Abstract
Lysine acetylation is tightly coupled to the nutritional status of the cell, as the availability of its cofactor, acetyl-CoA, fluctuates with changing metabolic conditions. Recent studies have demonstrated that acetyl-CoA levels act as an indicator of cellular nourishment, and increased abundance of this metabolite can block the induction of cellular recycling programs. Here we investigated the cross-talk between mitochondrial metabolic pathways, acetylation and autophagy, using chemical inducers of mitochondrial acetyl-CoA production. Treatment of cells with α-Lipoic Acid, a co-factor of the pyruvate dehydrogenase complex, led to the unexpected hyperacetylation of α-Tubulin in the cytosol. This acetylation was blocked by pharmacological inhibition of mitochondrial citrate export (a source for mitochondrial-derived acetyl-CoA in the cytosol), was dependent on the α-Tubulin acetyltransferase αTAT, and was coupled to a loss in function of the cytosolic histone deacetylase, HDAC6. We further demonstrate that α-Lipoic Acid slows the flux of substrates through autophagy-related pathways, and severely limits the ability of cells to remove depolarized mitochondria through PINK1-mediated mitophagy.
Keywords: α-Lipoic Acid, α-Tubulin acetylation, α-Tubulin acetyltransferase (αTAT), Autophagy, Histone Deacetylase 6 (HDAC6), Hyperacetylation, Mitochondria, Mitophagy
INTRODUCTION
During periods of nutrient deprivation, organisms can upregulate multiple catabolic pathways to maintain metabolite supply and cellular homeostasis. Autophagy, a cellular recycling process induced by nutrient stress, breaks down multiple subcellular components, allowing the cell to reutilize the biochemical structures contained within. To regulate autophagy induction, cells employ a number of mechanisms to monitor their energy status, with the best characterized being the AMPK-mTOR-ULK1 axis. Here, increases in the AMP:ATP ratio in the cytoplasm are sensed by AMP kinase (AMPK), which phosphorylates and inactivates the mammalian Target of Rapamycin (mTOR). Inactivation of mTOR removes its inhibition of the ULK1 kinase which, along with other upstream effectors, initiates the production of structures necessary to transport autophagy substrates to the lysosome for processing (reviewed in (1)).
Recent work has demonstrated that acetyl-CoA, like AMP/ATP, may act as a sensor molecule for cellular nutritional status (2). Using a number of approaches to manipulate cellular acetyl-CoA levels, it was shown that reductions in the cytosolic abundance of acetyl-CoA activated the cellular autophagic machinery. This process could be reversed using pharmacological agents that increase the availability of acetyl-CoA in the cytoplasm (2).
The final stages of these fuel metabolism pathways occur in the mitochondrion, and as such this organelle is one of the main regulators of acetyl-CoA abundance in the cell. This regulation occurs both within the mitochondria (by changing the levels of acetyl-CoA available for bioenergetic or biosynthetic pathways), and in the greater cellular environment (through the retrograde export of acetyl-CoA-transport molecules, such as citrate, to the cytoplasm; (2)).
We recently demonstrated that a reduction in mitochondrial protein lysine acetylation levels led to the increased turnover of mitochondrial proteins through mitophagy, an organelle-specific degradation pathway related to autophagy (3, 4). As acetyl-CoA is the cofactor for acetylation, there is a strong link between its availability and the acetylation status of mitochondrial proteins, as evidenced by the increased observation of this modification in models of nutrient excess (5, 6). We therefore hypothesized that increasing mitochondrial acetyl-CoA levels, using pharmacological inducers of pyruvate metabolism, would lead to the protection of mitochondria from degradation by mitophagy.
In this report we demonstrate that treatment of cells with α-Lipoic Acid, a co-factor in several mitochondrial metabolic pathways, including the pyruvate dehydrogenase complex, leads to the rapid and specific hyperacetylation of a single protein. Surprisingly, we show that this modification is localized outwith the mitochondria, and occurs on the K40 residue of α-Tubulin. Acetylation of this residue is dependent on the function of the tubulin acetyltransferase, αTAT, and is mediated by an inhibition of HDAC6 function. Finally, we demonstrate that α-Lipoic Acid treatment slows the flux of substrate processing by the autophagic machinery, and limits the removal of depolarized mitochondria through PINK1-related mitophagy pathways.
MATERIALS AND METHODS
Cell Culture and Treatments
African Green Monkey fibroblast COS-7 cells or Human hepatocarcinoma HepG2 cells were grown in high-glucose DMEM, supplemented with 10 % FBS and Anti-Anti (all Gibco), at 37 °C/5 % CO2. Cells were treated with the following chemicals (see figure legends for time and concentration): (±)-α-Lipoic Acid, sodium pyruvate (Pyr), dichloroacetate, N-acetylcysteine, 4-hydroxy-TEMPO, bafilomycin A1 and carbonyl cynanide 3-chlorophenolhydrazone, 6-BHA and BTC (all Sigma). Small-interfering RNAs for αTAT, along with non-targeting controls, were obtained from IDTDNA and transfected into cells using Lipofectamine 3000 (Life Technologies). GFP-Tubulin and GFP-K40R-Tubulin were obtained from Addgene and transfected using Lipofectamine 3000.
Cell Harvest, Subcellular Fractionation and Co-Immunoprecipitation
For standard western blots, cells were washed in ice-cold PBS and harvested by scraping. Cell pellets were lysed on ice for 30 min in CHAPS detergent buffer, and supernatents were recovered at 13000 g. For subcellular fractionation, cells were harvested as before and cell pellets lysed on ice using detergent-free sucrose buffer and a 25-gauge needle. After removal of cell nuclei and unlysed cells at 500 g, supernatents were centrifuged at 6000 g. The pellet represented the mitochondrial-rich heavy-membrane (HM) fraction, while the supernatent represented the cytosol. Both fractions were further lysed in CHAPS buffer prior to analysis by western blot. For co-immunoprecipitation experiments, cells were harvested in CHAPS buffer, and equal amounts of total protein were incubated at 4 °C overnight with the relevant antibody. Immunocaptured proteins were isolated using Protein-A agarose beads (Cell Signaling Technology), washed in CHAPS buffer and eluted at 95 °C in LDS sample buffer (Life Technologies). Acetyl-CoA levels were measured using a commercially available kit (Sigma).
Antibodies and Western Blotting
Protein lysates were prepared in LDS sample buffer and separated using the Bolt SDS-PAGE system on 12% Bis-Tris gels, before transfer to nitrocellulose membranes (all Life Technologies). Protein expression was analyzed using the following antibodies: mouse monoclonal acetyl-Lysine, mouse α-Tubulin, rabbit GAPDH, rabbit Glutamate Dehydrogenase (GDH), rabbit acetyl-Tubulin K40, rabbit HDAC6, rabbit LC3A/B, rabbit COX IV, rabbit Succinate Dehydrogenase A (SDHA), rabbit phospho-ACC from Cell Signaling Technologies; rabbit PINK1 from Novus Biochemicals. Fluorescent anti-mouse or anti-rabbit secondary antibodies (red, 700 nm; green, 800 nm) from LiCor were used to detect expression levels.
Confocal Microscopy and Analysis
HepG2 cells were grown on glass cover slips, fixed in 4% (v/v) paraformaldehyde and immunostained using antibodies for Tom20 (mitochondria), LC3 (autophagosomes), along with DAPI staining (nucleus). The figure shows representative images from the three experimental groups, and have been compressed to reduce size but retain resolution. All images were taken using a 60X (1.43NA) optic on a Nikon A1 with GASP detectors and NIS Elements software (Nikon Inc., Melville NY). 3D rendering on the confocal Z-stacks was performed to obtain measures of mitochondrial fragmentation using Imaris (Bitplane). Measurements were taken from each cell in 15 randomly chosen fields-of-view for each condition. Colocalization between Tom20 and LC3 was measured in voxels above pre-set thresholds, and Pearson Correlation Coefficients were generated to quantify colocalization of mitochondria and autophagosomes.
Image Preparation and Experimental Reproduction
Western blot images were captured using the Odyssey Fluorescent Imager (LiCor), which allows simultaneous analysis of two proteins on a single membrane. Images were processed and quantified using Odyssey software and assembled in Photoshop (Adobe). Alterations to brightness/contrast of figure panels were made using automatic software settings where necessary, and checked to ensure that no result was artificially manipulated or misrepresented. All experiments were conducted at least three times, and representative images from these repeats are presented.
RESULTS
α-Lipoic Acid promotes hyperacetylation of a single cytoplasmic protein
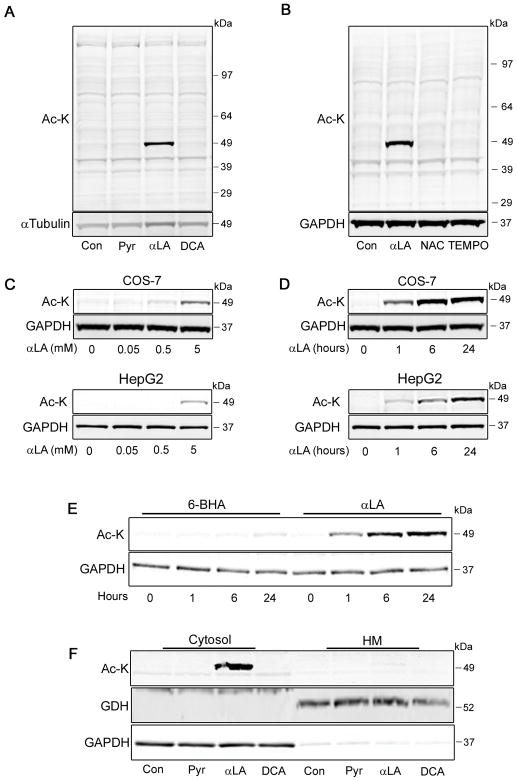
Recent research has demonstrated that chemical modulation of mitochondrial pathways related to acetyl-CoA metabolism regulates global autophagy pathways (2), and that changes in mitochondrial lysine acetylation levels are linked to fat uptake and usage (7). We first sought to investigate whether modulating mitochondrial acetyl-CoA levels through the pyruvate pathway would yield a similar effect on lysine acetylation. COS-7 cells were treated with 3 mM sodium pyruvate (Pyr; a concentration three-times higher than standard culture media), 5 mM αLA (a cofactor of the pyruvate dehydrogenase complex; (2)) or 20 mM DCA (an inhibitor of pyruvate dehydrogenase kinases; (8)), and total protein lysine acetylation was analyzed by western blot. After 4 h, there was a striking increase in the acetylation levels of a single ~50 kDa band in αLA-treated cells, which was absent in all other lanes (Fig. 1a).
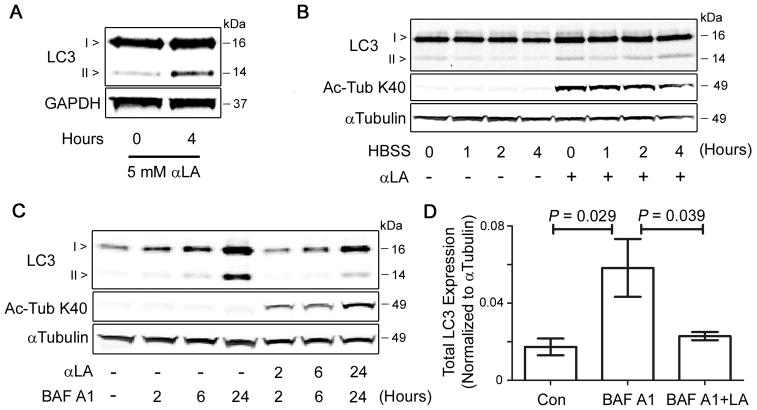
FIGURE 1. Treatment with αLA promotes lysine hyperacetylation of a single cytoplasmic protein.
(A) COS-7 cells were treated for 4 h with 3 mM Pyr (3x normal culture concentration), 5 mM αLA or 20 mM DCA to increase pyruvate-derived mitochondrial acetyl-CoA concentrations, followed by analysis of total protein lysine acetylation (Ac-K). (B) COS-7 cells were treated for 4 h with 5 mM αLA, 1 mM NAC or 1 mM 4-hydroxy-TEMPO, followed by analysis of total protein Ac-K. (C, D) COS-7 or HepG2 cells were treated with varying concentrations of αLA for 4 h (C), or for varying time-points with 5 mM αLA (D), followed by Ac-K analysis. (E) COS-7 cells were treated for varying time points with lipoic acid analogue 6-BHA (5 mM) or αLA (5 mM), followed by Ac-K analysis. (F) COS-7 cells were treated for 4 h with 3 mM Pyr, 5 mM αLA or 20 mM DCA, followed by harvest and separation into cytosolic and heavy-membrane (HM) fractions by differential centrifugation. Fractions were analyzed for Ac-K, with GDH (HM) and GAPDH (cytosolic) used as loading and fraction-purity controls. All immunoblots were performed on 12% Bis-Tris gels.
In addition to acting as a mitochondrial enzyme co-factor, αLA has been shown to have antioxidant properties (see e.g. (9, 10)). We therefore examined whether this hyperacetylation was linked to changes in cellular redox status. Treatment of cells with the broad-range antioxidants NAC or TEMPO had no effect on lysine acetylation (Fig. 1b), indicating that the changes seen in αLA-treated cells are not linked to the oxidation status of this protein.
We next tested whether the observed lysine hyperacetylation occurred dynamically, or whether it was caused by a non-biological process. COS-7 cells were treated over 4 h with a range of αLA concentrations (Fig. 1c), or at different time-points over the course of 24 h (Fig. 1d). Analysis by western blot showed that the acetylation status of this ~50 kDa protein changed dynamically in both a concentration- and time-dependent manner, beginning within an hour of treatment. Similar results were found in human hepatocarcinoma-derived HepG2 cells, indicating that the effects of αLA are not cell-line specific (Fig. 1c, d). Combined, these data suggest that the hyperacetylation induced by αLA treatment occurs in a controlled, biological manner.
To further demonstrate the specific effects of αLA, we repeated this time course using 6-BHA, an inactive analogue of lipoic acid (11). While robust hyperacetylation at ~50 kD was observed after 1 h in the αLA treated cells, there was negligible acetylation seen in 6-BHA treated cells from 1–24 h (Fig. 1e). These data further support that the observed hyperacetylation is specific to αLA.
Finally, we investigated the subcellular localization of this acetylation event. As the acetyl-CoA modulating chemicals were all proposed to work through mitochondrial pyruvate-utilization pathways, we hypothesized that the hyperacetylated protein would be localized in the mitochondrial compartment. To test this, we treated cells with Pyr, αLA and DCA, and then separated proteins into different cellular fractions using differential centrifugation. Surprisingly, the hyperacetylated protein was predominantly localized in the solulble cytoplasmic fraction, and not in the heavy membrame pellet containing mitochondria (Fig. 1f). This indicated that either αLA was acting predominantly in the cytoplasm, or that the pro-acetylation signals were being transmitted from the mitochondria to the cytosol, as previously proposed (2).
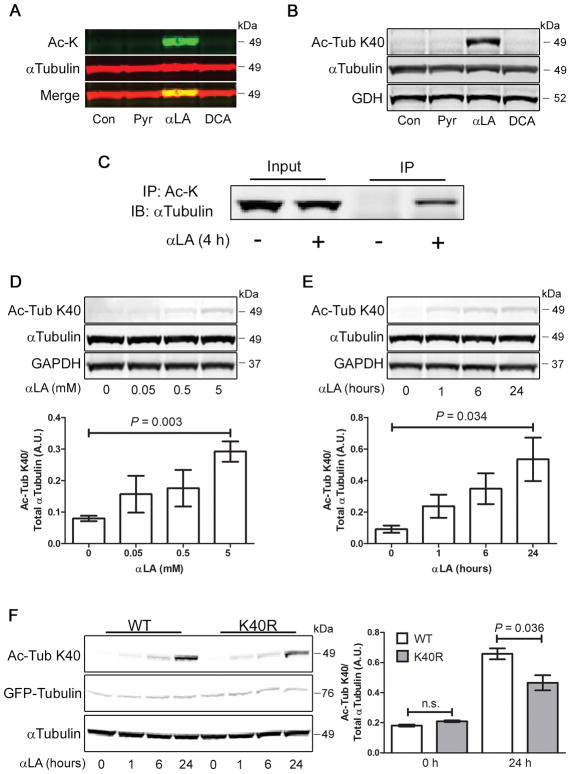
Identification of α-Tubulin Lysine-40 as the hyperacetylation substrate
The strength and rapidity of the hyperacetylation response induced by αLA suggested that the substrate may be an abundant cytosolic protein. As the size of the protein was assessed by SDS-PAGE to be around 50 kDa, we hypothesized that it may be a member of the tubulin family. To examine this more closely, we treated cells with Pyr, αLA and DCA for 4 h, then analyzed the lysates by western blot. Using the LiCor dual-color system, we simultaneously measured expression of α-Tubulin (red) and acetyl-lysine (green), and found that the αLA-induced hyperacetylated band overlapped α-Tubulin with high fidelity (Fig. 2a). Acetylation of α-Tubulin is a well characterized phenomenon, with the lysine residue at position 40 (K40) being the most common modification (12). To establish if αLA is promoting the hyperacetylation of this residue, we repeated the cell treatments and analyzed the lysates by western blot using an antibody specific for this modification. Following 4 h of αLA treatment, there was a marked increase in the abundance of acetylated-K40 α-Tubulin, which was absent in control, Pyr and DCA-treated cells (Fig. 2b). This indicates that K40 of α-Tubulin is the substrate for αLA-mediated hyperacetylation. To further confirm that α-Tubulin was the hyperacetylation substrate, we performed an immunoprecipitation analysis of its acetylation state. Cells treated with or without αLA for 4 h were immuoprecipitated with a global acetyl-lysine antibody, followed by immunoblotting for α-Tubulin. While we were able to detect a significant amount of acetylated α-Tubulin in αLA treated cells, this was absent in untreated cells (Fig. 2c). We next tested to see if the dynamic hyperacetylation seen using the global acetyl-lysine antibody was also observed at the K40 site. In keeping with the previous results, it was confirmed that acetylation of α-Tubulin at K40 occurs in a concentration- and time-dependent manner (Fig. 2d, e), indicating that it is potentially catalyzed through an enzymatic process. Finally, we tested to see if expression of a non-acetylatable mutant of α-Tubulin (K40R) would block the acetylation seen in αLA treated cells. Indeed, transient expression of GFP-WT-α-Tubulin, followed by αLA treatment, led to a robust hyperacetylation in α-Tubulin at K40, which was significantly attenuated in cells expressing the GFP-K40R mutant (Fig. 1f). Overall, these data suggest that the K40 residue of α-Tubulin is the substrate for αLA-induced hyperacetylation.
FIGURE 2. α-Tubulin is dynamically hyperacetylated at K40 in αLA treated COS-7 cells.
(A) Cells were treated for 4 h with 3 mM Pyr, 5 mM αLA or 20 mM DCA, followed by analysis by western blot. Simultaneous dual-color labeling indicates that lysine hyperacetylation occurs at the same molecular weight as α-Tubulin. (B) Cells treated with 5 mM αLA for 4 h using the same conditions show robust hyperacetylation of α-Tubulin at Lysine-40 (K40). (C) Cells were treated with αLA for 4 h, followed by immunoprecipitation with pan-Ac-K specific antibodies. Input and IP fractions were probed with an α-Tubulin antibody (D, E) α-Tubulin K40 acetylation induced by αLA occurs dynamically, and is both concentration- and time-dependent. (F) Cells were transfected with GFP-WT-α-Tubulin or GFP-K40R-α-Tubulin for 24 h, then treated with 5 mM αLA for 24 h. Acetylation of α-Tubulin was analyzed by Ac-K immunoblot. All immunoblots were performed on 12% Bis-Tris gels. Graphs show mean ± SE, with a Student’s t-test of P < 0.05 defined as significant.
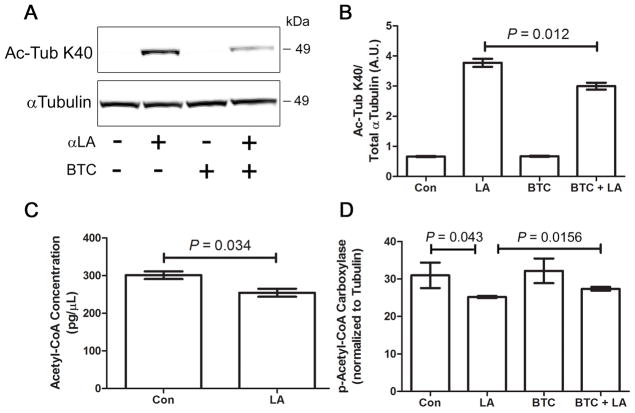
Mitochondrial citrate export is required for α-Lipoic Acid-mediated α-Tubulin acetylation
Previous work has shown that mitochondrial-derived citrate (an acetyl-CoA transport molecule) is one of the major sources of cytoplasmic acetyl-CoA required for lysine acetylation. Citrate is exported to the cytosol via the mitochondrial citrate carrier, where it is converted back to acetyl-CoA by ATP citrate lyase (reviewed in 2). To strengthen the link between αLA-mediated acetyl-CoA production in the mitochondria and cytosolic acetylation of α-Tubulin, we treated COS-7 cells with αLA in the presence or absence of BTC, a pharmacological inhibitor of mitochondrial citrate export (2). Loss of mitochondrial citrate export (and hence acetyl-CoA) in BTC/αLA co-treated cells led to a significant reduction in α-Tubulin acetylation, relative to cells treated with αLA alone (Fig. 3a,b).
FIGURE 3. Inhibition of mitochondrial citrate export inhibits α-tubulin hyperacetylation.
(A, B) COS-7 cells were treated with 5 mM αLA and/or 1.5 mM BTC for 24 h, followed by analysis for α-Tubulin K40 acetylation. (C) Measurement of cytosolic acetyl-CoA levels in COS-7 cells in the presence or absence of 5mM αLA for 24 h. (D) Analysis of acetyl-CoA carboxylase activation status in COS-7 cells treated with 5mM αLA and/or 1.5 mM BTC for 24 h. A reduction phospho-ACC levels indicates upregulated conversion of acetyl-CoA to malate. All immunoblots were performed on 12% Bis-Tris gels. Graphs show mean ± SE, with a Student’s t-test of P < 0.05 defined as significant.
As we hypothesized that αLA treatment would lead to increased free cytoplasmic acetyl-CoA, we measured the concentration of acetyl-CoA in cells treated with or without αLA for 24 h. Surprisingly, we found that there was a significant reduction in acetyl-CoA concentrations in αLA treated cells relative to the control (Fig. 3c). On investigating further, we found that αLA treatment significantly induced activity of acetyl-CoA carboxylase (ACC; measured as dephosphorylation of ACC), which converts free cytosolic acetyl-CoA into malonyl-CoA (Fig. 3d). To further implicate mitochondrial citrate in this process, we found that BTC could block the αLA-mediate induction of ACC (Fig. 3d). As such, we hypothesize that αLA treatment leads to a large increase in mitochondrial citrate transport, which provides sufficient acetyl-CoA to allow α-Tubulin hyperacetylation. Continued activity in this pathway leads to cellular homeostatic mechanisms in the cytosol to reduce free acetyl-CoA levels, resulting in ACC activation.
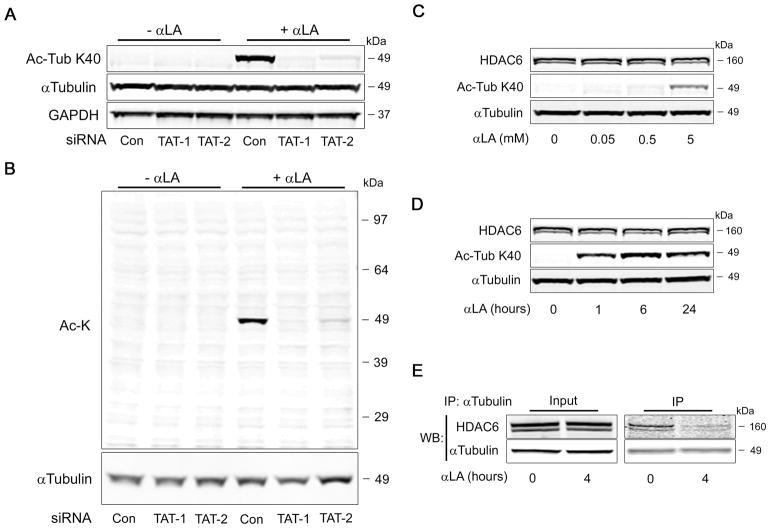
α-Lipoic Acid-mediated hyperacetylation is regulated by αTAT and HDAC6 activity
Lysine acetylation of α-Tubulin at K40 has previously been shown to be catalyzed by the α-Tubulin acetyltransferase protein αTAT (also known as MEC-17; (13, 14)). We therefore set out to establish if αTAT was responsible for the enzymatic hyperacetylation of α-Tubulin in response to αLA treatment. COS-7 cells were transfected for 48 h with non-targeting control siRNA (Con), or with two independent siRNAs targeting αTAT (TAT-1 and TAT-2), followed by treatment for 4 h with αLA. In agreement with earlier results, there was robust K40 acetylation of αLA-treated cells transfected with control siRNA. Strikingly, the hyperacetylation of α-Tubulin was largely abolished in cells transfected with siRNA targeting αTAT (Fig. 4a), suggesting that the effects of αLA are mediated by this acetyltransferase. We then repeated this analysis with a pan-acetylation antibody, to ensure that αTAT was responsible for the acetylation observed at ~50 kDa in previous experiments. As with the K40-specific antibody, loss of αTAT greatly diminished the observed hyperacetylation band (Fig. 4b), indicating that the previously observed modifications were indeed on α-Tubulin.
FIGURE 4. α-Tubulin hyperacetylation is dependent on αTAT activity and loss of HDAC6:α-tubulin interaction.
(A) COS-7 cells were transfected with two independent siRNAs (TAT-1 or TAT-2) to α-Tubulin acetyltransferase (αTAT), or a non-targeting control. After 48 h, cells were treated with or without 5 mM αLA for 4 h, followed by analysis for αTubulin K40 acetylation. (B) Analysis of total cellular lysine acetylation indicates that αTAT knockdown abolishes all hyperacetylation at ~50 kDa. (C, D) Effect of concentration- and time-dependent αLA treatment of cells on the expression of the α-Tubulin deacetylase protein, HDAC6. (E) Co-immunoprecipitation analysis of the interaction between α-Tubulin and HDAC6, after treatment with 5 mM αLA for 4 h. All immunoblots were performed on 12% Bis-Tris gels.
As this post-translational modification occurs through a reversible process, we next sought to investigate whether lysine deacetylation played a role in the acetylation status of α-Tubulin. In particular, we hoped to determine if the increased acetylation evident in αLA-treated cells was caused by a reduction in deacetylase abundance or activity. As before, we treated COS-7 cells with different concentrations of αLA for 4 h, or with 5 mM αLA at different time-points over 24 h. After harvesting the lysates, we analyzed the expression level of HDAC6, a cytosolic protein that has previously been shown to be the main α-Tubulin deacetylase (15, 16). However, there was neither a time- or concentration-dependent change in HDAC6 levels following treatment with αLA (Fig. 4c, d), indicating that this chemical has no effect on HDAC6 stability.
We next examined whether αLA could have a direct effect on ability of HDAC6 to deacetylate α-Tubulin. We treated COS-7 cells for 4 h with αLA, and utilized the cell lysates in co-immunoprecipitation experiments. Following immunocapture of α-Tubulin in untreated cells, we were able to detect a strong interaction between this protein and HDAC6. However, after 4 h of αLA treatment, we observed a significant decrease in the physical interaction between α-Tubulin and HDAC6 (Fig. 4e). We therefore conclude that αLA promotes the hyperacetylation of α-Tubulin by abolishing its interaction with HDAC6, thereby allowing unconstrained acetylation at K40 by αTAT.
α-Lipoic Acid treatment slows autophagic flux and blocks PINK1-mediated mitophagy
Finally, we set out to establish whether the hyperacetylation of α-Tubulin by αLA had an impact on cellular function. As the acetylation status of cytosolic proteins has recently been implicated in the regulation of global autophagy (2, 17), we examined the role of α-Tubulin acetylation in the induction of this cellular recycling program. Treatment of COS-7 cells for 4 h with αLA led to an increase in the lipidation of LC3-I to form LC3-II (Fig. 5a), a commonly-used marker of autophagy induction (18). However, the steady-state measurement of LC3-II levels offers an incomplete picture of the autophagy program, as it is unable to distinguish between the increased production of autophagic vacuoles (autophagosomes), and a decrease in the rate of autophagic flux (by a blockage in the autophagy pathway; (19)). As such, we performed time-course experiments to measure autophagic flux in control and αLA-treated cells. Firstly, we serum-starved COS-7 cells in HBSS for up to 4 h to induce autophagy, in the presence or absence of αLA. In cells treated with αLA there was a marked increase in LC3-II after extended serum withdrawal, which was not evident in untreated cells (Fig. 5b), indicating a potential reduction in the processing of autophagosomes. Secondly, treatment of cells with the lysosomal inhibitor bafilomycin A1, at various time-points over the course of 24 h, led to a steady increase in the expression of both forms of LC3. Co-treatment of cells with αLA diminished the rate of LC3-II accumulation following the addition of bafilomycin A1 (Fig. 5c, d), indicating that the hyperacetylation of α-Tubulin by αLA may slow the progression of substrates (such as LC3-II itself) through the autophagy pathway.
FIGURE 5. αLA treatment slows the flux of cellular autophagy.
(A) Activation status of the autophagy marker LC3, following treatment of COS-7 cells with 5 mM αLA for 4 h. Increases in LC3-II levels indicate either an increase in autophagy activity or decrease in autophagic flux. (B) COS-7 cells were serum starved in HBSS for 0–4 h, in the presence or absence of 5 mM αLA. Accumulation of LC3-II after several hours of starvation indicates a reduced rate of protein turnover by autophagy. (C) In time-course experiments, COS-7 cells treated with 5 mM αLA show diminished levels of LC3-II accumulation, relative to those treated with the lysosomal inhibitor BAF A1 (100 nM) alone, indicating that αLA slows the flux of substrates through the autophagy pathway. (D) Quantification of total LC3 expression in COS-7 cells after 24 h of treatment with BAF A1 and/or αLA. All immunoblots were performed on 12% Bis-Tris gels. Graphs show mean ± SE, with a Student’s t-test of P < 0.05 defined as significant.
Recent work has demonstrated that dysfunctional mitochondria are selectively targeted for removal, through an autophagy-related process called mitophagy (reviewed in (20)). We therefore sought to test if the hyperacetylation of α-Tubulin, and consequent disruption of the autophagic flux, would regulate the selective degradation of damaged mitochondria. We first took an imaging-based approach to analyze the impact of αLA on mitophagy. HepG2 cells were grown on coverslips and treated with 10 μM CCCP (an uncoupler of mitochondrial transmembrane potential), in the presence or absence of αLA for 4 h. Cells were fixed and stained for Tom20 (a mitochondrial marker), LC3 (an autophagsomal marker) and DAPI. In cells treated with CCCP alone, there was a large increase in mitochondrial:autophagosomal colocalization, which was attenuated by αLA co-treatment (Fig. 6a). This indicates that αLA may slow the delivery of damaged mitochondria to autophagosomes, or prevent CCCP-mediated damage upstream of this process. In addition, there was a significant increase in mitochondrial fragmentation in CCCP treated cells, which was greatly attenuated in αLA co-treated cells (Fig. 6a). This suggests that αLA can block mitochondrial fission, which is often observed prior to mitochondrial degradation in cells during upregulated mitophagy (20).
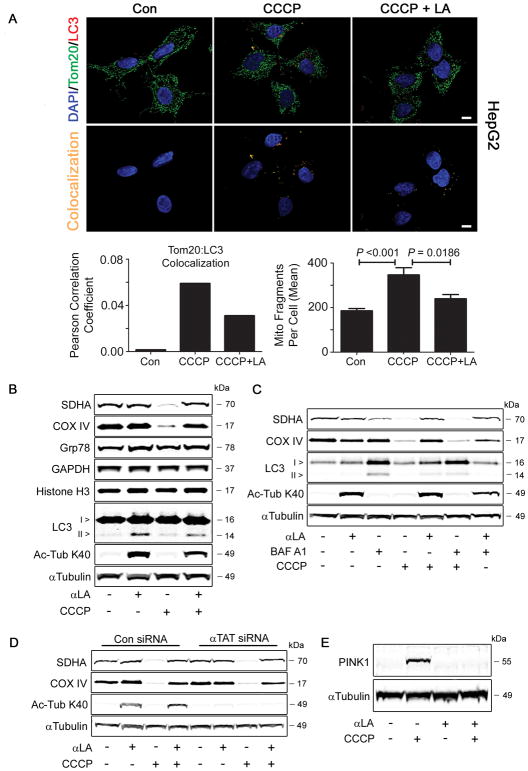
FIGURE 6. αLA treatment prevents the removal of depolarized mitochondria through PINK1-mediated mitophagy.
(A) HepG2 cells were treated with 10 μM CCCP in the presence or absence of 5 mM αLA for 4 h. Cells were fixed in 4% paraformaldehyde, then immunostained with the mitochondrial marker Tom20 and the autophagy marker LC3, along with nuclear DAPI. Confocal analysis and 3-D surface rendering of mitochondria from Z-stacks was used to measure Tom20:LC3 colocalization and mitochondrial fragmentation in response to treatment. (B) Treatment of COS-7 cells for 24 h with 10 μM CCCP led to the specific degradation of mitochondrial proteins COX IV and SDHA, without affecting the levels of cytosolic, microsomal or nuclear proteins. This was blocked by the simultaneous addition of 5 mM αLA. (C) αLA, but not bafilomycin A1, was able to prevent the degradation of mitochondrial proteins initiated by CCCP treatment. This suggests that the activity of αLA occurs prior to the lysosomal step of the autophagy pathway. (D) COS-7 cells were transfected for 48 h with either control or αTAT siRNA, followed by treatment with αLA (5 mM) or CCCP (10 μM). A loss of K40 hyperacetylation had no effect on the CCCP-mediated removal of mitochondrial proteins, indicating that the prevention of mitophagy by αLA is unrelated to its effect on α-Tubulin. (E) COS-7 cells treated with 10 μM CCCP showed stabilization of PINK1, a marker for mitophagy in the PINK1-Parkin pathway. This stabilization was abolished by concurrent treatment with 5 mM αLA, indicating that αLA acts upstream of this event. All immunoblots were performed on 12% Bis-Tris gels. Graphs show mean ± SE, with a Student’s t-test of P < 0.05 defined as significant. Scale bar is 10 μm.
We next analyzed whether the decrease in mitochondrial delivery to autophagosomes in αLA co-treated cells would have an effect on mitochondrial protein abundance. In COS-7 cells treated with CCCP alone, there was a marked reduction in mitochondrial protein content, as measured by western blotting for the mitochondrial proteins COX IV and SDHA. This reduction appeared to be specific to the mitochondrial compartment, as CCCP treatment had no effect on the abundance of the cytoplasmic GAPDH, nuclear histone H3 or the ER-localized Grp78 (Fig. 6b). Remarkably, co-treating cells with αLA completely blocked the removal of CCCP-damaged mitochondria, with levels of COX IV and SDHA expression remaining similar to those in untreated control cells (Fig. 6b). We next tested to see if this αLA-mediated block in mitophagy occurred before or after the delivery of substrates to the lysosome. While treatment of cells with bafilomycin A1 limited autophagic flux and led to accumulation of LC3-II, it failed to prevent the CCCP-induced degradation of mitochondrial proteins in a manner similar to αLA (Fig. 6c). This indicates that the lesion in the mitophagy pathway caused by αLA treatment occurs upstream of the delivery of mitochondrial substrates to the lysosome.
Following this result, we tested whether the hyperacetylation of α-Tubulin was responsible for the block in mitophagy, for example by affecting the transport of mitochondria along the cytoskeleton. COS-7 cells were transfected with control or αTAT siRNA for 48 h, then treated with αLA and/or CCCP for 24 h. Despite a robust reduction in K40 tubulin acetylation, there was no difference in the degradation of mitochondrial proteins between control or αTAT knockdown cells treated with CCCP, suggesting that the protective effect of αLA is independent of its pro-acetylation properties (Fig. 6d). Previous research had demonstrated that depolarization of mitochondria with CCCP stabilized levels of the mitochondrial kinase PINK1, which then recruits the ubiquitin ligase Parkin, leading to mitochondrial degradation (21, 22). We therefore tested to see if αLA had any effect on PINK1 stabilization, and found that it completely blocked the CCCP-mediated increase in this protein (Fig. 6e). As such, we conclude that αLA prevents the removal of mitochondria through mitophagy pathways by blocking the stabilization of PINK1 and recruitment of Parkin, rather than by its effects on tubulin and the cytoskeleton.
DISCUSSION
In this report we aimed to investigate the role of acetyl-CoA abundance and acetylation in the regulation of cellular recycling programs. We provide data suggesting that: (i) α-Lipoic Acid promotes the rapid hyperacetylation of α-Tubulin; (ii) that this acetylation was dependent on αTAT and required the inhibition of HDAC6 function; and (iii) α-Lipoic Acid treatment blocked the removal of mitochondria through PINK1-mediated mitophagy.
Our initial interest in examining this system lay in reports that changes in acetyl-CoA abundance, derived from mitochondrial sources, could act as a regulator of cellular autophagy pathways. Using an elegant set of nutritional and pharmacological interventions, Mariño et al. demonstrated that increasing the levels of acetyl-CoA in the mitochondria, followed by subsequent export to the cytosol of acetyl-CoA transport molecules such as citrate, led to a block on the induction of autophagy (2). In essence, acetyl-CoA derived from the metabolism of nutritional fuel sources was acting as a direct signal of cellular satiety, complementing pathways such as the AMPK-mTOR-ULK1 axis in regulating autophagic processes. Crucially, the regulation of autophagy by acetyl-CoA required the function of an acetyltransferase, EP300, indicating a link between nutritional status, the acetylation of cytosolic lysine residues (as a potential marker of fuel abundance) and the recycling of cellular components (2).
In order to regulate turnover through mitophagy, signaling mechanisms must exist between mitochondria and the cytoplasmic autophagy machinery, which allow specific organelles to be targeted for degradation. The PINK1-Parkin pathway has been shown to target dysfunctional organelles for removal via ubiquitination of mitochondrial proteins (21, 22), while BNIP3, Nix and FUNDC1 have been implicated in developmental and hypoxia-related mitophagy processes, acting as mediators autophagy-structure binding (23, 24, 25). We recently demonstrated that mitochondria displaying reduced mitochondrial lysine acetylation were targeted for degradation by mitophagy (3, 4), indicating that lysine acetylation – a proxy for acetyl-CoA levels (26) – may act as signaling mechanism for mitochondrial metabolic fitness.
In this report, we demonstrate that changing acetyl-CoA levels in the cell have a profound effect on α-Tubulin acetylation. In relation to studies on autophagy as a whole, the role of tubulin acetylation in the regulation of mitophagy has received little attention. Acetylation of α-Tubulin at K40 occurs on the luminal side of microtubules, and is catalyzed by the αTAT/MEC-17 acetyltransferase (13, 14). This reaction is counter-regulated by deacetylase enzymes, in particular HDAC6 (15, 16). Previous work, performed mainly in neurons, has shown that inhibiting HDAC6 leads to an increase in the velocity of mitochondrial movement (27). This would indicate that the tubulin hyperacetylation seen following treatment with αLA would not inhibit the transport of mitochondria to allow incorporation into autophagosomes, and indeed α-Tubulin acetylation had no effect on mitophagy (Fig. 6c). We hypothesize instead that the inhibition of HDAC6 function, caused by αLA treatment, may lead to impairment in maturation of autophagosome membranes, as seen previously (28). Further work will be required to determine how αLA treatment prevents mitochondrial removal upstream of autophagosome incorporation, leading to the loss of PINK1 stabilization and Parkin recruitment (Fig. 6d).
The role of αLA in biological systems has undergone a great deal of study, with several pharmaceutical applications being proposed. αLA is a co-factor in several mitochondrial enzymatic pathways, including the pyruvate dehydrogenase complex, the α-ketoglutarate dehydrogenase complex, the branched-chain oxoacid dehydrogenase complex and the glycine-cleavage system (29). Its enzyme-bound form, lipoamide, has been found to drive the pyruvate dehydrogenase complex to alter substrate usage and provide protection from damage to post-ischemic hearts (30). However, the majority of published research has focused on the role of αLA as an antioxidant, showing that it can mitigate oxidative stress in a number of systems (9, 10). In this study, we tested whether changing the redox environment would lead to α-Tubulin acetylation, but found that two other general antioxidants (NAC and TEMPO) were unable to induce this modification.
As an alternative, we postulate that the increase in mitochondria-derived acetyl-CoA after αLA treatment led to an increase in cytosolic acetyl-CoA levels, stimulating the acetyltransferase, EP300 (2). The active EP300 may then acetylate HDAC6 and disrupt its deacetylase activity (31), by causing it to physically disassociate from α-Tubulin (Fig. 4c). This results in the unconstrained hyperacetylation of α-Tubulin at K40 through αTAT. Work is actively underway in our laboratory to further elucidate the pathways by which αLA potently inhibits HDAC6, and to investigate whether this drug may be useful in constraining the induction of autophagy or mitophagy in situations where it may be potentially maladaptive (32).
Acknowledgments
We are greatly indebted to Dr. Michael N. Sack (National Heart, Lung and Blood Institute, NIH) for helpful discussions in the conception of this study. We would like to thank Jianxin Zeng (University of Pittsburgh) for assistance in immunofluorescence staining.
FUNDING
This work was funded by NHLBI HL116728 awarded to I.S.
ABBREVIATIONS
- 6-BHA
6-bromohexanoic acid
- Ac-K
acetyl-lysine
- αLA
α-Lipoic Acid
- ACC
acetyl-CoA carboxylase
- AMPK
AMP kinase
- αTAT
α-Tubulin acetyltransferase
- BTC
benzenetricarboxylate
- BAF A1
Bafilomycin A1
- CCCP
carbonyl cynanide 3-chlorophenolhydrazone
- DCA
dichloroacetate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GDH
glutamate dehydrogenase
- HDAC6
histone deacetylase 6
- HM
heavy membrane
- K40
lysine-40
- LC3
microtubule-associated light chain protein 3
- mTOR
mammalian Target of Rapamycin
- NAC
N-acetyl cysteine
- PINK1
PTEN-associated kinase 1
- Pyr
Sodium Pyruvate
- TEMPO
4-hydroxy-TEMPO
- ULK1
UNC-51-like kinase 1
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts to report
AUTHOR CONTRIBUTIONS
M.W.S., D.T., M.Z., G.G. and I.S. performed experiments, M.W.S., G.G., M.C., C.St.C. and I.S. analyzed data, I.S. and C.St.C. designed experiments, and I.S. wrote the manuscript.
References
- 1.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015;25:354–363. doi: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, Zamzami N, Scoazec M, Durand S, Enot DP, Fernandez AF, Martins I, Kepp O, Senovilla L, Bauvy C, Morselli E, Vacchelli E, Bennetzen M, Magnes C, Sinner F, Pieber T, Lopez-Otin C, Maiuri MC, Codogno P, Andersen JS, Hill JA, Madeo F, Kroemer G. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–725. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Webster BR, Scott I, Han K, Li JH, Lu Z, Stevens MV, Malide D, Chen Y, Samsel L, Connelly PS, Daniels MP, McCoy JP, Jr, Combs CA, Gucek M, Sack MN. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013;126:4843–4849. doi: 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott I, Webster BR, Chan CK, Okonkwo JU, Han K, Sack MN. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289:2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr, Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pougovkina O, te Brinke H, Ofman R, van Cruchten AG, Kulik W, Wanders RJ, Houten SM, de Boer VC. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Hum Mol Genet. 2014;23:3513–3522. doi: 10.1093/hmg/ddu059. [DOI] [PubMed] [Google Scholar]
- 8.Stacpoole PW. The pharmacology of dichloroacetate. Metabolism. 1989;38:1124–1144. doi: 10.1016/0026-0495(89)90051-6. [DOI] [PubMed] [Google Scholar]
- 9.Pilar Valdecantos M, Prieto-Hontoria PL, Pardo V, Modol T, Santamaria B, Weber M, Herrero L, Serra D, Muntane J, Cuadrado A, Moreno-Aliaga MJ, Alfredo Martinez J, Valverde AM. Essential role of NRF2 in the protective effect of lipoic acid against lipoapoptosis in hepatocytes. Free Radic Biol Med. 2015;84:263–278. doi: 10.1016/j.freeradbiomed.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Galilea M, Perez-Matute P, Prieto-Hontoria PL, Houssier M, Burrell MA, Langin D, Martinez JA, Moreno-Aliaga MJ. alpha-Lipoic acid treatment increases mitochondrial biogenesis and promotes beige adipose features in subcutaneous adipocytes from overweight/obese subjects. Biochim Biophys Acta. 2015;1851:273–281. doi: 10.1016/j.bbalip.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Selmi C, Mackay IR, Gershwin ME. The autoimmunity of primary biliary cirrhosis and the clonal selection theory. Immunol and Cell Biol. 2011;89:70–80. doi: 10.1038/icb.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janke C. The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467:218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davenport AM, Collins LN, Chiu H, Minor PJ, Sternberg PW, Hoelz A. Structural and functional characterization of the alpha-tubulin acetyltransferase MEC-17. J Mol Biol. 2014;426:2605–2616. doi: 10.1016/j.jmb.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 2003;22:1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morselli E, Marino G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Benit P, Rustin P, Criollo A, Kepp O, Galluzzi L, Shen S, Malik SA, Maiuri MC, Horio Y, Lopez-Otin C, Andersen JS, Tavernarakis N, Madeo F, Kroemer G. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 20.Saito T, Sadoshima J. Molecular Mechanisms of Mitochondrial Autophagy/Mitophagy in the Heart. Circ Res. 2015;116:1477–1490. doi: 10.1161/CIRCRESAHA.116.303790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rikka S, Quinsay MN, Thomas RL, Kubli DA, Zhang X, Murphy AN, Gustafsson AB. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 26.Weinert BT, Iesmantavicius V, Moustafa T, Scholz C, Wagner SA, Magnes C, Zechner R, Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Owens GC, Makarenkova H, Edelman DB. HDAC6 regulates mitochondrial transport in hippocampal neurons. PLoS One. 2010;5:e10848. doi: 10.1371/journal.pone.0010848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, Taylor JP, Cuervo AM, Yao TP. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassan BH, Cronan JE. Protein-protein interactions in assembly of lipoic acid on the 2-oxoacid dehydrogenases of aerobic metabolism. J Biol Chem. 2011;286:8263–8276. doi: 10.1074/jbc.M110.194191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sumegi B, Butwell NB, Malloy CR, Sherry AD. Lipoamide influences substrate selection in post-ischaemic perfused rat hearts. Biochem J. 1994;297:109–113. doi: 10.1042/bj2970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Y, Jeong HM, Jin YH, Kim YJ, Jeong HG, Yeo CY, Lee KY. Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochem Biophys Res Commun. 2009;383:88–92. doi: 10.1016/j.bbrc.2009.03.147. [DOI] [PubMed] [Google Scholar]
- 32.Ren J, Taegtmeyer H. Too much or not enough of a good thing - The Janus faces of autophagy in cardiac fuel and protein homeostasis. J Mol Cell Cardiol. 2015;84:223–226. doi: 10.1016/j.yjmcc.2015.03.001. [DOI] [PubMed] [Google Scholar]