Abstract
The zebrafish is a recent addition to animal models of human cancer, and studies using this model are rapidly contributing major insights. Zebrafish develop cancer spontaneously, after mutagen exposure and through transgenesis. The tumours resemble human cancers at the histological, gene expression and genomic levels. The ability to carry out in vivo imaging, chemical and genetic screens, and high-throughput transgenesis offers a unique opportunity to functionally characterize the cancer genome. Moreover, increasingly sophisticated modelling of combinations of genetic and epigenetic alterations will allow the zebrafish to complement what can be achieved in other models, such as mouse and human cell culture systems.
The cancer biology field is rapidly heading towards a post-genomic state, in which most human cancers will have been extensively sequenced. These sequencing projects have been integral to our understanding of the genetic abnormalities in human malignancy, and will be complemented by efforts in characterizing the epigenome of cancer. The data published thus far indicate that most malignancies are characterized by extensive genomic alterations, ranging from between 100 and 1,000 mutations in leukaemia1,2 to more than 70,000 point mutations in melanoma3. Perhaps more than at any other time, the field is now faced with an unprecedented opportunity to translate these findings into therapeutic advances; however, the tools required to place these abnormalities into a biological context have not yet been developed.
The next decade will witness a concerted effort to study the functional implications of this new data using human cell lines and animal models. No single model will fully capture the heterogeneous and evolving complexity of cancer, so we must rely on the strengths of various systems to contextualize this information. Although mouse models will remain a cornerstone of cancer research, the unique capabilities of the zebrafish have recently pointed towards it becoming a key model for helping us to understand cancer biology in vivo.
This Review highlights important insights that have been gained from studying cancer in zebrafish, how cancer biologists can take advantage of technologies that are unique to zebrafish and what the key roles of the zebrafish model will be in the coming decade.
Zebrafish as a cancer model
The zebrafish emerged as a model organism for developmental genetics in the 1960s, and was described by George Streisinger as a phage with a backbone. Its advantages for genetic studies are its high fecundity, the generation of transparent embryos that develop outside the mother and the conservation of vertebrate organs, which allows comparison with humans. The true usefulness of the model was established as a result of several large forward genetic screens4,5, which identified mutants in almost every organ or cell type, most of which are shared with mammals (FIG. 1), demonstrating that the fish could be used to identify genetic mutants for nearly any phenotype.
Figure 1 |. Zebrafish anatomy.
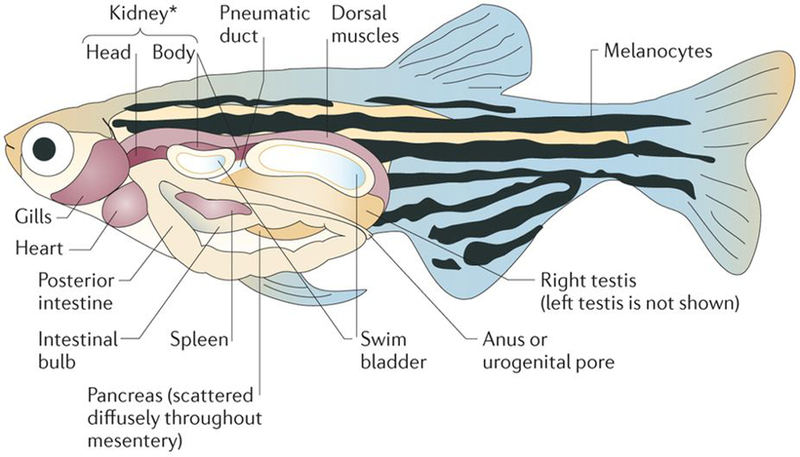
An adult zebrafish is shown with the anatomical structures labelled. Zebrafish share most of their organs with mammalian counterparts, including the brain, heart, liver, spleen, pancreas, gallbladder, intestines, kidney, testis and ovaries. *The kidney is also the site of haematopoiesis in zebrafish.
Fish have been known to develop cancer for more than a century. Xiphophorus species develop spontaneous melanoma, which was found to be due to activating mutations of the tyrosine kinase xiphophorus melanoma receptor kinase (xmrk)6. That zebrafish could be useful as a model for cancer was suggested in 1982, when it was found that exposure to carcinogens such as dimethylnitramine caused low-penetrance tumour formation in zebrafish7. By 2000, it was recognized that zebrafish exposed to more common mutagens such as ethylnitrosourea (ENU), DMBA and N-methyl-nitrosoguanadine (MNNG) develop various neoplasms, including skin papillomas, hepatic adenomas, rhabdomyosarcoma and seminoma8–10. However, it was with the emergence of rapid transgenic technology in the zebrafish that the field jumped forwards11–13. Langenau, Look and colleagues14 demonstrated that expression of the mouse oncogene Myc under the zebrafish recombinationactivating gene 2 (rag2) promoter resulted in the rapid onset of adult leukaemias that emerged from the thymus, quickly spread and that were fully transplantable, as seen by green fluorescent protein (GFP)-labelling of the tumour cells14. Since then, numerous zebrafish models of cancer using transgenic expression of dominant-acting oncogenes have been created (reviewed in REFS 15,16) (BOX 1). Furthermore, a zebrafish strain with mutant tp53 (which encodes the tumour suppressor p53) was shown to develop malignant peripheral nerve sheath tumours17, demonstrating that both oncogenes and tumour suppressors retain their evolutionarily conserved role in tumorigenesis. Increasing attention has recently focused on the role of immune and microenvironmental regulation of cancer, and it remains to be seen whether these aspects are also conserved in zebrafish.
Box 1 |. Positives and negatives of using zebrafish for cancer research.
Positives
Large numbers of transparent embryos that develop outside the female and that grow rapidly. A single adult mating pair can produce 200 embryos or more per week.
Embryonic phenotypes are strongly predictive of adult phenotypes in most organs, allowing for the screening of relevant adult phenotypes using space-efficient embryos.
As vertebrates, zebrafish share nearly all organs with mammals, including the brain, eyes, heart, intestines, pancreas, kidneys and liver.
Fish have a complex immune system with a full range of immune effectors, such as T cells and B cells, macrophages and monocytes.
Highly amenable to transgenic approaches. Mosaic (F0) transgenics can be created at a rate of 500–1,000 animals per day, and stable transgenic founders can be found in 50% of injected F0 animals using transposon-based systems.
Both forward genetic (using ethylnitrosourea) and reverse genetic (using TAL-like effector nuclease and CRISPRs) techniques are well characterized and highly scalable.
Transparent adult strains (that is, casper) allow for detailed in vivo imaging of tumour growth, migration and metastasis.
Large numbers of fluorescently tagged transgenic lines marking cells such as vascular endothelium, red and white blood cells, platelets and stroma are available.
There is high conservation of oncogenes such as BRAFV600E and NRASQ61K in zebrafish models of cancer. Microinjection of human genes under tissue-specific promoters leads to tumours that are similar to the human disease.
Tumours in zebrafish strongly resemble human tumours at the histological, gene expression and genomic levels.
Demonstrated success of chemical screening in zebrafish, which has led to clinical trials in patients with melanoma.
Negatives
Low incidence of spontaneous tumorigenesis, necessitating the use of mutagens and/or transgenic techniques.
Short-lived when compared with humans, which makes direct comparison of age-related cancer phenotypes limited.
Organs are typically simpler than mammalian counterparts. For example, the kidneys resemble the mesonephric rather than the metanephric stage.
Some mammalian organs are not conserved, including the mammary and prostate glands.
The genome size is approximately one-half the size of the human genome, making comparisons outside genic regions difficult.
The genome underwent a genome duplication event, so many genes have redundant copies, which complicates loss-of-function studies of tumour-suppressor genes.
Zebrafish grow at 28.5 °C, rather than at 37 °C, and are poikilothermic, limiting studies in which mammalian homeostatic temperature may be important to oncogenic phenotypes.
Limited range of antibody reagents, making protein-based analysis more difficult.
There are several technologies available in the zebrafish that have made it a unique contributor to the cancer research field (FIG. 2). In particular, cancer can be studied throughout the life of the animal, each stage with attendant experimental advantages that make zebrafish a powerful complement to other, more traditional, model systems. We highlight below some of the key techniques that have emerged using zebrafish, particularly those with direct relevance to human cancer pathogenesis.
Figure 2 |. Studying cancer in the zebrafish.
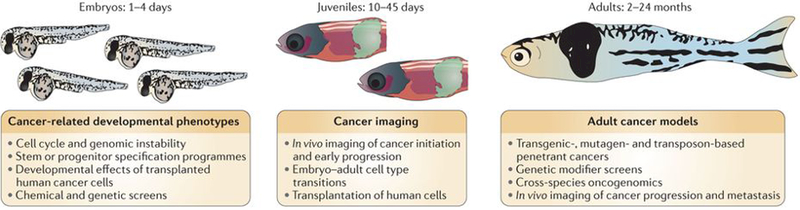
Differently aged animals each offers distinct advantages for cancer-relevant phenotypes. Embryos can be used to identify phenotypes that are highly relevant to cancer biology, such as defects in the cell cycle or genomic instability. Other embryo phenotypes may include stem or progenitor cells that act as cell of origin of the tumour or changes in embryo morphology on transplantation of human cells. Any of these phenotypes can then be used as the basis of chemical or genetic screens to find modifiers, which can be tested for their relevance to human cancer. Juvenile fish have the capacity for modelling early tumorigenesis and remain optically fairly translucent, lending themselves to detailed in vivo imaging. These cancers can be either from transgenic models or can arise via transplantation of tumour cells, and confocal imaging can be used to assess the tumour–stroma interaction at single-cell resolution. Adult fish develop fully penetrant and advanced cancers, both through transgenic techniques and through the transplantation of either zebrafish or human tumour cells. These animals are ideally suited to cross-species oncogenomics, either by directly testing candidate human genomic changes in the fish (by rapid transgenesis) or by comparing the profiles (DNA or RNA) of the mature tumour in the fish to that of the human to look for evolutionarily conserved events. Both the wild-type fish and the transparent casper model add improved capacities compared to mouse models for in vivo imaging and analysis of tumour stem cells and tumour progression and metastasis.
Cross-species oncogenomics
The zebrafish can be used to functionally characterize the large number of changes seen in human cancer, a major challenge that has emerged from projects such as The Cancer Genome Atlas. As recently highlighted18, the number of identified major-effect genes (that is, ‘drivers’) is limited, but the number of genes with unclear functional importance (which are presumably ‘passengers’) is vast. Functional testing of both frequently and infrequently altered genes can be carried out using zebrafish in two primary ways: by identifying changes in human cancer and then testing these candidate changes in transgenic zebrafish models (FIG. 3a); or by comparing genomic alterations found in human cancers to those found in zebrafish models of cancer to find evolutionarily conserved drivers (FIG. 3b–d; TABLE 1). Both approaches have been successfully applied, as outlined below.
Figure 3 |. Cross-species oncogenomics provides a powerful way to identify highly evolutionarily conserved events in tumorigenesis.
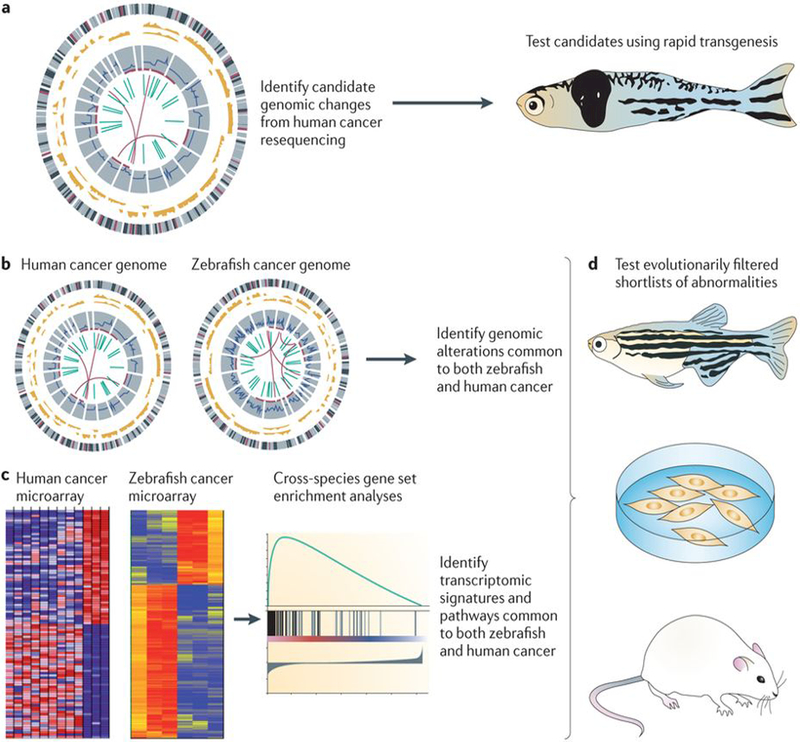
Candidate genomic changes in human cancers (part a) are being identified through consortiums such as The Cancer Genome Atlas and the International Cancer Genome Consortium (ICGC), which have revealed thousands of potential mutations, copy number changes and structural variants, most of which have not been functionally analysed in vivo. By testing a proportion of the high-confidence, recurrent events in transgenic zebrafish models, a direct readout of their effect on actual tumour biology can be rapidly achieved. This type of analysis can be carried out in the transient transgenic setting, allowing for thousands of animals or variants to be tested in a single experiment. Because there are now numerous models of cancer in the zebrafish (parts b,c), the genomic profile of these fish tumours can be directly compared with that of the human tumours to look for events that are common between the two species. This can be done either at the level of DNA (part b) by looking for common copy-number changes, mutations and structural variants, or at the RNA level (part c) by looking for transcriptomic commonalities. This will essentially act as a ‘filter’ to provide much shorter lists of highly penetrant changes that are conserved across millions of years of evolution. These abnormalities can then be efficiently tested in cell culture, zebrafish and mouse models (part d). Part c is reproduced, with permission, from REF. 28 © (2011) Macmillan Publishers Ltd. All rights reserved.
Table 1 |. Transgenic models of cancer in the zebrafish.
| Cancer | Oncogene | Tumour suppressor |
Use in cancer biology | Refs |
|---|---|---|---|---|
| Melanoma | mitfa–BRAFV600E | tp53−/− | Genetic and chemical modifier screens |
27, 30,31 |
| mitfa:EGFP:NRASQ61K | tp53−/− | |||
| kita–Gal4 × uas-HRAS | ||||
| Pancreatic | ptf1a–KRASG12V–GFP | Genetic modifier screens | 22,97 | |
| ptf1a:Gal4–VP16 × uas–KRASG12V–GFP | ||||
| T cell lymphoma or | rag2–myc | Cancer modelling and in vivo imaging | 14,98 | |
| leukaemia |
rag2–lox–dsRED2–lox–EGFP– mMyc × hsp70–cre |
Inducible cancer model | 99 | |
| rag2–NOTCH1 | NOTCH1 interaction with Bcl‑2 | 100,101 | ||
| rag2–myc × rag2–bcl2 | Mechanisms of leukaemia dissemination | 39 | ||
| B cell leukaemia |
Xenopus Spp. EF1α or zebrafish B actin– TEL–AML1 (ETV6–RUNX1) |
Initiating events in B cell leukaemia | 34 | |
| Numerous | b‑actin–lox–GFP–lox–KRASG12D × hsp70–cre | Inducible cancer model | 102 | |
| krt4:Gal4VP16;14 × uas:smoa1– |
Cooperation of hedgehog and AKT pathways | 103 | ||
| Rhabdomyosarcoma | rag2–KRASG12D | Identification of tumour-initiating cell populations | 29 | |
| Neuroblastoma | dβh:EGFP–MYCN | Cooperation of MYCN and ALK | 23 | |
| dβh:EGFP and dβh:ALKF1174L | Cooperation of MYCN and ALK | 23 | ||
| AML | pu1–MYST3/NCOA2–EGFP | First model of AML in zebrafish | 104 | |
| MPNST | tp53−/− | Conservation of tumour-suppressor pathways in zebrafish |
17 | |
| Major tumour type found in p53‑deficient zebrafish | ||||
| Lipoma | krt4–myrAKT1 | Platform for the study of drugs to treat lipoma and/or obesity |
105 | |
| Ewing’s sarcoma | hsp70 or β-actin–EWSR1–FLI1 | tp53−/− | Conserved function of EWS–FLI1 fusion protein from human to fish |
106 |
| Liver |
fabp10:LexPR; LexA:EGFP × cryB:mCherry; LexA:EGFP–krasG12V |
Inducible KRAS‑G12V hepatocellular cancer model | 36 | |
| fabp10:TA; TRE:xmrk; krt4:GFP | Inducible EGFR-homologue hepatocellular cancer model |
107 | ||
| Pancreatic neuroendocrine |
zmyod–MYCN | Pancreatic neuroendocrine model as a platform for downstream MYCN targets |
108 | |
| Myeloproliferative neoplasms |
sp1–NUP98–HOXA9 | NUP98–HOXA9‑induced oncogenesis from defects in haematopoiesis and aberrant DNA damage response |
109 | |
| Corticotroph adenoma and neoplasm |
POMC–PTTG | Identification of CDK inhibitors as possible treatment of corticotroph tumours |
110 | |
| Testicular germ cell tumour |
fugu flck–SV40 large T | Platform for modifier screens of testicular tumours | 35 |
ALK, anaplastic lymphoma kinase; AML, acute myeloid leukaemia; CDK, cyclin-dependent kinase; EF1α, elongation factor 1α; EGFP, enhanced green fluorescent protein; fabp10, fatty acid-binding protein 10, liver basic; GFP, green fluorescent protein; hsp70, heat shock protein 70; krt4, keratin 4; mitfa, microphthalmia-associated transcription factor a; MPNST, malignant peripheral nerve sheath tumour; myr, myristoylated; POMC, pro-opiomelanocortin; ptf1a, pancreas transcription factor 1 subunit a; PTTG, pituitary tumour-transforming gene; rag2, recombination-activating gene 2; smoa1, activated smoothened mutant; uas, upstream activating sequence.
Human melanoma is among the most genomically abnormal of all cancers, probably partly owing to the accumulation of large numbers of ultraviolet light-induced passenger mutations. Although common genetic alterations, such as BRAFV600E and NRASQ61K mutations, are well documented in human cancer, these are insufficient to explain the aggressive behaviour of the disease, and they probably extensively cooperate with other somatic changes during the course of tumorigenesis. Analyses of human melanoma samples using the GISTIC algorithm identified a number of regions of chromosomal amplification, and a subset of these were also overexpressed at the RNA level19, but there was no immediately available method to find which of these changes were biologically important. Focusing on a region of chromosomal gain at 1q21 comprising ~30 genes, Houvras, Ceol, Zon and colleagues developed an assay called miniCoopR to rapidly identify the key genes in this region20. Zebrafish harbouring a transgene overexpressing human BRAFV600E (under the melanocyte-specific microphthalmia-associated transcription factor a (mitfa) promoter and in the context of p53 loss of function) were bred to the nacre mutant, which encodes an inactivating mutation of mitfa. These animals are devoid of melanocytes and never develop melanoma. However, when embryos were injected with a rescuing miniCoopR plasmid encoding a mitfa minigene (again under the mitfa promoter), the resulting mosaic adults had partially restored melanocyte stripes and rapidly developed melanoma. This assay was then adapted so that the injected plasmid could contain not only the mitfa minigene but also any other gene of interest. Each of the 30 human genes in the 1q21 region was then systematically tested. More than 3,000 adult animals were screened, and a single gene, SET domain, bifurcated 1 (SETDB1), was found to cooperate with BRAFV600E in mediating melanoma growth. SETDB1 induces this effect partly by overcoming BRAFV600E-mediated senescence, and tumours co-expressing both BRAF and SETDB1 exhibited signs of increased aggressiveness. SETDB1, a histone methyltransferase, was found to target a broad range of transcriptional targets, particularly members of the HOX family. Other groups have since identified the 1q21 interval as a novel melanoma-susceptibility locus for familial melanoma21, solidifying the idea that SETDB1 is a bona fide oncogene in human melanoma.
This study provides one key example of how the zebrafish can be rapidly used to ‘filter’ the vast number of genetic alterations that are found by sequencing human tumour samples and can be used to identify the potential driver effects of such changes. Although similar approaches could be envisioned in human xenograft or mouse models (with short hairpin RNA (shRNA) or cDNA screens), the large number of zebrafish that can be observed in any one experiment provides a powerful incentive to use the fish as an initial screening tool. Furthermore, the power of this assay is that it provides a direct readout of tumour incidence, providing an immediate in vivo context for any genetic hit.
A similar approach was undertaken by Liu and Leach22 using a KRASG12V-driven pancreatic cancer model. In this system, an upstream activating sequence (UAS)–KRASG12V–GFP fusion protein is expressed in the exocrine pancreas, resulting in a 50% penetrance of pancreatic tumours by 5 months. By adapting this system to both transient and stable transgenic lines, almost any other cooperating oncogene can be tested. This work has been described in preliminary form, but the final results will allow both tumour initiation and progression to be assayed. Other work has delineated the role of NMYC and anaplastic lymphoma kinase (ALK) in the promotion of neuroblastoma using transgenic approaches23.
Aside from testing human genomic changes in zebrafish, it is also possible to directly compare the common genomic abnormalities in zebrafish and human tumours. The challenge in this approach is that even human cancers are markedly heterogeneous at the genetic level, so the likelihood of finding ‘common’ genetic drivers across species can be small. Nonetheless, Rudner and Trede24 carried out array comparative genomic hybridization on zebrafish T cell acute lymphoblastic leukaemia (T-ALL) and identified 893 genes that had significant copy number alterations. When multiple zebrafish and human tumours were analysed to look for recurrent events, ten genes were found to be repeatedly altered between the two species. Although this is a small number, these genes are likely to be functionally important, providing a focused set of genomic alterations that may be true drivers of T-ALL.
These types of analyses may also be carried out at the RNA level. Gong and colleagues25,26 compared zebrafish liver tumour signatures with human tumours and found a core set of 76 genes that are highly conserved between the species. These genes are enriched for members of the WNT–β-catenin and RAS–MAPK pathways, which is consistent with what is known about hepatocellular carcinoma (HCC) pathogenesis. Moreover, a subset of these genes was found to correlate with human HCC aggressiveness, demonstrating that the molecular changes found in zebrafish tumours could be used to predict genes that contribute to tumour progression. A similar approach to elucidating transcriptional similarities between zebrafish and human cancers has now been demonstrated for NRASQ61K- and BRAFV600E-driven melanomas27,28 and KRASG12D-driven rhabdomyosarcoma29. This approach suggests that the zebrafish tumour transcriptome can be used to filter down human expression data, but a concerted effort to comprehensively determine whether such filtered genes represent a true core functionality of these cancers awaits study.
Tumour transplantation and in vivo imaging
A core method of assessing tumorigenicity has been the use of tumour cell transplantation, in which tumour cells from a donor can be grown in a recipient either of the same species (allograft) or another species (xenograft). This is typically carried out by transplanting human cells into immunocompromised mice such as nu/nu, non-obese diabetic (NOD)–severe combined immunodeficient (SCID) or NOD interleukin-2 receptor, gamma chain (Il2rg)−/− (NOG)–SCID models. There are two major limitations to studying this process in mice. First, the limitation of animal numbers, as grafting more than ten to 12 recipients per group becomes physically and financially untenable. Second, imaging of the engrafted cells, often using GFP or luminescence, has limited resolution. Transplantation in zebrafish has emerged to overcome these limitations (FIG. 4).
Figure 4 |. Transplantation tools available in the zebrafish.
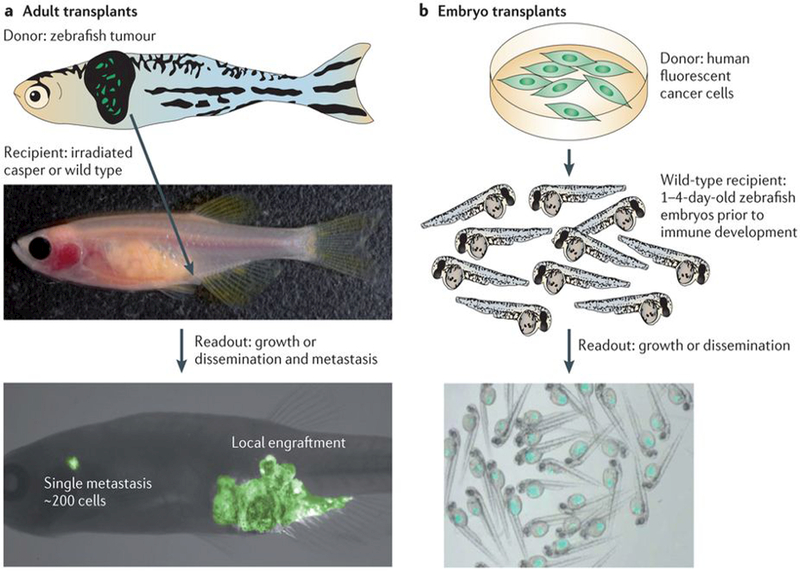
a | Transplantation of primary zebrafish tumour cells (top of the figure) into an irradiated, immunocompromised adult casper or wild-type recipient (middle) allows for detailed assessment of tumour growth and metastasis at single-cell resolution (bottom). b | Transplantation of well-characterized, fluorescently labelled human cell lines or primary human tumours (top) into the zebrafish embryo (middle) reliably leads to engraftment in recipients owing to a lack of immune system development at this time point. Hundreds to thousands of recipients (bottom) can be transplanted in this manner, which can be easily imaged and used for chemical or genetic screening approaches. Images in part a are reproduced, with permission, from REF. 38 © (2008) Cell Press.
Langenau and colleagues14 transplanted zebrafish T cell lymphoma cells, driven by MYC, into sublethally irradiated adult zebrafish recipients14 and found that these cells correctly homed to the thymus, suggesting a supportive microenvironment for these cells in this lymphoid-rich organ. Several other zebrafish models of cancer have also been shown to be transplantable, including models of melanoma27,30,31, pancreatic cancer, HCC32,33, B cell acute lymphoblastic leukaemia (driven by ets variant 6 (ETV6)–runt-related transcription factor 1 (RUNX1))34, SV40-driven germ cell tumours35 and Ewing’s sarcoma (driven by EWS RNA-binding protein 1 (EWSR1)–Friend leukaemia virus integration 1 (FLI1))36.
Detailed imaging of tumour cells in recipient zebrafish can also be achieved, which allows the study of engraftment and of tumour cell subpopulations. Building on work with a zebrafish model of embryonal rhabdomyosarcoma (ERMS)29, subpopulations of fluorescently labelled zebrafish ERMS cells were serially transplanted in limiting dilutions to identify the tumour-initiating cell37, and transgenic animals were then created in which various cell populations were fluorescently labelled. The animals were imaged with confocal microscopy, which revealed that late-stage ERMS showed distinct localization of the tumour-initiating cells away from more differentiated tumour cells. Importantly, the more differentiated tumour cell populations (which are incapable of tumour-initiating activity) were the most likely to intravasate blood vessels, the first key step in metastasis37, raising the intriguing hypothesis that tumour-initiating populations, metastatic populations and fully differentiated populations may all have distinct compartments and molecular identities.
Although the studies discussed above are useful for identifying cellular behaviour after transplantation, it has long been recognized that the opacity of the adult zebrafish provides a limit to cellular resolution during microscopy. Because one of the major strengths of the zebrafish system is the optically transparent embryos, we created an adult zebrafish that retained much of its transparency throughout adulthood, known as the casper model38 (FIG. 4a). The adult casper zebrafish line is translucent, allowing ready visualization of many internal organs, including the brain, heart, spinal cord, eggs, gut, liver and major blood vessels.
Feng, Look and colleagues39 used the casper system to investigate the mechanism of T cell lymphoma dissemination after transplantation39. Whereas transgenic expression of myc induces disseminated T-ALL in zebrafish, concomitant overexpression of bcl2 primarily led instead to localized T cell large lymphoblastic lymphoma. myc+ or myc+bcl2+ red fluorescent tumour cells were transplanted into a casper recipient in which all blood vessel endothelial cells were marked with GFP (the fli1a–GFP zebrafish line), and the myc+ cells could be seen to easily invade the blood vessels, whereas the myc+bcl2+ tumours lined up against the outer surface of those vessels but rarely, if ever, invaded through the vessel wall. By treating the recipient fish with chemicals, they found this effect was due to the overexpression of sphingosine 1-phosphate receptor 1 (S1PR1) by the myc+bcl2+ cells. These results were extended to observation of patient samples of T cell large lymphoblastic lymphoma, implicating a new pathway that regulates the transition between T cell lymphoma and T cell leukaemia and highlighting how in vivo imaging in the zebrafish can lead to novel insights into human cancer.
One important consideration in using the casper system is that the recipient is not wild type — its transparency results from mutations in mitfa and an unknown locus. For this reason, results obtained in casper must be validated by orthogonal methods to ensure that they are not artefacts of these defects. In addition, similar to studies using mice, all transplantation assays require immunosuppression of the recipient, a problem that has not completely been solved in the zebrafish. One advance has been to develop zebrafish tumours in isogenic, clonal lines such as that pioneered by Revskoy and co-workers32,33,40; this allows for the successful serial transplantation of T-ALL, HCC and pancreatic tumours. Although isogenic zebrafish can be difficult to maintain, this is an important advance that will probably be used by more groups in the future.
Xenotransplantation of human cells
The other method for overcoming the issues of immunosuppression is to transplant human cancer cells into transparent zebrafish embryos that do not yet have an intact immune system (FIG. 4b). These studies provide an important method that goes beyond what is typically achievable in mouse xenografts because they allow for detailed, three-dimensional analysis of single cells within an engrafted tumour. Because the embryo recipients are microscopic, thousands of recipients can be studied per day, so studying the effects of drugs on the transplanted cells is amenable to high-throughput approaches. Because the recipient zebrafish is not fully mature and is still undergoing organogenesis, the microenvironment is not fully static, but as a primary screening tool it offers speed and resolution not otherwise achievable.
Marques, Bagowski and colleagues41 fluorescently labelled fragments of human cancer explants derived from patients with gastrointestinal tumours and tracked their behaviour after transplantation into the yolk sac of 2-day-old fli1a–GFP transgenic recipients. Pancreatic cancer cells from these patients rapidly disseminated and formed micrometastases within 24 hours, with a preference for the liver, gut and intestines. When peritumoural pancreatic tissue, which was primarily composed of chronic pancreatitis, was transplanted, the cells failed to invade and migrate, indicating that this assay could be used to predict the ultimate biological behaviour of human tumour cells. The same group used this assay to show that small interfering RNA-mediated knockdown of LIM domain kinase 1 (LIMK1) and LIMK2 in human Panc-1 and Pa-Tu8988T pancreatic cancer cells decreased invasion and metastasis42. Determining whether the LIMKs are good antimetastatic therapeutic targets will take further study, but these results strengthen the in vivo evidence for the role of these proteins across species. Several other studies have since used this assay to examine primary and metastatic growth, angiogenesis and stem cell-like capacity43–47. The embryo assay has recently been adapted to a high-throughput approach using automated image analysis of recipients48, and proof of its usefulness as a tool for discovering therapies relevant to human cancer awaits further study.
One unexpected benefit of transplanting human cancer cells into zebrafish embryos is the availability of readouts of bidirectional crosstalk between tumour cells and the ‘environment’ of the embryo. The plastic developing embryo can act as an in vivo biosensor for factors that are produced by the tumour. Topczewska, Hendrix and colleagues49 used this to understand the role of Nodal signalling in melanoma pathogenesis49 by transplanting fluorescently labelled human melanoma cells into the developing blastula. The tumour cells disrupted embryogenesis, leading to a duplicated body axis that was highly reminiscent of embryonic defects induced by the secreted factor Nodal. This suggested that human melanoma cells secreted Nodal into the local microenvironment, which was subsequently confirmed by RNA and protein analysis. In this assay, the plastic microenvironment of the embryo can act as a readout for factors released by tumour cells that influence the microenvironment. This has not yet been repeated for other tumours but is clearly a unique assay that is achieved by xenografting human cancer cells into zebrafish. As metastasis is now well known to require many cell types within the microenvironment (reviewed in REF. 50), He and Snaar-Jagalska52,111 used the embryo assay to examine the role of neutrophils in this process. Fluorescent tumour cells were injected into the circulation of embryos, which led to widespread dissemination but with a clear concentration in the caudal haematopoietic tissue (CHT), which is known to be an important site of haematopoiesis and leukocyte differentiation53. This led to the hypothesis that leukocytes within that tissue are required for metastatic dissemination. Knockdown of the myeloid transcription factor Pu1 (also known as Spi1b) in recipient zebrafish led to a dramatic decrease in tumour (but not in normal) angiogenesis, indicating that these myeloid cells are required for angiogenesis and subsequent metastasis. This study provides an important link between the microenvironment and cancer cell metastasis, and points the way to future large-scale efforts at dissecting these complex interactions in vivo using zebrafish. Towards this goal, this assay has recently been applied to drug screening to find new inhibitors of tumour growth and metastasis54–56.
Stoletov, Klemke and colleagues57 transplanted fluorescent human breast (MDA-435) or sarcoma (HT1080) cells into the peritoneal cavity of 1-month-old zebrafish larvae, which had been treated with the immunosuppressant dexamethasone. Both cell types formed tumours at the injection site, but only the HT1080 sarcoma cells invaded and metastasized, as has previously been observed in mouse xenografts. They then injected these cells into the fli1a–GFP line and found that the MDA-435 cells were poorly angiogenic, probably accounting for their poor metastatic capacity. When the MDA-435 cells were transduced to overexpress vascular endothelial growth factor A (VEGFA) and transplanted into fli1a–GFP zebrafish, confocal microscopy revealed extensive vessel sprouting from the host vasculature and, most importantly, fenestration of the endothelium. However, these cells remained poorly metastatic because they failed to intravasate through vessel walls, but this could be induced by the overexpression of RHOC in MDA-435 cells. Co-expression of both VEGFA and RHOC allowed the cells to both recruit angiogenic vessels and to intravasate through the endothelial fenestrations. This study provides important in vivo mechanistic data regarding the role of RHOC in invasion and metastasis and is a logical follow-up of genomic studies from human cancer that had previously suggested the role of RHOC in metastasis58.
Chemical genetics comes into focus
The term chemical genetics encompasses two overlapping concepts. First is the idea of using small molecules to uncover a basic biological process, roughly analogous to what can be achieved using genetic mutants. Second is the use of in vivo capabilities of screening chemicals in a whole animal to find molecules with potential therapeutic relevance. Although a seemingly artificial separation, the choice of particular library, assay and downstream assessment of a chemical hit will generally determine the outcome of such approaches.
The zebrafish embryo is an ideal system in which to carry out such screens (FIG. 5). The embryo develops externally and one can obtain thousands of embryos per day, especially with the use of mass-mating chambers such as the iSpawn59. Phenotypes can be screened using bright-field, in situ hybridization or fluorescent microscopy approaches, and can be treated in 96-well format. Finally, one can vary the time of chemical exposure, so that modifiers of almost any cell type can be found if the chemical is applied at the correct developmental window. This is in contrast to genetic mutants, in which the expression of a given phenotype is controlled solely by the period in which maternal RNA and protein contribution runs out during embryonic development and the strength of the particular allele.
Figure 5 |. Chemical screening in the zebrafish.
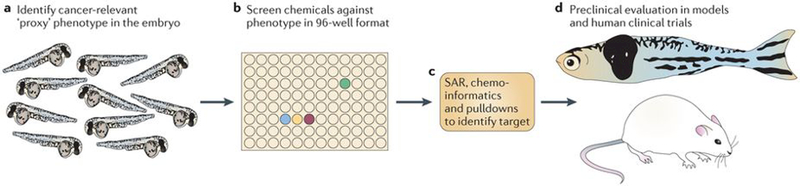
These types of screens are most efficiently carried out on embryos, given their amenability to large-scale, high-throughput manipulation and analysis. a | Identification of an embryonic phenotype that is highly relevant to cancer is a key step in this process. An embryo stained for the neural crest marker crestin is shown; neural crest cells give rise to pigmented melanocytes but also to melanoma in the zebrafish. b | After a relevant embryo phenotype is found, the embryos can be distributed in their chorions to plates, most typically the 96-well format. Each well will receive a distinct small molecule, either manually or with the aid of a liquid-handling robot. This method has been applied to screens ranging from 1,000 to 26,000 molecules. c | Identifying the mechanism of hits is shown. This will primarily depend on the nature of the library used. Molecules with unknown function may require methods such as structure–activity relationships (SARs), chemoinformatics (using algorithms and databases such as PubChem, ChemBank or DiscoveryGate) or pulldowns using tagged versions of drugs and mass spectrometry. For libraries biased towards chemicals with US Food and Drug Administration-approved or known mechanisms, this step can often be rapid, whereas for molecules of unknown function it can take up to 1 year or more. d | While mechanistic evaluation is ongoing, chemicals can be tested for their effects on cancer in multiple downstream assays, including zebrafish cancer models or mouse transgenic and xenograft models. Depending on potency and safety, some of these hits will be amenable to testing in clinical trials in humans.
The zebrafish has long been used to assess the teratogenic effects of chemicals60. However, it was the pioneering work of Schreiber, Peterson and colleagues61 that showed that careful regulation of the timing of drug administration and washout could affect specific organs61. They were the first to carry out an unbiased chemical screen, in which they tested several thousand small molecules for their effects on developmental end points, such as central nervous system, melanocyte, heart and ear development.
An early attempt to apply this approach to cancer was undertaken by Murphey, Zon and colleagues62, who screened for molecules that could rescue the cell cycle defect in a mutant called crash and burn (crb), in which the transcriptional regulator B-myb63 (also known as Mybl2) is mutated. They screened a library of 16,000 molecules (primarily of unknown function) and found a single hit, persynthamide, which almost completely normalized the mutant phenotypes. Unfortunately, the effects of this molecule could not be generalized to mammalian cancer cells.
Building on this experience, we carried out a chemical screen to identify suppressors of the neural crest progenitors that give rise to melanoma28. Transcriptional profiling of melanomas from the Mitfa–BRAFV600E tp53−/− model31 revealed an upregulation of genes such as crestin and SRY-box containing gene 10 (sox10) that are normally only expressed in the embryonic neural crest, the cell type that ultimately gives rise to pigmented melanocytes. Reasoning that these neural crest programmes are important in melanoma growth, we designed a screen using a library of ~2,000 chemicals in zebrafish embryos to find suppressors of the crestin+ lineage using in situ hybridization. We found that inhibitors of the metabolic enzyme dihydroorotate dehydrogenase (DHODH), such as leflunomide, which is used for patients with rheumatoid arthritis64, completely suppressed the expression of crestin and downstream genes such as sox10 and mitfa. Leflunomide primarily acts at the level of the neural crest stem cell and does not act on differentiated derivatives, and inhibition of DHODH was shown to interfere with the transcriptional elongation of key genes that are required for neural crest development (such as mitfa), as well as Myc-target genes that are known to be required for neural crest development65. Leflunomide inhibits the growth of human melanoma cells both in vitro and in mouse xenografts, and a Phase I/II clinical trial of leflunomide in combination with the BRAF inhibitor vemurafenib has been initiated in patients with melanoma (clinical trial ID number: NCT01611675). This is the first example of a zebrafish screen directly leading to a clinical trial in patients with melanoma, the results of which will be instructive for future screens in the zebrafish.
Because of developmental similarities between embryonic T cell lymphoblasts and T cell leukaemia blasts, Ridges, Trede and colleagues66 designed a screen to find suppressors of lymphoblast development that they posited could be therapeutics for T cell leukaemia66. Cross-testing against well-characterized human T cell leukaemia cell lines revealed one potent hit that they named lenaldekar, which is active in human T-ALL mouse xenografts, without substantial toxicity to the mice. Mechanistically, lenaldekar leads to decreased phosphorylation of downstream members of the PI3K–AKT–mTOR pathway, and identification of its direct target awaits further study. The inherent specificity of the screen combined with its potent activity makes this an exciting finding with potential for therapeutic use in patients. Given that only 5–7% of molecules that enter Phase I trials become useful therapeutics, screens such as this are likely to become models for future preclinical testing.
The field of chemical genetics in zebrafish is just emerging, but its usefulness will be proved by the results of clinical trials in humans, which will become increasingly common. The advantages of using zebrafish for chemical screens must be counterbalanced by the reality that chemicals can have very divergent effects in different species, and not everything that works in a zebrafish will work in the human context. Moreover, the fish are ‘bathed’ in the chemical, which does not allow for the establishment of tissue gradients that may be important for differential tissue effects. Finally, most chemical screens are done in zebrafish embryos, not in adult animals, and thus the effects may not be completely relevant to adult cancer phenotypes. The ability to carry out such screens in adults is limited by the larger size and complexity of handling thousands of adult zebrafish, and this has not yet been attempted in a systematic screening setting.
Forward genetic screens
Early experience with the treatment of adult fish with mutagens such as ENU8 suggested that forward genetics could be applied to finding novel, heritable cancer models in the zebrafish. Several studies have now shown this to be the case, each highlighting different technical strengths of this approach.
Because of its well-known role as a tumour suppressor, an effort was made to identify an ENU-based mutation in tp53, a reverse genetics technique known as TILLING. Several mutations in p53 have now been identified, including M214K17, N168K17 and I166T67. These zebrafish p53 mutants are susceptible to a number of heritable cancers, most prominently malignant peripheral nerve sheath tumours (MPNSTs), which are highly aneuploid68. In many respects, the p53-I166T mutation closely models Li–Fraumeni syndrome, and because p53 is inactivated or dysfunctional in most human cancers, this provides a genetic background on which to functionally test cooperation with candidate oncogenes.
A large-scale retroviral-based insertional mutagenesis screen to identify mutations that resulted in embryonic lethality (yielding ~300 mutant lines69) found that a considerable number of the mutants demonstrated inactivation of ribosomal proteins70,71 and heterozygous adults had a propensity to develop MPNSTs, reminiscent of the p53 mutants. Overall, 17 of 28 ribosomal protein mutants developed MPNSTs with varying penetrance, which correlated with overall growth impairment and translational defects. This raises the possibility that defects in the translation machinery might predispose to tumorigenesis, which awaits further validation in human cancer studies more globally, but is supported by the finding that the mTOR pathway, the deregulation of which is common to many human cancers, activates ribosome biogenesis and cap-dependent translation72. Furthermore, it was additionally discovered that four alleles of the Hagomoro insertional mutant were highly susceptible to neuroblastic tumours that have similarities to neuroblastoma73. The tumorigenic phenotype of these zebrafish is due to the upregulation of fibroblast growth factor 8a (fgf8a); this is the first time that this gene has been linked to the pathogenesis of neuroblastoma, but this finding awaits confirmation in human studies. Because genome instability is a hallmark of cancer74,75, Moore, Cheng and colleagues carried out an ENU-based screen to identify mutants that exhibited increased genomic instability and cancer susceptibility76. Spontaneous cancer development was seen in heterozygotes from 12 genome instability lines and in one highly penetrant line (gin10), which resulted in a 9.6-fold increase in spontaneous formation of tumours, including MPNSTs, testicular tumours, primitive neuro-ectodermal tumours, skin carcinoma and adenocarcinomas in the liver, pancreas and kidney. Although several of these mutants await identification, it is an important proof of principle that such methods can be used to find genes that are directly relevant to human cancer. Taking this approach further, mutants that exhibit cell cycle dysregulation, another hallmark of cancer, would also be expected to have alterations in cancer susceptibility. This was tested in an ENU screen identifying haploid zebrafish mutants with alterations in phospho-histone H3 (a marker of cellular proliferation), which revealed a total of seven altered phenotypes77,78. These included the crb-mutant phenotype78, as well as the cease and desist mutant phenotype, which is caused by a mutation in extra spindle poles like 1 (espl1; also known as separase)79. Both of these mutants have altered cancer phenotypes. When treated with the mutagen MNNG, the crb mutants showed a twofold increase in tumour formation, and the cease and desist mutants showed an increased rate of cancer and an eightfold increase in epithelial tumours. Taken together, these results suggest that both bmyb and espl1 act as tumour-suppressor genes in certain contexts and demonstrate how cellular defects elucidated in germline embryonic mutants are connected to cancer phenotypes.
The approaches discussed above have relied heavily on ‘proxy’ phenotypes, using a connection between an embryonic phenotype and tumour formation. To understand tumour formation more directly, several groups have used ENU screens to identify heritable cancer-associated alterations in specific organs. Neumann, Amatruda and colleagues77 identified a mutant fish phenotype named testicular germ cell tumour (tgct), which has a marked susceptibility to germ cell tumours both spontaneously and post-treatment with the mutagen MNNG77. This is due to a truncating mutation in bone morphogenetic protein receptor type 1bb (bmpr1bb; also known as alk6b)80, which causes a decrease in downstream bone morphogenetic protein (BMP) signalling, which is also seen in 60–90% of human well-differentiated germ cell tumours. Whether manipulation of BMP levels in patients with germ cell tumours has therapeutic relevance has yet to be established, but this screen helped to frame the unsuspected role of this pathway in the human condition. Similarly, zebrafish with mutations in leucine-rich repeat-containing protein 50 (Lrrc50; also known as Dnaaf1) unexpectedly developed seminomas, mutation of which was also found in human families with this disease23. This is a good example of how genes discovered in zebrafish can lead to the search for unexpected genotypes in human patients. Frazier, Trede and colleagues81 mutagenized transgenic p56lck–enhanced GFP (EGFP) zebrafish (in which thymocytes are marked with EGFP) and identified several mutant phenotypes with abnormal T cell development, three of which (shrek (srk), hulk (hlk) and oscar the grouch (otg)) then developed T cell malignancies that showed clonal T cell receptor rearrangements and were serially transplantable. The identification of the genes affected in these mutant fish awaits further study but will undoubtedly shed new light on the pathogenesis of T cell leukaemia, as these genes are sufficient, as single oncogenic hits, to cause adult leukaemia. Recent advances in using whole-genome or RNA sequencing approaches for the identification of mutant loci from forward genetic screens will make the utsefulness of this approach even more feasible for most laboratories82–84.
More recent approaches have capitalized on the mutational capacity offered by transposon-based approaches, especially those using the Sleeping Beauty system85. For example, 700–6,800 unique insertional events in individual zebrafish were mapped and 10% of the zebrafish developed tumours. Importantly, within the tumours, insertions tended to occur in genes that are known to be involved in cancer from mammalian systems, suggesting that this system can be adapted to forward genetic approaches for various tumour types.
The next decade of zebrafish in cancer biology
Many of the technologies described above are reaching technical maturity and are now widely available. The zebrafish is a unique model that has begun to shed light on cancer biology. The question for the field over the next 10 years is how does the zebrafish uniquely add to our knowledge of cancer biology in ways that are complementary, but distinct, from more mainstream models? We highlight below areas that are likely to be particularly fruitful and of great interest to cancer biologists.
Large-scale reverse genetics for identifying tumour suppressors
Most transgenic zebrafish models have relied on the overexpression of dominant-acting oncogenes. Comparatively few studies have interrogated tumour-suppressor genes, although recent work, such as with PTEN, points in the right direction51. However, many human cancer mutations occur in putative tumour suppressors or in genes with unknown functions. Complicating this is the fact that many genes can have both tumour suppressor and oncogenic functions, depending on cellular context.
Loss-of-function studies using shRNA approaches in human cell culture and mouse models have developed rapidly over the past few years but are hampered by the difficulties of studying a large number of candidate tumour-suppressor genes. This is one area in particular where the zebrafish model can contribute through two emerging technologies. First, through the use of TALlike effector nucleases (TALENs) and the CRISPR–Cas systems as extremely efficient and predictable methods of engineering genetic knockouts86,87. Second, through advances in RNA interference in the zebrafish88 using miR-30-based approaches, which are now commonly used in mouse shRNA studies89. This will allow tissue-specific and inducible knockdowns of almost any candidate gene. Capitalizing on these two technologies, along with the thousands of potentially injectable embryos generated per day in the zebrafish, comprehensively testing hundreds or thousands of potential tumour suppressors gleaned from human cancer-sequencing studies now seems possible. Because so many transgenic oncogene cancer models now exist, it will be straightforward to test these putative tumour-suppressor genes in those transgenic backgrounds.
Modelling multigenic changes in cancer
A second related issue is that cancer genomes harbour thousands or tens of thousands of mutations. The vast majority of mouse models study one to four genes at a time, although developments in creating targeted changes in embryonic stem cells and using these to generate chimeric animals holds promise for increasing this number89. Nonetheless, the simplicity of creating multigenic transgenic zebrafish models of cancer is unmatched. One can conceptualize creative extensions of the miniCoopR system to test not only two cooperating genes, but five, ten or more. This is a crucial missing piece of all current cancer models: the full range of human cancer is not a one- to four-gene disease but is a multigenic disease, and in vivo models must begin to address this reality and embrace this complexity. The zebrafish is particularly well suited to this challenge, and it will begin to allow for a rational assessment of how complex genetic changes function together to mediate cancer initiation and progression.
Epigenetic modifications in cancer
Rapid progress in analysing the epigenome of cancer is underway, assisted by the data that have emerged from the ENCODE consortium. There is little doubt that many cancers have an epigenetic component. Recent data in human leukaemia suggest that genetic and epigenetic abnormalities are linked: somatic mutations in epigenetic regulators such as TET2, isocitrate dehydrogenase 1 (IDH1), IDH2, additional sex combs-like protein 1 (ASXL1), enhancer of zeste homologue 2 (EZH2) and DNA methyltransferase 3A (DNMT3A) highlight these complex interdependencies (reviewed in REF. 90). The ability to carry out high-quality chromatin immunoprecipitation followed by deep sequencing91, as well as methylation profiling92,93, is rapidly emerging for zebrafish and will complement the work being carried out in mammalian systems. Moreover, the capacity for interrogating the phenotypic effects of knocking down or overexpressing epigenetic factors in transgenic cancer models will be a crucial way to dissect, in an unbiased manner, the roles of epigenomics in various cancers.
Immunological approaches
Recent success using antibody approaches against programmed cell death protein 1 (PD1) and cytotoxic T lymphocyte-associated protein 4 (CTLA4)94,95 have renewed the excitement in the tumour immunology field, offering the possibility of long-term remissions or cures by modulating the host immunological response to tumour cells. Although the zebrafish immune system clearly differs from the human system, fundamental principles of the way in which genetically distinct tumour cells are detected or immunoedited by the immune system will have direct relevance to immunotherapy of human cancer. This can be effectively studied through small-molecule screens or genetic screens to identify novel regulators of core immune functions.
Identifying cell-intrinsic and microenvironmental regulators of metastasis
The combination of zebrafish imaging, genetics, small-molecule screens and transgenics provides a novel method for interrogating both cell-intrinsic and microenvironmental regulators of metastasis. Metastasis is an evolving, multistep process, the study of which requires better animal models. The ability to easily manipulate tumours and their microenvironment on a large scale in zebrafish provides a unique opportunity to study this process in vivo. Many zebrafish transplant studies using the fli1a–GFP line have looked at how tumour cells interact with the vascular endothelium, but this cell type is only one of many in the microenviroment50. As transgenics that mark almost any cell type are now available, it is easy to envision many investigations into how tumour cells interact with fibroblasts, macrophages, keratinocytes, epithelial cells, haematopoietic stem cells, osteoblasts, and others. For example, Feng, Martin and colleagues96 showed that the induction of oncogenes such as HRAS in the melanocyte lineage leads to the recruitment of endogenous myeloperoxidase-positive neutrophils, indicating that one of the earliest host responses to oncogene activation is an inflammatory reaction. This type of study shows how transgenesis and in vivo imaging can come together in the zebrafish, and this approach will become even more powerful as such studies are layered onto genetic mutants with defects in any of these cell types in order to truly define which microenvironmental regulators are required for each step in metastasis.
Because of the heterogeneity of metastasis, our group has optimized a metastasis transplant assay using the casper zebrafish (FIG. 4a). Initial studies capitalized on the pigmented nature of zebrafish melanomas to visualize not only tumour growth at the transplant site but also single metastatic cells far from the transplant site. More recently, we have developed stable zebrafish melanoma cell lines marked by GFP that can be used for such transplant studies and that do not rely solely on pigmentation, which can vary widely among tumour cells (R.W., unpublished observations). Such lines can be used in high-throughput assays to probe metastatic factors in vivo.
Final conclusions
The concept of using zebrafish as a cancer model, proposed just over a decade ago, has now begun to yield results. We are optimistic that the community of researchers using this system will continue to grow robustly. It is incumbent on us to interact deeply with the mainstream cancer biology community who use human and mouse systems so that there is a bilateral recognition that each model system offers a unique set of tools for analysing tumour biology. Although the technologies described here are in varying stages of development, novel and unexpected tools will undoubtedly come to the forefront in the next decade, as the zebrafish community has a strong record of leading technological changes in biology research.
Key points.
Zebrafish develop cancer that is histologically and genetically similar to that of humans and it can develop spontaneously, after mutagen exposure or through transgenesis.
Large-scale transgenesis using hundreds to thousands of embryos per day allows for deep functional characterization of genetic abnormalities that have been discovered in human cancer studies, such as The Cancer Genome Atlas.
Chemical screens have successfully been used in zebrafish embryos to find molecules and pathways that are directly relevant to human malignancy, and drugs from these screens have entered clinical trials in patients.
Large-scale forward genetic screens offer a unique opportunity to identify cancer phenotypes in an unbiased manner.
Transparent embryos and adults allows for in vivo visualization of cancer growth and progression at single-cell resolution.
Xenotransplantation of human cancer cells into zebrafish larvae provides opportunities for personalized cancer screens.
Major areas of growth in the coming decade include modelling multigenic changes in cancer, epigenetics and the dissection of metastasis.
Glossary
- GISTIC
(Genomic identification of significant targets in cancer). An algorithm for the identification of copy number gains and losses in human cancer, using genome-wide data such as high-density single nucleotide polymorphism arrays.
- Caudal haematopoietic tissue
(CHT). A region of the zebrafish embryo that acts as an intermediate site of embryonic blood development.
- TILLING
(Targeting induced local lesions in genomes). A technique to identify specific mutations in a gene of interest. Unbiased mutagenesis of the genome using ethylnitrosourea is carried out, followed by PCR-based sequencing of the region of interest to identify zebrafish with the preferred mutation.
- Li-Fraumeni syndrome
An autosomal-dominant condition caused by an inherited mutation in p53 that renders humans highly susceptible to a number of cancers, including breast cancer, leukaemia and sarcoma.
- CRISPR-Cas
Clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated (Cas).
- miR-30 based approaches
A technique for improving the efficiency of in vivo RNA interference. A short hairpin RNA against a gene of interest is embedded within an endogenous microRNA 30 (miR-30) backbone, which allows for high-level expression from RNA polymerase II promoters.
Footnotes
Forward genetic screens
An approach in which germline mutations are induced by mutagens such as and offspring are scored for phenotype of interest. The causal mutation is then identified in mutants with interesting phenotypes.
Competing interests statement
The authors declare competing financial interests: see Web version for details.
References
- 1.Zhang J et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger MF et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485, 502–506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driever W et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37–46 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Haffter P et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123, 1–36 (1996). References 4 and 5 are landmark papers describing the use of the zebrafish in a phenotypic forward genetic screen. [DOI] [PubMed] [Google Scholar]
- 6.Dimitrijevic N et al. Activation of the Xmrk proto-oncogene of Xiphophorus by overexpression and mutational alterations. Oncogene 16, 1681–1690 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Pliss GB, Zabezhinski MA, Petrov AS & Khudoley VV Peculiarities of N-nitramines carcinogenic action. Arch. Geschwulstforsch 52, 629–634 (1982). [PubMed] [Google Scholar]
- 8.Beckwith LG, Moore JL, Tsao-Wu GS, Harshbarger JC & Cheng KC Ethylnitrosourea induces neoplasia in zebrafish (Danio rerio). Lab Invest 80, 379–385 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Spitsbergen JM et al. Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz[a] anthracene by two exposure routes at different developmental stages. Toxicol. Pathol. 28, 705–715 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Spitsbergen JM et al. Neoplasia in zebrafish (Danio rerio) treated with N-methyl- Nʹ-nitro-N-nitrosoguanidine by three exposure routes at different developmental stages. Toxicol. Pathol 28, 716–725 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Culp P, Nusslein-Volhard C & Hopkins N High-frequency germ-line transmission of plasmid DNA sequences injected into fertilized zebrafish eggs. Proc. Natl Acad. Sci. USA 88, 7953–7957 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S, Yang S & Hopkins N lacZ expression in germline transgenic zebrafish can be detected in living embryos. Dev. Biol 161, 77–83 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Lin S et al. Integration and germ-line transmission of a pseudotyped retroviral vector in zebrafish. Science 265, 666–669 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Langenau DM et al. Myc-induced T cell leukemia in transgenic zebrafish. Science 299, 887–890 (2003). The first description of a transgenic cancer model in the zebrafish. [DOI] [PubMed] [Google Scholar]
- 15.Berghmans S et al. Making waves in cancer research: new models in the zebrafish. Biotechniques 39, 227–237 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Stoletov K & Klemke R Catch of the day: zebrafish as a human cancer model. Oncogene 27, 4509–4520 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Berghmans S et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl Acad. Sci. USA 102, 407–412 (2005). The first description of a conserved tumour-suppressor function of p53 in zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin WM et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res 68, 664–673 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceol CJ et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–517 (2011). An in vivo genetic overexpression screen that used human oncogenomic data with in vivo melanoma modelling in the adult zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macgregor S et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nature Genet 43, 1114–1118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S & Leach SD Screening pancreatic oncogenes in zebrafish using the Gal4/UAS system. Methods Cell Biol 105, 367–381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu S et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 21, 362–373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudner LA et al. Shared acquired genomic changes in zebrafish and human T-ALL. Oncogene 30, 4289–4296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam SH et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nature Biotech 24, 73–75 (2006). A comprehensive analysis demonstrating the conservation of transcriptomic signatures between zebrafish and human liver cancer. [DOI] [PubMed] [Google Scholar]
- 26.Lam SH & Gong Z Modeling liver cancer using zebrafish: a comparative oncogenomics approach. Cell Cycle 5, 573–577 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Dovey M, White RM & Zon LI Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish 6, 397–404 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White RM et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518–522 (2011). A chemical genetic screen in zebrafish to identify small-molecule suppressors of melanoma progenitors, the results of which have led to human clinical trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langenau DM et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev 21, 1382–1395 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santoriello C et al. Kita driven expression of oncogenic HRAS leads to early onset and highly penetrant melanoma in zebrafish. PLoS ONE 5, e15170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patton EE et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol 15, 249–254 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Mizgireuv IV & Revskoy SY Transplantable tumor lines generated in clonal zebrafish. Cancer Res 66, 3120–3125 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Mizgirev I & Revskoy S Generation of clonal zebrafish lines and transplantable hepatic tumors. Nature Protoc 5, 383–394 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Sabaawy HE et al. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc. Natl Acad. Sci USA 103, 15166–15171 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill JA et al. Enforced expression of Simian virus 40 large T-antigen leads to testicular germ cell tumors in zebrafish. Zebrafish 7, 333–341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen AT et al. An inducible krasV12 transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis. Model. Mech 5, 63–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ignatius MS et al. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell 21, 680–693 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White RM et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 (2008). A paper describing the development of the transparent casper transplant model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng H et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell 18, 353–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith AC et al. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood 115, 3296–3303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marques IJ et al. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer 9, 128 (2009). A demonstration of the feasibility of transplanting human cancer cells into embryonic zebrafish as a readout for metastatic capacity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vlecken DH & Bagowski CP LIMK1 and LIMK2 are important for metastatic behavior and tumor cell-induced angiogenesis of pancreatic cancer cells. Zebrafish 6, 433–439 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Rouhi P et al. Pathological angiogenesis facilitates tumor cell dissemination and metastasis. Cell Cycle 9, 913–917 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Stoletov K et al. Visualizing extravasation dynamics of metastatic tumor cells. J. Cell Sci 123, 2332–2341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao C et al. A novel xenograft model in zebrafish for high-resolution investigating dynamics of neovascularization in tumors. PLoS ONE 6, e21768 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao C et al. Distinct contributions of angiogenesis and vascular co-option during the initiation of primary microtumors and micrometastases. Carcinogenesis 32, 1143–1150 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Eguiara A et al. Xenografts in zebrafish embryos as a rapid functional assay for breast cancer stem-like cell identification. Cell Cycle 10, 3751–3757 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Ghotra VP et al. Automated whole animal bio-imaging assay for human cancer dissemination. PLoS ONE 7, e31281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Topczewska JM et al. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nature Med 12, 925–932 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Joyce JA & Pollard JW Microenvironmental regulation of metastasis. Nature Rev. Cancer 9, 239–252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choorapoikayil S, Kuiper RV, de Bruin A & den Hertog J Haploinsufficiency of the genes encoding the tumor suppressor Pten predisposes zebrafish to hemangiosarcoma. Dis. Model. Mech 5, 241–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patton EE Live imaging in zebrafish reveals neu(trophil) insight into the metastatic niche. J. Pathol 227, 381–384 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Bertrand JY et al. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 134, 4147–4156 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murphy EA et al. Disruption of angiogenesis and tumor growth with an orally active drug that stabilizes the inactive state of PDGFRβ/B-RAF. Proc. Natl Acad. Sci. USA 107, 4299–4304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corkery DP, Dellaire G & Berman JN Leukaemia xenotransplantation in zebrafish--chemotherapy response assay in vivo. Br. J. Haematol 153, 786–789 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Wang C et al. Rosuvastatin, identified from a zebrafish chemical genetic screen for antiangiogenic compounds, suppresses the growth of prostate cancer. Eur. Urol 58, 418–426 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Stoletov K, Montel V, Lester RD, Gonias SL & Klemke R High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc. Natl Acad. Sci. USA 104, 17406–17411 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clark EA, Golub TR, Lander ES & Hynes RO Genomic analysis of metastasis reveals an essential role for RhoC. Nature 406, 532–535 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Adatto I, Lawrence C, Thompson M & Zon LI A new system for the rapid collection of large numbers of developmentally staged zebrafish embryos. PLoS ONE 6, e21715 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Leeuwen CJ, Grootelaar EM & Niebeek G Fish embryos as teratogenicity screens: a comparison of embryotoxicity between fish and birds. Ecotoxicol. Environ. Saf 20, 42–52 (1990). [DOI] [PubMed] [Google Scholar]
- 61.Peterson RT, Link BA, Dowling JE & Schreiber SL Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc. Natl Acad. Sci. USA 97, 12965–12969 (2000). The first systematic chemical screen in zebrafish. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stern HM et al. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nature Chem. Biol 1, 366–370 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Saville MK & Watson RJ B-Myb: a key regulator of the cell cycle. Adv. Cancer Res 72, 109–140 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Golicki D et al. Leflunomide in monotherapy of rheumatoid arthritis: meta-analysis of randomized trials. Pol. Arch. Med. Wewn. 122, 22–32 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Hong SK, Tsang M & Dawid IB The mych gene is required for neural crest survival during zebrafish development. PLoS ONE 3, e2029 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ridges S et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood 119, 5621–5631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parant JM, George SA, Holden JA & Yost HJ Genetic modeling of Li-Fraumeni syndrome in zebrafish. Dis. Model. Mech 3, 45–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang G et al. Highly aneuploid zebrafish malignant peripheral nerve sheath tumors have genetic alterations similar to human cancers. Proc. Natl Acad. Sci. USA 107, 16940–16945 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amsterdam A et al. Identification of 315 genes essential for early zebrafish development. Proc. Natl Acad. Sci. USA 101, 12792–12797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amsterdam A et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2, E139 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai K et al. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev. Dyn 238, 76–85 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guertin DA & Sabatini DM Defining the role of mTOR in cancer. Cancer Cell 12, 9–22 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Amsterdam A et al. Zebrafish Hagoromo mutants up-regulate fgf8 postembryonically and develop neuroblastoma. Mol. Cancer Res 7, 841–850 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanahan D & Weinberg RA The hallmarks of cancer. Cell 100, 57–70 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Moore JL, Rush LM, Breneman C, Mohideen MA & Cheng KC Zebrafish genomic instability mutants and cancer susceptibility. Genetics 174, 585–600 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neumann JC, Dovey JS, Chandler GL, Carbajal L & Amatruda JF Identification of a heritable model of testicular germ cell tumor in the zebrafish. Zebrafish 6, 319–327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shepard JL et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc. Natl Acad. Sci. USA 102, 13194–13199 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shepard JL et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev 21, 55–59 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neumann JC et al. Mutation in the type IB bone morphogenetic protein receptor Alk6b impairs germ-cell differentiation and causes germ-cell tumors in zebrafish. Proc. Natl Acad. Sci. USA 108, 13153–13158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frazer JK et al. Heritable T-cell malignancy models established in a zebrafish phenotypic screen. Leukemia 23, 1825–1835 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miller AC, Obholzer ND, Shah AN, Megason SG & Moens CB RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res 23, 679–686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hill JT et al. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res 23, 687–697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bowen ME, Henke K, Siegfried KR, Warman ML & Harris MP Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics 190, 1017–1024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McGrail M et al. Somatic mutagenesis with a Sleeping Beauty transposon system leads to solid tumor formation in zebrafish. PLoS ONE 6, e18826 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bedell VM et al. In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dahlem TJ et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 8, e1002861 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Rienzo G, Gutzman JH & Sive H Efficient shRNA-mediated inhibition of gene expression in zebrafish. Zebrafish 9, 97–107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Premsrirut PK et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell 145, 145–158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shih AH, Abdel-Wahab O, Patel JP & Levine RL The role of mutations in epigenetic regulators in myeloid malignancies. Nature Rev. Cancer 12, 599–612 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Ganis JJ et al. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev. Biol 366, 185–194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu SF et al. DNA methylation profiling in zebrafish. Methods Cell Biol 104, 327–339 (2011). [DOI] [PubMed] [Google Scholar]
- 93.Goll MG & Halpern ME DNA methylation in zebrafish. Prog. Mol. Biol. Transl. Sci 101, 193–218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wolchok JD et al. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med 369, 122–133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hamid O et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med 369, 134–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feng Y, Santoriello C, Mione M, Hurlstone A & Martin P Live imaging of innate immune cell sensing of transformed cells in zebrafish larvae: parallels between tumor initiation and wound inflammation. PLoS Biol 8, e1000562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park SW et al. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology 134, 2080–2090 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Langenau DM et al. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA 102, 6068–6073 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feng H et al. Heat-shock induction of T‑cell lymphoma/ leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br. J. Haematol 138, 169–175 (2007). [DOI] [PubMed] [Google Scholar]
- 100.Chen J et al. NOTCH1‑induced T‑cell leukemia in transgenic zebrafish. Leukemia 21, 462–471 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Blackburn JS et al. Notch signaling expands a pre-malignant pool of T‑cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia 26, 2069–2078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Le X et al. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc. Natl Acad. Sci. USA 104, 9410–9415 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ju B, Spitsbergen J, Eden CJ, Taylor MR & Chen W Co‑activation of hedgehog and AKT pathways promote tumorigenesis in zebrafish. Mol. Cancer 8, 40 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhuravleva J et al. MOZ/TIF2‑induced acute myeloid leukaemia in transgenic fish. Br. J. Haematol 143, 378–382 (2008). [DOI] [PubMed] [Google Scholar]
- 105.Chu CY et al. Overexpression of Akt1 enhances adipogenesis and leads to lipoma formation in zebrafish. PLoS ONE 7, e36474 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leacock SW et al. A zebrafish transgenic model of Ewing’s sarcoma reveals conserved mediators of EWS‑FLI1 tumorigenesis. Dis. Model. Mech 5, 95–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Z et al. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet‑on xmrk transgenic zebrafish. J. Hepatol 56, 419–425 (2012). [DOI] [PubMed] [Google Scholar]
- 108.Yang HW et al. Targeted expression of human MYCN selectively causes pancreatic neuroendocrine tumors in transgenic zebrafish. Cancer Res 64, 7256–7262 (2004). [DOI] [PubMed] [Google Scholar]
- 109.Forrester AM et al. NUP98‑HOXA9‑transgenic zebrafish develop a myeloproliferative neoplasm and provide new insight into mechanisms of myeloid leukaemogenesis. Br. J. Haematol 155, 167–181 (2011). [DOI] [PubMed] [Google Scholar]
- 110.Liu NA et al. Targeting zebrafish and murine pituitary corticotroph tumors with a cyclin-dependent kinase (CDK) inhibitor. Proc. Natl Acad. Sci. USA 108, 8414–8419 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.He S et al. Neutrophil-mediated experimental metastasis is enhanced by VEGFR inhibition in a zebrafish xenograft model. J. Pathol 227, 431–445 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]


