Abstract
The asymmetric outcome of female meiosis I, whereby an entire set of chromosomes are discarded into a polar body, presents an opportunity for selfish genetic elements to cheat the process and disproportionately segregate to the egg. Centromeres, the chromosomal loci that connect to spindle microtubules, could potentially act as selfish elements and “drive” in meiosis. We review the current understanding of the genetic and epigenetic contributions to centromere identity and describe recent progress in a powerful model system to study centromere drive in mice. The progress includes mechanistic findings regarding two main requirements for a centromere to exploit the asymmetric outcome of female meiosis. The first is an asymmetry between centromeres of homologous chromosomes, and we found this is accomplished through massive changes in the abundance of the repetitive DNA underlying centromeric chromatin. The second requirement is an asymmetry in the meiotic spindle, which is achieved through signaling from the oocyte cortex that leads to asymmetry in a posttranslational modification of tubulin, tyrosination. Together, these two asymmetries culminate in the biased segregation of expanded centromeres to the egg, and we describe a mechanistic framework to understand this process.
Sexual reproduction in eukaryotes depends on a haploid–diploid life cycle. Meiosis, the process by which haploids are generated, provides an opportunity for genetic elements to compete for transmission to the offspring because each gamete carries only one of the two alleles of a gene. According to Mendel’s Law of Segregation (First Law), alleles are transmitted with equal probability, but it is increasingly clear that this law can be violated, and segregation can be manipulated by selfish genetic elements through meiotic drive. The impact of meiotic drive on many aspects of evolution and genetics is now recognized, with examples widespread across eukaryotes (Werren 2011; Rice 2013; Helleu et al. 2015; Lindholm et al. 2016), but the underlying cell biological mechanisms are largely unknown. Selfish elements can drive by eliminating competing gametes (e.g., sperm killing or spore killing) or by increasing their transmission to the egg in female meiosis, which is the focus of this review.
Because of its inherent asymmetry, female meiosis provides a clear opportunity for selfish elements to cheat: Only chromosomes that segregate to the egg can be transmitted to offspring, whereas the rest are degraded in polar bodies (Sandler and Novitski 1957; Pardo-Manuel de Villena and Sapienza 2001). The centromere drive hypothesis (Fig. 1A–C; Henikoff et al. 2001) proposes that a centromere, as the locus that directs chromosome segregation, can act as a selfish element by increasing its own transmission through female meiosis at the expense of the homologous chromosome. The hypothesis was formulated to explain the paradox that although centromere function is essential for eukaryotic cell division and highly conserved, both repetitive centromere DNA and centromere-binding proteins have evolved rapidly. Because the basic mechanisms of kinetochore assembly at centromeres and interactions with spindle microtubules (MTs) are similar across many eukaryotes, the expectation is that purifying selection would minimize amino acid changes. Key centromere proteins such as CENP-A and CENP-C, however, show strong signatures of positive selection based on the ratio of synonymous to nonsynonymous substitutions in their coding sequences (Malik and Henikoff 2001; Talbert et al. 2004; Schueler et al. 2010; Zedek and Bureš 2016).
Figure 1.
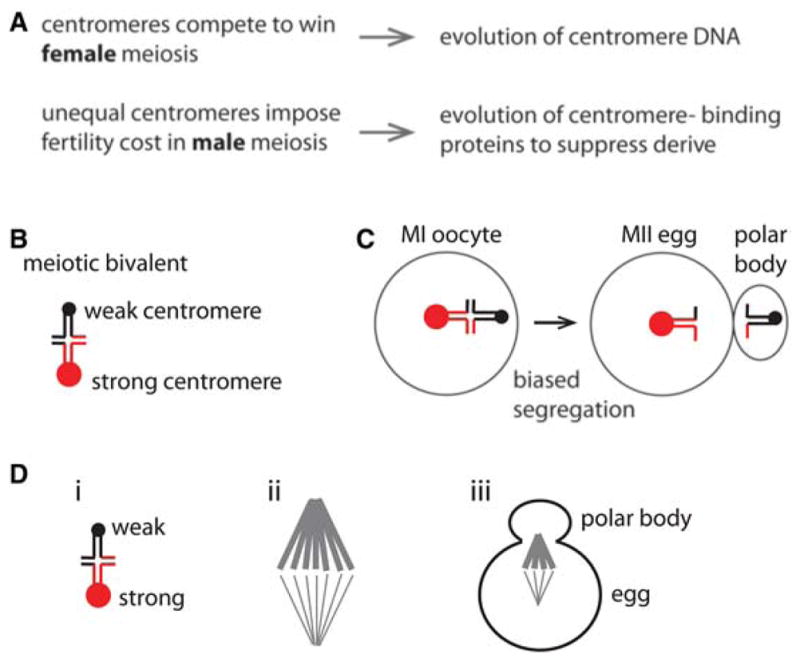
The centromere drive hypothesis. (A) Hypothesis for the evolution of centromere DNA and proteins. (B) A meiotic bivalent with unequal centromeres (red and black circles). (C) Biased segregation with the stronger centromere remaining in the egg. (D) A meiotic bivalent with weaker and stronger centromeres (i) preferentially orients on an asymmetric spindle (ii), which orients relative to the cortex where the polar body forms (iii).
The centromere drive hypothesis has two parts. First, evolution of centromere DNA is driven by competition to orient toward the spindle pole that will remain in the egg, and expansion of repetitive sequences (or other changes to these sequences) at a centromere somehow leads to preferential orientation. The second part explains the evolution of centromere proteins through conflict between individual centromeres, which expand to gain an advantage in female meiosis, and the rest of the genome. Differences between centromeres of homologous chromosomes, which lead to biased segregation in female meiosis, may also impose a fitness cost such as reduced male fertility. This cost would provide selective pressure favoring alleles of centromere-binding proteins that equalize centromeres and suppress drive by binding independent of sequence.
This review focuses on cell biological and molecular mechanisms for centromere drive in female meiosis, based on recent work in a mouse model system. Conceptually, drive depends on three conditions (Fig. 1D). The first is asymmetry in female meiotic cell division and cell fate, which is well established and a universal feature of sexual reproduction in animals (Gorelick et al. 2016). The second is a difference between the centromeres of homologous chromosomes that influences their segregation, discussed in Part 1 below. The third is asymmetry in the meiotic spindle that can be exploited by selfish centromeres if they preferentially attach to the egg side, discussed in Part 2.
PART 1: CENTROMERES
A classic example of selfish chromosome behavior involves the heterochromatic knob locus in maize (Rhoades 1942; Yu et al. 1997). In other situations, centromeres can represent the selfish element (Fishman and Saunders 2008; Iwata-Otsubo et al. 2017). For both noncentromeric and centromeric selfish elements in meiotic chromosome drive, the unifying concept is that they direct advantageous molecular interactions with the meiotic spindle that positively bias the transmission of the chromosome in which they reside. Knobs generate a new structure that interacts directly with spindle MTs (Yu et al. 1997) using a special minus-end-directed kinesin motor protein (K Dawe, pers commun), bypassing the typical connection that occurs through a centromere-localized kinetochore. Centromeric selfish elements could, in principal, function by biasing connections to the egg-oriented side of an asymmetric spindle.
Centromeres have long been considered the “black box” of the chromosome because in animals and most eukaryotes the underlying DNA is highly repetitive. Thus, the balance of genetic and epigenetic contributions to centromere identity and function remains difficult to nail down (Fig. 2). For instance, does the DNA sequence at centromeres even matter? As mentioned above, centromeric DNA is very rapidly evolving (Henikoff et al. 2001; Dumont and Fachinetti 2017). In addition, it has long been appreciated that the repetitive DNA at human centromeres is neither necessary nor sufficient for centromere function (Earnshaw and Migeon 1985; Depinet et al. 1997; du Sart et al. 1997; Warburton et al. 1997; Eichler 1999). On the other hand, human artificial chromosomes (HACs) have been reported to require specific forms of higher-order structures of human centromeric repeats (Harrington et al. 1997; Schueler et al. 2001) or artificial amplification of non-HAC forming higher-order centromere repeats (Hayden et al. 2013). Further, there is a requirement in HAC formation for the only known DNA sequence specific centromere-binding protein in metazoans, CENP-B (Ohzeki et al. 2002; Okada et al. 2007), arguing for an important genetic contribution. CENP-B also has direct physical connections to CENP-A and CENP-C at the centromere that contribute to centromere function (Fachinetti et al. 2015; Hoffmann et al. 2016). Strong data support a major epigenetic contribution to centromere identity, where the histone H3 variant, CENP-A, forms nucleosomes that are fundamental to specifying centromere location (for review, see Black and Cleveland 2011). To this point, it is possible to seed a new functional centromere capable of epigenetic propagation by initially directing a local high density of CENP-A nucleosome assembly (Barnhart et al. 2011; Mendiburo et al. 2011; Ohzeki et al. 2012; Hori et al. 2013; Chen et al. 2014; Logsdon et al. 2015; Tachiwana et al. 2015). It is attractive to think that the balance of genetic and epigenetic forces at the centromere could be at the center of a molecular “tug-of-war” that drives rapid centromere evolution (Henikoff et al. 2001). At a bare minimum, the strong evidence for both genetic and epigenetic contributions to centromere function requires that both should be considered when trying to decipher the molecular mechanisms of centromere drive in any particular branch of the eukaryotic evolutionary tree.
Figure 2.
Foundations for our understanding of a balance between genetic and epigenetic contributions to human centromere identity. (A) Example of centromere silencing and de novo formation on the same chromosome. Anti-centromere antisera (ACA) recognize both CENP-A at the neocentromere (arrowhead) and CENP-B at the silenced centromere at the original location (asterisk). Scale bar, 2 μm. (B) Example of a HAC assay for functional centromeric DNA. The small arrow denotes the HAC formed by functional X chromosome centromere DNA, whereas the arrowhead indicates the centromere from a natural copy of the X chromosome. X chromosome centromere DNA FISH is in red. CENP-E immunofluorescence is in green. (C) HAC formation results indicating a requirement for both CENP-B and the CENP-B box within α-satellite DNA. WTR, wild type repeat; MTR, mutant repeat. (D) Example of HAC formation (artificial chromosome, AC) in mouse cells (left), and instances where a HAC failed to form, integrating (Int) into a natural chromosome instead (center and right). Scale bar, 2 μm. (E) Seeding centromeric chromatin that can form a functional kinetochore. Tethering a Lac repressor (LacI) fusion with the CENP-A binding domain (HJURPScm3) of HJURP to a Lac repressor (LacO) array assembles CENP-A nucleosomes, leading to formation of functional centromeric chromatin that can recruit kinetochore components such as the microtubule-binding protein, Ndc80. Scale bar, 5 μm.
(A, Adapted, with permission, from Bassett et al. 2010; B, adapted from Schueler et al. 2001, with permission from The American Association for the Advancement of Science; C,D, adapted from Okada et al. 2007, with permission from Elsevier; E, adapted, with permission, from Barnhart et al. 2011.)
The mouse model has emerged as an exciting system to reveal the balance of epigenetic and genetic influences in the molecular arms race at centromeres. It has already provided a strong system to interrogate the cell biological basis for meiotic drive. A key early finding by one of our laboratories (Lampson’s), was that different natural and laboratory strains of mice have “stronger” and “weaker” centromeres. Two key specific findings were that (1) stronger centromeres accumulated more of the kinetochore protein, Hec1/Ndc80, and (2) the imbalanced centromeres between homologous chromosomes caused aberrant alignment between the poles of the metaphase spindle of meiosis I (Chmátal et al. 2014). A hypothesis to emerge from these studies was that the larger kinetochore of the stronger centromere strains was built on centromeric chromatin that was somehow more attractive to recruiting kinetochore components than their weaker centromere strain counterparts.
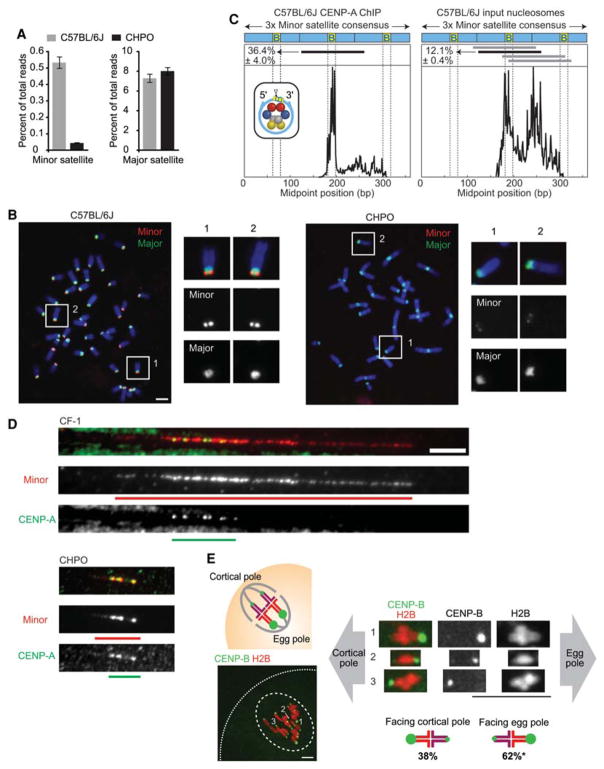
With the CENP-A nucleosome as the candidate to serve this centromeric chromatin role, we initially considered three possible ways in which this chromatin could be altered: one implicating the CENP-A protein and two implicating the DNA that wraps the CENP-A-containing histone octamer. For the CENP-A protein, because it is so extremely long-lived that it has no measurable turnover at mouse oocyte centromeres (Smoak et al. 2016), we considered that it could have substitutions in its primary sequence that might affect centromere strength through the germline of a hybrid animal. However, the strong centromere strains (CF-1 and C57BL/6J) and weak centromere strain (CHPO) all have identical protein sequences (Iwata-Otsubo et al. 2017). Thus, we focused on the CENP-A nucleosomal DNA (Fig. 3), and we considered both the sequence and the abundance of the repeating monomeric unit of mouse centromere DNA (termed “minor satellite”). Biochemical isolation of CENP-A nucleosomes and nuclease (MNase) digestion of total chromatin coupled to sequencing (CENP-A native chromatin immunoprecipitation [ChIP]-seq and MNase-seq, respectively) indicated that the sequences of minor satellite monomers were very homogeneous within a strain and also very similar between strains (Iwata-Otsubo et al. 2017). On the other hand, the sequencing experiments and complementary fluorescence in situ hybridization (FISH) experiments revealed that stronger centromeres contain six- to 10-fold more minor satellite DNA than do weaker centromeres (Fig. 3A,B; Iwata-Otsubo et al. 2017). This increase leads to a similarly large increase of the CENP-B protein on stronger centromeres because the minor satellite monomer contains its recognition element, the CENP-B box (Masumoto et al. 1989). CENP-A abundance at a centromere, and downstream centromere components (e.g., its direct binding partner, CENP-C), is limited by the expression levels of itself and proteins, such as its chromatin assembly factor, HJURP, in the epigenetic pathway for centromeric chromatin assembly (Zasadzińska and Foltz 2017). Nonetheless, CENP-A and CENP-C are increased on stronger centromeres (Iwata-Otsubo et al. 2017) to a similar extent, as is Hec1/Ndc80 (Chmátal et al. 2014). The massive differential in CENP-B levels facilitated tracking stronger and weaker centromeres of bivalent chromosomes in meiosis I. Remarkably, we observed biased orientation of the stronger centromeres toward the egg pole, with the weaker centromeres oriented toward the cortex and destined to be discarded into the polar body (Fig. 3E; Iwata-Otsubo et al. 2017). Thus, our findings explain the molecular basis for strengthening centromeres through expansion of the assembly site for CENP-A nucleosomes.
Figure 3.
Amplified mouse minor satellite repeats act as selfish elements in female meiosis. (A) Quantitation of the MNase-seq reads from a stronger centromere strain (C57BL/6J) and weaker centromere strain (CHPO). Weaker centromeres have a much smaller amount of minor satellite DNAwhile maintaining a similar level of major satellite DNA relative to stronger centromeres. (B) FISH analysis also shows that weaker centromeres have only very low levels of minor satellite DNA. Scale bar, 5 μm. (C) CENP-A nucleosomes are specifically phased with one primary position within the minor satellite repeat monomer unit. CENP-A ChIP-seq analysis of midpoints of CENP-A nucleosomes shows a striking enrichment for a single primary assembly site within the monomer unit of mouse minor satellite (three tandem monomers are shown in the diagram at top; the horizontal black line indicates the primary CENP-A nucleosome assembly site). Notably, the midpoint (dyad axis of symmetry of the nucleosome, marked by a triangle in the inset nucleosome schematic) is within the CENP-B box. Canonical nucleosomes are the major form of nucleosome on minor satellite DNA in stronger centromere mouse strains, and the input “bulk” nucleosomes are not nearly as well-phased compared to CENP-A nucleosomes. The CENP-A specific phasing suggests a specific connection between the genetic and epigenetic factors involved in specifying mouse centromeres. (D) CENP-A nucleosomes fill the minor satellite region at weaker centromeres. Images of CENP-A and minor satellite localized by immunofluorescence and FISH, respectively, on extended chromatin fibers from stronger (CF1, top images) or weaker (CHPO, bottom images) centromere strains. Green and red bars show the length of CENP-A and minor satellite signals, respectively. Scale bar, 5 μm. CENP-A nucleosomes fill the minor satellite region at weaker centromeres (bottom panels) but not stronger centromeres (top panels). (E) Stronger centromeres orient preferentially to the egg in meiosis I. Schematic shows bivalents in CF-1 × CHPO oocytes, with CF-1 centromeres facing the egg. Image shows a CF-1 × CHPO oocyte expressing CENP-B-EGFP and H2B-mCherry, shortly before anaphase onset; dashed white lines show cortex and spindle outline. The orientation of each bivalent was determined using CENP-B-EGFP intensity to distinguish CF-1 (brighter) and CHPO (dimmer) centromeres. Asterisk indicates that it is significantly different from 50%. Scale bar, 10 μm. (Adapted, with permission, from Iwata-Otsubo et al. 2017.)
Two other important findings emerged from our genomic analysis of centromeric chromatin in the stronger and weaker centromere strains. First, nucleosomes containing CENP-A, but not those containing its canonical counterpart, histone H3, are faithful to a single nucleosome assembly site within the monomer minor satellite sequence (Fig. 3C; Iwata-Otsubo et al. 2017). Outside of the strictly genetically defined centromere of budding yeast (Clarke and Carbon 1980; Furuyama and Biggins 2007), this is the first example of which we are aware to indicate such a strong and specific positioning of CENP-A nucleosomes. Further, the center of the nucleosomal DNA (the so-called nucleosomal dyad position) falls within the CENP-B box (Fig. 3C). It is now important to test the possibility that the CENP-B protein plays an important role in positioning CENP-A nucleosomes and in mediating centromere drive. Such tests are made feasible by the fact that CENP-B is nonessential in mice (Hudson et al. 1998), unlike other centromere proteins (e.g., CENP-A and CENP-C) (Kalitsis et al. 1998; Howman et al. 2000). Second, we found that a substantial fraction of CENP-A nucleosomes shift to the adjacent major satellite DNA but only in weaker centromeres (Iwata-Otsubo et al. 2017). It is not clear if this weak centromere strain-specific shift has any functional consequence in major satellite DNA, because there it remains >100-fold more dilute per unit length of DNA compared to its enrichment on minor satellite DNA. For the minor satellite positions where the functional centromere resides, our cytological (Fig. 3D) and genomic findings that weaker centromere strains have a far higher density of CENP-A nucleosomes (Iwata-Otsubo et al. 2017) (as high as half of all nucleosomes on minor satellite DNA), compared to stronger centromere strains where CENP-A is only a very minor chromatin component (found in <8% of nucleosomes), suggest that minor satellite nucleosome assembly sites account for centromere strength.
Very broadly, our findings add an important genetic contribution on top of epigenetic contributions known to be important for centromere identity and strength. Our favored model is that the abundance of minor satellite plays a central role in determining centromere strength, leading to preferential orientation on the spindle of the meiotic oocyte and retention in the egg. It should be noted, however, that evidence in flies supports the notion that simply altering the levels of CENP-A in the male germline in one generation can influence the amount of CENP-A in the next generation (Raychaudhuri et al. 2012). A pressing issue to resolve in the future is the extent to which such epigenetic forces shape the strength of centromeric chromatin in the female germline in flies, in our mouse model for drive, and in other eukaryotes. When thinking broadly about eukaryotic evolution, it is important to note that such a fundamental chromosomal process as chromosome inheritance through the germline could be biased through multiple pathways. Sorting out the molecular basis (genetic and/or epigenetic) of centromere selfishness between closely related strains/species will likely require direct experimental interrogation in every case. Thinking back to the classic example of meiotic chromosome drive in maize where noncentromeric heterochromatic knobs direct connections to the spindle (Rhoades 1942; Yu et al. 1997), one must also always consider potential mechanisms of biased chromosome inheritance that bypass the centromere altogether.
PART 2: MEIOTIC SPINDLE OF THE OOCYTE
The biased orientation observed with hybrid bivalents in the CHPO × CF-1 model system (Fig. 3E) implies some asymmetry within the spindle that is exploited by stronger centromeres to increase their transmission to the egg. Spindle asymmetry has been reported in grasshopper (Hewitt 1976), and there are examples in other organisms that were not analyzed in depth (Crowder et al. 2015). In mouse oocytes we showed that the MI spindle is asymmetric for a specific posttranslational modification of tubulin (Janke 2014), tyrosinated α-tubulin, with more tyrosinated MTs oriented toward the cortex (Fig. 4A; Akera et al. 2017). The cortex near the spindle is enriched for CDC42 and RAC GTPases, and this cortical polarization is established by RANGTP generated around the chromosomes (Li and Albertini 2013). Inhibition of either RAN or CDC42 prevents both spindle asymmetry and biased orientation of hybrid bivalents in our model system (Fig. 4B,C), which has two important implications (Akera et al. 2017). First, spindle asymmetry is established by localized CDC42 activity at the cortex (Fig. 5A). Second, centromeres can interact with the asymmetric spindle to bias their orientation. Because the asymmetry is generated by a signal from the cortex, it has a consistent orientation with the tyrosinated side toward the cortex, and therefore provides spatial cues that can be exploited by selfish centromeres. One important outstanding question arising from this work is how CDC42 regulates tubulin tyrosination, but here we will focus on a different question: how bivalents with weaker and stronger centromeres interact with the asymmetric spindle to bias their transmission to the egg.
Figure 4.
Spindle asymmetry and biased orientation of selfish centromeres. (A) Oocytes fixed and stained at metaphase I show asymmetry within the spindle for tyrosinated α-tubulin, which is enriched on the cortical side of the spindle, whereas β-tubulin is symmetric. (B) Oocytes expressing dominant negative mutants of RAN or CDC42 were fixed and stained for tyrosinated α-tubulin. Both mutants prevent spindle asymmetry. Images A and B show the whole oocyte (left) or a magnified view of the spindle (right); dashed line, cortex; scale bars, 10 μm. Graphs are line scans of tubulin intensity across the spindle. (C) Bivalent orientation was measured in CHPO × CF-1 hybrid oocytes as in Fig. 3, either shortly after spindle migration to the cortex (early meta I), or shortly before anaphase onset (late meta I). The fraction of bivalents with the stronger centromere oriented toward the egg is shown (*indicates significant deviation from 50%, P < 0.005). Biased orientation is lost if spindle asymmetry is prevented by expression of a constitutively active RAN mutant or a dominant negative CDC42 mutant. (D) Schematic showing initially unbiased orientation of hybrid bivalents immediately after spindle migration to the cortex. The bias is observed later in metaphase I. (Portions reproduced from Akera et al. 2017, with permission from AAAS.)
Figure 5.
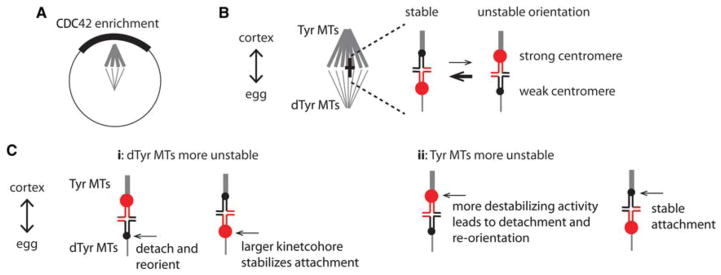
Model for biased chromosome orientation. (A) Spindle asymmetry and cortical CDC42 enrichment. The cortical side of the spindle has higher levels of tyrosinated α-tubulin compared to the egg side. (B) The orientation with the stronger centromere binding to more tyrosinated (Tyr) MTs and the weaker centromere to more detyrosinated (dTyr) MTs is labile and will tend to reorient. The opposite configuration is more stable and persists. (C) Models for how one bivalent configuration is selectively stabilized relative to the other. If dTyr MTs are more unstable (i), weaker centromeres may detach to initiate reorientation on the egg side. Conversely, stronger centromeres stabilize interactions on that side of the spindle to prevent reorientation. Alternatively, if Tyr MTs are more unstable (ii), stronger centromeres may initiate reorientation by destabilizing interactions with the cortical side of the spindle.
One possible model is that MTs from the egg side of the spindle preferentially capture stronger centromeres, and these attachments are maintained until anaphase. If MTs on the egg side were more dynamic, for example, they might initially interact with centromeres that present a larger kinetochore target with more MT-binding proteins (Chmátal et al. 2017). Weaker centromeres would then capture MTs from the cortical side, leading to biased orientation. A second model is that bivalents sample both configurations, and the one with stronger centromeres toward the more tyrosinated MTs on the cortical side is labile and will tend to reorient, whereas the opposite configuration is more stable (Fig. 5B). This trial-and-error mechanism, in which the preferred configuration is selectively stabilized, is analogous to the long-standing model for how correct, bi-oriented attachments are stabilized by tension (Nicklas 1997; Lampson and Grishchuk 2017).
The timing of meiotic events in our model system supports the second model. The MI spindle forms around the chromosomes, which are initially positioned in the center of the oocyte, and initial kinetochore-MT attachments are established at this time (Kitajima et al. 2011). The chromosomes and spindle then migrate together to the cortex to allow the highly asymmetric cell division that preserves most of the cytoplasm in the egg while extruding a relatively small polar body. We find that spindles are asymmetric only when positioned near the cortex. Furthermore, orientation of hybrid bivalents is unbiased immediately after migration to the cortex, and the bias is established later (Fig. 4C,D; Akera et al. 2017). These findings indicate that bivalents preferentially reorient on the asymmetric spindle to bias stronger centromeres toward the egg side.
Preferential reorientation indicates both differences between stronger and weaker centromeres in how they interact with spindle MTS and a difference between the two sides of the spindle, such that one configuration is selectively stabilized relative to the other. One model is that detyrosinated MTs on the egg side of the spindle tend to make more unstable kinetochore attachments, but stronger centromeres stabilize interactions with the egg side by building kinetochores with more MT-binding activity (Fig. 5Ci). The other configuration, with weaker centromeres connected to these unstable MTs, would be labile and likely to reorient. In this model, stronger centromeres win by preventing reorientation after reaching their preferred configuration. Supporting this model, stronger centromeres do build kinetochores with increased levels of kinetochore proteins such as CENP-A and CENP-C and the major MT-binding protein, HEC1 (Chmátal et al. 2014; Iwata-Otsubo et al. 2017). Conversely, an alternative model is that tyrosinated MTs on the cortical side make more unstable kinetochore attachments (Fig. 5Cii). In this case stronger centromeres can win by preferentially destabilizing the configuration where they face the cortical side, triggering reorientation toward the egg side. The configuration with weaker centromeres toward the cortical side would persist if these centromeres have less destabilizing activity.
Our findings are consistent with the second model, in which stronger centromeres destabilize their interactions with the cortical side of the spindle rather than stabilizing interactions with the egg side. Briefly, we showed that kinetochore MTs attached to stronger centromeres are more unstable than those of weaker centromeres, based on sensitivity to low temperature, and the attachments are most unstable when facing the cortical side. Furthermore, manipulating tubulin tyrosination levels showed that more tyrosinated MTs are more unstable (Akera et al. 2017). These results suggest that increased MT destabilizing activity at stronger centromeres drives preferential reorientation when facing the cortical side, which is more tyrosinated. Predictions of this model, for example that reorientation events are initiated by the stronger centromere, still need to be tested directly.
Overall, our results provide a cell biological framework for centromere drive. Chromosome positioning near the cell cortex is crucial for the highly asymmetric division in female meiosis (Li and Albertini 2013). The chromosomes direct cortical polarization by producing RANGTP, and the resulting enrichment of CDC42 on the polarized cortex generates asymmetry in α-tubulin tyrosination in the spindle. Spindle asymmetry is thus inherent to female meiosis, at least in this system, and can be exploited by centromeres or other selfish elements to bias their transmission to the egg. Observations in our model system indicate that centromeres can bias their orientation to the egg side of the spindle by preferentially destabilizing their interactions with tyrosinated MTs. Whether this strategy represents a broader theme in female drive systems is currently unknown because of the paucity of cell biological studies. One prominent example is knobs that drive in maize meiosis, which are not centromere linked and have adopted a different strategy based on recruiting a motor that positions them favorably on the spindle (Dawe 2009).
A natural question that follows from our model is the molecular identity of the MT destabilizing activity. Candidates include Aurora B kinase, which localizes to centromere chromatin and phosphorylates kinetochore substrates that bind MTs to destabilize these interactions, and kinesin-13 family members such as MCAK (mitotic centromere-associated kinesin), which catalyze MT depolymerization. Intriguingly, MCAK prefers tyrosinated MTs as a substrate (Peris et al. 2009; Sirajuddin et al. 2014). If stronger centromeres are enriched for MCAK activity, they would preferentially destabilize interactions with the cortical side of the spindle, which is more tyrosinated, driving reorientation. Future work will address this molecular interpretation of the destabilization model.
Progress in our model system for centromere drive raises additional pressing questions for future research. Our findings suggest that natural selection favors centromeres that increase destabilizing activity, specifically acting on tyrosinated spindle MTs. What are the underlying mechanisms that determine this activity, and what is the molecular link to increased levels of centromere chromatin at stronger centromeres, as indicated by CENP-A and CENP-C levels? Our molecular understanding of centromere assembly and function has advanced rapidly in recent years, and integrating this knowledge into models of centromere drive and selective pressures shaping centromere evolution promises to be an exciting research direction.
CONCLUSION
The centromere drive hypothesis originated as an explanation for why signatures of positive selection are detected in centromere proteins. To briefly restate the hypothesis, natural selection favors changes in centromere DNA that increase transmission to the egg in female meiosis, but there is a fitness cost associated with unbalanced centromeres, so selection also favors changes in centromere proteins that equalize centromeres and suppress drive. Work in our mouse model system so far has focused on the first part: understanding how stronger centromeres win in female meiosis. If there is a fitness cost associated with such drive, it has not yet been revealed, and how centromere proteins may have evolved to minimize this cost is unknown. Thus, the primary question underlying the drive hypothesis—whether suppression of drive is the selective pressure responsible for rapid evolution of centromere proteins—remains unanswered. Based on our results, such suppression might reduce the influence of DNA sequence on centromere function (e.g., by minimizing the contribution of CENP-B, which is the only protein known to bind directly to minor satellite sequences). Negating any sequence dependence of interactions between DNA and either CENP-A or CENP-C would also suppress drive, although such dependence has yet to be demonstrated. Alternatively, if centromeres win by increasing MT destabilizing activity, drive could be suppressed by weakening the link between centromere expansion and recruitment of those activities. Testing these models is an important challenge for future investigations of centromere drive.
Acknowledgments
We thank K. Dawe for sharing unpublished data, H. Willard, H. Masumoto, and D. Foltz for permission to reproduce data in Fig. 2, T. Akera and J. Dawicki-McKenna for comments on the manuscript, and members of the Black and Lampson labs for helpful discussions. Work in our labs is supported by the National Institutes of Health (NIH) (GM082989 to B.E.B. and GM122475 to M.A.L.).
References
- Akera T, Chmátal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, Lampson MA. Spindle asymmetry drives non-Mendelian chromosome segregation. Science. 2017;358:668–672. doi: 10.1126/science.aan0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Wood S, Salimian KJ, Ajith S, Foltz DR, Black BE. Epigenetic centromere specification directs Aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol. 2010;190:177–185. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol. 2014;204:313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Schultz RM, Black BE, Lampson MA. Cell biology of cheating—Transmission of centromeres and other selfish elements through asymmetric meiosis. Prog Mol Sub-cell Biol. 2017;56:377–396. doi: 10.1007/978-3-319-58592-5_16. [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Crowder ME, Strzelecka M, Wilbur JD, Good MC, von Dassow G, Heald R. A comparative analysis of spindle morphometrics across metazoans. Curr Biol. 2015;25:1542–1550. doi: 10.1016/j.cub.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RK. Maize centromeres and knobs (neocentromeres) In: Bennetzen JL, Hake S, editors. Handbook of maize genetics and genomics. Springer; New York: 2009. pp. 239–250. [Google Scholar]
- Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, et al. Characterization of neo-centromeres in marker chromosomes lacking detectable alpha-satellite DNA. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- Dumont M, Fachinetti D. DNA sequences in centromere formation and function. Prog Mol Subcell Biol. 2017;56:305–336. doi: 10.1007/978-3-319-58592-5_13. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Repetitive conundrums of centromere structure and function. Hum Mol Genet. 1999;8:151–155. doi: 10.1093/hmg/8.2.151. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev Cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R, Carpinone J, Derraugh LJ. No universal differences between female and male eukaryotes: Anisogamy and asymmetrical female meiosis. Biol J Linn Soc. 2016;120:1–21. [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF. Sequences associated with centromere competency in the human genome. Mol Cell Biol. 2013;33:763–772. doi: 10.1128/MCB.01198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleu Q, Gérard PR, Montchamp-Moreau C. Sex chromosome drive. Cold Spring Harb Perspect Biol. 2015;7:a017616. doi: 10.1101/cshperspect.a017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Meiotic drive for B-chromosomes in the primary oocytes of Myrmeleotettix maculatus (Orthopera: Acrididae) Chromosoma. 1976;56:381–391. doi: 10.1007/BF00292957. [DOI] [PubMed] [Google Scholar]
- Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, McMahon MA, Hervé S, Cleveland DW, Fachinetti D. CENP-A is dispensable for mitotic centromere function after initial centromere/kinetochore assembly. Cell Rep. 2016;17:2394–2404. doi: 10.1016/j.celrep.2016.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang W-H, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol. 2017;27:2365–2373e8. doi: 10.1016/j.cub.2017.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C. The tubulin code: Molecular components, readout mechanisms, and functions. J Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Earle E, Hill J, Choo KH. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc Natl Acad Sci. 1998;95:1136–1141. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- Lampson MA, Grishchuk EL. Mechanisms to avoid and correct erroneous kinetochore-microtubule attachments. Biology (Basel) 2017;6:1. doi: 10.3390/biology6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Albertini DF. The road to maturation: Somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- Lindholm AK, Dyer KA, Firman RC, Fishman L, Forstmeier W, Holman L, Johannsesson H, Knief U, Kokko H, Larracuente AM, et al. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol Evol. 2016;31:315–326. doi: 10.1016/j.tree.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J Cell Biol. 2015;208:521–531. doi: 10.1083/jcb.201412011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H. Breaking the HAC barrier: Histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31:2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Nonrandom segregation during meiosis: The unfairness of females. Mamm Genome. 2001;12:331–339. doi: 10.1007/s003350040003. [DOI] [PubMed] [Google Scholar]
- Peris L, Wagenbach M, Lafanechère L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, Andrieux A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol. 2009;185:1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol. 2012;10:e1001434. doi: 10.1371/journal.pbio.1001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MM. Preferential segregation in maize. Genetics. 1942;27:395–407. doi: 10.1093/genetics/27.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Nothing in genetics makes sense except in light of genomic conflict. Annu Rev Ecol Evol Syst. 2013;44:217–237. [Google Scholar]
- Sandler L, Novitski E. Meiotic drive as an evolutionary force. Am Nat. 1957;91:105–110. [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Green ED NISC Comparative Sequencing Program. Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol. 2010;27:1585–1597. doi: 10.1093/molbev/msq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoak EM, Stein P, Schultz RM, Lampson MA, Black BE. Long-term retention of CENP-A nucleosomes in mammalian oocytes underpins transgenerational inheritance of centromere identity. Curr Biol. 2016;26:1110–1116. doi: 10.1016/j.cub.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Müller S, Blümer J, Klare K, Musacchio A, Almouzni G. HJURP involvement in de novo CenH3 (CENP-A) and CENP-C recruitment. Cell Rep. 2015;11:22–32. doi: 10.1016/j.celrep.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Werren JH. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci. 2011;108(Suppl):10863–10870. doi: 10.1073/pnas.1102343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H-G, Hiatt EN, Chan A, Sweeney M, Dawe RK. Neo-centromere-mediated chromosome movement in maize. J Cell Biol. 1997;139:831–840. doi: 10.1083/jcb.139.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadzińska E, Foltz DR. Orchestrating the specific assembly of centromeric nucleosomes. Prog Mol Subcell Biol. 2017;56:165–192. doi: 10.1007/978-3-319-58592-5_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Bureš P. CenH3 evolution reflects meiotic symmetry as predicted by the centromere drive model. Sci Rep. 2016;6:33308. doi: 10.1038/srep33308. [DOI] [PMC free article] [PubMed] [Google Scholar]