Abstract
Objectives:
The method of colorectal cancer (CRC) tissue sampling would affect its molecular profile and the downstream processing. In this study, we described the impact of CRC tissue sampling procedures on the quality control (QC) metrics of cytoscan HD array.
Methods:
We employed a high-resolution cytoscan HD microarray platform to investigate the chromosomal aberrations that could be associated with CRC. We compared the tissue extraction procedures and their impact on the QC parameters from the cytoscan HD array determined by chromosome analysis software (Suite3.1). Median of absolute values of all pairwise differences (MAPD), waviness-standard deviation (Waviness-SD), and single nucleotide polymorphism QC (SNPQC) were the QC parameters that were analyzed.
Results:
From 67 patients, we collected 843 colorectal tissues. Of these, 65.7% were obtained through endoscopic procedures, and the rest was after surgical resections. The mean transit time between tissue excision and preservation was 26 ± 15.5 and 74.6 ± 24.8 min, respectively. The tissues extracted from the surgical procedure showed mean MAPD of 0.28 ± 0.06 compared to 0.24 ± 0.06, for endoscopy, P = 0.005, degree of waviness-SD of 0.20 ± 0.1 compared to 0.2 ± 0.1, P = 0.64, and SNPQC of 9.6 ± 4.2 compared to 11.1 ± 4.6, P = 0.23.
Conclusions:
This report provides objective results that can help in tissue sampling intended to be used for DNA based molecular studies. Tissue collection protocol should be optimized to support microarray-analysis methods. Tissue extraction from endoscopic procedures had faster transit time and relatively better quality metrics outcome than surgical procedures. However, surgical procedures have less refusal rate, higher tissue quantity, and less negative results for malignancy.
Keywords: Colorectal cancer, cytogenetic, quality control metrics, sampling procedure, tissue collection
Introduction
Colorectal cancer (CRC) is the second most common cancer in female and the third most common cancer among male worldwide.[1] It was estimated that around 1.4 million people were diagnosed with CRC in 2012, which represents 10% of all total cancers, and 693,900 estimated deaths from this disease in the same year.[1] In the United States, CRC is the third most common cancer in both genders with estimated 8% incidence.[2] The estimated cases of CRC in 2016 was 134,490, and around 49,190 deaths were reported.[2] In the Kingdom of Saudi Arabia (KSA), the incidence of CRC is found to be increasing,[3,4] with overall survival of 44.6%.[4] It is the most common cancer among male and the third most common among female in KSA.[4]
Development and progression of CRC are a multistep process and may be related to genetic alterations, which involve loss/gain of chromosomes, loss of heterozygosity and uniparental disomy.[5,6] Studying CRC tissue samples from patients are a vital need to find out clinically relevant diagnostic, therapeutic, prognostic, and predictive markers of CRC. These studies are gaining prominence with the mounting evidence about the role of genetic factors in the development of CRC.[7]
The most common source of CRC tissue for experimental research is from patients following diagnostic or therapeutic procedures through endoscopy or surgical resections. The methods of tissue extraction and processing would affect its molecular profile, which in turn could affect the downstream processing.[8] To minimize tissue desiccation, contamination and to prevent biochemical, structural and environmental changes that could affect the integrity of extracted nucleic acids, it is necessary to optimize tissue collection protocol that suits the need of scientific experiments. The proper handling of tissue samples is crucial for getting reliable results from downstream applications. These tissues should be resected cut, submerged, transported, stored, thawed and analyzed in a proper way using appropriate and specific equipment and fixation/storage reagents. These aspects of sample collection may affect the results generated using these tissues. Hence, there is a need to study the effect of the tissue sampling methods on the results emanating from the downstream processes like cytogenetic studies.[9] Cytogenetic studies using microarray technology relies on the purity and quality of genomic DNA extracted from the tissue which in turn may get affected by the sampling and storage protocol. Cancer samples are known to have a complex genomic structure due to major changes in the chromosomal composition. The objective of this study is to describe the impact of CRC sample collection procedures on the quality control (QC) metrics of a cytogenetic microarray-analysis based study. This study will pave the way for benchmarking the QC metrics for CRC samples obtained from biopsies and surgical resections and will be helpful in designing and standardizing the sample collection procedures in future studies.
Methods
In the current study, we compared the colorectal tissue sampling methods through two different approaches (endoscopic procedure and surgical resection). We analyzed the impact of sample collection procedures on the QC metrics of cytoscan HD microarray.
In addition, we described the sample collection process (tissue contamination, resection, fixation, handling, transporting, storage, and extraction of nucleic acids), number of extracted tissues, histopathology results, the used fixation/storage reagents, the transit time, the cycle time, informed consent process and patients’ recruitment rate.
“Transit time” is defined as the time from tissue extraction to the time of completed tissue fixation procedures; while “cycle time” refers to the time of arrival to the sample extraction site (endoscopy unit or operation theater) until departure.
Setting
The study was conducted in a 1000-bed teaching hospital in Riyadh, KSA. The hospital carries the Joint Commission International and the College of American Pathology accreditations. Samples were collected through endoscopic procedures in the endoscopy unit and after surgical resections in the operation theater. The samples were processed at the central anatomic pathology laboratory in the hospital and the cancer research laboratory at King Abdullah International Medical Research Center located in King Abdul Aziz Medical City (KAMC), Riyadh, KSA. The study was approved by the Institutional Review Board of KAMC.
Sample size
This study aimed to collect 80 sets of samples from patients who are diagnosed with or suspected to have CRC. Each sample will necessarily comprise a set of colorectal neoplastic and adjacent normal tissues.
Inclusion and exclusion criteria
Adult patients with or suspected to have a clinical diagnosis of CRC were included in the study. Patients who had received prior radiation or chemotherapy were excluded.
Sample collection process
After confirming the eligible patients through convenient sampling technique, a team member (surgeon or gastroenterologist) discusses the purpose and procedure of the study to the patient. The informed consent form (ICF) was signed by each patient before starting the procedure and sample collection.
The normal and neoplastic tissues were identified and resected from patients by a gastroenterologist in the endoscopy unit or by the gastroenterology surgeon in the operation theater. Subsequently, the proper size of required tissue (0.5 × 0.5 × 0.5 cm or smaller) was determined and cut by trained study team. The tissues were submerged into appropriate fixation reagent according to the priority of tissue collection in the following order: 1-RNA stabilizing reagent (RNAlater) 2-flash freezing in liquid nitrogen 3-Optimal Cutting Temperature (OCT) compound embedded sample (for cryosectioning) 4-formalin fixation. Multiple samples were collected from patients as appropriate for greater use of tissues. This collection pattern is a modified form of the standard operating procedures for biospecimen collection from National Cancer Institutes (NCI), National Institute of Health, USA.[10]
Tissues contamination
To avoid cross-contamination between normal and neoplastic tissues, separated sterile suture sets were used to cut and submerge the tissues. All normal and neoplastic samples were packed and sealed separately in hazard material bags.
Storage temperatures
Cold preservation is crucial as a stabilizing factor for integrity of the tissues. For the purpose of this study, samples were stored under the following temperatures:
The tissues that were fixed in RNA stabilizing reagent were stored at 4°C overnight and then at −20°C for long time storage. Cryovials were kept immediately in Dewar containing liquid nitrogen and then stored in a cryogenic storage vessel at or below −150°C. The submerged tissues on cryomold filled with OCT were placed inside a tightly closed Petri dish and place it on the mouth of liquid nitrogen transport Dewar for 1 min to let it solidify, and then store at −80°C. Tissues fixed in formalin were stored at room temperature.
Time of sample collection and processing
The tissues were submerged immediately into storage reagent after excision and cutting. All samples were sent immediately to the molecular and pathology lab for storing. The transit and cycle time was calculated.
Extraction of nucleic acids
All samples stored in RNAlater were used for extracting DNA and RNA in a single protocol as described before.[11] Briefly, Macherey-Nagel Tri Prep kit (Germany) was used to extract DNA followed by RNA from a maximum of 30 mg samples obtained from endoscopic biopsies or surgical resections. Homogenization was carried out by Tissuelyzer (Qiagen, Cat#85600) using 5 mm stainless steel beads (Qiagen, Cat#69989). Purified DNA and RNA were checked for purity and quantity using Nanodrop (Thermo scientific).
Cytoscan HD Microarray studies
DNA extracted from these tissues were used to carry out the cytoscan HD microarray analysis (Affymetrix) using the protocol as described before.[11,12] A maximum of eight samples were processed in a batch. Samples from neoplastic-normal pairs from the same patient were run in the same batch to avoid batch effect within the patient comparison.
QC metrics
For QC metrics, we used the parameters from the cytoscan HD array as determined by chromosome analysis software (Suite3.1) available from Affymetrix.[13] The software reports three QC metrics to evaluate the quality of the generated data: Median of the absolute values of all pairwise differences (MAPD), waviness standard deviation (SD), and single nucleotide polymorphism QC (SNPQC). The MAPD is a per chip estimate of variability to assess the variation in microarray probes across the genome. It represents the median of the distribution of changes in log2 ratio between adjacent probes. The waviness-SD measures genome-wide long-range variation of probes. The SNPQC estimates the distributions of homozygous and heterozygous alleles and calculates the distance between them.[14]
These QC metrics represent measurements of signal-to-noise, which have a direct association to the functional resolution, sensitivity, and specificity which in turn is directly associated with quality of PCR product.[14] The higher number of SNPQC and the lower number of MAPD and waviness-SD represent better quality results.
Statistical analyses
Continuous variables were described as the mean and SD. Categorical variables were expressed as absolute and relative frequencies. Computations were performed by IBM SPSS Statistics V23 (IBM Corp., Armonk, NY).
The test of normality using skewness coefficient indicated that MAPD, Waviness-SD and SNPQC were not normally distributed. Therefore, the Mann–Whitney test was used as a test of significance to compare the impact of sample collection methods on the QC metrics. P < 0.05 was considered as significant.
Results
Samples characteristics
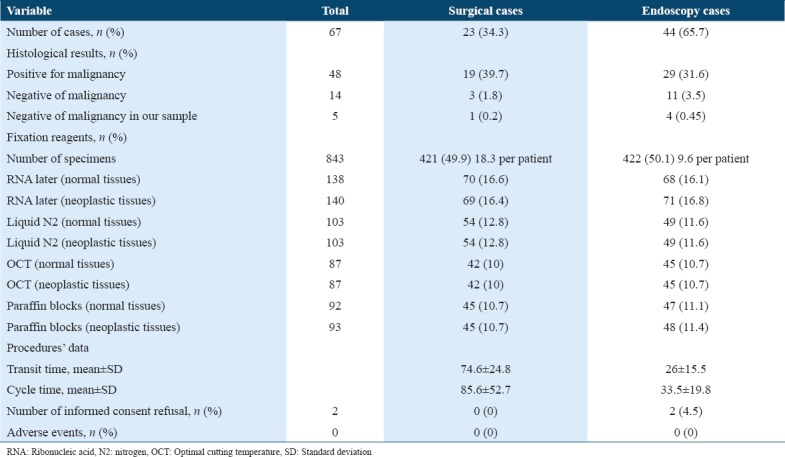
A total of 74 patients were screened to be a part of this study. Of these, 5 patients were excluded because they received chemotherapy or radiotherapy and 2 cases refused to sign the ICF. A total of 67 patients were included in this study, in which 843 colorectal tissues were collected. Of these 420 samples were from normal colorectal tissues and 423 from neoplastic/suspected neoplastic colorectal tissues.
Majority of sample collection was through endoscopic procedures (n = 44) (65.7%). From all cases, 48 (71.6%) showed positive results for malignancy, 14 (20.9%) cases were negative for malignancy, and 5 (7.5%) cases were negative for malignancy in our samples despite the malignancy being confirmed in patients’ diagnostic samples. Majority of negative cases came from the endoscopic procedures.
As presented in Table 1, more patients were consented in the endoscopy unit for biopsies. However, due to larger tissue mass the numbers of samples collected through surgical resection in the operation theater were more than the endoscopic procedures (18 tissues per patient vs. 10 tissues per patient, respectively). The transit time for sample collection was longer in the operation theater 74.6 ± 24.8.
Table 1.
Sample characteristics
Comparison of cytoscan HD microarray quality metrics shows higher short-range noise in surgical resections
As shown in Table 2, the SNPQC reading was higher in the tissues collected after endoscopic procedures (11.1 ± 4.6) than the surgical resections (9.6 ± 4.2), with no significant statistical difference, P = 0.23. The MAPD was lower in the tissues collected after endoscopic procedure (0.24 ± 0.06 vs. 0.28 ± 0.06 for surgical resections, P = 0.005, respectively). The Waviness-SD was similar in both collected tissues after endoscopic procedures and surgical resections 0.2 ± 0.1.
Table 2.
Quality metrics results
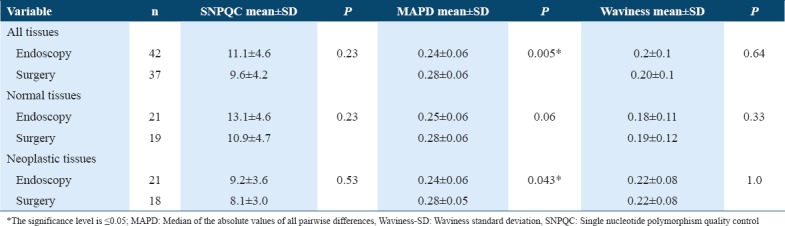
Similar results were revealed when we compared the QC metrics for the neoplastic tissues alone. The MAPD was lower in the tissues collected after endoscopic procedures compared with the surgical resections 0.24 ± 0.06 and 0.28 ± 0.05, respectively, P = 0.043. However, no statistically significant difference observed for the normal tissue collected from the endoscopic procedures when compared with the normal tissue collected from surgical resections [Table 2].
QC metrics based on empirical dataset may not affect the biological interpretation of data from cytoscan HD array
The values of the QC metrics described by Affymetrix are derived from an empirical data set. Our values for these QC metrics show a wide range as described in Table 3. We applied publicly available bioinformatics tools like patchwork to analyze our data and the information obtained was biologically relevant. Despite QC metrics below the recommended values, we could detect chromosomal aberrations. As shown in Figure 1a (normal) and 1b (tumor) representing one of the samples sets there is an observed uniparental disomy/loss of heterozygosity in chromosome 4 and 10. The data obtained from the cytoscan HD platform were used to identify novel genes associated with CRC in the Saudi population.[11,15]
Table 3.
Quality metrics values
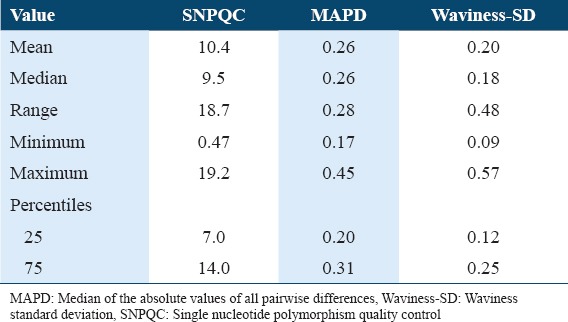
Figure 1.
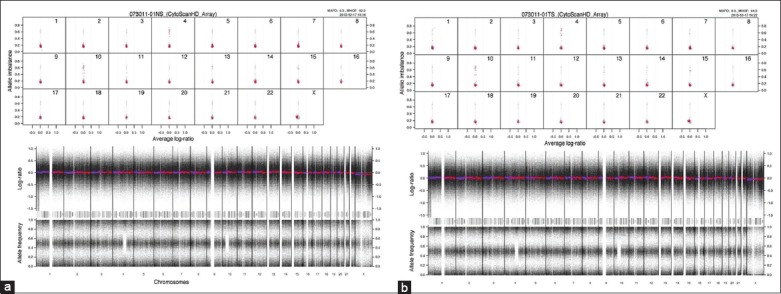
Allele frequency pattern of one set of representative samples from a patient. Normal (a) and tumor samples (b) surgically resected from a patient are depicted here. Bottom panel represents allelic frequency and log ratio of the complete genotype of the sample. Top panel shows inferred allelic imbalance in chromosome 4 and 10
Discussion
This study aimed to analyze the sample collection procedure for CRC patients and its effect on the quality metrics of a downstream application. In this report, we focused on cytoscan HD array. This array has been used for cytogenetic analyses to detect copy number variations, loss of heterozygosity and uniparental disomy.
Our observations indicate high compliance of Saudi patients to participate in studies involving genomic analyses. Only 2 (3%) patients refused to participate in this study. This value is much lower than that reported in previous national studies.[16,17] This result suggests that including a larger sample size of Saudi subjects in genomic studies could be feasible.
To obtain good quality nucleic acids and minimize the potential damage of the samples for microarray and similar studies, we modified the specimen collection criteria of NCI standard operating procedures for biospecimen collection. RNA later samples were our first priority due to better stability of the samples at room temperature and proven integrity of the extracted nucleic acid.[18-21] The usual practice in the endoscopy and operation theater is to store the samples in the formalin for pathological examination.[8] This is a standard procedure for pathological analyses but would be detrimental for the integrity of nucleic acids. We, therefore, adopted the storage scheme that utilized RNA later (stabilizing agent)[18-21] along with flash freezing as storage methods.
Studies that compare the feasibility, procedures and QC metrics of colorectal sampling methods through endoscopy or surgical resection are limited. In this study, sampling through the surgical resections allowed collection of more quantities of neoplastic and normal tissues than the endoscopic procedures without any negative impact on patients’ health status as there was no adverse event reported. Unlike the tissues taken from surgically resected masses, usually, the tissues extracted from endoscopic procedures are limited for diagnostic purposes. This reduced the chances of getting more quantities of tissue for research purposes. In addition, sampling through surgical resections would provide more accurate and confirmed malignant tissues. This is reflected in our results where more negative samples were from endoscopy than from surgical resections. This could be due to established criteria for classifying a sample to be tumor, which necessitates at least 60% cells to be cancerous.[10] From the consent rate, we can suggest that patients undergoing surgical resections were more willing to participate in research studies that patients undergoing endoscopy. This is expected since patients undergoing endoscopic procedure may feel anxiety, fear of pain or bleeding as potential complications due to extra tissues for research purposes;[22] however, the overall refusal rate was very low in this study.
We used QC metrics parameters from the cytoscan HD array determined by chromosome analysis software (Suite3.1). This program used an external reference set derived from phenotypically healthy samples to be used as a baseline. Neoplastic colorectal samples may introduce different sources of variability relative to the default reference set included in the software. The existing literature that determines the QC metrics to measure the cutoffs for colorectal tissues is lacking. Hence, the available quality metrics thresholds may not be suitable for tissues from CRC patients. In this study, we provide evidence that SNPQC, MAPD, and Waviness-SD values could range beyond recommended values without compromising the accuracy of the obtained information. Thus, the effect of sampling method is reflected in the MAPD values only irrespective of the nature of the sample. However, the genetic composition for tumor and normal cells is significantly different.[23] This difference can cause a shift in the QC metric values.
Several factors may affect the quality of the sample tissue,[9,24-26] the reasons behind the MAPD variation between sampling collection methods could not be definite. However, the large difference in the transit time between these sampling methods might contribute to this variation.[9] The transit time of sample collection is an important factor to get high-quality samples.[9] Tissue ischemia, delay of tissue fixation and freezing may influence the integrity of the molecular features.[9,24-26]
Taken together, the results suggest endoscopy to be a better procedure for extraction as reflected in the MAPD values which are significantly different irrespective of the nature of the sample. More studies are needed to ascertain the exact details of this variation.
To our knowledge, this is the first study that comprehensively describes the impact of sampling methods on the QC metrics of a cytogenetic microarray-analysis based study. Our evidence suggests that QC cutoff values should be decided based on observed copy number changes as reflected in allelic imbalance. However, the study has potential limitations; the study was conducted in a single center, relatively small sample size, and the analysis was limited only to the tissues stored in RNA later.
Conclusion
Colorectal tissue collection method protocol should be optimized to suit the downstream analyses. Sampling through endoscopic procedures had faster transit time and relatively better quality metrics outcomes than colorectal tissue sampling through surgical resections. However, colorectal tissue samplings through surgical resections have less refusal rate, higher tissue quantity, and less negative results for malignancy. The QC metrics determined by Affymetrix are not binding for inferring the cytogenetic information required from microarray-based studies. Further analysis is needed to determine the factors affecting the quality of tissue in both approaches; including comparing the molecular profile of fresh frozen colorectal tissues with the same tissues fixed in other fixation reagents.
Acknowledgment
This study was supported by research grant RC10/083 awarded to MAA by King Abdullah International Medical Research Center.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Mosli MH, Al-Ahwal MS. Colorectal cancer in the kingdom of saudi arabia:Need for screening. Asian Pac J Cancer Prev. 2012;13:3809–13. doi: 10.7314/apjcp.2012.13.8.3809. [DOI] [PubMed] [Google Scholar]
- 4.Alsanea N, Abduljabbar AS, Alhomoud S, Ashari LH, Hibbert D, Bazarbashi S, et al. Colorectal cancer in saudi arabia:Incidence, survival, demographics and implications for national policies. Ann Saudi Med. 2015;35:196–202. doi: 10.5144/0256-4947.2015.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen CL, Wiuf C, Kruhøffer M, Korsgaard M, Laurberg S, Ørntoft TF, et al. Frequent occurrence of uniparental disomy in colorectal cancer. Carcinogenesis. 2007;28:38–48. doi: 10.1093/carcin/bgl086. [DOI] [PubMed] [Google Scholar]
- 6.Loo LW, Tiirikainen M, Cheng I, Lum-Jones A, Seifried A, Church JM, et al. Integrated analysis of genome-wide copy number alterations and gene expression in microsatellite stable, cpG island methylator phenotype-negative colon cancer. Genes Chromosomes Cancer. 2013;52:450–66. doi: 10.1002/gcc.22043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valle L. Genetic predisposition to colorectal cancer:Where we stand and future perspectives. World J Gastroenterol. 2014;20:9828–49. doi: 10.3748/wjg.v20.i29.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werner M, Chott A, Fabiano A, Battifora H. Effect of formalin tissue fixation and processing on immunohistochemistry. Am J Surg Pathol. 2000;24:1016–9. doi: 10.1097/00000478-200007000-00014. [DOI] [PubMed] [Google Scholar]
- 9.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–71. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NIH. Fresh Frozen Tissue Protocol-Appendices. [[Last cited on 2017 Oct 18]]. Available from: https://www.epi.grants.cancer.gov/CFR/bio-fresh-tissue-app.html .
- 11.Eldai H, Periyasamy S, Al Qarni S, Al Rodayyan M, Muhammed Mustafa S, Deeb A, et al. Novel genes associated with colorectal cancer are revealed by high resolution cytogenetic analysis in a patient specific manner. PLoS One. 2013;8:e76251. doi: 10.1371/journal.pone.0076251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aziz MA, Periyasamy S, Al Yousef Z, AlAbdulkarim I, Al Otaibi M, Alfahed A, et al. Integrated exon level expression analysis of driver genes explain their role in colorectal cancer. PLoS One. 2014;9:e110134. doi: 10.1371/journal.pone.0110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Affymetrix. Chromosome Analysis Suite (ChAS) Software-Support Materials. 2015. [[Last cited on 2017 Oct 09]]. https://www.affymetrix.com/support/technical/byproduct.affx?product=chas .
- 14.Affymetrix. Unlocking Formalin-Fixed Paraffin-Embedded (FFPE) Samples with CytoScan®Cytogenetics Solution. 2013. https://tools.thermofisher.com/content/sfs/brochures/cytoscan_hd_solution_ffpe_appnote.pdf .
- 15.Aziz MA, Periyasamy S, Yousef Z, Deeb A, AlOtaibi M. Colorectal cancer driver genes identified by patient specific comparison of cytogenetic microarray. Genom Data. 2014;2:29–31. doi: 10.1016/j.gdata.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Jumah M, Abolfotouh MA, Alabdulkareem IB, Balkhy HH, Al-Jeraisy MI, Al-Swaid AF, et al. Public attitude towards biomedical research at outpatient clinics of king abdulaziz medical city, riyadh, saudi arabia. East Mediterr Health J. 2011;17:536–45. [PubMed] [Google Scholar]
- 17.Al-Qadire MM, Hammami MM, Abdulhameed HM, Al Gaai EA. Saudi views on consenting for research on medical records and leftover tissue samples. BMC Med Ethics. 2010;11:18. doi: 10.1186/1472-6939-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dekairelle AF, Van der Vorst S, Tombal B, Gala JL. Preservation of RNA for functional analysis of separated alleles in yeast:Comparison of snap-frozen and RNA Later solid tissue storage methods. Clin Chem Lab Med. 2007;45:1283–7. doi: 10.1515/CCLM.2007.281. [DOI] [PubMed] [Google Scholar]
- 19.Mutter GL, Zahrieh D, Liu C, Neuberg D, Finkelstein D, Baker HE, et al. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics. 2004;5:88. doi: 10.1186/1471-2164-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez GE, Mondala TS, Head SR. Assessing a novel room-temperature RNA storage medium for compatibility in microarray gene expression analysis. Biotechniques. 2009;47:667–8, 670. doi: 10.2144/000113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatzis C, Sun H, Yao H, Hubbard RE, Meric-Bernstam F, Babiera GV, et al. Effects of tissue handling on RNA integrity and microarray measurements from resected breast cancers. J Natl Cancer Inst. 2011;103:1871–83. doi: 10.1093/jnci/djr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greiner KA, Born W, Nollen N, Ahluwalia JS. Knowledge and perceptions of colorectal cancer screening among urban african americans. J Gen Intern Med. 2005;20:977–83. doi: 10.1111/j.1525-1497.2005.00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palumbo E, Russo A. Chromosome imbalances in cancer:Molecular cytogenetics meets genomics. Cytogenet Genome Res. 2016;150:176–84. doi: 10.1159/000455804. [DOI] [PubMed] [Google Scholar]
- 24.Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, et al. Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5 nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn. 2000;2:84–91. doi: 10.1016/S1525-1578(10)60621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visvikis S, Schlenck A, Maurice M. DNA extraction and stability for epidemiological studies. Clin Chem Lab Med. 1998;36:551–5. doi: 10.1515/CCLM.1998.094. [DOI] [PubMed] [Google Scholar]
- 26.Berger A, Bruschek M, Grethen C, Sperl W, Kofler B. Poor storage and handling of tissue mimics mitochondrial DNA depletion. Diagn Mol Pathol. 2001;10:55–9. doi: 10.1097/00019606-200103000-00009. [DOI] [PubMed] [Google Scholar]


