Abstract
Synapse associated protein of 97KDa (SAP97) belongs to a family of scaffolding proteins, the membrane-associated guanylate kinases (MAGUKs), that are highly enriched in the postsynaptic density of synapses and play an important role in organizing protein complexes necessary for synaptic development and plasticity. The Dlg-MAGUK family of proteins are structurally very similar, and an effort has been made to parse apart the unique function of each Dlg-MAGUK protein by characterization of knockout mice. Knockout mice have been generated and characterized for PSD-95, PSD-93, and SAP102, however SAP97 knockout mice have been impossible to study because the SAP97 null mice die soon after birth due to a craniofacial defect. We studied the transcriptomic and behavioral consequences of a brain-specific conditional knockout of SAP97 (SAP97-cKO). RNA sequencing from hippocampi from control and SAP97-cKO male animals identified 67 SAP97 regulated transcripts. As large-scale genetic studies have implicated MAGUKs in neuropsychiatric disorders such as intellectual disability, autism spectrum disorders, and schizophrenia (SCZ), we analyzed our differentially expressed gene (DEG) set for enrichment of disease risk-associated genes, and found our DEG set to be specifically enriched for SCZ-related genes. Subjecting SAP97-cKO mice to a battery of behavioral tests revealed a subtle male-specific cognitive deficit and female-specific motor deficit, while other behaviors were largely unaffected. These data suggest that loss of SAP97 may have a modest contribution to organismal behavior. The SAP97-cKO mouse serves as a stepping stone for understanding the unique role of SAP97 in biology.
Introduction
Intellectual disabilities and neuropsychiatric behavioral disorders affect about 17.9% of individuals over their lifetime and interfere with the ability of people to experience a fulfilling and productive life (nimh.nih.gov). Some of these disorders are clearly developmental. For example, autism spectrum disorders (ASD) are characterized by impairments in social interaction and communication, and by restricted, repetitive behaviors and about 1% of children show signs and symptoms that lead to the diagnosis of ASD [1,2]. Schizophrenia (SCZ) is another mental disorder that is characterized by disordered thought processes and disturbed emotional responsiveness [3]. The symptoms of SCZ usually appear during young adulthood, with an overall prevalence of about 0.7% [3,4]. Technological advances have brought unprecedented insights into the genetic architecture of these and many other neuropsychiatric disorders [4–7].
Exome-sequencing technology has allowed us to systematically scan genes for de novo mutations at the single-base resolution, potentially offering insights into risk-determining genes [4,8]. Whole-exome sequencing results from patients with ASD or SCZ reveal significantly enriched copy number variant (CNV) mutations in the synaptic gene set [4]. Among the most prevalent synaptic genes that have been uncovered in large-scale genomic studies have been alterations in the neurexins/neurolignins along with the ProSAP/Shank family. Various genetically manipulated mice of these gene families recapitulate some of the behavioral features of ASD, SCZ, and intellectual disability [9–12]. However, none of these models completely phenocopy disease in humans, consistent with the polygenic origin of these disorders.
Another important group of synaptic genes that has been implicated to be involved in ASD or SCZ is the Discs-large (Dlg) family of membrane associated guanylate kinases (MAGUKs) [7,13–16]. The Dlg family is the most comprehensively studied family of MAGUKs, and is comprised of PSD-95, PSD-93, SAP102, and SAP97. They share a common domain structure comprised of three PDZ domains, along with an SH3 and GUK domain. The Dlg-MAGUK family directly binds to many proteins in the postsynaptic density (i.e. glutamate receptor subunits, TARPS, and neurexin/neuroligin clusters), and regulates synaptic nanoscale structure and synaptic transmission [17–19] . Mice with a targeted deletion of PSD-95, PSD-93, and SAP102 show a range of phenotypes also displayed by individuals with psychiatric disorders [15,16,20].
A variety of evidence implicates SAP97 in the etiology of ASD and SCZ: 1) single nucleotide polymorphisms in SAP97 have been linked to an increased risk of schizophrenia in males [21], 2) a more recent genetic association analysis detected an association between schizophrenia and single nucleotide polymorphisms located within a newly identified primate-specific exon of SAP97 [22], 3) individuals with 3q29 microdeletions spanning the SAP97 locus display autism and intellectual disability [23], 4) a meta-analysis demonstrated the 3q29 deletion confers a 40-fold increased risk for SCZ [24], and 5) a study of expression levels of multiple postsynaptic density proteins found a specific decrement in the level of SAP97 in post mortem frontal lobe from schizophrenic patients [25]. SAP97 is also the only member of the Dlg-MAGUK family that directly binds to the extreme C-terminus of the GluA1 AMPA receptor (AMPAR), a subunit that promotes dendritic growth and patterned synaptic innervation [26,27]. Thus, it is plausible that defects in these SAP97-dependent mechanisms contribute to a ASD and SCZ phenotype. While these findings advocate for the participation of SAP97 in the etiology of neuropsychiatric disorders, the issue has been difficult to study because SAP97 knockout mice die a few days after birth from a craniofacial defect [28].
In this study, we chose to pursue a broad behavioral and transcriptomic characterization of SAP97 deficient mice to gain an overall understanding of the role of SAP97 in brain function and organismal level. We generated mice that have a conditional knockout of SAP97 targeted to neurons using the Cre-loxP system. We then characterized these mice at the behavioral and biochemical level. Overall, our results suggest that loss of SAP97 results in mild sex-specific behavioral abnormalities as well as regulates transcripts of SCZ risk-related genes. This study provides us with the framework to conduct subsequent studies that can address whether SAP97 directly contributes to the pathophysiology of SCZ.
Materials and methods
Animals
All animal procedures were approved by the Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee. Mice were housed (2–5 animals per cage) with a 12/12 hr light/dark cycle and with ad libitum access to food and water. Approximately 100 total animals were used for this study. The Cre-loxP system was used to generate a SAP97 conditional knockout (cKO) mouse. SAP97fl/- mice were generated as previously described. Nestin-cre+/- mice on a C57Bl/6 background were purchased from Jackson Labs (stock number 003771). Nestin-cre+/-; SAP97fl/- mice were generated by crossing male Nestin-cre+/- with female SAP97fl/- mice. Nestin-cre+/-; SAP97fl/- male mice were then crossed with female SAP97fl/- mice to generate Nestin-cre+/-; SAP97fl/fl (SAP97-cKO) and littermate control animals. Littermate control animals (genotype: Nes-cre+/-, SAP97fl/fl, and wild-type) were averaged and compared to cKO animals. Genomic DNA was extracted from tail snips (obtained at 10 days of age) using the Phenolcholoroform acetate method to confirm genotypes. The primers used for genotyping were as follows: SAP97 flox fwd- AGAGTATGCTCTATGTGATGTTGTGTG rev-TAAGAAGGATCAACTGGCAAAGGTG; CRE fwd- ACCTGATGGACATGTTCAGG rev-CGAGTTGATAGCTGGCTGG
Behavioral experiments
Open field
Assessment of general exploratory behavior was evaluated using the open field paradigm. Mice were placed in a white, opaque plexiglass box (40cm x 40cm) and were given 15 minutes to explore the apparatus. Exploratory locomotor activity (total distance traveled and average speed) was scored using the Any-MAZE tracking software (San Diego Instruments, San Diego, CA).
Elevated plus maze
Assessment of anxiety-like behaviors was evaluated using an elevated plus maze (Coulbourn Instruments, Whitehall, PA). The mouse was initially placed in the center “free zone”, and was allowed to freely explore the apparatus for the 5-minute trial time. Time in the open arms versus the closed arms, as well as number of entries to these arms, was measured using the Any-MAZE tracking software.
Accelerating rotarod
Assessment of motor learning and motor coordination was evaluated using the accelerating rotarod (Ugo Basile, Varese, Italy). The starting acceleration was 4 rpm, and accelerated to 40 rpm over a 5-minute trial time. Mice underwent 3 trials per day for 4 consecutive days, for a total of 12 trials. Latency to fall from the rod was manually measured and compared across the 4 days.
Novel Object Recognition (NOR)
Assessment of cognition was evaluated using the NOR paradigm. The testing apparatus was a white, opaque plexiglass box (40cm x 40cm). On day 1, mice were habituated to the testing apparatus for 15 minutes. On day 2, the mice were reintroduced to the testing apparatus and allowed to explore two identical objects equally spaced from the walls of the apparatus (objects A and A’) for 5 minutes and the animal was then removed. Any-MAZE tracking software was used to measure the time spent investigating each object, and a preference index (PI) was calculated by dividing time spent investigating A’ by time spent investigating A (A’/A). One hour later after identical object exploration, the mouse was placed back in the testing apparatus where one of the identical objects had been replaced with a novel object that differed in shape, color, and texture (object B). Again, the mouse was given 5 minutes to explore the two objects, and preference index for the novel object was calculated by dividing time spent investigating B by time spent investigating A (B/A). Significant preference for the novel object was assessed by comparing the PI from the training phase to the PI from the testing phase.
Three chambered social choice
Assessment of sociability was evaluated using the standard three-chambered social choice paradigm. A white, opaque plexiglass rectangular box was used, with three partitions (each 20cm x 40cm). The mouse was first given 5 minutes to habituate to the empty apparatus. After habituation, into the left and right compartments was placed either with an inanimate object (nonsocial zone) or an age and sex-matched C57Bl/6 mouse (social zone). The object and mouse were placed under clear, plexiglass cylinders with perforations to allow odor detection. During the testing phase, the test mouse was allowed five minutes to explore either zone. Time in each zone was measured using the Any-MAZE tracking software. Zone preference index was calculated by dividing specific zone time (either social or nonsocial) by total zone exploration time.
Biochemistry
Mice were anesthetized with a pentobarbital solution and decapitated. The brain was removed, and each hemisphere of the cerebellum, cerebral cortex, and hippocampus was dissected. One hemisphere was rapidly transferred to a mortar and pestle prechilled on dry ice, and ground into a fine powder to be processed for RNA extraction by the RNeasy mini kit (Qiagen, Catalogue #74134) according to the manufacturer’s instructions. Once RNA extraction was complete, conversion to cDNA was done using the iScript Supermix (Bio-Rad, Catalogue #1708841, Hercules, CA). The other hemisphere was transferred to a dounce prechilled on ice, and lysed in 1% Triton-X lysis buffer (150mM NaCl, 50mM Tris-HCl pH 8.0, 1mM EGTA, 5mM EDTA) with protease and phosphatase inhibitors for generation of protein lysates.
Antibodies
The following antibodies were used in this study as follows: immunoblotting of SAP97 (Thermo Fisher Scientific, catalogue # PA1-741); immunoblotting of PSD95 (NeuroMab, catalogue # 75–348); immunoblotting of PSD93 (NeuroMab, catalogue # 75–284); immunoblotting of SAP102 (NeuroMab, catalogue # 75–058); immunoblotting beta-actin (Cell Signaling Technology, catalogue # 3700 (mouse), or Sigma-Aldrich, catalogue # A2066 (rabbit)). Secondary antibodies for immunoblots (IRDye) were purchased from Li-COR (Catalogue # 925–32210, and Catalogue # 925–68021).
Western blot
Western blot was performed according to standard procedures [29–31].
Quantitative PCR
Quantitative real-time PCR (qPCR) was carried out as previously described using the delta delta Ct method to calculate relative gene expression levels [32]. Ribosomal S17 and S18 (RS17, RS18) were used as reference genes. Each reaction consisted of cDNA, primers, and Power SYBR Green PCR Master Mix (Applied Biosystems, Catalogue # 4367659, Waltham, MA) with a total 25uL reaction volume. Melting curve analysis of the target sequences showed that all primers used in this study generated amplification of a single peak, without primer-dimer artifacts. Primer and cDNA concentrations were optimized prior to use in qPCR experiments. Each qPCR experiment consisted of 4–6 biological replicates, as well as three technical replicates per sample. The primers used for qPCR included: GLUA1 fwd- CCCTGAGAGGTCCCGTAAAC rev- GCTCAGAGCACTGGTCTTGT; GLUA3 fwd- CCATGCTCTTGTCAGCTTCG rev- AGTCCACCTATGCTGATGGT; GLUA4 fwd- TGAATGAACAAGGCCTCTTGGA rev- AGGCACTCGTCTTGTCCTTG; NRCAM fwd- AAGACCCGCTGGACTTTGAA rev- GGCTTGCCATTGCCTTCTTA; HUWE1 fwd- GTTGGGATTTCCCACCAGGA rev- CAGTCTGCAGGAGCTTCAGT; PTEN fwd- CCTGCAGAAAGACTTGAAGGTG rev- CTGTGCAACTCTGCAGTTAAA; ADAM10 fwd- GGCTGGGAGGTCAGTATGGA rev- CTCGTGTGAGACTGCTCGTT; WAS fwd- TCAGCTGAACAAGACCCCTG rev- CATGCATCAGGGCACCTACT; ERBB4 fwd- ACCCAGGGGTGTAACGGT rev- TGGTAAAGTGGAATGGCCCG; SEMA4C fwd- GGTGGCCGGAGTCAAACG rev- TTCAGTCCAGCAGCCCTCTTT; KCNA3 fwd- TCCGAAAAGCCCGGAGTAAC rev- CTGTGGAGTTGCCCGTTTTG; KCNA4 fwd- CACTTGCTGGGAATGGTGAAGT rev- GAGAAGGTGGTAGACGCAGT; KCNA5 fwd- TAGGACACTGGCTGACCCAT rev- ACGCACAAGCAGCTCAAAAG; GNG13 fwd- TTGCTGTCTCCTCCAAAACCTC rev- TCCCTCTTGAAGGCCAGTTG; FZD7 fwd- AGAACCTCGGCTACAACGTG rev- ACCGAACAAAGGAAGAACTGC; DLGAP4 fwd- TTTGCTTCTCTGCCCGATCC rev- TGATGAACATTGCTTCAAGAGC; CTNNA1 fwd- CAGTTCGCTGCAGAAATGAC rev- ACCTGTGTAACAAGAGGCTCC; CALM3 fwd- GAGTAACCTCGATCCCCGAG rev- GAAGGCTTCCTTGAACTCTGC; KCNC1 fwd- CTACGCGCGGTATGTGGC rev- TCGGTCTTGTTCACGATGGG; S18 fwd- CAGCTCCAAGCGTTCCTGG rev-GGCCTTCAATTACAGTCGTCTTC; S17 fwd- GATTCAGAGAGGGCCTGTGAG rev-CTGAGACCTCAGGAACGTAGT
RNA sequencing
RNA was isolated from four control and four SAP97-cKO male hippocampi, quality evaluated by Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA), and sequenced with an Illumina HiSeq 4000 High-Throughput Sequencing System. The RNA-seq reads were aligned to the mouse genome mm10.GRCm38.p5 using STAR version 2.5.3a [33]. Next, normalization and quantification were performed with the PORT version 0.8.2a-beta pipeline (http://github.com/itmat/Normalization) which first removes reads that map to ribosomal RNA sequences or mitochondrial DNA and then uses a read re-sampling strategy for normalization to account for batch effects and differences in sequencing depth among the samples. After the normalization procedure, the gene level quantification was done by PORT with respect to the Ensemblv90 annotation. The normalized count of reads mapping to exon 10 of SAP97 showed almost 30-fold reduction from an average of 242 in the control samples to an average of 8 in the SAP97-cKO samples affirming the efficacy of the knockdown procedure. The differential expression analysis was performed using the R Bioconductor package limma-voom [34,35]. The top 566 genes with FDR < 0.5 and fold-change of greater than 1.6 were used for general pathway enrichment analyses, which were performed using Ingenuity IPA.
Analysis of overlap between differentially expressed genes and risk-associated disease genes
The significance of overlap between the set of differentially expressed genes (DEGs) and risk-associated ASD, SCZ, and other neuronal disorder genes was analyzed using non-parametric analysis. The ASD gene list was chosen from research by Silvia De Rubeis et al., while the SCZ and ataxia gene lists were chosen from online resources (szdb.org, genedx.com). Details of the lists chosen and overlap analysis are discussed in results section. The mean and variance of the corresponding hypergeometric distribution were calculated. The p value of the significance of the overlap was estimated using the hypergeometric probability test.
Statistics
Data were analyzed using Prism (GraphPad Software, La Jolla, CA). Significant differences within groups were determined using either Student’s t-test, one-way ANOVA followed by Tukey’s test for multiple comparisons, or repeated-measures two-way ANOVA followed by Tukey’s test for multiple comparisons. For all tests except for RNAseq, the significance threshold was set to p<0.05. The significance threshold for DEGs in the RNAseq experiment was set to FDR <0.25.
Results
Targeted deletion of SAP97 to neurons
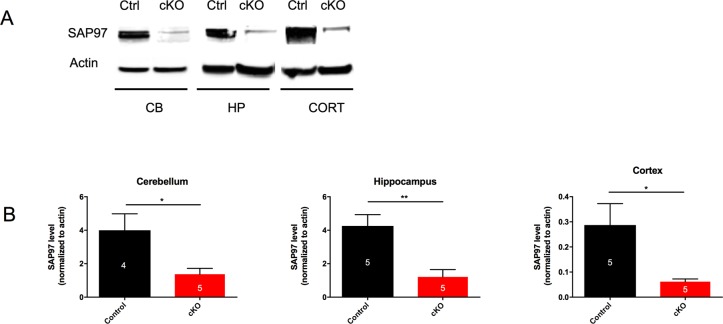
Global SAP97 knockout mice have been generated, but die soon after birth due to a craniofacial defect. In order to study the effect of loss of SAP97 on neuronal development and behavior, we conditionally knocked out SAP97 by crossing Nestin-cre mice with SAP97 floxed mice (see Methods). SAP97-cKO mice were born at Mendelian ratios and were grossly normal. At two months of age, we harvested tissue from the cerebellum, hippocampus, and cortex from control and SAP97-cKO animals of both sexes and prepared protein lysates for western blot analysis. In all three brain regions, the abundance of SAP97 was significantly reduced (Cerebellum: Ctrl 3.998 ± 0.9833, n = 4; SAP97-cKO 1.373 ± 0.348, n = 5, p = 0.0279; Hippocampus: Ctrl 4.252 ± 0.6751, n = 5; SAP97-cKO 1.215 ± 0.4314, n = 5, p = 0.0053; Cortex: Ctrl 0.287 ± 0.08557, n = 5; SAP97-cKO 0.06179 ± 0.01072, n = 5, p = 0.0311), indicating that we successfully generated cKO animals (Fig 1).
Fig 1. SAP97 protein is sufficiently knocked down in SAP97-cKO animals.
(A) Western blots showing reduced SAP97 band intensity in cerebellum, hippocampus, and cortex. (B) Quantification of western blot analysis. *P < .05, **P < .01 (two-tailed Student’s t test). Data are presented as mean ± SEM.
No apparent compensation by other Dlg-MAGUK family members
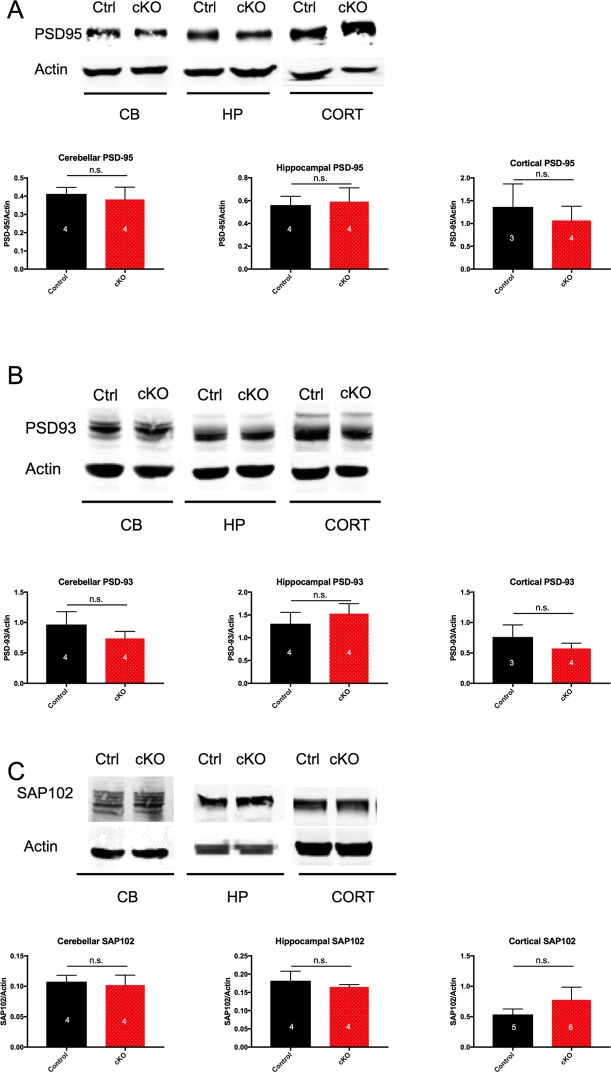
Previous work shows that the Dlg-MAGUKs (PSD-95, PSD-93, SAP102, and SAP97) can have redundant functions in electrophysiological assays. In order to examine whether loss of SAP97 led to compensatory changes in the abundance of the other Dlg-MAGUK family members, we measured total protein levels in the cerebellum, hippocampus, and cortex. There was no significant difference in the abundance of any other Dlg-MAGUK at the protein level in control versus SAP97-cKO male animals (Fig 2). These data suggest that if members of the Dlg-MAGUK family compensate for the lack of SAP97, they do so without a change in overall abundance.
Fig 2. No compensation by Dlg-MAGUK family abundance in SAP97-cKO animals.
(A) Western blots and quantification showing no significant change in abundance of PSD-95 in cerebellum, hippocampus, and cortex. (B) Western blots and quantification showing no significant change in abundance of PSD-93 in cerebellum, hippocampus, and cortex. (C) Western blots and quantification showing no significant change in abundance of SAP102 in cerebellum, hippocampus, and cortex. n.s., no significance (two-tailed Student’s t test). Data are presented as mean ± SEM.
No changes in gene expression level of known SAP97 binding partners or interactors
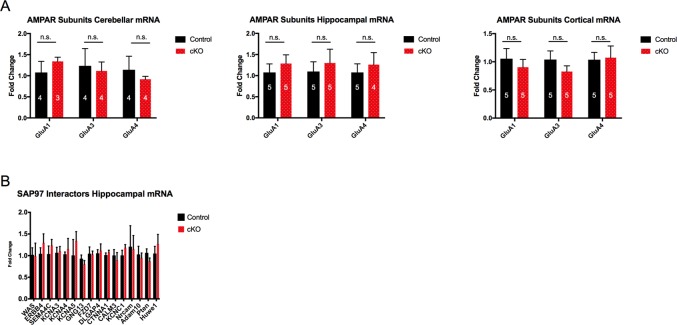
SAP97 is a scaffolding protein that allows for a large number of protein-protein interactions. Thus, the absence of SAP97 could potentially affect the expression level of numerous proteins. To determine whether loss of SAP97 contributes to changes in expression levels of other identified members of the postsynaptic density, we conducted a directed qPCR screen. We measured mRNA levels firstly of all AMPAR subunits, as SAP97 is known to be the only Dlg-MAGUK to directly bind GluA1. mRNA levels of GluA1, GluA3, and GluA4 remained unchanged in the three brain regions that were probed (cerebellum, hippocampus, and cortex) (Fig 3A). Results from GluA2 were highly variable and thus removed from the study.
Fig 3. No change in mRNA expression level of AMPAR subunits and selected SAP97 interactor genes in SAP97-cKO animals.
(A) qPCR results showing no significant change in abundance of GluA1, GluA3, or GluA4 mRNA transcripts in selected brain regions. (B) qPCR results showing no significant change in abundance of mRNA levels of selected SAP97 interactor genes in hippocampus. n.s., no significance (two-tailed Student’s t test). Data are presented as mean ± SEM.
We next sought to determine whether the levels of other proteins known to interact with SAP97 were affected by loss of SAP97. We measured mRNA levels of 16 genes in the hippocampus. From our selection of 16 genes, we observed no differences in the mRNA expression level between control and SAP97-cKO animals (Fig 3B). These results would suggest that the abundance of genes from our selection is not significantly regulated by SAP97 expression.
Identification of SAP97-regulated transcripts in the hippocampus
Given that we observed no group differences in our directed qPCR screen, we sought a broader, unbiased approach by performing RNAseq analysis on hippocampi from SAP97-cKO and control mice (n = 4 per group). For each animal, we verified the presence or absence of SAP97 by western blot on brain tissue before submitting hippocampal samples for sequencing.
A total of 66 genes were found to be significantly downregulated in the SAP97-cKO animals as compared to control hippocampi (FDR < 0.25) (Fig 4A, Table 1). In contrast, only one gene was upregulated in the hippocampi of SAP97-cKO animals as compared to control hippocampi. Gene ontology analysis of the DEGs revealed enrichment for numerous cellular and molecular functional categories, including those related to “Cell Morphology,” “Cellular Development,” and “Cell-To-Cell Signaling and Interaction” (Tables 2 and 3). Additionally, the top enriched ID Associated Network Functions included “Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function,” and “Developmental Disorder, Embryonic Development, Organ Development” (Table 4). Gene ontology terms to describe gene products known to be associated with ASD or the neurexin-neuroligin-SHANK complex in mice frequently include “Cell Communication” and “Nervous System Development”, which overlaps with the findings in our RNAseq study [36]. Previous studies that have conducted RNAseq on SCZ patients and performed gene ontology analysis on the resulting DEGs have identified regulation of the actin cytoskeleton as a key pathway [6]. While the actin cytoskeleton was not directly implicated by our RNAseq study, it is essential for many of the gene ontology analysis terms listed in our data set. As proper arrangement of the actin cytoskeleton is essential for neuronal cell maturation and migration, neurite outgrowth, and maintenance of synaptic density and plasticity, dysregulation of these pathways in the nervous system could have severe consequences in psychiatric disorders such as SCZ.
Fig 4. Loss of SAP97 leads to downregulation of DEGs and enrichment of SCZ risk-related genes.
(A) Heat map representation of downregulation of DEGs in SAP97-cKO hippocampus. (B) DEGs are specifically enriched for SCZ risk-related genes. *P < .05 (two-tailed Student’s t test), **P < .01 (hypergeometric probability test).
Table 1. List of genes with significant expression differences between control and SAP97-cKO mice.
| Ensemble Genome ID | Gene Symbol | Log Fold Change |
|---|---|---|
| ENSMUSG00000019970 | Sgk1 | 0.604982801 |
| ENSMUSG00000022770 | Dlg1 | 0.920172279 |
| ENSMUSG00000023224 | Serping1 | 1.092300119 |
| ENSMUSG00000029304 | Spp1 | 2.086028102 |
| ENSMUSG00000031431 | Tsc22d3 | 0.662945543 |
| ENSMUSG00000021390 | Ogn | -1.36634343 |
| ENSMUSG00000026728 | Vim | -0.73795218 |
| ENSMUSG00000022769 | Sdf2l1 | 0.813450163 |
| ENSMUSG00000020467 | Efemp1 | 0.785266079 |
| ENSMUSG00000033227 | Wnt6 | 1.860240362 |
| ENSMUSG00000055128 | Cgrrf1 | 0.544558433 |
| ENSMUSG00000037254 | Itih2 | -1.44816667 |
| ENSMUSG00000070691 | Runx3 | 2.433336132 |
| ENSMUSG00000032575 | Manf | 0.581728345 |
| ENSMUSG00000031289 | Il13ra2 | 1.998579379 |
| ENSMUSG00000029661 | Col1a2 | 1.062558267 |
| ENSMUSG00000067038 | Rps12-ps3 | 2.467941625 |
| ENSMUSG00000054619 | Mettl7a1 | -0.39870621 |
| ENSMUSG00000024650 | Slc22a6 | 1.704915114 |
| ENSMUSG00000015090 | Ptgds | 1.253057418 |
| ENSMUSG00000026043 | Col3a1 | 1.300397001 |
| ENSMUSG00000030357 | Fkbp4 | 0.343944084 |
| ENSMUSG00000071005 | Ccl19 | 2.363646432 |
| ENSMUSG00000105843 | Gm42644 | -1.52239332 |
| ENSMUSG00000030711 | Sult1a1 | 0.875568486 |
| ENSMUSG00000079293 | Clec7a | 2.007730582 |
| ENSMUSG00000070436 | Serpinh1 | 0.658564574 |
| ENSMUSG00000030154 | Klrb1f | 2.174662835 |
| ENSMUSG00000004951 | Hspb1 | 1.133328217 |
| ENSMUSG00000036777 | Anln | 0.750523147 |
| ENSMUSG00000027248 | Pdia3 | 0.350126593 |
| ENSMUSG00000030218 | Mgp | 1.053034845 |
| ENSMUSG00000030108 | Slc6a13 | 1.258734628 |
| ENSMUSG00000057836 | Xlr3a | 2.154616401 |
| ENSMUSG00000000303 | Cdh1 | 1.987938463 |
| ENSMUSG00000024087 | Cyp1b1 | -0.95956574 |
| ENSMUSG00000032231 | Anxa2 | 0.831693618 |
| ENSMUSG00000060591 | Ifitm2 | -0.82147731 |
| ENSMUSG00000049241 | Hcar1 | 1.730353275 |
| ENSMUSG00000032060 | Cryab | 0.561687665 |
| ENSMUSG00000026638 | Irf6 | 1.238846117 |
| ENSMUSG00000022548 | Apod | 0.868596841 |
| ENSMUSG00000013584 | Aldh1a2 | 1.291353747 |
| ENSMUSG00000019539 | Rcn3 | 0.615255839 |
| ENSMUSG00000040055 | Gjb6 | -0.57564983 |
| ENSMUSG00000040310 | Alx4 | 1.225356858 |
| ENSMUSG00000031367 | Ap1s2 | 0.377838987 |
| ENSMUSG00000023272 | Creld2 | -0.51269519 |
| ENSMUSG00000038155 | Gstp2 | -1.96171475 |
| ENSMUSG00000107215 | Gm43197 | 2.106413397 |
| ENSMUSG00000074896 | Ifit3 | 0.830338737 |
| ENSMUSG00000020484 | Xbp1 | 0.420933711 |
| ENSMUSG00000034435 | Tmem30b | 2.348757156 |
| ENSMUSG00000022766 | Serpind1 | 1.521272379 |
| ENSMUSG00000005125 | Ndrg1 | 0.558146377 |
| ENSMUSG00000038393 | Txnip | 0.584362558 |
| ENSMUSG00000041548 | Hspb8 | 0.559621993 |
| ENSMUSG00000034165 | Ccnd3 | 0.399017781 |
| ENSMUSG00000027048 | Abcb11 | -2.19248413 |
| ENSMUSG00000073418 | C4b | 0.516645555 |
| ENSMUSG00000025823 | Pdia4 | -0.55418986 |
| ENSMUSG00000031070 | Mrgprf | 1.766073987 |
| ENSMUSG00000032179 | Bmp5 | 1.738362368 |
| ENSMUSG00000043795 | Prr33 | 2.024615333 |
| ENSMUSG00000048368 | Omd | 2.520720092 |
| ENSMUSG00000066861 | Oas1g | 2.341011247 |
| ENSMUSG00000024371 | C2 | 1.517189655 |
Table 2. List of top diseases identified through IPA that were affected in hippocampus of SAP97-cKO animals.
| Name | p-value | Genes Affected |
|---|---|---|
| Organismal Injury and Abnormalities | 2.58E-02–1.30E-05 | 37 |
| Respiratory Disease | 2.52E-02–1.30E-05 | 10 |
| Endocrine System Disorders | 2.52E-02–1.42E-05 | 16 |
| Gastrointestinal Disease | 2.21E-02–1.42E-05 | 21 |
| Immunological Disease | 2.52E-02–1.42E-05 | 14 |
Table 3. List of top molecular and cellular functions identified through IPA that were affected in hippocampus of SAP97-cKO animals.
| Name | p-value | Genes Affected |
|---|---|---|
| Cell Death and Survival | 2.83E-02–5.79E-05 | 21 |
| Cellular Movement | 2.52E-02–6.58E-05 | 18 |
| Cell Morphology | 2.52E-02–2.70E-04 | 19 |
| Cellular Development | 2.52E-02–2.70E-04 | 25 |
| Cell-To-Cell Signaling and Interaction | 2.47E-02–3.32E-04 | 17 |
Table 4. List of top networks identified through IPA that were affected in hippocampus of SAP97-cKO animals.
| ID Associated Network Functions | Score |
|---|---|
| Organismal Injury and Abnormalities, Respiratory Disease, Cellular Movement | 35 |
| Cellular Development, Cellular Growth and Proliferation, Hematological System Development and Function | 22 |
| Ophthalmic Disease, Organismal Injury and Abnormalities, Hereditary Disorder | 22 |
| Cell Cycle, Gene Expression, Skeletal and Muscular System Development and Function | 5 |
| Developmental Disorder, Embryonic Development, Organ Development | 2 |
Schizophrenia risk enrichment in DEG set
Given that SAP97, along with the other members of the Dlg-MAGUK family, have been strongly implicated in psychiatric disorders such as ASD and SCZ, we chose to examine whether the DEG set was enriched for disease risk-associated genes. In order to determine whether the DEG set had a significant overlap with genes implicated in psychiatric disorders such as ASD and SCZ, we compared our DEG set with disease-related gene databases. For determining overlap with ASD-related genes, we used the gene set previously generated from the transmission and de novo association test (TADA), which consists of 107 genes. When we matched our DEG list to the TADA ASD gene list, we did not find the match percentage to be significant based on the hypergeometric distribution (Distribution mean = 0.30, standard deviation = 0.30; SAP97-cKO DEG 0.0) (Fig 4B). We next chose to compare our DEG list to SCZ risk-related genes found from SZDB: A Database for Schizophrenia Genetic Research (szdb.org). The distilled list of genes from this database gives a score for each gene based on criteria such as convergent functional genomics, copy number variation, differential expression, genome wide association study, and linkage and association studies. The more categories a certain gene is implicated in, the higher the score for that gene. Based on this model, we chose the top 1,000 genes from this database to match to our SAP97-cKO DEG list. Interestingly, we found the SAP97-cKO DEG list to have a significant amount of overlap to the SZDB list based on the hypergeometric test (Distribution mean = 2.79, standard deviation = 1.63; SAP97-cKO DEG 13.43, p = 0.0018) (Fig 4B). Finally, we matched our SAP97-cKO DEG list to ataxia risk-related genes as a negative control, as ataxia is not classified as a neuropsychiatric disorder and SAP97 has not previously been implicated in ataxia. We used a list of ataxia risk-related genes compiled from GeneDx, whose clinical team compiled using multiple sources, including Online Mendelian Inheritance in Man (OMIM), Human Gene Mutation Database (HGMD), and Human Phenotype Ontology (HPO) terms. The total number of genes in this list was 993, which would also allow us to control for the size of the SCZ gene list used. When we compared our SAP97-cKO DEG list to the GeneDx ataxia gene set, we found no significant match percentage (Distribution mean = 2.77, standard deviation = 2.65; SAP97-cKO DEG 4.48) (Fig 4B). Together, these results suggest that SAP97-cKO DEGs are specifically enriched for SCZ risk-related genes.
Behavioral analysis of SAP97-cKO mice
Next, we performed a battery of behavioral tests to screen for behavioral deficits in the SAP97-cKO mice.
Mild exploratory behavior deficit in female SAP97-cKO animals
We first performed the open field test to examine general ambulation. In the males, we observed no change in the total distance traveled (Ctrl 64.8 ± 2.102, n = 29; SAP97-cKO 61.76 ± 3.763, n = 19) (Fig 5A) or the speed of the animals (Ctrl 0.07214 ± 0.002259, n = 29; SAP97-cKO 0.06847 ± 0.004183, n = 19) (Fig 5B). This indicates that the male SAP97-cKO mice do not have a basic impairment in movement. However, female SAP97-cKO animals exhibited decreases in total distance traveled (Ctrl 64.39 ± 3.495, n = 24; SAP97-cKO 52.1 ± 4.519, n = 12, p = 0.0446) (Fig 5A) and speed (Ctrl 0.0717 ± 0.004071, n = 24; SAP97-cKO 0.058 ± 0.005053, n = 12, p = 0.0497) (Fig 5B). These observations may indicate either a motor deficit or an impairment in exploratory behavior that is specific to female SAP97-cKO animals.
Fig 5. Open field behavior indicates female-specific deficit in exploratory motor behavior of SAP97-cKO animals.
(A) No group differences seen in average distance traveled in male animals. Female SAP97-cKO animals display significantly less distance traveled. (B) No group differences seen in average speed in male animals, while female SAP97-cKO show decreased speed. n.s., no significance, *P < .05, **P < .01 (two-tailed Student’s t test). Data are presented as mean ± SEM.
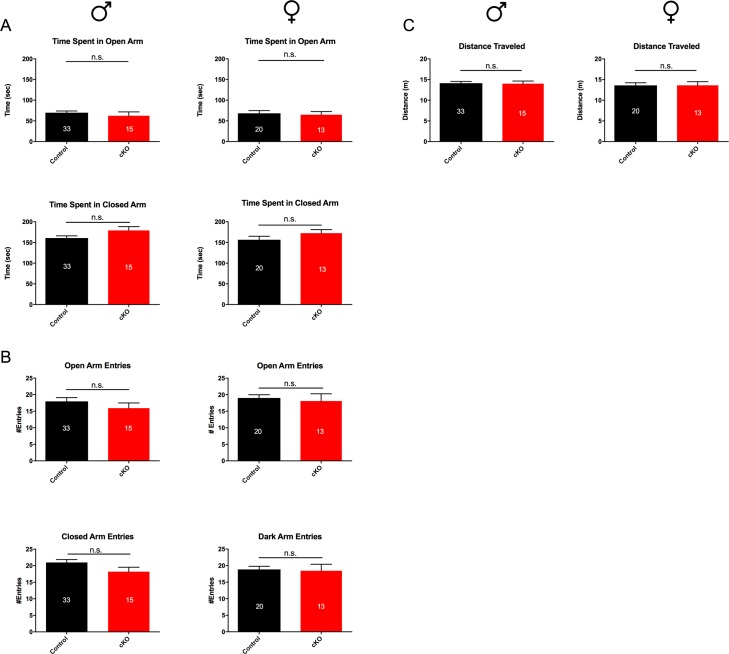
No changes in anxiety levels in SAP97-cKO animals
We next chose to examine anxiety-like behavior. In order to gauge whether SAP97-cKO mice had alterations in anxiety-like behavior, we performed the standard elevated plus maze. When comparing time spent in open arms versus the closed arms, we saw no significant differences between genotypes in both males (open arms: Ctrl 69.75 ± 4.098, n = 33; SAP97-cKO 62.47 ± 9.331, n = 15; closed arms: Ctrl 160.8 ± 5.339, n = 33; SAP97-cKO 179.3 ± 8.893, n = 15) and females (open arms: Ctrl 68.17 ± 6.734, n = 20; SAP97-cKO 64.85 ± 7.72, n = 13; closed arms: Ctrl 156.6 ± 8.515, n = 20; SAP97-cKO 172.5 ± 8.559, n = 13) (Fig 6A). Likewise, the number of entries into the open versus closed arms was similar between genotypes of both sexes (Fig 6B). Total distance traveled in the maze was also measured and compared between Ctrl and SAP97-cKO animals to ensure no significant differences in overall exploration of the maze (Fig 6C). These observations indicate no obvious anxiety-like phenotype in the SAP97-cKO animals.
Fig 6. Comparison of elevated plus maze behavior between control and SAP97-cKO animals.
(A) No group differences seen in average total time spent in open arms vs closed arms of maze. (B) No group differences seen in total open arm entries or closed arm entries. (C) No group differences seen in average distance traveled in elevated plus maze apparatus. n.s., no significance (two-tailed Student’s t test). Data are presented as mean ± SEM.
Mild male-specific cognitive deficit in SAP97-cKO mice
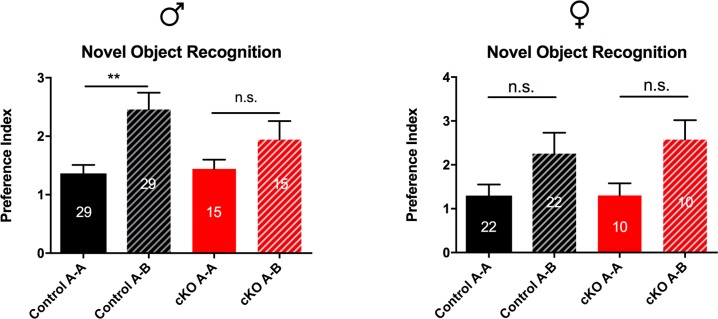
We next chose to examine cognitive behavior in the SAP97-cKO mice. The novel object recognition task is a standard test for cognition that measures ability to recall an object previously observed, as indicated by preference for a novel object (see Methods). During the training phase of this task, we observed no significant differences between the ratio of time spent investigating the two identical objects A and A’ for both males and females (Male: Ctrl 1.364 ± 0.1438, n = 29; SAP97-cKO 1.441 ± 0.1578, n = 15; Female: Ctrl 1.3 ± 0.2506, n = 22; SAP97-cKO 1.301 ± 0.2775, n = 10) indicating that the animals had no prior bias. During the testing phase, control male mice displayed a marked increase in the preference index for the novel object, while SAP97-cKO male mice showed no significant increase in novel object preference index (Ctrl A-A 1.364, Ctrl A-B 2.457; SAP97-cKO A-A 1.441, SAP97-cKO A-B 1.941, F (3, 79) = 5.311, p = 0.0022) (Fig 7). When we examined this behavior in the females, we observed a trending, but not significant, increase in preference index for the novel object in both control and SAP97-cKO animals (Ctrl A-A 1.3, Ctrl A-B 2.255; SAP97-cKO A-A 1.301, SAP97-cKO A-B 2.575, F (3, 57) = 2.59, p = 0.0616) (Fig 7). These findings suggest a modest male-specific cognitive deficit in the SAP97-cKO animals.
Fig 7. Comparison of novel object recognition behavior indicates male-specific cognitive deficit.
Control male animals exhibit preference for novel object (Ctrl A-A vs Ctrl A-B), while SAP97-cKO male animals do not show preference. Both control and SAP97-cKO female animals show trend for preference of novel object, but did not reach significance. n.s., no significance, **P < .01 (ordinary one-way ANOVA with Tukey’s test for multiple comparisons). Data are presented as mean ± SEM.
Female-specific motor learning deficit in SAP97-cKO mice
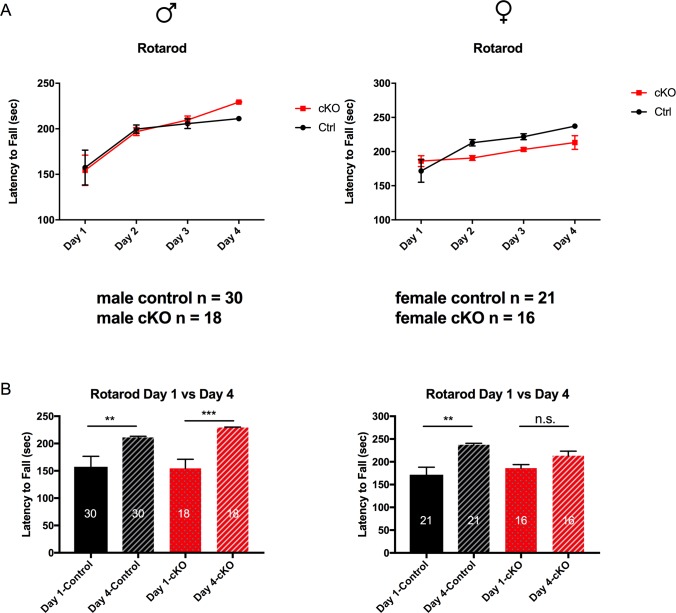
We next chose to examine motor learning behavior in the SAP97-cKO mice. In order to determine whether this behavioral change is present in the SAP97-cKO mice, we performed the standard rotarod task (see Methods). Analysis of both sexes showed significant time effects (Male: F (3, 6) = 12.31, p = 0.0057; Female: F (3, 6) = 9.126, p = 0.0118), while only female animals showed a trend for genotype effects and significant time and genotype interaction effects (genotype effect: F (1, 2) = 11.53, p = 0.0769; time x genotype effect: F (3, 6) = 5.099, p = 0.0434) (Fig 8A). Control animals of both sexes showed a significant increase in latency to fall from the rod from day 1 to day 4 (Male: Day 1 157.5 ± 19.12, Day 4 211.1 ± 2.074, n = 30, p = 0.0010; Female: Day 1 171.6 ± 16.53, Day 4 237.3 ± 3.439, n = 21, p = 0.0021), indicating learning of the task (Fig 8A and 8B). However, while male SAP97-cKO mice showed no learning impairment (Day 1 154.4 ± 16.64, Day 4 229.4 ± 0.8372, n = 18, p = 0.0002), female SAP97-cKO mice showed no significant learning over the timecourse of this task (Day 1 186 ± 7.927, Day 4 213.3 ± 9.988, n = 16) (Fig 8A and 8B). These results suggest that there is a female-specific motor learning or coordination deficit present in the SAP97-cKO mice.
Fig 8. Comparison of rotarod behavior indicates female-specific motor learning deficit.
(A) Both control and SAP97-cKO male animals show learning over the 4-day course of rotarod paradigm. Control female animals show increased motor learning over course of 4 days, while female SAP97-cKO show no significant increase in motor learning. (B) Plots showing comparison of Day 1 versus Day 4 rotarod data for control and SAP97-cKO animals indicates female-specific motor learning deficit. n.s., no significance, **P < .01, ***P < .001 (repeated-measures two-way ANOVA with Tukey’s test for multiple comparisons). Data are presented as mean ± SEM.
No social deficits present in SAP97-cKO mice
The last behavior we chose to examine in the SAP97-cKO mice was sociability. We looked for changes in this behavioral domain by using the three-chambered social choice paradigm (see Methods). During the testing phase of this paradigm, we measured preference for the social target zone versus the nonsocial target zone. Control and SAP97-cKO animals of both sexes exhibited a strong preference for spending time in the social target zone (Male: Ctrl-Nonsocial 0.334, Ctrl-Social 0.666, n = 15; SAP97-cKO-Nonsocial 0.4164, SAP97-cKO-Social 0.5836, n = 11, F (3, 48) = 24.67, p<0.0001; Female: Ctrl-Nonsocial 0.3641, Ctrl-Social 0.6359, n = 8; SAP97-cKO-Nonsocial 0.3045, SAP97-cKO-Social 0.6955, n = 8, F (3, 20) = 14.53, p<0.0001) (Fig 9). These results suggest no social deficit in the SAP97-cKO mice.
Fig 9. Comparison of social choice behavior between control and SAP97-cKO animals.
No significant differences observed between control and SAP97-cKO male or female animals in preference for social target. **P < .01, ***P < .001, ****P < .0001 (ordinary one-way ANOVA with Tukey’s test for multiple comparisons). Data are presented as mean ± SEM.
Discussion
SAP97 is a member of the Dlg-MAGUK family, a family of genes that plays an important role in synapse biology and has repeatedly been implicated in neuropsychiatric disorders [7,21,23,37]. In this study, we aim to address the direct role of SAP97 in overall behavior and its potential for contributing to pathology. We generated and studied mice that were null for SAP97 in the nervous system and make three principal observations. First, there are no compensatory changes in expression levels of other Dlg-MAGUKS or AMPARs in the SAP97-cKO versus controls. Second, loss of SAP97 is associated with moderate changes in gene transcripts related to SCZ. And third, SAP97-cKO animals display a mild behavioral phenotype, consisting of a male-specific cognitive deficit and female-specific motor deficit. While future behavioral experiments will be necessary to understand whether SAP97 regulates SCZ-specific behaviors, our results indicate that SAP97 probably plays a limited role in organismal behavior under standard laboratory conditions and its absence leads to transcriptomic changes associated with SCZ.
ASD and SCZ as polygenic disorders
Investigations of ASD, SCZ, and other related psychiatric disorders indicate a highly polygenic architecture with small effect sizes of each implicated risk variant. Mouse modeling of these disorders by targeting one such risk variant typically demonstrates a moderate, or incomplete manifestation of the human disorder. This is well illustrated by human and mouse studies of the ProSAP/SHANK family member SHANK3. Human genetic studies link mutations in SHANK3 to a broad range of neuropsychiatric disorders. For example, deletions of exons 1–9 or exons 1–17 of SHANK3 have been found in patients exhibiting severe language delay and significant intellectual disability. Mice generated to mimic these deletions were generated by Peca et al. and the main behavioral effects were repetitive grooming and deficits in social interaction [9]. Jiang et al. used a different targeting strategy to mimic the human deletions and the mice displayed repetitive behaviors, deficits in social interaction, abnormal ultrasonic communication patterns and learning and memory deficits [38]. In a second well-studied family, affected individuals displayed ASD-features and this was linked to a deletion of SHANK3 exon 21 (an exon that included the Homer binding domain). Mice generated to mimic this genetic lesion were created by Kouser and Speed et al. and ~2.5 month old animals exhibit defects in spatial learning and memory, motor-coordination deficits, hypersensitivity to heat, novelty avoidance, but minimal social abnormalities and no repetitive grooming behavior [11]. Together, this work demonstrates that creating a mouse with a genetic lesion that closely mimics, or is identical, to the gene defect in humans with neuropsychiatric disease only partially recapitulates the human behavioral phenotypes.
This disparity between genetic lesions associated with psychiatric phenotypes and mice created to mimic the human condition also extends to the Dlg-MAGUK family. Nonsynonymous missense mutations in the Dlg-MAGUK family members have been found in ASD and SCZ patients, and decreased protein expression of PSD-95, PSD-93, and SAP97 has been observed in the cortex of postmortem SCZ patients. To model with in mice, null alleles of PSD-95, PSD-93, and SAP102 have been created—PSD-95 and SAP102 knockout animals share spatial learning memory deficits [20,39], while animals null for PSD-95 or PSD-93 share a hyper-social phenotype [15]. PSD-95 and SAP102 knockout animals display a mild, and PSD-93 knockout animals display a severe, motor function defect [15,20]. The PSD-95 null mouse has been the most extensively investigated animal. Feyder et al. characterized the PSD-95 knockout mice and the mice exhibit increased repetitive behaviors, abnormal communication, hyper-social behavior, impaired motor coordination, and increased stress-reactivity and anxiety-related responses [16]. The extent to which the PSD-95 null mice faithfully report on the contribution of PSD-95 to psychiatric disease is an open question. Our behavioral characterization of the SAP97-cKO animals was conducted broadly as a first-pass assessment of behavioral changes. We are unable to conclude that the behavioral abnormalities observed in our study are disease specific, and further behavioral tests that are specific to ASD and SCZ will be necessary to claim whether SAP97 directly contributes to a partial manifestation of disease phenotype. Nevertheless, our behavioral results provide a framework for understanding the role of SAP97 in organismal behavior and brain function.
SAP97 splice variants and their differing roles in the nervous system
SAP97 has wide molecular diversity, which is created by extensive alternative splicing. The two most well-studied SAP97 splice variants are SAP97α and SAP97β. In SAP97α, the prototypic N-terminal L27 domain is replaced with a putative palmitoylation motif. Overexpression of SAP97α (but not SAP97β) was shown to enhance the synaptic levels of AMPARs and to compensate for the shRNA-mediated loss of PSD-95 in organotypic slices [40]. SAP97 isoform-specific biology may also extend into human SCZ data. Uezato and colleagues identified a new SAP97 splicing variant that is transcribed from a previously unreported 95-base-pair exon (exon 3b). In post-mortem prefrontal cortices of patients with SCZ, mRNA expression of exon 3b was significantly reduced, specifically in patients with early-onset SCZ [37]. A more recent genetic association analysis detected an association between SCZ and single nucleotide polymorphisms located within exon 3b [22]. How reduced levels of the SAP97 3b transcript may be involved in the susceptibility and pathophysiology of early-onset SCZ is unknown. While our study provides a broad characterization of the effect of SAP97 on brain function, it will be necessary to conduct future studies aimed at addressing the individual roles of prominent splice variants.
The role of the serpin family as a molecular module in SCZ
The RNAseq study we conducted on the hippocampi of SAP97-cKO animals indicated 67 DEGs, which were specifically enriched for SCZ-related risk genes. The specific SCZ-related risk genes we identify in our data are SERPING1, RUNX3, CLEC7A, SERPINH1, CDH1, AP1S2, XBP1, SERPIND1, and C4B. These observations lead us to hypothesize that SAP97 is a component of a “molecular module” of gene products that together subserve aspects of normal behavior. Further, we hypothesize that abnormalities in the operation of this molecular module give rise to select behavioral alterations. Defects in many molecular modules in aggregate manifest as the complex psychiatric disorder we recognize as SCZ. The components of this module may interact physically, functionally, developmentally, or in terms of localization. Future work will be required to elucidate: 1) how the components of this hypothesized molecular module mechanistically interact, and 2) how this impacts brain function and behavior.
Our attention is drawn to three genes that were differentially expressed in the hippocampus of SAP97-cKO mice versus controls—serine peptidase inhibitors (serpins), as this group of genes has previously been reported in the literature to be associated with SCZ [41–46]. SERPING1 was found to be upregulated in postmortem brain tissue from SCZ patients [44,45]. Additionally, a study of adult Swedish twins enriched for SCZ showed an association between gene expression level of SERPING1 and thickness across the cortex, a characteristic that is potentially involved in the pathogenesis of SCZ [41]. Polymorphisms in the promoter regions of genes on 22q11, a chromosomal region that has been associated with various psychiatric illnesses including SCZ, resulted in activity differences in the gene SERPIND1 [42]. Another well-studied member of the serpin family previously implicated in SCZ, but not directly by our RNAseq data, is neuroserpin (SERPINI1). SERPINI1 is restricted to regions in the brain where synaptic changes are associated with learning and memory (cortex, hippocampus, amygdala, and olfactory bulb) [46]. SERPINI1 has also been implicated in dendrite growth, as overexpression studies in primary neurons leads to increased dendritic arborization and altered dendritic spine shape [47]. Additionally, mice with dysregulated expression of SERPINI1 show selective reduction of locomotor activity in novel environments, anxiety-like responses, and neophobic response to novel objects [43]. These behavioral phenotypes in the SERPINI1 deficient mice are reminiscent of the defects we see in the SAP97-cKO animals. SERPINI1 is also a known inhibitor of the extracellular protease tissue-type plasminogen activator (tPA). Conditions that affect the activity of tPA have consistently been described in drug-naïve cases of SCZ [48–51]. Interestingly, psychotic patients on chronic warfarin therapy for deep-vein thrombosis showed remission of psychotic symptoms, indicating that defective modulation of the coagulation pathway might contribute to the pathogenesis of SCZ [52]. C4B, or complement component b, is another gene directly listed from our RNAseq study that has known roles in the coagulation pathway and is an important cofactor to the serine protease family. The strongest genetic association of SCZ at a population level involves variation in the Major Histocompatibility Complex (MHC) locus, where the association of SCZ with the MHC locus arises substantially from many diverse alleles of the C4 genes [41,53,54]. These prior observations along with our findings from the SAP97-cKO RNAseq study may highlight a potential mechanism by which SAP97 contributes to the etiology of SCZ.
Sex-specific differences in psychiatric disease
Psychiatric disorders are characterized by substantial sex-differences in their prevalence, symptomology, and treatment response [55]. Women are more likely than men to develop dementia, panic disorder, post-traumatic stress disorder, and major depression [56,57]. Conversely, the incidence of neurodevelopmental disorders such as ASD and SCZ is higher in males [58,59]. In our study, we conducted RNAseq on male hippocampal tissue from SAP97-cKO tissue and found the resulting DEGs to be specifically enriched for SCZ risk-related gene sets. However, our behavioral screen was undertaken on both male and female SAP97-cKO animals and identified interesting sex-specific differences. This raises the possibility that the RNAseq profile of female SAP97-cKO mice may be at least partially distinct from the male SAP97-cKO dataset.
One potential limitation of our study of female behavior is the lack of assessment of the estrous cycle. Female mice in distinct stages of the estrous cycle have been previously shown to perform differently in behavioral tasks related to anxiety and cognition. Furthermore, it is thought that oestrogens play a protective role against SCZ [60]. It will be vital to perform behavioral testing at different stages of the female estrous cycle, as well as corroborate behavioral findings with RNAseq data in order to have a complete understanding of the role of SAP97 in the female brain. Additionally, results from these future experiments may address the behavioral sex differences we observed in the SAP97-cKO animals.
Conclusion
Our study provides the first broad behavioral and transcriptomic characterization of SAP97 in the mouse nervous system. Despite study limitations, we show that loss of SAP97 contributes to moderate behavioral abnormalities in both male and female animals, as well as an enrichment of SCZ related genes in the male animals. Our findings are a first step to understanding the direct role of SAP97 in overall organismal behavior, and potentially provide a stepping-stone for understanding the molecular mechanism by which SAP97 contributes to neuropsychiatric disorders.
Acknowledgments
We thank the High-throughput Sequencing (HTS) Core at the Children’s Hospital of Philadelphia for technical assistance in generating the cDNA libraries. We greatly appreciate the help by Komal S. Rathi and Dr. Deanne M. Taylor early in the project with the design and implementation of the RNAseq experiments. We also thank Dr. Ted Abel, Dr. Minghong Ma, Dr. Marc V. Fucillo, and Dr. Zhaolan Zhou for ongoing insightful feedback on the project.
Data Availability
Data are available from the figshare database (DOI 10.6084/m9.figshare.5942476; https://figshare.com/s/00d5f26e315c997fddff).
Funding Statement
This work was supported by Public Health Service grants NS095746 and NS093439 (to R.G.K.), the NIH training grant 5T32MH017168-33 (to P.G.), and by The National Center for Advancing Translational Sciences grant 5UL1TR000003. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493: 327–337. doi: 10.1038/nature11860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchino S, Waga C. SHANK3 as an autism spectrum disorder-associated gene. Brain and Development. The Japanese Society of Child Neurology; 2013;35: 106–110. doi: 10.1016/j.braindev.2012.05.013 [DOI] [PubMed] [Google Scholar]
- 3.Grabrucker S, Proepper C, Mangus K, Eckert M, Chhabra R, Schmeisser MJ, et al. The PSD protein ProSAP2/Shank3 displays synapto-nuclear shuttling which is deregulated in a schizophrenia-associated mutation. Experimental Neurology. Elsevier Inc; 2014;253: 126–137. doi: 10.1016/j.expneurol.2013.12.015 [DOI] [PubMed] [Google Scholar]
- 4.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506: 179–184. doi: 10.1038/nature12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515: 209–215. doi: 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu S-A, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry. 2014;20: 563–572. doi: 10.1038/mp.2014.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing J, Kimura H, Wang C, Ishizuka K, Kushima I, Arioka Y, et al. Resequencing and Association Analysis of Six PSD-95-Related Genes as Possible Susceptibility Genes for Schizophrenia and Autism Spectrum Disorders Nature Publishing Group; Nature Publishing Group; 2016;: 1–8. doi: 10.1038/srep27491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh A, Michalon A, Lindemann L, Fontoura P, Santarelli L. Drug discovery for autism spectrum disorder: challenges and opportunities. Nat Rev Drug Discov. 2013;12: 777–790. doi: 10.1038/nrd4102 [DOI] [PubMed] [Google Scholar]
- 9.Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472: 437–442. doi: 10.1038/nature09965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han K, Holder JL Jr, Schaaf CP, Lu H, Chen H, et al. SHANK3 overexpression causesmanic-like behaviour with uniquepharmacogenetic properties. Nature. Nature Publishing Group; 2014;503: 72–77. doi: 10.1038/nature12630 24153177 [Google Scholar]
- 11.Kouser M, Speed HE, Dewey CM, Reimers JM, Widman AJ, Gupta N, et al. Loss of Predominant Shank3 Isoforms Results in Hippocampus-Dependent Impairments in Behavior and Synaptic Transmission. Journal of Neuroscience. 2013;33: 18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Human Molecular Genetics. 2011;20: 3093–3108. doi: 10.1093/hmg/ddr212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristiansen LV, Beneyto M, Haroutunian V, Meador-Woodruff JH. Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Mol Psychiatry. 2006;11: 737–747. doi: 10.1038/sj.mp.4001844 [DOI] [PubMed] [Google Scholar]
- 14.Funk AJ, Rumbaugh G, Harotunian V, McCullumsmith RE, Meador-Woodruff JH. Decreased expression of NMDA receptor-associated proteins in frontal cortex of elderly patients with schizophrenia. NeuroReport. 2009;20: 1019–1022. doi: 10.1097/WNR.0b013e32832d30d9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler D, Daher F, Wüstefeld L, Hammerschmidt K, Poggi G, Seelbach A, et al. Hypersocial behavior and biological redundancy in mice with reduced expression of PSD95 or PSD93. Behavioural Brain Research. Elsevier B.V; 2017;: 1–11. doi: 10.1016/j.bbr.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 16.Feyder M, Karlsson R-M, Mathur P, Lyman M, Bock R, Momenan R, et al. Association of Mouse Dlg4(PSD-95) Gene Deletion and Human DLG4Gene Variation With Phenotypes Relevant to Autism Spectrum Disorders and Williams' Syndrome. AJP. 2010;167: 1508–1517. doi: 10.1176/appi.ajp.2010.10040484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bats C, Groc L, Choquet D. The Interaction between Stargazin and PSD-95 Regulates AMPA Receptor Surface Trafficking. Neuron. 2007;53: 719–734. doi: 10.1016/j.neuron.2007.01.030 [DOI] [PubMed] [Google Scholar]
- 18.Mondin M, Labrousse V, Hosy E, Heine M, Tessier B, Levet F, et al. Neurexin-Neuroligin Adhesions Capture Surface-Diffusing AMPA Receptors through PSD-95 Scaffolds. Journal of Neuroscience. 2011;31: 13500–13515. doi: 10.1523/JNEUROSCI.6439-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannone G, Mondin M, Grillo-Bosch D, Tessier B, Saint-Michel E, Czöndör K, et al. Neurexin-1β Binding to Neuroligin-1 Triggers the Preferential Recruitment of PSD-95 versus Gephyrin through Tyrosine Phosphorylation of Neuroligin-1. CellReports. The Authors; 2013;3: 1996–2007. doi: 10.1016/j.celrep.2013.05.013 [DOI] [PubMed] [Google Scholar]
- 20.Cuthbert PC, Stanford LE, Coba MP, Ainge JA, Fink AE, Opazo P, et al. Synapse-Associated Protein 102/dlgh3 Couples the NMDA Receptor to Specific Plasticity Pathways and Learning Strategies. Journal of Neuroscience. 2007;27: 2673–2682. doi: 10.1523/JNEUROSCI.4457-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uezato A, Kimura-Sato J, Yamamoto N, Iijima Y, Kunugi H, Nishikawa T. Further evidence for a male-selective genetic association of synapse-associated protein 97 (SAP97) gene with schizophrenia. Behavioral and Brain Functions. BioMed Central Ltd; 2012;8: 2 doi: 10.1186/1744-9081-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uezato A, Yamamoto N, Jitoku D, Haramo E, Hiraaki E, Iwayama Y, et al. Genetic and molecular risk factors within the newly identified primate-specific exon of the SAP97/DLG1gene in the 3q29 schizophrenia-associated locus. Am J Med Genet. 2017;174: 798–807. doi: 10.1002/ajmg.b.32595 [DOI] [PubMed] [Google Scholar]
- 23.Quintero-Rivera F, Sharifi-Hannauer P, Martinez-Agosto JA. Autistic and psychiatric findings associated with the 3q29 microdeletion syndrome: Case report and review. Am J Med Genet. 2010;152A: 2459–2467. doi: 10.1002/ajmg.a.33573 [DOI] [PubMed] [Google Scholar]
- 24.Mulle JG. The 3q29 deletion confers >40-fold increase in risk for schizophrenia Nature Publishing Group; 2015;20: 1028–1029. doi: 10.1038/mp.2015.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyooka K, Iritani S, Makifuchi T, Shirakawa O, Kitamura N, Maeda K, et al. Selective reduction of a PDZ protein, SAP-97, in the prefrontal cortex of patients with chronic schizophrenia. 2002;: 1–10. [DOI] [PubMed] [Google Scholar]
- 26.Zhou W, Zhang L, Guoxiang X, Mojsilovic-Petrovic J, Takamaya K, Sattler R, et al. GluR1 Controls Dendrite Growth through Its Binding Partner, SAP97. Journal of Neuroscience. 2008;28: 10220–10233. doi: 10.1523/JNEUROSCI.3434-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Jablonski AM, Mojsilovic-Petrovic J, Ding H, Seeholzer S, Newton IP, et al. SAP97 Binding Partner CRIPT Promotes Dendrite Growth in Vitroand in Vivo. eNeuro. 2017;: ENEURO.0175–17.2017–61. doi: 10.1523/ENEURO.0175-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caruana G, Bernstein A. Craniofacial Dysmorphogenesis Including Cleft Palate in Mice with an Insertional Mutation in the discs large Gene. Molecular and Cellular Biology. 2001;21: 1475–1483. doi: 10.1128/MCB.21.5.1475-1483.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David S, Kalb RG. Serum/glucocorticoid-inducible kinase can phosphorylate the cyclic AMP response element binding protein, CREB. FEBS Letters. 2005;579: 1534–1538. doi: 10.1016/j.febslet.2005.01.040 [DOI] [PubMed] [Google Scholar]
- 30.Kim C-H, Takamiya K, Petralia RS, Sattler R, Yu S, Zhou W, et al. Persistent hippocampal CA1 LTP in mice lacking the C-terminal PDZ ligand of GluR1. Nat Neurosci. 2005;8: 985–987. doi: 10.1038/nn1432 [DOI] [PubMed] [Google Scholar]
- 31.Mojsilovic-Petrovic J. Protecting Motor Neurons from Toxic Insult by Antagonism of Adenosine A2a and Trk Receptors. Journal of Neuroscience. 2006;26: 9250–9263. doi: 10.1523/JNEUROSCI.1856-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 33.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29: 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43: e47–e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. 2014;: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel S, Roncaglia P, Lovering RC. Using Gene Ontology to describe the role of the neurexin-neuroligin-SHANK complex in human, mouse and rat and its relevance to autism. BMC Bioinformatics. BMC Bioinformatics; 2015;: 1–18. doi: 10.1186/s12859-014-0430-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uezato A, Yamamoto N, Iwayama Y, Hiraoka S, Hiraaki E, Umino A, et al. Reduced cortical expression of a newly identified splicing variant of the DLG1 gene in patients with early-onset schizophrenia Nature Publishing Group; 2015;5: e654–8. doi: 10.1038/tp.2015.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y-H, Ehlers MD. Modeling Autism by SHANK Gene Mutations in Mice. Neuron. Elsevier Inc; 2013;78: 8–27. doi: 10.1016/j.neuron.2013.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. 1998;: 1–7. [DOI] [PubMed] [Google Scholar]
- 40.Waites CL, Specht CG, Hartel K, Leal-Ortiz S, Genoux D, Li D, et al. Synaptic SAP97 Isoforms Regulate AMPA Receptor Dynamics and Access to Presynaptic Glutamate. Journal of Neuroscience. 2009;29: 4332–4345. doi: 10.1523/JNEUROSCI.4431-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allswede DM, Zheutlin AB, Chung Y, Anderson K, Hultman CM, Ingvar M, et al. Complement Gene Expression Correlates with Superior Frontal Cortical Thickness in Humans Nature Publishing Group; Nature Publishing Group; 2017;: 1–9. doi: 10.1038/npp.2017.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O'Donovan MC, Buckland PR. Functional analysis of polymorphisms in the promoter regions of genes on 22q11. Hum Mutat. 2004;24: 35–42. doi: 10.1002/humu.20061 [DOI] [PubMed] [Google Scholar]
- 43.Madani R, Kozlov S, Akhmedov A, Cinelli P, Kinter J, Lipp H-P, et al. Impaired explorative behavior and neophobia in genetically modified mice lacking or overexpressing the extracellular serine protease inhibitor neuroserpin. Molecular and Cellular Neuroscience. 2003;23: 473–494. doi: 10.1016/S1044-7431(03)00077-0 [DOI] [PubMed] [Google Scholar]
- 44.Saetre P, Emilsson L, Axelsson E, Kreuger J, Lindholm E, Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7: 551–10. doi: 10.1186/1471-244X-7-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang X, Liu Y, Hahn C-G, Gur RE, Sleiman PMA, Hakonarson H. RNA-seq analysis of amygdala tissue reveals characteristic expression profiles in schizophrenia Nature Publishing Group; Nature Publishing Group; 2017;7: 1–8. doi: 10.1038/tp.2017.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reumann R, Vierk R, Zhou L, Gries F, Kraus V, Mienert J, et al. The serine protease inhibitor neuroserpin is required for normal synaptic plasticity and regulates learning and social behavior. Learn Mem. 2017;24: 650–659. doi: 10.1101/lm.045864.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borges VM, Lee TW, Christie DL, Birch NP. Neuroserpin regulates the density of dendritic protrusions and dendritic spine shape in cultured hippocampal neurons. J Neurosci Res. 2010;88: n/a–n/a. doi: 10.1002/jnr.22428 [DOI] [PubMed] [Google Scholar]
- 48.Song X, Fan X, Li X, Kennedy D, Pang L, Quan M, et al. Serum levels of BDNF, folate and homocysteine: In relation to hippocampal volume and psychopathology in drug naïve, first episode schizophrenia. Schizophrenia Research. Elsevier B.V; 2014;159: 51–55. doi: 10.1016/j.schres.2014.07.033 [DOI] [PubMed] [Google Scholar]
- 49.Halacheva K, Dimova S, Tolev T, Dimov D, Nikolova M. Elevated anticardiolipin antibodies in schizophrenic patients before and during neuroleptic medication. Psychiatry Research. Elsevier Ltd; 2009;169: 51–55. doi: 10.1016/j.psychres.2008.05.011 [DOI] [PubMed] [Google Scholar]
- 50.Gris J-C, Nobile B, Bouvier S. Neuropsychiatric presentations of antiphospholipid antibodies. Revista Brasileira de Hematologia e Hemoterapial. Elsevier Masson SAS; 2015;135: S56–S59. doi: 10.1016/S0049-3848(15)50445-3 [DOI] [PubMed] [Google Scholar]
- 51.Delluc A, Rousseau A, Le Galudec M, Canceil O, Woodhams B, Etienne S, et al. Prevalence of antiphospholipid antibodies in psychiatric patients users and non-users of antipsychotics. Br J Haematol. 2013;164: 272–279. doi: 10.1111/bjh.12627 [DOI] [PubMed] [Google Scholar]
- 52.Hoirisch-Clapauch S, Amaral OB, Mezzasalma MAU, Panizzutti R, Nardi AE. Dysfunction in the coagulation system and schizophrenia Nature Publishing Group; 2015;6: e704–8. doi: 10.1038/tp.2015.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schizophrenia Working Group of the Psychiatric Genomics Consortium, Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530: 177–183. doi: 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rezende SM. Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex. Blood. 2003;103: 1192–1201. doi: 10.1182/blood-2003-05-1551 [DOI] [PubMed] [Google Scholar]
- 55.Kokras N, Dalla C. Sex differences in animal models of psychiatric disorders. Br J Pharmacol. 2014;171: 1–25. doi: 10.1111/bph.12291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittchen HU, Jacobi F, Rehm J, Gustavsson A, Svensson M, Jönsson B, et al. The size and burden of mental disorders and other disorders of the brain in Europe 2010. European Neuropsychopharmacology. Elsevier B.V; 2011;21: 655–679. doi: 10.1016/j.euroneuro.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 57.Kessler RC. The global burden of anxiety and mood disorders: Putting ESEMeD findings into perspective. J Clin Psychiatry. 2008;: 1–19. [PMC free article] [PubMed] [Google Scholar]
- 58.Häfner H. Gender differences in schizophrenia. Psychoneuroendocrinology. 2003;28: 17–54. doi: 10.1016/S0306-4530(02)00125-7 [DOI] [PubMed] [Google Scholar]
- 59.Fombonne E. The Prevalence of Autism. JAMA 2003;: 1–3. [DOI] [PubMed] [Google Scholar]
- 60.Kulkarni J, Gavrilidis E, Worsley R, Van Rheenen T, Hayes E. The Role of Estrogen in the Treatment of Men with Schizophrenia. Int J Endocrinol Metab. 2013;11: 1–8. doi: 10.5812/ijem.7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the figshare database (DOI 10.6084/m9.figshare.5942476; https://figshare.com/s/00d5f26e315c997fddff).