda M, Zhu H, Zhang Z, Guo X, Li T, Li Z, Peng X, Nakajima K, Duan L et al. 2015. Spatiotemporal sequestration of miR165/166 by Arabidopsis Argonaute10 promotes shoot apical meristem maintenance. Cell Reports 10: 1819–1827. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X. 2011. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhou Y, Castillo-Gonzalez C, Lu A, Ge C, Zhao YT, Duan L, Li Z, Axtell MJ, Wang XJ et al. 2013. Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nature Structural & Molecular Biology 20:1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielezinski A, Dolata J, Alaba S, Kruszka K, Pacak A, Swida-Barteczka A, Knop K, Stepien A, Bielewicz D, Pietrykowska H et al. 2015. MirEX 2.0 – an integrated environment for expression profiling of plant microRNAs. BMC Plant Biology 15: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]