Abstract
Anti-vascular endothelial growth factor (VEGF) therapy has demonstrated efficacy in treating human metastatic cancers, but therapeutic resistance is a practical limitation and most tumors eventually become unresponsive. To identify microenvironmental factors underlying the resistance of cancer to antiangiogenesis therapy, we conducted genomic analyses of intraperitoneal ovarian tumors in which adaptive resistance to anti-VEGF therapy (B20 antibody) developed. We found that expression of the microseminoprotein, prostate-associated (MSMP) gene was substantially upregulated in resistant compared with control tumors. MSMP secretion from cancer cells was induced by hypoxia, triggering MAPK signaling in endothelial cells to promote tube formation in vitro. Recruitment of the transcriptional repressor CCCTC-binding factor (CTCF) to the MSMP enhancer region was decreased by histone acetylation under hypoxic conditions in cancer cells. MSMP siRNA, delivered in vivo using the DOPC nanoliposomes, restored tumor sensitivity to anti-VEGF therapy. In ovarian cancer patients treated with bevacizumab, serum MSMP concentration increased significantly only in non-responders. These findings imply that MSMP inhibition combined with the use of antiangiogenesis drugs may be a new strategy to overcome resistance to antiangiogenesis therapy.
Keywords: MSMP, CCR2, anti-VEGF antibody, human ovarian cancer, bevacizumab, adaptive resistance
Introduction
Angiogenesis is an essential step in tumor growth and metastasis.1 Successful inhibition of human tumor xenograft growth by a monoclonal antibody specific for vascular endothelial growth factor (VEGF)2 has contributed to the development of antiangiogenesis therapy for cancer. In 2004, bevacizumab became the first anti-VEGF agent approved by the U.S. Food and Drug Administration for cancer patients. In phase 3 clinical trials, bevacizumab therapy has been effective for treatment of patients with metastatic ovarian,3, 4, 5 colorectal,6 lung,7 or renal8 cancers, and glioblastoma.9 However, improvement in clinical outcomes of cancer patients using this therapy has been modest. Despite initial response, most tumors became resistant to such drugs, and survival benefits for patients receiving antiangiogenesis therapy for gynecological malignancies are usually measured in months.
Microseminoprotein, prostate-associated (MSMP) is a highly conserved, secreted protein containing a signal peptide sequence that lacks the classical structure of chemokines and is strongly expressed in the hormone-insensitive prostate cancer cell line PC3. It is abundantly expressed in benign and malignant human prostate and breast tumors and generally expressed to a lesser extent in ovarian and lung tumors.10, 11, 12 Previous reports suggested that MSMP is a chemoattractant protein acting as a C–C chemokine receptor 2 (CCR2) ligand to recruit peripheral blood monocytes that may influence inflammation and cancer development,11 but its functions are incompletely understood.
Here, we searched for mechanisms involved in the emergence of tumor resistance to anti-VEGF antibody-based therapy. We found that MSMP expression is induced with prolonged anti-VEGF therapy, specifically under hypoxia, and has an important proangiogenic role in treatment-resistant ovarian tumors.
Results
MSMP expression in ovarian tumors is upregulated by anti-VEGF antibody therapy in vivo
To identify mechanisms of escape from antiangiogenesis therapy, we first carried out genomic analyses of ovarian tumors in which VEGF was blocked in vivo by treatment with the anti-VEGF antibody B20 (targets both murine and human VEGF). Specifically, we gave this antibody to mice with established intraperitoneal SKOV3ip1 human ovarian or IG10 murine ovarian tumors. Following initial response, we used bioluminescence imaging to identify tumors that grew despite treatment (adaptive resistance). We collected resistant tumors from the mice and isolated their RNA for gene expression studies. This analysis identified 17 genes with at least three-fold higher or 70% lower expression in B20-resistant tumors than in control tumors (Figure 1A). Chromogranin A (CHGA) and MSMP were the most upregulated genes (7.7- and 4.6-fold, respectively) in the resistant tumors and IGFBP1 (insulin like growth factor binding protein 1), SST (somatostatin), SLC40A1 (solute carrier family 40 member 1), AKR1C2 (aldo-keto reductase family 1 member C2) and BBOX1 (gamma-butyrobetaine hydroxylase 1) followed these two genes. To confirm these findings, we analyzed gene expression in the SKOV3ip1 and IG10 adaptive resistance models by using quantitative reverse transcriptase–PCR (qRT–PCR). Chromogranin A expression did not validate, but MSMP expression was increased significantly in the B20-resistant tumors (P=0.04) (Figure 1B) compared with the B20-sensitive tumors. IGFBP5, SST, SLC40A1, AKR1C2 and BBOX1 were also increased in SKOV3ip1 in the B20-resistant tumors compared with the B20-sensitive tumors (Supplementary Figure S1). To determine the localization of MSMP expression, we used immunofluorescence staining of ovarian tumors from the mice and found that MSMP was predominantly expressed in the cancer cell cytoplasm (P<0.01) and was absent from stromal cells (Figure 1C). These results suggested that cancer cells are the main source of MSMP in B20-resistant tumors.
Figure 1.
Anti-VEGF antibody therapy and hypoxia induce MSMP expression in ovarian tumors in vivo and in vitro. A, SKOV3ip1 models were established from athymic nude mice. Tumor-bearing mice were given the anti-VEGF antibody B20. The heat map shows the expression of 15 upregulated and 2 downregulated genes in B20-resistant SKOV3ip1 tumors compared with that in control tumors in biological triplicates. B, Real-time RT-PCR analysis of chromogranin A (CHGA) and MSMP expression in whole SKOV3ip1 and IG10 tumors in vivo. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. C, Immunohistochemical (IHC) and immunofluorescence (IF) analysis of MSMP expression in sections of B20-resistant and control ovarian tumors. Left: representative images of MSMP staining in SKOV3ip1 tumors and adjacent tissue (red, MSMP; green, CD31 in IF; magnification, ×20). Right: ratios of MSMP-positive cells per total cells per high-power field. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. D, Secreted MSMP levels in media conditioned with SKOV3ip1, OVCAR8, or RF24 cells under hypoxic conditions assessed using ELISA. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. E, Ratios of MSMP to B-actin expression in OVCAR8 and RF24 cells incubated under normoxic and hypoxic conditions. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. F, Real-time RT-PCR analysis of MSMP expression in SKOV3ip1, OVCAR8, and RF24 cells incubated under normoxic or hypoxic conditions. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays.
Hypoxia increases in vitro MSMP levels in cancer cells
To identify the mechanisms underlying increased MSMP expression, we searched for genes near the MSMP locus in the 9p13 chromosomal region and potential transcription factors (TFs) recruited to this region in human cells13. Analysis of the related canonical pathways of 120 genes and 30 TFs using Ingenuity Pathway Analysis (QIAGEN, Hilden, Germany) revealed that genes found in this region primarily belong to the hypoxia signaling pathway (Supplementary Table S1). Because hypoxia is an important consequence of anti-angiogenesis therapy, we next examined whether hypoxia could augment MSMP expression to promote angiogenesis using enzyme-linked immunosorbent assay (ELISA), Western blotting, and qRT-PCR with human cancer cell lines incubated under hypoxic conditions. OVCAR8 and SKOV3ip1 cells secreted 78 ng/ml and 62 ng/ml of MSMP at the highest levels under these conditions (1% O2) for 48 hours (P = 0.04 and 0.04, respectively) (Figure 1D). Intracellular MSMP expression levels were low in ovarian, lung, and colon cancer cell lines under normoxic conditions and high in SKOV3ip1, OVCAR8, A549, and DLD1 cells under hypoxic conditions (Figure 1E; Supplementary Figure S2A). Both secreted and intracellular MSMP levels remained unchanged under hypoxic conditions in RF24 endothelial cells (Figure 1D and 1E; Supplementary Figure S2B). Similarly, MSMP mRNA expression levels increased significantly under hypoxic conditions in OVCAR8 and SKOV3ip1 cells (P = 0.04 and 0.01, respectively) (Figure 1F). When HIF1a was induced by CoCl2, MSMP protein expression did not increase (Supplementary Figure S2C), suggesting HIF1a is not involved in the induction of MSMP.
The transcriptional repressor CCCTC-binding factor (CTCF) regulates MSMP expression
We next determined potential mechanisms underlying increased MSMP expression with anti-angiogenesis therapy using human ovarian cancer cell lines. To determine whether transcriptional or post-transcriptional mechanisms are involved in MSMP expression, we treated SKOV3ip1 and OVCAR8 cells with the transcriptional inhibitor actinomycin D. MSMP mRNA expression levels were not significantly different under normoxic or hypoxic conditions with this treatment (Supplementary Figure S3A), suggesting that MSMP expression is mainly regulated transcriptionally. Our search of the University of California, Santa Cruz genome browser for TFs that could bind with the MSMP promoter and enhancer regions14 identified about 2,500 base pairs upstream of the MSMP transcription start site, a putative enhancer region exhibiting enhancer properties such as DNAse I hypersensitivity clusters and an open chromatin structure. This region exhibited recruitment of TFs, including CTCF, POLR2A, MAX, RAD2, JUNB, and JUND. A search of the ENCODE chromatin immunoprecipitation (ChIP) sequencing database demonstrated that among these TFs, CTCF binding with this region was most prominent and conserved in different cell lines (Supplementary Figure S3B).
Because CTCF is a multifunctional TF that can function as either an activator or repressor of transcription, we first confirmed the role of CTCF in regulation of MSMP expression using siRNA. CTCF silencing by two different siRNAs significantly increased MSMP mRNA (P < 0.01) (Figure 2A) and protein levels under hypoxic conditions (Figure 2B). In addition, ectopically expressed CTCF decreased MSMP expression under hypoxic conditions in SKOV3ip1 cells (Figure 2C).
Figure 2.
MSMP expression in ovarian cancer cells is regulated by histone acetylation under hypoxic conditions. A, Real-time qRT-PCR analysis of CTCF and MSMP expression in OVCAR8 and SKOV3ip1 cells transfected with 1 of 2 CTCF siRNAs or control siRNAs. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. B, Immunoblot of CTCF, MSMP, and B-actin expression in SKOV3ip1 cells transfected with control siRNA or CTCF siRNA. C, Left: immunoblot of MSMP, CTCF, and B-actin expression in SKOV3ip1 cells treated with an ectopic CTCF expression vector. Right: real-time qRT-PCR analysis of CTCF expression in SKOV3ip1 cells treated with ectopic CTCF protein. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. D and E, ChIP assay data. OVCAR8, SKOV3ip1, and RF24 cells were cultured under normoxic or hypoxic conditions. Chromatin in the cells was pulled down using (D) anti-CTCF and anti-rabbit IgG antibodies or (E) anti-H3ac and anti-rabbit IgG antibodies. Left: representative images of PCR products. Right: fold enrichment of CTCF binding to the MSMP enhancer region. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. F, Real-time RT-PCR analysis of MSMP expression in SKOV3ip1 cells treated with 1 of 2 HDAC inhibitors. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays.
We then studied whether CTCF expression was altered under hypoxic conditions using human ovarian cancer cell lines. Western blotting demonstrated that CTCF expression did not change in SKOV3ip1, OVCAR8, or RF24 cells under hypoxic conditions (Supplementary Figure S3C). Because CTCF was equally expressed under normoxic and hypoxic conditions, we next asked whether recruitment of CTCF to the MSMP enhancer differed for normoxia and hypoxia. Because CTCF is a DNA methylation-sensitive repressor 15, we next treated cells with 5-azacytidine to determine whether methylation regulated expression of MSMP in ovarian tumors. Under hypoxic conditions, however, treatment with 5-azacytidine for 48 hours did not change MSMP mRNA expression levels in SKOV3ip1 cells (Supplementary Figure S3D). Because inhibiting methylation did not change expression of MSMP, methylation at the CTCF binding site is unlikely to be responsible for differences in CTCF binding to the MSMP enhancer. Therefore, we asked whether hypoxia-induced changes to the conformation of chromatin in this region were responsible for the differences in CTCF binding. A ChIP assay demonstrated that CTCF binding to the MSMP enhancer decreased significantly under hypoxic conditions after 48 hours in OVCAR8 and SKOV3ip1 cells (P < 0.01 and 0.02, respectively), but did not demonstrate this binding in RF24 cells under either normoxic or hypoxic conditions (Figure 2D). To determine whether changes in CTCF binding to the MSMP enhancer resulted from alterations in chromatin structure, we investigated histone markers at the MSMP enhancer in ovarian cancer cells in the context of hypoxia. ChIP assays with an antibody against H3ac, a marker associated with open chromatin structures where TFs can be recruited, demonstrated that H3ac’s association with the MSMP enhancer decreased significantly in OVCAR8 and SKOV3ip1 cells after 48 hours under hypoxic conditions (P < 0.01), but demonstrated no binding in RF24 cells under either normoxic or hypoxic conditions (Figure 2E). We next measured MSMP mRNA expression in SKOV3ip1 cells treated with the pan-histone deacetylase (HDAC) inhibitor panobinostat or suberoylanilide hydroxamic acid, both of which inhibit the removal of histone acetylation from the MSMP enhancer and therefore keep the MSMP enhancer region in an open configuration. Under hypoxic conditions, MSMP expression in these cells decreased significantly after 12 hours of treatment with either panobinostat (P = 0.01 and P < 0.01 at 10 nM and 25 nM, respectively) or suberoylanilide hydroxamic acid (P = 0.02 and P < 0.01 at 10 nM and 25 nM, respectively) (Figure 2F).
MSMP expression stimulates angiogenesis
CCR2 has previously been considered a primary receptor for MSMP 11. To identify cells targeted by secreted MSMP, we compared CCR2 expression in SKOV3ip1, OVCAR8, and RF24 cells under both normoxic and hypoxic conditions. Both mRNA and protein expression for CCR2 were highest in RF24 cells under hypoxic conditions (Supplementary Figure S4A), suggesting that the effects of MSMP are recognized primarily by endothelial cells. To assess the angiogenic effects of MSMP in endothelial cells, we performed a tube formation assay. We treated RF24 cells with media depleted of MSMP via conditioning with SKOV3ip1, OVCAR8, or A549 cells transfected with siRNAs against MSMP. Cultured media from cells transfected with MSMP siRNAs resulted in lower extent of tube formation by the RF24 cells compared to the control cells (by 50% P = 0.04, by 89% P < 0.01, and by 75% P < 0.01 in OVCAR8, SKOV3ip1, and A549 cells, respectively). Moreover, recombinant MSMP (rMSMP) rescued tube formation by RF24 cells treated with MSMP-depleted media (Figure 3A). rMSMP also had a direct and robust proangiogenic effect on RF24 cells even with no or low levels of other growth factors (with 0% and 2.5% fetal bovine serum [FBS]; P < 0.01) (Figure 3B). We observed this proangiogenic effect in the presence of an anti-VEGF antibody, as well (Supplementary Figure S4B). CCR2 silencing in RF24 cells significantly decreased tube formation (P < 0.01), and rMSMP did not overcome inhibition of tube formation by CCR2-silenced RF24 cells (Figure 3C). In addition, ectopically expressed CCR2 significantly increased tube formation (P < 0.01, Figure 3D, Supplementary Figure S4C).
Figure 3.
Secreted MSMP has a proangiogenic effect on endothelial cells. A, Effects of a medium conditioned with ovarian and lung cancer cells in which MSMP expression was or was not depleted by siRNA on tube formation by RF24 cells with or without the addition of rMSMP. Left: representative images of tube formation assays, magnification, ×20. Right: numbers of tubes in each assay in representative high-power fields. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. CM, conditioned medium. B, Direct effects of rMSMP on RF24 tube formation with no or low concentrations of FBS with growth factors. Left: representative images of tube formation assays. Right: numbers of tubes in each assay in representative high-power fields. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. C, Effects of CCR2 silencing on RF24 tube formation with or without added rMSMP. Left: representative images of tube formation assays, magnification, ×100. Right: numbers of tubes in each assay in representative high-power fields. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. D, Effects of CCR2 expression on RF24 tube formation. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays. E, Left: immunoblot of phosphorylated ERK (pERK), ERK, and B-actin expression in RF24 cells in which CCR2 was or was not silenced with or without the addition of rMSMP. Si-Control, control siRNA; si-CCR2, CCR2 siRNA. Right: effects of GSK1120212 on RF24 tube formation. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD of triplicate assays.
To identify the mechanisms underlying promotion of angiogenesis by MSMP, we employed Ingenuity Pathway Analysis to discover proangiogenic factors regulated by CCR2 signaling and angiogenesis pathway-related molecules. The results suggested that the MAPK pathway caused proangiogenic effects downstream from CCR2 (Supplementary Figure S4D). Treatment of control siRNA-transfected RF24 cells with rMSMP resulted in increased phosphorylation of ERK compared to the CCR2 siRNA-transfected cells (Figure 3E). The MAPK inhibitor GSK1120212 significantly decreased tube formation by the RF24 cells compared to the control cells (P < 0.05), and CCR2 expression rescued tube formation by RF24 cells treated with GSK1120212 (Figure3E, Supplementary Figure S4E).
Sensitization of tumors to treatment with an anti-VEGF antibody by MSMP silencing in vivo
To examine the biological effects of MSMP silencing on human ovarian tumors in vivo, we used a B20-resistant SKOV3ip1 athymic mouse model. For these experiments, siRNA was packaged in our well-characterized 1, 2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) nanoliposomes (Figure 4A). The tumors removed from mice given B20 and MSMP siRNA-DOPC weighed 89% less than did those from mice given B20 and control siRNA-DOPC (P < 0.01) (Figure 4B). Also, the number of ovarian tumor nodules was 78% lower in the MSMP siRNA-DOPC group (P < 0.01) (Figure 4B). Immunostaining using CD31 antibody on SKOV3ip1 in vivo samples showed that microvessel density was decreased in si-MSMP treated group compared with control siRNA treated group (P = 0.04, Figure 4C).
Figure 4.
Sensitization of ovarian tumors to an anti-VEGF antibody via MSMP silencing in vivo. A, Experimental schedule for the B20-resistant SKOV3ip1 model and representative IVIS images of intraperitoneal tumors. B, Tumor weights, numbers of tumor nodules, and representative images of the abdominal cavity in the control siRNA and MSMP siRNA groups of mice (white border, tumor in a representative control siRNA mouse). The Mann-Whitney U test and an unpaired t-test with Welch’s correction were used for statistical analysis of tumor weight and number of nodules, respectively. C, The mean vessel density and representative images of immunofluorescence analysis of CD31 expression in sections of the control siRNA and MSMP siRNA groups of mice (red, CD31; magnification, ×20). Mean vessel density was calculated by averaging vessel counts from five random fields per slide. Data are shown as means ± SD of triplicate assays.
Serum MSMP levels in patients with ovarian cancer resistant to bevacizumab
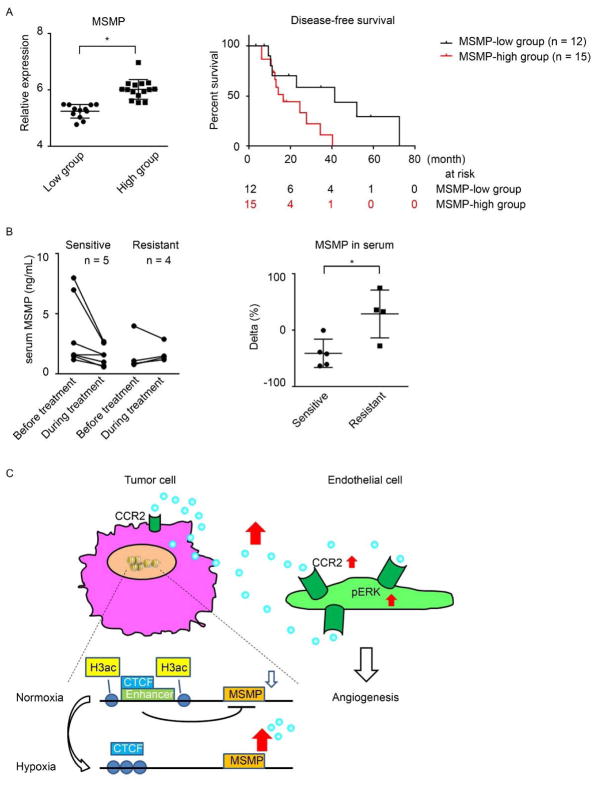
To determine the role of MSMP expression in ovarian cancer patient outcomes, we used data from The Cancer Genome Atlas project on cancer cases for which treatment with bevacizumab was prescribed (Supplementary Table S2). We analyzed MSMP mRNA expression levels in 27 tumor samples from ovarian cancer patients. Based on these samples, we classified the patients as either MSMP-low or MSMP-high using a cutoff value for MSMP expression determined according to basal levels of MSMP expression (P < 0.01). Increased expression of MSMP was associated with significantly reduced disease-free survival in ovarian cancer patients given bevacizumab (P = 0.04) (Figure 5A).
Figure 5.
MSMP overexpression confers clinical adaptive resistance to anti-angiogenesis therapy to ovarian cancer. A, Left: MSMP expression in ovarian cancer patients in the MSMP-low and MSMP-high groups. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD. Right: Kaplan-Meier disease-free survival curves for patients with ovarian cancer treated with bevacizumab according to stratified tumoral expression of MSMP. Data were taken from The Cancer Genome Atlas data sets. A log-rank test was used for statistical analysis. B, Left: serum concentrations of MSMP measured in 12 ovarian cancer patients using ELISA before and during treatment with a bevacizumab-containing regimen. The concentrations in patients whose disease was resistant to the regimen were compared with those in patients whose disease was sensitive to the regimen. Right: fluctuations in serum MSMP levels during anti-angiogenesis treatment in the 2 patient groups. A 2-tailed unpaired Student t-test was used for statistical analysis. Data are shown as means ± SD. The same data set was used in both graphs. C, A proposed model for regulation of MSMP expression in cancer cells under hypoxic conditions and the pro-angiogenic role of MSMP in ovarian tumors.
To identify adaptive changes in MSMP expression in cancer patients in response to anti-angiogenesis therapy, we evaluated serum concentrations of MSMP in patients with FIGO stage III or IV high-grade epithelial ovarian, primary peritoneal, or fallopian tube cancer treated with the combination of bevacizumab and weekly paclitaxel and carboplatin as first-line chemotherapy after surgery in a phase II clinical trial. We selected 9 patients who provided paired blood samples before and during treatment. We stratified the subjects by treatment response according to the Response Evaluation Criteria in Solid Tumors. We defined complete and partial responses as sensitive and stable and progressive disease as resistant. Although the baseline concentration was variable in both sensitive and resistant groups, when we compared the levels of serum MSMP concentration before and during treatment for each patient, we found that the concentration increased significantly only in patients with resistant disease (P = 0.02) (Figure 5B).
Discussion
In the present study, we demonstrated that MSMP is an important mediator of adaptive resistance of ovarian cancer to anti-angiogenesis therapy. MSMP expression was upregulated mainly in cancer cells by the hypoxic microenvironment induced by treatment with anti-angiogenesis drugs. Induction of MSMP expression in the context of hypoxia occurred in ovarian cancer cells, but not in endothelial cells, a distinction owing to inhibition of recruitment of the transcriptional repressor CTCF to the MSMP enhancer by cancer-specific chromatin remodeling (Figure 5C). A previous study demonstrated that histone deacetylase activity was markedly upregulated by hypoxia in tumor cells.16 The present study demonstrated that under hypoxic conditions, histone deacetylation directly regulated the transcription of proangiogenic genes in ovarian cancer cells. This finding is important for explaining the diversity of adaptive resistance-related genes in resistant tumors, and has implications for design of additional clinical trials.17–19
Our results implicate a role for CTCF in adaptive resistance of human ovarian cancers to anti-angiogenesis therapy. Previous studies demonstrated that CTCF controlled local epigenetic modifications of histones by erasing the H3K27me3 histone marker as an insulator of target genes. Under that scenario, CTCF silencing should decrease expression of genes located next to the CTCF boundary flanking the H3k27me3 domain.20 However, our results demonstrated that CTCF silencing increased MSMP expression in human cancer cell lines, suggesting that hypoxia-inducible histone modification was the principal mechanism of epigenetic regulation of MSMP and that CTCF acted as a transcription repressor.
In summary, changes in MSMP enhancer acetylation and protein expression in ovarian cancer cells in response to anti-angiogenesis therapy provide new knowledge related to adaptive changes in the tumor microenvironment. Therefore, targeting of MSMP in combination with the use of anti-angiogenesis drugs may provide an important avenue for improving efficacy of anti-VEGF therapy and blocking adaptive resistance.
Materials and Methods
Materials
SiRNA sequences were designed by and purchased from Sigma-Aldrich (St. Louis, MO) (Supplementary Materials). For western blotting, primary antibodies against MSMP (1:500, ab185471; Abcam, Cambridge, UK), β-actin (1:5000, A5316; Sigma-Aldrich), CTCF (1:1000, 3418S; Cell Signaling Technology, Danvers, MA), p44/42 MAPK (Erk1/2; 1:1000, 9102S; Cell Signaling Technology), and phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204; 1:1000, 9101S; Cell Signaling Technology) were used. For ELISAs, a rabbit polyclonal anti-MSMP antibody (1:200; Abcam) and goat polyclonal anti-MSMP antibody (1:400, D-15; Santa Cruz Biotechnology, Santa Cruz, CA) were used.
Cell cultures
Cell lines were purchased from The University of Texas MD Anderson Cancer Center Characterized Cell Line Core Facility or the ATCC21–25, and maintained in 5% CO2 at 37°C. We cultured ovarian (A2780, SKOV3ip1, OVCAR8, and HeyA8), lung (A549), and colon (DLD1 and SW480) cancer cell lines in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10–15% FBS and 0.1% gentamicin sulfate (Gemini Bio-Products, Calabasas, CA). We cultured the endothelial cell line RF24 in modified essential medium supplemented with 10% FBS, sodium pyruvate, modified essential medium vitamins, L-glutamine, and modified essential medium nonessential amino acids. We also maintained the prostate cancer cell line PC3 in Dulbecco’s modified Eagle’s medium supplemented with 15% FBS. All the cell lines were routinely tested to confirm the absence of Mycoplasma and cultured for fewer than 6 months after resuscitation.
Microarray analysis
We extracted total RNA from whole control and B20-treated SKOV3ip1 tumors using a mirVana RNA isolation kit (Ambion, Austin, TX). NanoDrop spectrophotometer (Thermo Fisher Scientific) was used for the assessment of both RNA quality and quantity. Total RNA (700 ng) from each tumor was labeled and hybridized to Bead Chips according to the manufacturer’s protocols (Illumina; San Diego, CA). We scanned Bead Chips with an Illumina BeadArray Reader, and the resulting data were normalized and expressed as log2 values. Genes expressed in resistant tumors with P values less than 0.05 compared with those in the control group were considered significantly different.
Cell treatment
MSMP, CTCF, and CCR2 genes in human cancer or endothelial cells were silenced using forward or reverse transfection protocols with the Lipofectamine RNAiMAX transfecting agent (Thermo Fisher Scientific) and specific human siRNAs according to the agent manufacturers’ recommendations. Briefly, 40 nM siRNA was mixed with RNAiMAX and added to culture medium for 6 to 12 hours. We added fresh medium to the mixture, and cells were allowed to grow in it.
We purchased rMSMP protein from OriGene (Rockville, MD). The nonselective HDAC inhibitor panobinostat was purchased from Selleckchem (S1030; Houston, TX). The HDAC inhibitor suberoylanilide hydroxamic acid (vorinostat), actinomycin D, and 5-azacytidine were purchased from Sigma-Aldrich.
RNA isolation and qRT-PCR
We isolated RNA from cultured ovarian and lung cancer cell lines and IG10-induced murine tumors using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. RNA quality and quantity were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific). We synthesized cDNA from 1000 ng of RNA using a Verso cDNA kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. mRNA expression in the cells was analyzed using qRT-PCR with a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) and SYBR Green Real-Time PCR Master Mix. Each of the 40 PCR cycles consisted of 15 seconds of denaturation at 95°C and 1 minute of annealing and extension at 60°C. We calculated the level of expression of the target gene in the cells using the ΔΔCt method and normalized according to 18S mRNA expression.
In vivo models
All animal protocols for this study were approved by the Institutional Animal Care and Use Committee at MD Anderson. We performed all animal experiments using 8- to 12-week-old female athymic nude mice obtained from Taconic Farms (Hudson, NY). Murine VEGF-A–targeted monoclonal antibody B20 was obtained from Genentech (South San Francisco, CA). We injected SKOV3ip1 human ovarian cancer cells or IG10 murine ovarian cancer cells (1×106) into the peritoneal cavities of nude mice. Three weeks later, we gave mice B20 (5 mg/kg body weight twice per week). Two weeks after B20-based treatment began, the mice were placed in B20-sensitive and B20-resistant groups based on the results of IVIS imaging (Xenogen, Alemeda, CA). The B20-resistant group of mice in the SKOV3ip1 intraperitoneal model was randomly divided into 2 groups and given either control siRNA (n = 9) or MSMP siRNA (n = 8) in 1, 2-dioleoyl-sn-glycero-3-phosphocholine nanoparticles (5 μg intraperitoneally twice per week) along with B20 until they became moribund. Investigators killed the mice via cervical dislocation, and their tumor weights, tumor nodule numbers, and metastasis distributions were noted. The mice were housed 5 per cage in standard acrylic glass cages in a room maintained at a constant temperature and humidity. They were fed a regular autoclaved chow diet with water ad libitum. For animal studies, investigators performing the necropsy were blinded to the specific treatment groups.
Western blotting
We prepared protein lysates from cultured cancer cells by exposing them to a modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton, 0.5% deoxycholate) with 25 mg/mL leupeptin, 10 mg/mL aprotinin, 2 mM ethylenediaminetetraacetic acid, and 1 mM sodium orthovanadate. For each lysate, the protein concentration was determined using a BCA Protein Assay Reagent Kit (Thermo Fisher Scientific). Protein lysates were separated using 8% or 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and blocked with 5% bovine serum albumin in Tris-buffered saline with Tween 20 for 1 hour at room temperature. Membranes were probed with primary antibodies in 5% bovine serum albumin and Tris-buffered saline with Tween 20 overnight at 4°C. The bands in the resulting gels were then incubated with a horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody (GE Healthcare, Little Chalfont, UK) for 1 hour at room temperature. We developed blots with an enhanced chemiluminescence detection kit (Pierce Biotechnology, Waltham, MA) and assessed in a semi-quantitative manner using the ImageJ software program (National Institutes of Health, Bethesda, MD).
Immunostaining
Immunostaining was performed with formalin-fixed, paraffin-embedded SKOV3ip1 tumor sections (5 μm thick) or optimal cutting temperature compound-embedded frozen sections of the SKOV3ip1 and IG10 tumors. After deparaffinization, rehydration, and antigen retrieval or fixation, we treated sections with 3% H2O2 for 10 minutes to block the endogenous peroxidase activity. Nonspecific epitopes were blocked with 4% fish gelatin in a phosphate-buffered saline solution for 20 minutes. Slides were incubated with primary antibodies against MSMP (rabbit polyclonal anti-human, 1:200; Abcam) and CD31 (rat monoclonal anti-mouse, 1:800; Pharmingen, San Jose, CA) overnight at 4°C. For immunohistochemical analysis, the sections were washed with phosphate-buffered saline solution, the appropriate amount of horseradish peroxidase-conjugated secondary antibody was added to the sections with 3,3′-diaminobenzidine chromogen for visualization, and the sections were counterstained with Gill’s hematoxylin #3. For immunofluorescence, the secondary antibody stain was either Alexa 488 (CD31) or Alexa 594 (MSMP; Jackson ImmunoResearch, West Grove, PA). We used Hoechst 33342 for nuclear staining (1:10,000; Molecular Probes, Waltham, MA). We defined cells with strongly stained MSMP in cytoplasm which are similar to positive control of prostate cancer cells as positive. For analysis of proportion of immunohistochemical positivity, we checked ten high power field for each samples.
Enzyme-linked immunosorbent assay
We determined MSMP expression levels using an MSMP ELISA Kit (Mybiosource, San Diego, CA) according to the manufacturer’s recommendations for human blood samples. For in vitro samples of cell culture media, a rabbit polyclonal anti-MSMP antibody was used as the capture antibody (1:300), and a goat polyclonal anti-MSMP antibody (1:400) was used as the detection antibody. SKOV3ip1, OVCAR8, and RF24 cells were grown to 90% confluence under normoxic or hypoxic (1% O2) conditions for 48 hours.
CTCF and CCR2 expression plasmid
Full-length CTCF cDNA was amplified using PCR and inserted into the EcoRI and BamHI sites of the pcDNA3 vector (Addgene, Cambridge, MA) to generate a CTCF expression vector. CCR2 expression plasmid was obtained from Addgene (CCR2-Tango).
ChIP assay
Human cancer cell lines were subjected to hypoxia (1% O2) for 48 hours. We performed ChIP assays to confirm binding of TF to the genome enhancer region using a Chip-it Express Kit (Active Motif, Carlsbad, CA) as recommended by the manufacturer. In brief, cultured cells were collected from cell culture dishes and subjected to lysis, sonication, and immunoprecipitation with an anti-H3ac antibody (1:10; Active Motif), anti-CTCF antibody (1:50), or normal rabbit IgG as a control (1:50). Immunocomplexes of chromatin and TF were collected from sonicated samples with protein G magnetic beads and eluted. Cross-links between proteins and chromatins were reversed via incubation of immunocomplexes at 95°C. Fold enrichment of the binding between the MSMP enhancer and CTCF or that between the MSMP enhancer and H3ac was quantified using a PCR.
Tube formation assay
For evaluation of the indirect effects of rMSMP on tube formation by endothelial cells, we treated SKOV3ip1, OVCAR8, and A549 cells with either control siRNA or MSMP siRNA for 48 hours. RF24 cells (1×104) were plated and incubated for 6 hours at 37°C with or without bevacizumab in the presence of a 50% conditioned medium. Tube formation by the cells was assayed using a Matrigel-coated μ-Slide Angiogenesis ibiTreat kit (ibidi, Madison, WI). We assessed direct effects of CCR2 silencing on tube formation 48 hours after transfection of RF24 cells with siRNAs. For tube formation rescue experiments, rMSMP was added to the wells.
Patient samples
Serum samples were obtained from 9 ovarian cancer patients who were enrolled in an Institutional Review Board-approved phase 2 clinical trial (NCT01097746) conducted at MD Anderson and collaborating institutions. Informed consent was obtained from all subjects. All cases were diagnosed histologically as epithelial ovarian cancer, peritoneal primary carcinoma, or fallopian tube cancer; were FIGO stage III or IV; were defined surgically at the completion of initial abdominal surgery; and had sufficient tissue available for histologic evaluation.
The Cancer Genome Atlas Data analysis
For measurement of MSMP expression in human ovarian cancer, MSMP RNA expression in cancer tissue was analyzed using RNASeqV2 data available from the open-access database of The Cancer Genome Atlas (http://www.cbioportal.org/public-portal/).
Statistics
For in vitro studies, continuous variables were compared using the Student t-test if normally distributed. We compared differences in variables that were not normally distributed using the nonparametric Mann-Whitney U test. Data were presented as means ± SD. For animal experiments, tumor weights and nodule numbers were presented as medians and means ± SD, respectively. Statistical comparisons of experimental groups were analyzed using the nonparametric Mann-Whitney U test and an unpaired t-test with Welch’s correction, respectively. We estimated sample sizes to ensure adequate power to detect a pre-specified effect size based on previously generated data. The Student t-test was used to classify the patients as either MSMP-low or MSMP-high and compare their serum MSMP concentrations. Kaplan-Meier survival curves and log-rank tests were used to examine the association between MSMP expression and overall survival in The Cancer Genome Atlas data. All statistical tests were 2-sided, and P values less than 0.05 were considered significant. Only 2-tailed values were reported. For every figure, the variance is similar between the groups.
Supplementary Material
Acknowledgments
Financial support: S.P. is supported by a Foundation for Women’s Cancer grant and Ovarian Cancer Research Fund Alliance. S.W. is supported by Ovarian Cancer Research Fund Alliance, Foundation for Women’s Cancer, Texas Center for Cancer Nanomedicine, and Cancer Prevention and Research Institute of Texas training grants (RP101502 and RP101489, respectively). K.N. is supported by the Ovarian Cancer Research Fund Alliance (grant 292015) and KRIBB Research Initiative Program. T.H. is supported by Uehara Memorial Foundation Research Fellowships for Research Abroad. Portions of this work were supported by the National Institutes of Health grants (CA016672, CA109298, P50 CA083639, P50 CA098258, and UH3 TR000943), Ovarian Cancer Research Fund, Inc. (Program Project Development Grant), American Cancer Society Research Professor Award, Blanton-Davis Ovarian Cancer Research Program, RGK Foundation, and the Frank McGraw Memorial Chair in Cancer Research.
Footnotes
Conflict of interest.
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Hillen F, Griffioen AW. Tumour vascularization: sprouting angiogenesis and beyond. Cancer Metastasis Rev. 2007;26:489–502. doi: 10.1007/s10555-007-9094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim KJ, Li B, Winer J, Armanini M, Gillett N, Phillips HS, et al. Inhibition of Vascular Endothelial Growth Factor-Induced Angiogenesis Suppresses Tumor-Growth Invivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 3.Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 4.Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365:2484–2496. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 5.Pujade-Lauraine E, Hilpert F, Weber B, Reuss A, Poveda A, Kristensen G, et al. Bevacizumab combined with chemotherapy for platinum-resistant recurrent ovarian cancer: The AURELIA open-label randomized phase III trial. J Clin Oncol. 2014;32:1302–1308. doi: 10.1200/JCO.2013.51.4489. [DOI] [PubMed] [Google Scholar]
- 6.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 7.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 8.Rini BI, Halabi S, Rosenberg JE, Stadler WM, Vaena DA, Ou SS, et al. Bevacizumab plus interferon alfa compared with interferon alfa monotherapy in patients with metastatic renal cell carcinoma: CALGB 90206. J Clin Oncol. 2008;26:5422–5428. doi: 10.1200/JCO.2008.16.9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman HS, Prados MD, Wen PY, Mikkelsen T, Schiff D, Abrey LE, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27:4733–4740. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- 10.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 11.Pei X, Sun Q, Zhang Y, Wang P, Peng X, Guo C, et al. PC3-secreted microprotein is a novel chemoattractant protein and functions as a high-affinity ligand for CC chemokine receptor 2. J Immunol. 2014;192:1878–1886. doi: 10.4049/jimmunol.1300758. [DOI] [PubMed] [Google Scholar]
- 12.Valtonen-Andre C, Bjartell A, Hellsten R, Lilja H, Harkonen P, Lundwall A. A highly conserved protein secreted by the prostate cancer cell line PC-3 is expressed in benign and malignant prostate tissue. Biol Chem. 2007;388:289–295. doi: 10.1515/BC.2007.032. [DOI] [PubMed] [Google Scholar]
- 13.Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MS, Kwon HJ, Lee YM, Baek JH, Jang JE, Lee SW, et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat Med. 2001;7:437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 17.Drappatz J, Lee EQ, Hammond S, Grimm SA, Norden AD, Beroukhim R, et al. Phase I study of panobinostat in combination with bevacizumab for recurrent high-grade glioma. J Neurooncol. 2012;107:133–138. doi: 10.1007/s11060-011-0717-z. [DOI] [PubMed] [Google Scholar]
- 18.Lee EQ, Reardon DA, Schiff D, Drappatz J, Muzikansky A, Grimm SA, et al. Phase II study of panobinostat in combination with bevacizumab for recurrent glioblastoma and anaplastic glioma. Neuro Oncol. 2015;17:862–867. doi: 10.1093/neuonc/nou350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strickler JH, Starodub AN, Jia J, Meadows KL, Nixon AB, Dellinger A, et al. Phase I study of bevacizumab, everolimus, and panobinostat (LBH-589) in advanced solid tumors. Cancer Chemother Pharmacol. 2012;70:251–258. doi: 10.1007/s00280-012-1911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weth O, Paprotka C, Gunther K, Schulte A, Baierl M, Leers J, et al. CTCF induces histone variant incorporation, erases the H3K27me3 histone mark and opens chromatin. Nucleic Acids Res. 2014;42:11941–11951. doi: 10.1093/nar/gku937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bast RC, Jr, Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest. 1981;68:1331–1337. doi: 10.1172/JCI110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buick RN, Pullano R, Trent JM. Comparative properties of five human ovarian adenocarcinoma cell lines. Cancer Res. 1985;45:3668–3676. [PubMed] [Google Scholar]
- 23.Landen CN, Kim TJ, Lin YG, Merritt W, Kamat AA, Han L, et al. Tumor-Selective Response to Antibody-Mediated Targeting of alpha(v)beta(3) Integrin in Ovarian Cancer. Neoplasia. 2008;10:1259–1267. doi: 10.1593/neo.08740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau DH, Lewis AD, Ehsan MN, Sikic BI. Multifactorial mechanisms associated with broad cross-resistance of ovarian carcinoma cells selected by cyanomorpholino doxorubicin. Cancer Res. 1991;51:5181–5187. [PubMed] [Google Scholar]
- 25.Yoneda J, Kuniyasu H, Crispens MA, Price JE, Bucana CD, Fidler IJ. Expression of angiogenesis-related genes and progression of human ovarian carcinomas in nude mice. J Natl Cancer Inst. 1998;90:447–454. doi: 10.1093/jnci/90.6.447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.