Abstract
The BRCA-Fanconi anemia (FA) pathway preserves the genome and suppresses cancer and is a main determinant of chemotherapeutic efficacy. The hereditary breast cancer genes BRCA1 and BRCA2 function in DNA double-strand break repair mediating distinct steps of homologous recombination (HR). More recently, independent of DNA repair, functions in the replication stress response have come to light, providing insight as to how the BRCA-FA pathway also balances genome preservation with proliferation. The BRCA-FA proteins associate with the replisome and contribute to the efficiency and recovery of replication following perturbations that slow or arrest DNA replication. Although the full repertoire of functions in the replication stress response remains to be elucidated, the function of BRCA1 and BRCA2 in protecting stalled replication forks contributes along with HR to the sensitivity of BRCA-associated tumors to chemotherapy. Moreover, chemoresistance evolves from restoration of either HR and/or fork protection. Although mechanisms underlying the restoration of HR have been characterized, it remains less clear how restoration of fork protection is achieved. Here, we outline mechanisms of “rewired” fork protection and chemotherapy resistance in BRCA cancer. We propose that mechanisms are linked to permissive replication that limits fork remodeling and therefore opportunities for fork degradation. Combating this chemoresistance mechanism will require drugs that inactivate replication bypass mechanisms.
BRCA-FA PATHWAY FUNCTIONS BEYOND DNA REPAIR
Deficiency in the BRCA-Fancomi anemia (FA) pathway has widespread physiological consequences. Germ-line mutations in the hereditary breast cancer genes such as BRCA1 and BRCA2 are highly penetrant and predispose 20%–80% of carriers to breast and ovarian cancer (Apostolou and Fostira 2013). In addition, biallelic inactivation of BRCA genes causes FA, a rare multigenic disease in which loss of any of the 21 distinct complementation groups, FANC-A, FANC-B, FANC-C, …, FANC-T, confers disease (for reviews, see Bhattacharjee and Nandi 2017; Cheung and Taniguchi 2017). Patients are characterized by progressive bone marrow failure, developmental defects, and cancer predisposition. FA patient cells are also exquisitely sensitive to agents that induce DNA interstrand cross-links (ICLs). Consequently, research has focused on understanding the function of the BRCA-FA pathway in ICL repair. In particular, given the role of BRCA proteins in fixing double-strand DNA breaks by homologous recombination (HR), this function was thought to extend to the repair of ICL-induced breaks. However, essential functions in preserving replication have emerged that are independent of HR. This role was not readily apparent because BRCA-FA-deficient cells are not universally sensitive to fork slowing or stalling drugs. Recent advances in studying the biology of DNA replication and associated machinery have clarified roles in replication and the replication stress response and provide a new perspective for understanding BRCA-FA disease and the function of BRCA-FA proteins in cell survival.
BRCA-FA PROTEINS PROTECT STALLED REPLICATION FORKS FROM DEGRADATION
A key initial finding linking the BRCA-FA proteins to the replication stress response was the observation that BRCA2 protects stalled replication forks from break formation following treatment with hydroxyurea (HU) that decreases the generation of deoxyribonucelotides through inhibition of ribonucleotide reductase (Lomonosov et al. 2003). More recently, BRCA1, BRCA2, FANCD2, and FANCA were shown to protect nascent DNA strands of replication forks stalled by HU. Mechanistically, BRCA1 and BRCA2 protect nascent DNA at stalled forks from MRE11-mediated degradation by loading RAD51 on ssDNA exposed at stalled forks (Schlacher et al. 2011; Ying et al. 2012; Chaudhuri et al. 2016). Correspondingly, in the absence of BRCA1 or BRCA2 nascent DNA strands of stalled replication forks undergo extensive nucleolytic degradation leading to long stretches of single-stranded DNA (ssDNA) (Schlacher et al. 2011, 2012; Kolinjivadi et al. 2017b). Consistent with this mode of action, a degradation phenotype also underlies a RAD51 mutation in patients with an FA-like phenotype (Ameziane et al. 2015; Wang et al. 2015; Zadorozhny et al. 2017). Thus, RAD51 is a key mediator of BRCA function in HR at DNA breaks and in protecting nascent DNA at stalled forks. In both settings, RAD51 loading on ssDNA could restrict MRE11 resection to initiate recombination-based mechanisms to promote repair at breaks and restart at stalled forks. The mechanistic overlap of HR proteins operating in DNA repair and DNA replication were recently reviewed (Kolinjivadi et al. 2017a).
Synchrony in fork processing is established in part by the fact that factors regulating DNA end resection also contribute to fork slowing and subsequent restart mechanisms. Consequently, loss of several BRCA-FA factors causes not only fork degradation phenotypes but also slowing and restart defects following replication stress. In particular, RAD51, FANCD2, FANCM, FAN1, and the FANCD2–FAN1 interaction ensure that replication elongation is slowed in response to HU (Luke-Glaser et al. 2010; Lossaint et al. 2013; Zellweger et al. 2015; Lachaud et al. 2016). BRCA-FA-deficient cells also display defects in slowing replication in response to ICLs that were proposed to underlie the pronounced G2/M arrest and sensitivity (Sala-Trepat et al. 2000). Even during unperturbed replication, RAD51 loss leads to the accumulation of ssDNA gaps that are visible by electron microscopy (EM) indicating its role in normal replication (Lopes et al. 2006; Hashimoto et al. 2010). Gaps could result from discontinuous replication if DNA synthesis skips over barriers and reinitiates downstream by employing the primase, PrimPol (Guilliam and Doherty 2017). Whatever the source of gaps, they could lead to extensive resection that underlies fork degradation phenotypes that characterize loss of BRCA-FA proteins. Gaps could be initiating sites for remodelers that mediate aberrant processing in BRCA-FA cells (Kolinjivadi et al. 2017a). Future work will ideally reveal how fork slowing, reversal, protection, and recombination are ultimately coordinated and/or linked with other BRCA-FA pathway functions including replication restart (Petermann et al. 2010; Schwab et al. 2013; Raghunandan et al. 2015; Lemacon et al. 2017; Rondinelli et al. 2017), regulation of new origin firing (Thompson et al. 2017), and traversing the replisome past replication blocking lesions (Huang et al. 2013).
DNA REPAIR AND FORK PROTECTION UNDERLIES BRCA1/2 GENOME STABILITY AND TUMOR SUPPRESSION FUNCTIONS
These recently identified roles for BRCA-FA pathway in replication fork stability raise the possibility that maintenance of replication fork stability may contribute to its genome preservation and tumor suppression functions. The idea that replication dysfunction independent of HR drives cancer formation is in part supported by analysis of mammary epithelial cells from BRCA1 mutation carriers that have not yet developed cancer. These premalignant cells were found to show functional HR and DNA damage checkpoint signaling but defects in the protection of stalled replication forks (Pathania et al. 2014). Moreover, cells carrying BRCA2 heterozygous truncating mutations showed extensive replication stress–induced fork degradation (Tan et al. 2017). Thus, loss of DNA repair may not be the only crucial factor in the etiology of BRCA cancer. More likely, it could be a combined loss of and/or coordination between DNA repair and DNA replication that undermines the maintenance of genome integrity and tumor suppression.
Until recently, restoration of the HR pathway was the only described mechanism by which BRCA1/2-mutant cancers survive genotoxins, such as cisplatin or inhibitors of poly(ADP-ribose) polymerase (PARPi) (Edwards et al. 2008; Sakai et al. 2008, 2009). In BRCA1-mutant cells, restoration of HR is achieved by several mechanisms including reversion mutations or loss of the nonhomologous end joining (NHEJ) factor, 53BP1 (Bunting et al. 2010, 2012). Conversely, in BRCA2-mutant cells, restoration of HR is solely achieved by reversion mutations (Edwards et al. 2008; Sakai et al. 2008, 2009). The finding that approximately half of BRCA2-mutant cancers develop chemoresistance in the absence of restored HR suggested that reversion-independent resistance mechanisms awaited identification (Norquist et al. 2011). To define genetic determinants of cisplatin resistance, we performed a genome-wide short-hairpin RNA (shRNA) screen and found that loss of the chromatin remodeler, CHD4, mediates resistance to cisplatin and PARPi in BRCA2-mutated cancer (Guillemette et al. 2015). CHD4 depletion did not restore HR but reduced the levels of chromosomal aberrations in BRCA2-mutant cells exposed to cisplatin (Guillemette et al. 2015). The elevated resistance and genomic stability correlated with reduced MRE11 chromatin association and increased fork protection (Chaudhuri et al. 2016). A series of recent publications concur that rewired fork protection confers chemoresistance independent of HR. This is achievable in BRCA1/2-deficient cells by several mechanisms, including loss of the chromatin modifier complex MLL3–4/PTIP/MRE11, the fork remodelers, SMARCAL1, HLTF, or ZRANB3, PARP1, the methyltransferase EZH2, and the negative regulator of RAD51, RADX (Chaudhuri et al. 2016; Ding et al. 2016; Dungrawala et al. 2017; Kolinjivadi et al. 2017b; Taglialatela et al. 2017; Vujanovic et al. 2017), as well as through up-regulation of FANCD2 (Kais et al. 2016; Michl et al. 2016). The clinical significance of this mechanism is confirmed by the poor patient response and survival outcomes observed upon restored fork protection (Guillemette et al. 2015; Chaudhuri et al. 2016; Rondinelli et al. 2017). Thus, these factors may prove useful as potential biomarkers of BRCA1/2-deficient tumor response to chemotherapy.
DEFINING DEGRADATION FACTORS AND FORK DYNAMICS IN BRCA1/2-DEFICIENT CELLS
A common feature of rewired fork protection is that nuclease activity is tempered by various mechanisms. In particular, in BRCA1/2-mutant cells, suppressing MRE11-nucleolytic degradation restores fork protection. This can be done with the MRE11 inhibitor mirin or by reestablishing Rad51 filament formation at stalled forks through expression of a RAD51 mutant lacking ATPase activity or by depletion of the anti-RAD51 factor, RADX (Schlacher et al. 2011; Dungrawala et al. 2017). In addition, loss of factors that facilitate the recruitment of MRE11 to stalled forks such as PARP1, PTIP, or CHD4 restores fork protection in BRCA1/2-deficient cells (Table 1; Chaudhuri et al. 2016; Ding et al. 2016). Given that MRE11 has limited nucleolytic activity (Cannavo and Cejka 2014), other nucleases likely contribute to fork degradation. In particular, CtIP initiates the MRE11-dependent degradation that is extended by EXO1 (Lemacon et al. 2017). Although it remains debated whether DNA2 contributes to degradation in BRCA1/2-deficient cells (Chaudhuri et al. 2016; Spies et al. 2016; Kolinjivadi et al. 2017b; Lemacon et al. 2017), DNA2 degrades forks in cells deficient in Abro1, a paralog of the BRCA1-interacting protein, Abraxas (Xu et al. 2017). An MRE11-independent mechanism has also been described in which the nuclease MUS81 contributes to fork degradation in a pathway with EZH2 (Table 1; Rondinelli et al. 2017).
Table 1.
Simplified overview of factors whose loss restores fork protection and/or chemoresistance in BRCA1/2-deficient cells
| Factor | Function | Fork Protection | Chromosome stability | Chemoresistance | Clinical Relevance | Publication(s) |
|---|---|---|---|---|---|---|
| MRE11 | Nuclease that degrades nascent DNA in stalled replication forks | Inhibition restores fork protection in BRCA2- depleted (V-C8), BRCA1- depleted U2OS and BRCA1- mutant (UWB1) cells. | Schlacher et al. 2011 and 2012; Ying et al. 2012; Lemacon et al. 2017; Mijic et al. 2017 | |||
| CHD4 | Recruits MRE11 to stalled fork | Depletion restores fork protection in BRCA2- mutant (PEO1) cells. | Depletion protects against chromosomal aberrations in BRCA2-mutant (PEO1) cells (cisplatin). | Depletion increased resistance in BRCA2-mutant (PEO1, Capanl, FA- D1) cells (cisplatin/PARPi). | Low CHD4 correlated with reduced progression-free survival (PFS) in BRCA2- mutant ovarian cancers | Guillemette et al. 2015; Chaudhuri et al. 2016 |
| PARP1 | Recruits MRE11 to stalled fork, fork reversal activity | Deletion restores fork protection in Brcat1−/− or Brca2−/−B cells and Brca2−/− mESC. | Deletion protects against chromosomal aberrations in Brca2−/− mESC and Brca1−/−B cells (cisplatin, camptothecin). | PARP1 deletion reduces tumor- free survival in Brca2−/− mouse model. | Chaudhuri et al. 2016; Ding et al. 2016 | |
| PTIP | Recruits MRE11 to stalled fork | Depletion restores fork protection in Brca1−/− or Brca2−/−B cells. | Deletion protects against chromosomal aberrations in Brca1−/− or Brca2−/−B cells (HU, cisplatin, camptothecin). | Deletion increased resistance in BRCA1−/−B cells (HU). | Low PTIP correlated with reduced PFS in BRCA2-mutant ovarian cancer | Chaudhuri et al. 2016 |
| HLTF | Fork reversal activity, E3 ubiquitin ligase for PCNA | Depletion restores fork protection in BRCA1- or BRCA2- depleted MCF10A cells. | Taglialatela et al. 2017 | |||
| SMARCAL1 | Fork reversal activity | Depletion restores fork protection in BRCA1- or BRCA2- depleted MCF10A cells or in BRCA2- depleted Xenopus extracts. | Depletion protects against chromosomal aberrations in BRCA1- and BRCA2-depleted MCF10A cells (camptothecin). | Depletion increased resistance in BRCA1-mutant (MDA-MB-436) cells (cisplatin/PARPi). | Low SMARCAL1 correlated with reduced overall survival (OS) in BRCA1-mutant breast cancer | Kolinjivadi et al. 2017b; Taglialatela et al. 2017 |
| ZRANB3 | Fork reversal activity | Depletion restores fork protection in BRCA1- or BRCA2- depleted MCF10A cells. | Depletion protects against chromosomal aberrations in BRCA1- and BRCA2-depleted MCF10A cells (camptothecin), Deletion causes increased chromatid breaks/gaps in BRCA2- depleted U2OS cells (HU). | Vujanovic et al. 2017; Taglialatela et al. 2017; Mijic et al. 2017 | ||
| RAD52 | Fork reversal activity, loads Rad51 onto ssDNA, recruits MRE11 to stalled fork | Depletion restores fork protection in BRCA2- depleted U2OS cells | Depletion protects against chromosomal breakage in BRCA2-depleted U2OS cells (HU). | Mijic et al. 2017 | ||
| RAD51 | Fork reversal activity, binds and protects ssDNA | Depletion restores fork protection in BRCA2- depleted RPE-1 cells | Mijic et al. 2017 | |||
| EXO1 | Nuclease that extends MRE11 fork degradation | Depletion restores fork protection in BRCA2- mutant (PEO1) or BRCA1- mutant (UWB1) cells, and BRCA1- or BRCA2- depleted U2OS cells. | Depletion protects against chromosomal aberrations in BRCA2-depleted U2OS (HU). | Lemacon et al. 2017 | ||
| MLL4 | Induces histone H3 methylation at Lysine 4 (H3K4) to recruit MRE11 to stalled fork | Deletion restores fork protection in Brca1−/− or Brca2−/− B cells. | Deletion protects against chromosomal aberrations in Brca2−/−B cells (PARPi, cisplatin). | Chaudhuri et al. 2016 | ||
| RADX | Inhibits the accumulation of Rad51 at forks | Depletion restores fork protection in BRCA2- depleted U2OS cells, BRCA2 mutant Capanl cells | Depletion increased resistance in BRCA2-depleted U2OS cells (PARPi). | Dungrawala et al. 2017 | ||
| EZH2 | Induces trimethylation of histone H3 at Lysine 27 (H3K27me3) to recruit Mus81 to stalled fork | Depletion restores fork protection in BRCA2- mutant (VU423) cells. | Inhibition protects against chromosomal aberrations in BRCA2-depleted Hela or BRCA2- mutant (VU423) cells (mitomycin C). | Depletion increases resistance in BRCA2-depleted HeLa cells (PARPi, cisplatin). | Low EZH2 correlated with reduced PFS in BRCA2-mutant ovarian cancer EZH2 inhibition promotes relapse in Brca2−/− breast tumor mouse model (PARPi). | Rondinelli et al. 2017 |
Replication fork structures also play a prominent role in nucleolytic degradation. In particular, a reversed replication fork, a so-called “chicken foot,” is the target of MRE11 digestion in BRCA1/2-deficient cells (Mijic et al. 2017). Accordingly, fork reversal has been shown to be a prerequisite for fork degradation in BRCA1/2-deficient cells (Kolinjivadi et al. 2017b). In addition to the remodelers HLTF, SMARCAL1, and ZRANB3 (Kolinjivadi et al. 2017b; Taglialatela et al. 2017), RAD52, PARP1 and RAD51 drive fork reversal–dependent degradation in BRCA1/2-deficient cells (Table 1; Lemacon et al. 2017; Mijic et al. 2017; Ray Chaudhuri et al. 2012). There are numerous proteins known to generate reversed forks such as DNA helicases FBH1, BLM, WRN, RECQL5 and DNA translocases FANCM and RAD54 (Blastyák et al. 2010; Bétous et al. 2012; Ciccia et al. 2012; Fugger et al. 2015; Kile et al. 2015; Neelsen and Lopes 2015). The relevance of these factors to fork degradation in BRCA cancer remains to be investigated. How loss of each remodeler can individually restore fork protection in BRCA1/2-deficient cancer also awaits further analysis. Fork remodelers may act in a common pathway; however, HLTF, ZRANB3, and SMARCAL1 do not form a complex (Taglialatela et al. 2017). It also remains to be determined how fork remodelers interact with the other pathways of chemoresistance such as EZH2 or MLL3–4/PTIP/MRE11, both of which act independently of each other (Rondinelli et al. 2017). Given that the reversed fork structure has exposed DNA ends that mimic DNA double-stranded break ends, it will also be important to determine if in addition to MRE11, these ends are an entry point for NHEJ factors. If so, the reported elevated NHEJ activity that causes genomic instability in BRCA-FA cells (Adamo et al. 2010; Bunting et al. 2012; Pace et al. 2010) may be suppressed by depletion of fork remodelers. However, as described below, preventing fork reversal and subsequent degradation or NHEJ reactions is not always sufficient for chemoresistance in BRCA1/2-deficient tumors.
COMPLEXITY OF REWIRED FORK PROTECTION MECHANISMS
The emerging literature indicates that rewired fork protection mechanisms and their relationship to cell viability, genome stability, and chemoresistance are complex. Rewired fork protection in BRCA1/2-deficient cells is achieved by loss of either of three remodelers (HLTF, SMARCAL1, and ZRANB3), PTIP, or PARP1, but loss of CHD4 or EZH2 is restricted to BRCA2-mutant cells (Guillemette et al. 2015; Ding et al. 2016; Ray Chaudhuri et al. 2016; Rondinelli et al. 2017; Taglialatela et al. 2017). These findings suggest that stalled fork structures or compensating pathways in BRCA1 and BRCA2 cells may be distinct, but the overall strategy of rewired fork protection remains consistent; forks that escape nucleases are protected from degradation. However, not all routes that escape degradation will confer chemotherapy resistance. RAD51, MRE11, or RAD52 are required for viability of BRCA1/2-deficient cells, so their loss will not result in chemoresistance but rather synthetic lethality (Fig. 1; Feng et al. 2011; Ying et al. 2012; Lok et al. 2013). The balance between lethality and survival/chemoresistance also appears dependent on the sequence by which a fork degradation factor is lost, before or after BRCA deficiency. Depletion of PARP1 in BRCA2-deficient cells leads to synthetic lethality (Feng and Jasin 2017), whereas deletion of PARP1 before BRCA2 depletion provides some protective effects (Ding et al. 2016).
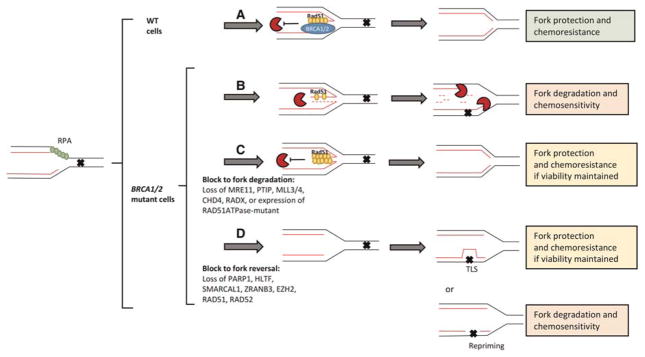
Figure 1.
Model of the mechanisms and potential consequences of fork protection in BRCA1/2 -deficient cells. (A) In wild-type (WT cells), when replication stress (represented by X) is encountered in the course of cancer therapy, there is a reversal of replication forks, and the protection of nascent DNA in a BRCA1-, BRCA2-, and Rad51-dependent manner, that limits resection by nucleases such as MRE11, CtIP, and EXO1. Following TS or recombination-based restart, the replication fork is restarted, thereby conferring chemo-resistance. In absence of BRCA1/2, (B) Rad51 is no longer stabilized on reversed forks, allowing access of nucleases to nascent DNA, resulting in extensive degradation and chemosensitivity. (C) Extensive nascent cell degradation can be avoided by either loss of fork degradation factors or gain in stabilization factors that will allow forks to restart via template switch (TS) and confer chemotherapy resistance if viability is not also compromised. (D) Loss of fork reversal will limit fork degradation if translesion synthesis (TLS) is active at the fork and gaps generated by repriming reactions are avoided.
An additional level of complexity derives from the fact that some proteins possess multiple functions in the replication stress response. For example, RAD51 participates in both the formation and protection of reversed forks (Hashimoto et al. 2010; Zellweger et al. 2015). Therefore, depending on which RAD51 function is disrupted, fork protection or degradation could be altered. Further findings show the importance of cellular context, specifically mutational status, for therapy resistance. For example, CHD4 depletion conferred cisplatin resistance in BRCA2-mutant cell lines harboring truncated BRCA2, but did not improve cisplatin resistance in BRCA2-depleted cells, indicating resistance requires the maintenance of a residual BRCA2-mutant species (SB Cantor, unpubl.). As different cell types show differing levels of reversed forks (Ahuja et al. 2016), this may explain the findings that SMARCAL1 depletion resulted in chemoresistance in the MCF10A cancer cell line but not in nonmalignant mammary epithelial cells, despite both cell lines being BRCA1-deficient (Taglialatela et al. 2017). Likewise, Ptip depletion in Brca2-null mouse B cells improves genome stability and resistance to HU, but in BRCA2-null primary human cells fails to restore viability (Chaudhuri et al. 2016; Feng and Jasin 2017). Ideally future work will unravel these distinctions so that expectations for therapy are better understood.
Finally, it is important to realize that mechanisms of fork protection may vary for different genotoxic agents; mechanisms revealed following treatment of cells with HU may not be informative for mechanism following treatment of cells with cisplatin or PARPi, as these agents have distinct modes of disrupting replication. For example, EZH2 and MUS81 are required for the restart of replication in BRCA2-deficient cells following their release from HU (Lemacon et al. 2017; Rondinelli et al. 2017). HU treatment promotes replication fork reversal (Zellweger et al. 2015), and reversed forks are extensively resected by nucleases in BRCA2-deficient cells. MUS81 cleaves the over-resected reversed DNA forks in BRCA2-deficient cells and promotes POLD3-dependent fork rescue, explaining their dependence on MUS81 for replication fork progression and resistance to HU (Lai et al. 2017; Lemacon et al. 2017). Conversely, following treatment with PARPi, it is less clear how MUS81 depletion achieves PARPi resistance in BRCA2-deficient cells (Rondinelli et al. 2017). In particular, MUS81 is probably not required for fork restart upon treatment with PARPi because PARPi suppresses fork slowing and reversal depriving cells of a MUS81 substrate (Sugimura et al. 2008; Ray Chaudhuri et al. 2012). Likewise, it remains to be determined if similar to depletion of SMARCAL1, ZRANB3 depletion confers cisplatin/PARPi resistance. ZRANB3 depletion restores fork protection in distinct cell systems; however, genomic instability is increased or decreased in BRCA1/2-deficient cells depending on the source of replication stress (Mijic et al. 2017; Taglialatela et al. 2017).
REWIRED FORK PROTECTION AND REPLICATION PROGRESSION WITHOUT FORK REVERSAL
When rewired fork protection confers therapy resistance in BRCA1/2-mutant cells, the mechanism of replication should be quite different than as found in BRCA1/2-proficient cells. In this regard, it is important to note that upon replication stress, replication forks typically slow and reverse (Zellweger et al. 2015). As described above, rewired fork protection achieved through suppression of HLTF, SMARCAL1, ZRANB3, PARP1, and RAD51 is expected to suppress fork reversal. Even though reversed forks are dangerous species that present DNA ends for resection, ssDNA generation, and fork degradation, they also serve to liberate DNA lesions from replisome entanglements, allowing more efficient DNA repair processing. DNA ends in reversed forks also provide DNA substrates for recombination. In addition, the reversed fork is thought to facilitate other strategies for repair or bypass of DNA lesions, such as template switch (TS). TS is an error-free DNA damage tolerance pathway that uses the newly synthesized daughter strand DNA for homology-directed repair to bypass lesions (for review, see Lovett 2017). Access to recombination, TS, and other repair mechanisms through fork remodeling activities is an important genome stabilizing response to stress that limits fork breakage (Ray Chaudhuri et al. 2012; Neelsen and Lopes 2015). Thus, rewired fork protection that limits replication fork slowing and reversal should compromise genomic stability. In BRCA-deficient cancer cells, however, fork reversal could have dire consequences given that its loss provides genomic stability and chemoresistance.
Clues to replication progression mechanisms that confer therapy resistance come from how other activities are altered in BRCA1/2-deficient cells that have rewired fork protection. In particular, depletion of HLTF not only results in loss of fork reversal, but TS is also disrupted. This stems from the role of HLTF as an E3 ubiquitin ligase responsible for the addition of ubiquitination chains on PCNA, an essential step in TS (Branzei 2011; Lin et al. 2011). Rewired fork protection established through ZRANB3 depletion will also alter the replication stress response. Polyubiquitinated PCNA recruits ZRANB3 to sites of replication stress, both of which are critical for fork reversal (Ciccia et al. 2012; Vujanovic et al. 2017) and proposed to mediate TS. In addition to catalyzing the regression of stalled replication forks, ZRANB3 also prevents inappropriate recombination (Ciccia et al. 2012). Therefore, loss of ZRANB3 could elevate aberrant recombination as well as interfere with a polyUb-PCNA-dependent axis required for engagement of TS. Furthermore, loss of ZRANB3-PCNA interaction may allow access of de-ubiquitination enzymes thereby decreasing polyUb-PCNA and in turn increase monoUb-PCNA. If so, loss of ZRANB3, similar to loss of HLTF, could enhance the overall monoUb-PCNA. This result could have great significance as monoUb-PCNA serves as a molecular switch for the error-prone mechanism of translesion synthesis (TLS), a DNA tolerance mechanism that allows the timely bypass of DNA lesions using error-prone TLS polymerases (Choe and Moldovan 2017).
REWIRED FORK PROTECTION VIA TLS AT THE FORK
Could TLS operate at the fork to limit gap formation, maintain replication, and promote chemoresistance in BRCA1/2-deficient cells (Fig. 1)? TLS is best described for filling in gaps postreplication and operates effectively when restricted from S phase (Karras and Jentsch 2010). However, TLS may operate at the fork, as the replisome is passing through the site of DNA damage or other barrier to replication (Sale et al. 2012). Indeed, TLS polymerases enable replication to continue despite DNA damage or other barriers induced by chemotherapy because they have low-fidelity, which allows the insertion of nucleotides opposite to bulky DNA lesions that block high-fidelity replicative DNA polymerases. Moreover, when nucleotide pools are reduced as in HU-treated cells, TLS could also tolerate insertion of mismatched nucleotide (Edmunds et al. 2008; Quinet et al. 2014). Consistent with the role of TLS reactions occurring at the elongating fork, replication fork stalling is observed in cells depleted of TLS polymerases (Quinet et al. 2014, 2016). TLS activity at the fork may also be restricted to a subset of polymerases. Whereas Rev3L operates in postreplicative gap filling, Rev1 and Polη are required for TLS at stalled forks (Quinet et al. 2016).
In considering the role of TLS in rewired fork protection, it is important to note that TLS may be compromised in BRCA-FA cells. Indeed, in some respects, loss of the BRCA-FA pathway phenocopies loss of TLS and vice versa. Sensitivity to ICL-inducing agents is a phenotype attributed to loss of either pathway. Moreover, FA-like phenotypes in the hematopoietic stem cells result from loss of Ub-PCNA (Pilzecker et al. 2017). Pathway interconnections between BRCA-FA and TLS are established by interactions and the employment of common ubiquitin-modifying enzymes (for reviews, see Kim and D’Andrea 2012; Kim et al. 2012; Fu et al. 2013; Tian et al. 2013; Boisvert and Howlett 2014; Choe and Moldovan 2017). Notably the pathways are also genetically linked. Biallelic inactivation of Rev7(FANCV), the regulatory subunit of the TLS polymerase Polζ, underlies the genetic defect in the FA-V complementation group l (Bluteau et al. 2016). One could speculate that TLS defects in BRCA-FA-deficient cells selects for compensatory mechanisms that activate TLS downstream from the BRCA-FA pathway. This could underlie how BRCA-FA cells overcome proliferation defects due to endogenous lesions such as aldehyde-induced damage and R-loops, and/or how bone marrow failure transforms into leukemia in FA patients (García-Rubio et al. 2015). Moreover, a gain in TLS could eliminate mitotic DNA synthesis that compensates for under-replication (Bhowmick et al. 2016) especially in BRCA2-deficient cells (Lai et al. 2017). Indeed, an investigation of the mutational signature in cells deficient in BRCA1 suggests a compensatory up-regulation of TLS (Zamborszky et al. 2017).
Although it may be easier to tip the balance toward TLS in BRCA-FA cancer cells, the rewired replisome may take this a step further. In particular, TLS could suffice to replace replicative polymerases during replication if gaps are very short, which could be possible if nuclease-dependent resection at the fork is blocked or limited. In addition, the simultaneous loss of RAD51 and associated Pol α (Kolinjivadi et al. 2017b) could enable other gap-filling pathways to compensate as the replisome stalls. Indeed, monoUb-PCNA and RAD51 have nonredundant functions in gap filling (Hashimoto et al. 2010). A switch to TLS through a series of modifications on PCNA (Kannouche and Lehmann 2004) and increased chromatin access to stalled forks may underlie the mechanism by which CHD4 loss confers therapy resistance. Indeed, CHD4 loss in BRCA2-mutant cells reduced chromatin bound MRE11 and RAD51, while also elevating focal accumulation of the E3 ubiquitin ligase RAD18 that is necessary for PCNA ubiquitination (Guillemette et al. 2015; Chaudhuri et al. 2016). Notably, chromatin-bound RAD51 in BRCA2-mutant cells is already very low, but CHD4 depletion reduces this further (Guillemette et al. 2015). Thus, by disrupting the residual RAD51 that operates independent of BRCA2 to promote fork reversal (Kolinjivadi et al. 2017b) and/or that captures ssDNA for gap-filling reactions postreplication (González-Prieto et al. 2013), CHD4 loss could block fork reversal and degradation as well as liberate ssDNA for TLS reactions at the fork. Other routes to elevated TLS through changes in chromatin access or RAD18 complexes have been described (Kim et al. 2014; Yamada et al. 2014). We also found that the FANCJ DNA helicase has the ability to aberrantly promote TLS and suppress HR when its interaction with BRCA1 or its carboxy-terminal acetylation are disrupted (Cantor and Nayak 2016; Xie et al. 2010a, 2012). Conceivably, when not properly regulated by BRCA1, FANCJ disrupts fork-remodeling pathways and/or improves TLS efficiency by unfolding DNA secondary structures that interfere with replication. Interestingly, the mismatch repair protein MSH2, which binds secondary structures formed at stalled replication forks, blocks TLS pathways in FANCJ-deficient cells (Peng et al. 2014).
Further characterization will be required to decipher the contribution of TLS to chemoresistance and rewired fork protection in BRCA cancers. Nevertheless, it is an important therapeutic target (Yamanaka et al. 2017) given the growing body of evidence illustrating a causative role for TLS in the development of chemoresistance (Doles et al. 2010; Xie et al. 2010b; Srivastava et al. 2015). Importantly, significant progress has been made in the development of TLS inhibitors (Actis et al. 2013; Korzhnev and Hadden 2016; Sail et al. 2017; Sanders et al. 2017). TLS inhibitors may be efficacious as a first-line combination chemotherapy or prevent chemoresistance via restored fork protection when used in combination with cisplatin or PARPi. It will also be important to determine whether alternative strategies to block HR and/or fork protection mechanisms of chemoresistance such as inhibitors of ATR (Yazinski et al. 2017) disrupt permissive replication mediated by TLS or other pathways at the replication fork.
CONCLUSION
The BRCA-FA pathway displays indispensable roles in maintaining genome stability, suppressing tumors, and mediating chemoresistance. It does this, not only through integral roles in DNA repair, but growing evidence indicates the BRCA/FA pathway contributes to the maintenance of replication forks during times of replication stress. In the absence of replication fork maintenance, BRCA1/2-mutant cancers are capable of “rewiring” the replication fork to allow replication to proceed. We propose that the TLS bypass pathway plays an important part in the mechanism by which replication forks proceed through replication stress. The role of the FA-BRCA pathway in maintaining replication forks and the newly established role for rewired replication forks in chemoresistance provide exciting new possibilities for the development of new chemotherapeutic interventions.
Acknowledgments
We thank the members of the Cantor laboratory for helpful discussions. Special thanks to Dr. George-Lucien Moldovan for a critical reading of the review. This work was supported by National Institutes of Health (NIH) grant R01 CA176166-01A as well as charitable contributions from the Lipp Family Foundation.
References
- Actis M, Inoue A, Evison B, Perry S, Punchihewa C, Fujii N. Small molecule inhibitors of PCNA/PIP-box interaction suppress translesion DNA synthesis. Bioorg Med Chem. 2013;21:1972–1977. doi: 10.1016/j.bmc.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Ahuja AK, Jodkowska K, Teloni F, Bizard AH, Zellweger R, Herrador R, Ortega S, Hickson ID, Altmeyer M, Mendez J, et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat Commun. 2016;7:10660. doi: 10.1038/ncomms10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameziane N, May P, Haitjema A, van de Vrugt HJ, van Rossum-Fikkert SE, Ristic D, Williams GJ, Balk J, Rockx D, Li H, et al. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat Commun. 2015;6:8829. doi: 10.1038/ncomms9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou P, Fostira F. Hereditary breast cancer: The era of new susceptibility genes. Biomed Res Int. 2013;2013:747318. doi: 10.1155/2013/747318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Nandi S. DNA damage response and cancer therapeutics through the lens of the Fanconi Anemia DNA repair pathway. Cell Commun Signal. 2017;15:41. doi: 10.1186/s12964-017-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R, Minocherhomji S, Hickson ID. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol Cell. 2016;64:1117–1126. doi: 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- Blastyák A, Hajdú I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluteau D, Masliah-Planchon J, Clairmont C, Rousseau A, Ceccaldi R, Dubois d’Enghien C, Bluteau O, Cuccuini W, Gachet S, Peffault de Latour R, et al. Biallelic inactivation of REV7 is associated with Fanconi anemia. J Clin Invest. 2016;126:3580–3584. doi: 10.1172/JCI88010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert RA, Howlett NG. The Fanconi anemia ID2 complex: Dueling saxes at the crossroads. Cell Cycle. 2014;13:2999–3015. doi: 10.4161/15384101.2014.956475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D. Ubiquitin family modifications and template switching. FEBS Lett. 2011;585:2810–2817. doi: 10.1016/j.febslet.2011.04.053. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Kozak ML, Kim JM, Wong N, López-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- Cantor SB, Nayak S. FANCJ at the FORK. Mutat Res. 2016;788:7–11. doi: 10.1016/j.mrfmmm.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri AR, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung RS, Taniguchi T. Recent insights into the molecular basis of Fanconi anemia: Genes, modifiers, and drivers. Int J Hematol. 2017;106:335–344. doi: 10.1007/s12185-017-2283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KN, Moldovan GL. Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol Cell. 2017;65:380–392. doi: 10.1016/j.molcel.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell. 2012;47:396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Ray Chaudhuri A, Callen E, Pang Y, Biswas K, Klarmann KD, Martin BK, Burkett S, Cleveland L, Stauffer S, et al. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat Commun. 2016;7:12425. doi: 10.1038/ncomms12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT. Suppression of Rev3, the catalytic subunit of Polζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci. 2010;107:20786–20791. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Bhat KP, Le Meur R, Chazin WJ, Ding X, Sharan SK, Wessel SR, Sathe AA, Zhao R, Cortez D. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol Cell. 2017;67:374–386. e375. doi: 10.1016/j.molcel.2017.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- Feng W, Jasin M. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat Commun. 2017;8:525. doi: 10.1038/s41467-017-00634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Dudimah FD, Zhang J, Pickering A, Paneerselvam J, Palrasu M, Wang H, Fei P. Recruitment of DNA polymerase η by FANCD2 in the early response to DNA damage. Cell Cycle. 2013;12:803–809. doi: 10.4161/cc.23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger K, Mistrik M, Neelsen KJ, Yao Q, Zellweger R, Kousholt AN, Haahr P, Chu WK, Bartek J, Lopes M, et al. FBH1 catalyzes regression of stalled replication forks. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.028. [DOI] [PubMed] [Google Scholar]
- García-Rubio ML, Pérez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, Aguilera A. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Prieto R, Muñoz-Cabello AM, Cabello-Lobato MJ, Prado F. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 2013;32:1307–1321. doi: 10.1038/emboj.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette S, Serra RW, Peng M, Hayes JA, Konstantinopoulos PA, Green MR, Cantor SB. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 2015;29:489–494. doi: 10.1101/gad.256214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliam TA, Doherty AJ. PrimPol-prime time to reprime. Genes (Basel) 2017;8:20. doi: 10.3390/genes8010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kais Z, Rondinelli B, Holmes A, O’Leary C, Kozono D, D’Andrea AD, Ceccaldi R. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 2016;15:2488–2499. doi: 10.1016/j.celrep.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–1013. [PubMed] [Google Scholar]
- Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Kile AC, Chavez DA, Bacal J, Eldirany S, Korzhnev DM, Bezsonova I, Eichman BF, Cimprich KA. HLTF’s ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol Cell. 2015;58:1090–1100. doi: 10.1016/j.molcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Gene Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yang K, Dejsuphong D, D’Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol. 2012;19:164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Dejsuphong D, Adelmant G, Ceccaldi R, Yang K, Marto JA, D’Andrea AD. Transcriptional repressor ZBTB1 promotes chromatin remodeling and translesion DNA synthesis. Mol Cell. 2014;54:107–118. doi: 10.1016/j.molcel.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinjivadi AM, Sannino V, de Antoni A, Técher H, Baldi G, Costanzo V. Moonlighting at replication forks—A new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 2017a;591:1083–1100. doi: 10.1002/1873-3468.12556. [DOI] [PubMed] [Google Scholar]
- Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, Técher H, Baldi G, Shen R, Ciccia A, Pellegrini L, et al. Smarcal1-mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol Cell. 2017b;67:867–881. e867. doi: 10.1016/j.molcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev DM, Hadden MK. Targeting the translesion synthesis pathway for the development of anti-cancer chemotherapeutics. J Med Chem. 2016;59:9321–9336. doi: 10.1021/acs.jmedchem.6b00596. [DOI] [PubMed] [Google Scholar]
- Lachaud C, Moreno A, Marchesi F, Toth R, Blow JJ, Rouse J. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science. 2016;351:846–849. doi: 10.1126/science.aad5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X, Broderick R, Bergoglio V, Zimmer J, Badie S, Niedzwiedz W, Hoffmann JS, Tarsounas M. MUS81 nuclease activity is essential for replication stress tolerance and chromosome segregation in BRCA2-deficient cells. Nat Commun. 2017;8:15983. doi: 10.1038/ncomms15983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemacon D, Jackson J, Quinet A, Brickner JR, Li S, Yazinski S, You Z, Ira G, Zou L, Mosammaparast N, et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat Commun. 2017;8:860. doi: 10.1038/s41467-017-01180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Zeman MK, Chen JY, Yee MC, Cimprich KA. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell. 2011;42:237–249. doi: 10.1016/j.molcel.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–3558. doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Décaillet C, Gari K, Constantinou A. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell. 2013;51:678–690. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- Lovett ST. Template-switching during replication fork repair in bacteria. DNA Repair (Amst) 2017;56:118–128. doi: 10.1016/j.dnarep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010;29:795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl J, Zimmer J, Buffa FM, McDermott U, Tarsounas M. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat Struct Mol Biol. 2016;23:755–757. doi: 10.1038/nsmb.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, Mutreja K, Ursich S, Ray Chaudhuri A, Nussenzweig A, Janscak P, et al. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat Commun. 2017;8:859. doi: 10.1038/s41467-017-01164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W, Karlan BY, Taniguchi T, Swisher EM. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008–3015. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- Pathania S, Bade S, Le Guillou M, Burke K, Reed R, Bowman-Colin C, Su Y, Ting DT, Polyak K, Richardson AL, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Xie J, Ucher A, Stavnezer J, Cantor SB. Crosstalk between BRCA-Fanconi anemia and mismatch repair pathways prevents MSH2-dependent aberrant DNA damage responses. EMBO J. 2014;33:1698–1712. doi: 10.15252/embj.201387530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilzecker B, Buoninfante OA, van den Berk P, Lancini C, Song JY, Citterio E, Jacobs H. DNA damage tolerance in hematopoietic stem and progenitor cells in mice. Proc Natl Acad Sci. 2017;114:E6875–E6883. doi: 10.1073/pnas.1706508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet A, Vessoni AT, Rocha CR, Gottifredi V, Biard D, Sarasin A, Menck CF, Stary A. Gap-filling and bypass at the replication fork are both active mechanisms for tolerance of low-dose ultraviolet-induced DNA damage in the human genome. DNA Repair (Amst) 2014;14:27–38. doi: 10.1016/j.dnarep.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Quinet A, Martins DJ, Vessoni AT, Biard D, Sarasin A, Stary A, Menck CF. Translesion synthesis mechanisms depend on the nature of DNA damage in UV-irradiated human cells. Nucleic Acids Res. 2016;44:5717–5731. doi: 10.1093/nar/gkw280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunandan M, Chaudhury I, Kelich SL, Hanenberg H, Sobeck A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi anemia core complex. Cell Cycle. 2015;14:342–353. doi: 10.4161/15384101.2014.987614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondinelli B, Gogola E, Yucel H, Duarte AA, van de Ven M, van der Sluijs R, Konstantinopoulos PA, Jonkers J, Ceccaldi R, Rottenberg S, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol. 2017;19:1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- Sail V, Rizzo AA, Chatterjee N, Dash RC, Ozen Z, Walker GC, Korzhnev DM, Hadden MK. Identification of small molecule translesion synthesis inhibitors that target the Rev1-CT/RIR protein–protein interaction. ACS Chem Biol. 2017;12:1903–1912. doi: 10.1021/acschembio.6b01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, et al. Secondary mutations as a mechanism of cis-platin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, Wurz K, Higgins J, Villegas E, Taniguchi T. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 2009;69:6381–6386. doi: 10.1158/0008-5472.CAN-09-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat M, Rouillard D, Escarceller M, Laquerbe A, Moustacchi E, Papadopoulo D. Arrest of S-phase progression is impaired in Fanconi anemia cells. Exp Cell Res. 2000;260:208–215. doi: 10.1006/excr.2000.4994. [DOI] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MA, Haynes B, Nangia-Makker P, Polin LA, Shekhar MP. Pharmacological targeting of RAD6 enzyme-mediated translesion synthesis overcomes resistance to platinum-based drugs. J Biol Chem. 2017;292:10347–10363. doi: 10.1074/jbc.M117.792192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shin-Ya K, Niedzwiedz W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J Cell Biol. 2013;201:33–48. doi: 10.1083/jcb.201208009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies J, Waizenegger A, Barton O, Sürder M, Wright WD, Heyer WD, Löbrich M. Nek1 regulates Rad54 to orchestrate homologous recombination and replication fork stability. Mol Cell. 2016;62:903–917. doi: 10.1016/j.molcel.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Han C, Zhao R, Cui T, Dai Y, Mao C, Zhao W, Zhang X, Yu J, Wang QE. Enhanced expression of DNA polymerase η contributes to cisplatin resistance of ovarian cancer stem cells. Proc Natl Acad Sci. 2015;112:4411–4416. doi: 10.1073/pnas.1421365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J Cell Biol. 2008;183:1203–1212. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan R, et al. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol Cell. 2017;68:414–430. e418. doi: 10.1016/j.molcel.2017.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SLW, Chadha S, Liu Y, Gabasova E, Perera D, Ahmed K, Constantinou S, Renaudin X, Lee M, Aebersold R, et al. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017;169:1105–1118. e1115. doi: 10.1016/j.cell.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EL, Yeo JE, Lee EA, Kan Y, Raghunandan M, Wiek C, Hanenberg H, Scharer OD, Hendrickson EA, Sobeck A. FANCI and FANCD2 have common as well as independent functions during the cellular replication stress response. Nucleic Acids Res. 2017;45:11837–11857. doi: 10.1093/nar/gkx847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Sharma S, Zou J, Lin SY, Wang B, Rezvani K, Wang H, Parvin JD, Ludwig T, Canman CE, et al. BRCA1 promotes the ubiquitination of PCNA and recruitment of trans-lesion polymerases in response to replication blockade. Proc Natl Acad Sci. 2013;110:13558–13563. doi: 10.1073/pnas.1306534110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, Taglialatela A, Huang JW, Holland CL, Zwicky K, et al. Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol Cell. 2017;67:882–890. doi: 10.1016/j.molcel.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, et al. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Litman R, Wang S, Peng M, Guillemette S, Rooney T, Cantor SB. Targeting the FANCJ-BRCA1 interaction promotes a switch from recombination to polη-dependent bypass. Oncogene. 2010a;29:2499–2508. doi: 10.1038/onc.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci. 2010b;107:20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Peng M, Guillemette S, Quan S, Maniatis S, Wu Y, Venkatesh A, Shaffer SA, Brosh RM, Jr, Cantor SB. FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response. PLoS Genet. 2012;8:e1002786. doi: 10.1371/journal.pgen.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Wu X, Wu L, Castillo A, Liu J, Atkinson E, Paul A, Su D, Schlacher K, Komatsu Y, et al. Abro1 maintains genome stability and limits replication stress by protecting replication fork stability. Genes Dev. 2017;31:1469–1482. doi: 10.1101/gad.299172.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Masai H, Bartek J. Regulation and roles of Cdc7 kinase under replication stress. Cell Cycle. 2014;13:1859–1866. doi: 10.4161/cc.29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Chatterjee N, Hemann MT, Walker GC. Inhibition of mutagenic translesion synthesis: A possible strategy for improving chemotherapy? PLoS Genet. 2017;13:e1006842. doi: 10.1371/journal.pgen.1006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, Ho CK, Todorova Kwan T, Morris R, Lauffer S, Nussenzweig A, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31:318–332. doi: 10.1101/gad.290957.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72:2814–2821. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- Zadorozhny K, Sannino V, Belan O, Mlcouskova J, Spirek M, Costanzo V, Krejci L. Fanconi-anemia-associated mutations destabilize RAD51 filaments and impair replication fork protection. Cell Rep. 2017;21:333–340. doi: 10.1016/j.celrep.2017.09.062. [DOI] [PubMed] [Google Scholar]
- Zamborszky J, Szikriszt B, Gervai JZ, Pipek O, Poti A, Krzystanek M, Ribli D, Szalai-Gindl JM, Csabai I, Szallasi Z, et al. Loss of BRCA1 or BRCA2 markedly increases the rate of base substitution mutagenesis and has distinct effects on genomic deletions. Oncogene. 2017;36:746–755. doi: 10.1038/onc.2016.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]