Abstract
Synthetic biologists use engineering principles to design and construct genetic circuits for programming cells with novel functions. A bottom-up approach is commonly used to design and construct genetic circuits by piecing together functional modules that are capable of reprogramming cells with novel behavior. While genetic circuits control cell operations through the tight regulation of gene expression, a diverse array of environmental factors within the extracellular space also has a significant impact on cell behavior. This extracellular space offers an addition route for synthetic biologists to apply their engineering principles to program cell-responsive modules within the extracellular space using biomaterials. In this review, we discuss how taking a bottom-up approach to build genetic circuits using DNA modules can be applied to biomaterials for controlling cell behavior from the extracellular milieu. We suggest that, by collectively controlling intrinsic and extrinsic signals in synthetic biology and biomaterials, tissue engineering outcomes can be improved.
1. Introduction
Synthetic biology has transformed how cells can be reprogrammed, providing a means to reliably and predictably control cell behavior with the assembly of genetic parts into more complex gene circuits. This bottom-up approach enables the combination of characterized pieces of DNA, or modules, with specific functions to create genetic programs to engineer cells with desired behavior. Traditionally, these functional modules have come from orthogonal organisms to minimize crosstalk between the engineered circuits and the host’s cell machinery, which allows robust and tunable gene expression in response to a specified input. Examples of these programs include switches [1–12], oscillators [13–19], timers and counters [20, 21], amplifiers [22, 23], logic gates [24–27], biosensors [28–40], tight gene control [41–51], enhanced protein production [52, 53], and for various therapeutic applications [54–66].
Gene expression in mammalian cells can generally be described as starting with the recruitment of transcription factors to DNA regulatory sites within the nucleus to activate or repress the transcription of target genes. This controls the production of pre-messenger RNA (mRNA) transcripts that undergo post-transcriptional modifications including splicing introns out of the transcript and adding a 5’ cap and a 3’ poly (A) tail. These mature mRNAs are exported into the cytoplasm where they bind to ribosomes and undergo translation [67, 68]. The control of all of these processes is essential for cells to control gene expression to maintain their viability and to execute cell fate decisions. In higher organisms, any disruption to this regulation can lead to serious consequences including cancer [69–71], underdeveloped or weak hypertrophic tissues [72–74], and/or metabolic diseases [75–80]. Therefore, it is important that genes are only expressed when needed, and that the correct level of expression is maintained. Mammalian synthetic biologists have intervened in the molecular processes of gene expression by implementing novel gene expression patterns using genetic circuits capable of controlling gene expression by maintaining tight off states [81, 3], tuning gene expression levels [82–84, 3, 85], and implementing Boolean logic functions [86–89, 27, 90], making approaches in synthetic biology ideal for applications in tissue engineering.
The primary goal of tissue engineering is to repair and regenerate damaged and diseased tissues [91–94]. The central thrust of many approaches in tissue engineering is to deliver stem cells and biochemical factors within material scaffolds, ultimately to direct stem cell differentiation and promote the production of new, viable tissue. All tissues are composed of an extracellular matrix (ECM) as well as lineage committed cells that secrete and maintain the ECM. Stem cells are an imperative cell population for tissue maintenance, regeneration, and repair because they have the ability to replenish their own population, and to differentiate into a number of cell types to restore tissue function. Both intrinsic (transcription factor expression) as well as extrinsic (environmental) mechanisms have been shown to be involved with the regulation of stem cell self-renewal and differentiation, and this interplay between intrinsic and extrinsic cues has made it difficult to direct an entire population of stem cells toward a desired lineage [95–98]. This begs the question of whether a combined approach that incorporates cues from both inside and outside of the cell might yield more robust stem cell differentiation outcomes. A variety of biomaterials have been explored as three-dimensional (3D) scaffolds for tissue engineering approaches [99–113]. Similarly, recent efforts in synthetic biology have focused on the precise orchestration of gene expression that can be used to augment stem cell self-renewal and differentiation [114–119, 63]. This review focuses on how bottom-up approaches used in synthetic biology to build genetic circuits from individual functional modules can be applied to biomaterials for building 3D microenvironments with programmed ECMs that contain extracellular domains/modules to direct cell behavior for tissue engineering applications. Specifically, we examine both intrinsic and extrinsic efforts to guide cell behavior and discuss various target areas to manipulate and control cell function (Fig. 1). Synthetic biologists build genetic circuits and intervene at the various steps in gene expression to control cell behavior, and tissue engineers focus on creating biomaterials with custom domains to control cell behavior. In both cases, functional modules are pieced together to build genetic circuits and environments for obtaining desired cell outcomes. Altogether, we view this bottom-up approach in both synthetic biology and biomaterials to be a promising means to enhance desired cell behavior that can be used for tissue-engineering applications.
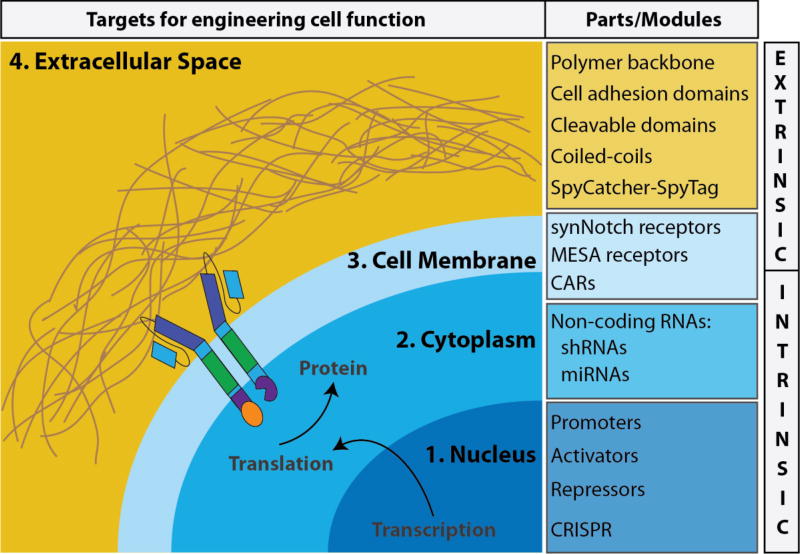
Figure 1. Bottom-up approaches for controlling cell behavior.
Synthetic biologists use interchangeable and well-characterized genetic parts, or modules, (listed in blue boxes) of DNA to build complex genetic circuits to program cells with robust and tunable gene expression for obtaining desired cell behavior. Synthetic biologists use a variety of genetic tools to target: (1) the nucleus, (2) the cytoplasm, and (3) the cell membrane to control gene expression that ultimately control cell behavior. The extracellular space (4) is another area for synthetic biologists to consider using a bottom-up approach to construct a cell-instructive microenvironment using well-characterized extracellular matrix (ECM) protein domains/modules (listed in yellow box). Together, controlling the intrinsic and extrinsic signals that cells are exposed to will likely improve our ability to guide the growth and behavior of cells.
2. Targeting the cell – modules in synthetic biology
Advances in DNA manipulation have enabled the construction of complex genetic circuits in mammalian cells that have improved our ability to probe and program mammalian cells. These advances make it possible to envision novel therapeutic interventions for many therapeutic applications, including treating damaged and diseased tissue. Spatial and temporal control of gene expression is critical for applications in tissue engineering and regenerative medicine. Engineering approaches in synthetic biology have benefited from the increased number of available genetic modules to enable the reprogramming of cells with novel biological behavior [114, 120, 121, 118, 122], dynamic gene control [82–84, 3, 85], and Boolean logic capabilities [86–89, 27, 90], which has led to an improved ability to control mammalian cell behavior. These genetic tools can be used toward one of the goals in tissue engineering, which is to direct stem cell differentiation to create new tissues. In this case, tools for regulating gene expression are essential, and in this section, we provide an overview of how advances in mammalian synthetic biology have led to a new generation of novel genetic tools that can be used for exciting new directions in tissue engineering.
2.1 Transcriptional control
Transcription factors are proteins that bind to regions of DNA to control the expression of downstream genes. These DNA-binding domains can function as transcriptional activators or repressors to engineer synthetic promoters to be used as modules for building genetic circuits. Altogether, synthetic promoters that contain corresponding transcription factor binding sequences can be used to control the rate of transcription of a gene of interest. The activity of each synthetic promoter can be engineered to strongly influences important cell processes such as proliferation, apoptosis, and differentiation by regulating the expression of genes in either ‘on’ or ‘off states [123]. Synthetic biologists have capitalized on this by isolating transcriptional regulatory parts from other species and transferring them to mammalian cells. For example, the binding domains of the bacterial repressor proteins, LacI and TetR, have been placed within promoter regions to enable downstream gene expression to be controlled [124, 125]. The effects of LacI and TetR can be reversed by adding the small molecule inducers, isopropyl β-D-1-thiogalactopyranoside (IPTG) and tetracycline, respectively. When moving these transcriptional regulators to mammalian systems, they can be fused to a transcriptional activator domain (e.g. VP16) or a transcriptional repressor domain (e.g. KRAB) to employ desired control over gene expression [126, 127]. More recently, gene regulatory modules from Neurospora crassa were coupled to the Lac repressor to create a new orthogonal genetic tool, called LacQ, for regulating gene expression in mammalian cells [4]. Many of these transcription factor genetic circuits have been extensively reviewed elsewhere [128–137, 55, 138, 139, 47, 140, 59, 141–146, 66].
The engineering of transcription factor genetic circuits has demonstrated how cells can be programmed with novel gene expression patterns, which have already shown utility in stem cell and tissue engineering applications. Recently, the LacQ switch was shown to function in mouse embryonic stem cells to tightly control gene expression in the pluripotent state [4]. In a different study, a light activated genetic switch was engineered to turn gene expression on after exposure to blue light [10]. This light induced genetic switch was used to control the expression of the master myogenic factor, MyoD, to direct mesenchymal stem cells to differentiate into skeletal muscle cells. The same optogenetic switch was also used to control the expression of Vascular Endothelial Growth Factor (VEGF) and Angiopoietin 1 (ANG1) to induce new blood vessel formation, which is a critical occurrence for tissue regeneration and wound healing applications. Another study showed that pulsing the expression of GATA6 in human induced pluripotent stem (iPS) cells initiated the development of all three germ layers to form liver bud-like tissue in vitro, an important step for recapitulating the formation of organs and tissues [147]. While traditional differentiation protocols require a mixture of growth factors and cytokines to be administered over successive temporal windows, the authors’ GATA-6-induced differentiation methods did not require any exogenous biologic aside from those to maintain iPS cell pluripotency. When examining individual cells in culture, differentiation outcomes were found to be a result of both cellular GATA-6 expression levels as well as neighboring cell types. This demonstrated that cell niches can be spontaneously developed using genetically programmed differentiation regimens. Importantly, emerging tissue function was apparent with the development of blood-like cells that produced hemoglobin subunits. Lastly, a genetic circuit that functions as a band-pass filter was engineered to temporally control the expression of the three different essential transcription factors, Ngn3, Pdx1, and MafA for the development of pancreatic β cells [148]. This genetic circuit was shown to temporally control the expression of these three transcription factors to effectively direct human iPS cells differentiation into glucose-sensitive insulin secreting like β cells.
2.2 Genome editing
In addition to transcription factor genetic circuits, mammalian synthetic biologists have utilized enzymes from bacteria and yeast that bind to site-specific DNA sequences and function to alter the sequence through irreversible excisions, inversions, and integrations, depending on the orientation and architecture of the DNA recognition sites [129]. This recombinase technology has been utilized in many organisms to program cells with Boolean logic functions [149, 86, 88], cellular computation [150, 20], and cellular memory [151–155, 89]. Recently, recombinases were used to engineer mammalian cells with multiple programmed inputs and outputs to allow for more targeted complex manipulation of cell behavior [156]. This study introduced a new platform called “BLADE”, or Boolean Logic and Arithmetic through DNA Excision, to generate multiple genetic circuits that can be customized based on the desired cellular outcomes. This is a significant advancement in mammalian synthetic biology because it provides exciting opportunities to design custom-made complex genetic programs that can be used for directing stem cell differentiation.
In addition to recombinases, the discovery of programmable DNA nucleases, zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), and clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9, have made it possible to target specific locations within the genomic DNA and introduce site-specific gene modifications by cleaving the DNA at specific target sequences [132, 157]. These nucleases are able to perform efficient, precise genetic modifications by creating double stranded DNA breaks, which allows for gene deletion, mutated sequence corrections, and gene insertion [26, 158]. The double stranded breaks stimulate cellular DNA repair mechanisms, including nonhomologous end joining (NHEJ) and homology-directed repair (HR), which enables the directed reprogramming of cells using these genetic modules [159–161, 117, 162–165]. Genome targeting technologies have shown great promise in reprogramming stem cells for directing their differentiation, in addition to correcting disease causing DNA mutations [166–171]. For example, efforts are underway to study the effects of correcting DNA sequences that cause diseases such as Duchenne muscular dystrophy (DMD), which is an inherited X-linked disease caused by mutations in the gene encoding for dystrophin, a protein required for muscle fiber integrity [172, 167, 169]. Patients with mutated dystrophin protein typically experience muscle degeneration that results in death at a young age, typically from a weakening heart and breathing complications. Studies correcting this mutation within the genome are promising for future genome editing approaches [173, 168].
Altogether, the ability to specifically target the cell genome using genome targeting genetic tools enable the placement of landing pads for the direct insertion of larger genetic circuits into the genome [174, 175]. These important advancements make it possible to program stem cells with genetic circuits capable of tightly regulating gene expression for directing their differentiation.
2.3 Noncoding RNAs
Many genes produce small RNAs that do not encode for any protein. These noncoding RNAs make up a diverse set of transcripts that function as regulatory components within cells. With advances in sequencing technologies, many of these noncoding RNAs have been identified [176]. The discovery of sequence-specific RNA-mediated pathways for turning off gene expression [177] has empowered synthetic biologists with genetic tools to target transcripts before they are translated [119]. For example, early in mammalian synthetic biology, the RNAi pathway was utilized to tightly regulate gene expression by controlling the production of short hairpin RNA (shRNA) using a TetR-regulated promoter [3]. This study demonstrated how coupling repressor protein and RNAi modules lead to tight gene control that behaves like a rheostat, enabling tightly regulated and graded levels of gene expression. More recent investigations have leveraged noncoding RNA modules to correlate miRNA with gene expression levels to create a model for predicting protein-responsive miRNAs [178]. This work also enabled the intracellular measurement of the nuclear protein concentration of β-catenin. Several synthetic RNA-based control circuits have also been engineered to recognize unique microRNA identifiers of cancer cells, enabling genetically programmed cells to distinguish between healthy and cancerous states by classifying specific microRNA levels of each cell [62]. Once these microRNAs are detected, the genetic circuit induces apoptosis of the cancerous cells. In a different study, negative feedback control was obtained from an RNAi-based genetic circuit to maintain homeostatic protein levels in mammalian cells [81]. Overall, cell recognition-response genetic tools offer significant value in building tissues that are composed of multiple cell types.
2.4 Receptors
In addition to engineering genetic circuits that respond to small diffusible molecules and light, cell membrane receptors have been engineered to link extrinsic signals to control intracellular molecular events. A typical mammalian cell is exposed to hundreds of different signals in its environment. These signals can be soluble, bound to the ECM, or located on the surface of neighboring cells, and they can act in many combinations to regulate stem cell behavior and cell fate decisions. Rewiring receptors to connect with genetic circuits and thereby capture receptor activation has the potential to monitor what cells experience in their native environments (e.g. changes in the ECM, presence/absence of other cells, levels of proteins, etc.). This has the exciting potential to provide insights into the extracellular triggers that direct cells down particular differentiation pathways. To capitalize on the natural sensing ability of cell receptors, synthetic biologists have rewired receptor activation to connect to genetic circuits that trigger the expression of reporter genes or therapeutic proteins [30, 179]. The Modular Extracellular Sensor Architecture (MESA) system pioneered this effort using bio-orthogonal membrane receptors [31, 36]. The engineered receptors consisted of two transmembrane chains, each with a single variable fragment fused to either an intracellular protease chain or a target chain containing the protease ligand, as well as a synthetic transcription factor. The authors more recently demonstrated that MESA could be multiplexed to implement cellular logic in response to extracellular cues [180]. Upon binding to the target ligand, the two receptor fragments dimerize and trigger proteolytic cleavage of the synthetic transcription factor, which initiates an intracellular transcriptional response. This strategy enabled the development of logic functions as a means to reduce deleterious off-target receptor activation [180]. In a similar approach, the Notch receptor has been rewired to trigger synthetic gene modules (synNotch) [181, 182]. synNotch receptors capitalize on the proteolytic release of the Notch intracellular domain upon binding of the Notch receptor to its ligand. This intracellular domain translocates to the nucleus and initiates transcriptional activation of the nuclear reporter module or in the case of synNotch, activates a synthetic gene module [182]. These novel technologies are exciting from a tissue engineering perspective because they can be used as a means to rewire extracellular-to-intracellular transduction pathways for enhanced cell differentiation outcomes.
Similarly, T cells have been engineered for tumor targeting using a wide range of surface receptors with distinct intracellular functions [183]. These chimeric antigen receptors (CARs) were used to create T cells that have synthetic receptors consisting of an antigen-specific single-chain variable fragment, which is derived from a monoclonal antibody fused to the core intracellular signaling domain. Upon T cell activation, the host’s immune system is stimulated to attack the tumor [184]. Notably, CARs allow for flexible user-defined retargeting simply by swapping the ligand-binding domain, while the intracellular processing and signal transduction are limited to endogenous signaling cascades.
3. Targeting the extracellular space – modules in biomaterials
All cells naturally interact with their ECM to maintain healthy tissues, and any changes in the ECM can lead to significant changes in cell behavior. The adhesion of cells to the ECM has a profound influence on cellular behavior, affecting cell migration, differentiation, and proliferation. Differentiation outcomes of stem cells are significantly impacted by the mechanical properties (e.g. stiffness and elasticity) of their surrounding environment [185–191]. Aspects regarding the mechanical environment and its influence on stem cell behavior have been extensively reviewed elsewhere [192, 187, 193–195]. Additionally, when growth factors are added to the media, studies have shown that they act synergistically with the mechanical properties of the material when both target the same differentiation outcome [185]. For example a stiff matrix combined with bone (osteogenic) media results in robust differentiation of stem cells into osteoblasts, the cells that secrete matrix for bone formation. On the other hand, mixed environments produce varied differentiation outcomes. A soft matrix combined with osteogenic media, was found to produce a mixed population of cells, and resulted in a low efficiency of differentiation into osteoblasts. Therefore, both the biochemical and mechanical properties within the cellular environment are essential mediators of differentiation, and their synergy is paramount to promoting robust outcomes.
Maintaining a stem cell’s ability to differentiate into more specialized cell types, or “stemness”, is essential to tissue engineering applications because it allows for the expansion of a stem cell population to generate enough cell numbers to form a functioning tissue. Recently, it was shown that the ability of neural progenitor stem cells to differentiate into multiple neural cell types was dependent upon the capacity of the ECM to undergo matrix remodeling via material degradation [196]. This remodeling was demonstrated to occur in the presence of protease-degradable domains as well as physical crosslinks within the biomaterial, suggesting that neural progenitor stem cells require degradable microenvironments to maintain stemness. This study suggests that biomaterial degradation can be programmed to first promote the proliferation of stem cells, followed by their differentiation into more specialized cells. Altogether, studies have illustrated that the mechanical and biochemical characteristics within the cellular milieu have a significant impact on stem cell behavior. Therefore, most tissue engineering approaches focus on engineering biomaterials to mimic aspects of the native tissue ECM.
While synthetic polymer chemistry has traditionally been a means to create biomaterials for growing cells in 3D scaffolds, these materials generally lack the cues necessary to control cell attachment and differentiation for tissue production. This has motivated efforts to modify 3D scaffolds with bioactive sequences and factors throughout the material [197]. Recombinant strategies for producing peptide scaffolds permit modularity in the design of protein products that are capable of forming 3D materials to grow cells. Because the design of these materials is encoded within DNA, they are more amenable to be programmed for interacting with other synthetic or biological systems [198]. We therefore discuss how the same bottom-up approaches used in synthetic biology to build complex gene circuits out of genetic modules can be applied to engineer protein-based biomaterials that harbor functional modules. Specifically, we examine how these domains/modules can build instructive environments for stem cells to receive timely extrinsic signals for improving differentiation outcomes. Many reviews have been published summarizing how biomaterial scaffolds have been used for tissue engineering applications [199, 93, 200, 201, 113]. Here, we discuss how taking a bottom-up approach to assemble modular protein domains within biomaterial scaffolds will enable the construction of complex cell-responsive extracellular matrices for directing cell fate decisions. These scaffolds have the ability to assemble either through non-covalent or covalent interactions, while remaining biologically orthogonal within the mammalian context. While not all of these materials have been directly assembled in the extracellular space of a mammalian culture, we predict that their translation into this space is imminent. Furthermore, we highlight how these components can be utilized to control the mechanical and chemical features of biomaterials and to endow extracellular spaces with sensing and bioactive functionalities.
Hydrogels are common tissue engineering scaffolds that provide 3D environments for cell growth, mimicking in vivo settings with facile assembly. These materials are hydrophilic in nature, which provides an optimal environment for molecules and nutrients to move freely throughout the material. Hydrogels can be made from many different polymer chains (e.g. collagen, polyethylene glycol (PEG), alginate, etc.) that crosslink to form gels, however many additional domains are required for cell attachment [102, 92, 202, 203, 111, 204–206]. Furthermore, hydrogels typically lack the proper signals to induce robust differentiation for tissue production, resulting in a need for their modification of bioactive sequences and factors [207–212]. From a synthetic biology perspective, elastin-like polypeptides (ELPs) are an attractive platform of materials because they can be easily modified and recombinantly expressed in E. coli to produce high yields of tailor-made programmed materials for encapsulating mammalian cells [210]. Below is a non-exhaustive list of significant functional modules that can be built into biomaterials for directing cell function.
3.1 Cell adhesion domains
Improving engineering approaches of ECMs by programming in functional domains/modules to better control cell behavior has the potential to improve how tissue substitutes can be engineered from stem cells. A common modification to materials is based upon the observation that cells require integrin receptors to bind to ECM ligands for cell attachment, which has been shown to influence and regulate cell migration, growth, differentiation, and apoptosis. One of these ligands is the RGD (R, arginine; G, glycine; and D, aspartic acid) peptide sequence, which is an integrin-binding domain present in fibronectin. RGD can be attached to various materials, or programmed into the DNA of recombinant proteins. This cell-ECM binding module was taken from the fibronectin ECM protein [94, 213], and has been shown to transform an otherwise non-cell adhesive biomaterial into a microenvironment that mimics native tissues [214–217]. Overall, the addition of cell adhesion domains within biomaterial scaffolds has shown to significantly impact cell behavior in the areas of cell attachment, cell spreading, cytoskeletal reorganization, the formation of focal adhesions, as well as proliferation and cell motility [218].
3.2 Cleavable domains
Matrix metalloproteinases (MMPs) are secreted by cells, which function in the extracellular environment to degrade ECM proteins. MMPs have shown to play a major role in tissue repair and remodeling in response to injury and, for this reason, have been popular targets in disease treatments [219]. The inclusion of MMP-cleavable domains within a biomaterial has been demonstrated to promote stem cell viability and differentiation [111]. These MMP-degradable sites allow surrounding cells to permit matrix remodeling by making room for newly synthesized tissue. In a recent publication, it was shown that the incorporation of MMP-7-cleavable RGDS (RGD plus serine) into a biomaterial system, along with chondroitin sulfate and heparin, two bioactive molecules known to enhance chondrogenesis (the formation of cartilage) of human mesenchymal stem cells [220]. MMPs are therefore a module that enables matrix remodeling that can also be used to release bioactive molecules from the scaffold in a spatial and temporal fashion to positively influence stem cell differentiation and tissue remodeling.
3.3 Non-covalent modules in biomaterials
3.3.1 Peptide cross-linking
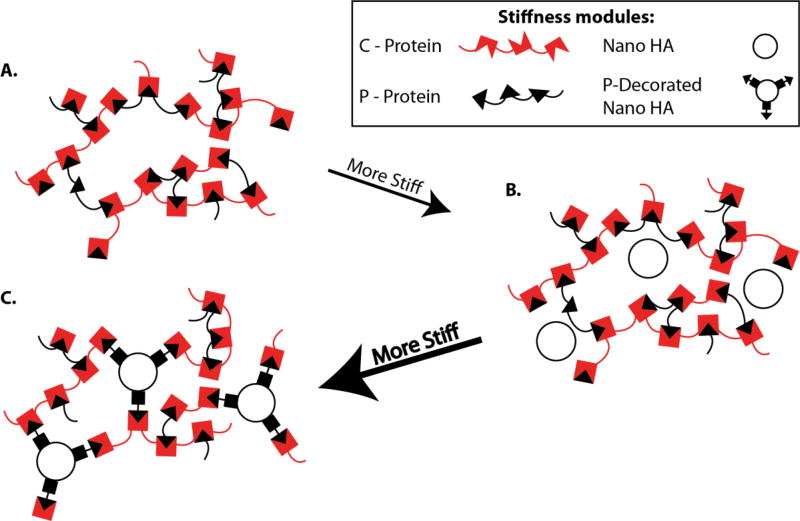
Capitalizing on protein-protein interactions, it was shown that peptide cross-linking groups could be added to various modules using recombinant technologies to customize cellular microenvironments. In one such example, specific protein domains containing two tryptophan residues, called WW motifs, fold with stable triple-stranded β-sheets that recognize and physically cross-links with proline-containing ligands [221]. Many WW motifs together (called a “C” domain) have been shown to create a soft hydrophilic hydrogel that provides a scaffold for cells to grow [222]. This hydrogel environment can be decorated with bioactive domains to impact cell behavior by adding proline-rich ligands (called a “P” domain) that have been engineered to contain other functional modules. For example, to create tailor-made materials for bone regeneration, hydrogels made from C and P domains with functional peptides (i.e. RGD and hydroxyapatite (HA), a mineral naturally found in bone) genetically fused to them [203]. Upon mixing these peptides, the system forms a hydrogel (Fig. 2A) that achieved a material stiffness determined by the ratio of the three constituents, with a significantly greater stiffness being achieved with the inclusion of the nano-HA (Fig. 2B), and an even greater stiffness with the P-decorated nano-HA (Fig. 2C). Upon gelation, the hydrogel demonstrated self-healing properties and recovered its viscosity following mechanical perturbation, an important property to consider in applications with mechanical forces playing a significant role in the outcome of tissue development. This study exemplifies how using recombinant technologies can be used to modify the mechanical properties of the extracellular microenvironment with the presence of different functional modules that can be used for directing cell fate decisions.
Figure 2. Controlling matrix stiffness with peptide crosslinking modules.
The stiffness of ECM scaffolds can be controlled by using modules that contain repeat motifs of either a “P” (proline rich – black triangles) or “C” (WW – red broken boxes). These modules can be mixed to control the stiffness of the matrix. A. When P and C domains are mixed, they form a soft hydrogel. B. When Nano HA modules are added to the P and C gels, the matrix stiffens. C. The addition of P-Decorated Nano HA modules enables an even stiffer matrix to be formed.
3.3.2 Coiled-coils
Coiled-coil interactions provide a means to tailor the binding associations between two peptide domains that specifically associate. Coiled-coils are two or more alpha-helices that combine in a parallel or anti-parallel fashion to create a superhelix [223]. The individual helices of coiled-coils are composed of heptad repeats and precise placement of amino acids within the coils produce high degrees of specificity between different constituents of the coils. Coiled-coil domains are found throughout nature, including within ECM that contains proteins such as collagen [224] and laminin [225]. Due to their specificity and ease of customization, coiled-coils have been employed in recombinant material design to create orthogonal protein linkages in the extracellular space. For example, it was shown that the dynamic mechanical properties of a hydrogel can be controlled by functionalizing the polymer backbone chains with various coiled-coil domains, and adjusting the coil primary structure to obtain desired matrix outcomes (Fig. 3A and B) [226]. Specifically, these properties allow the elasticity and the rates of dissociation/reassociation to dynamically change based on the design of the coiled-coils used in the matrix. Altogether, this work demonstrates that coiled-coil domains can be used to engineer mechanically dynamic hydrogels by controlling the amino acid structure of the coils.
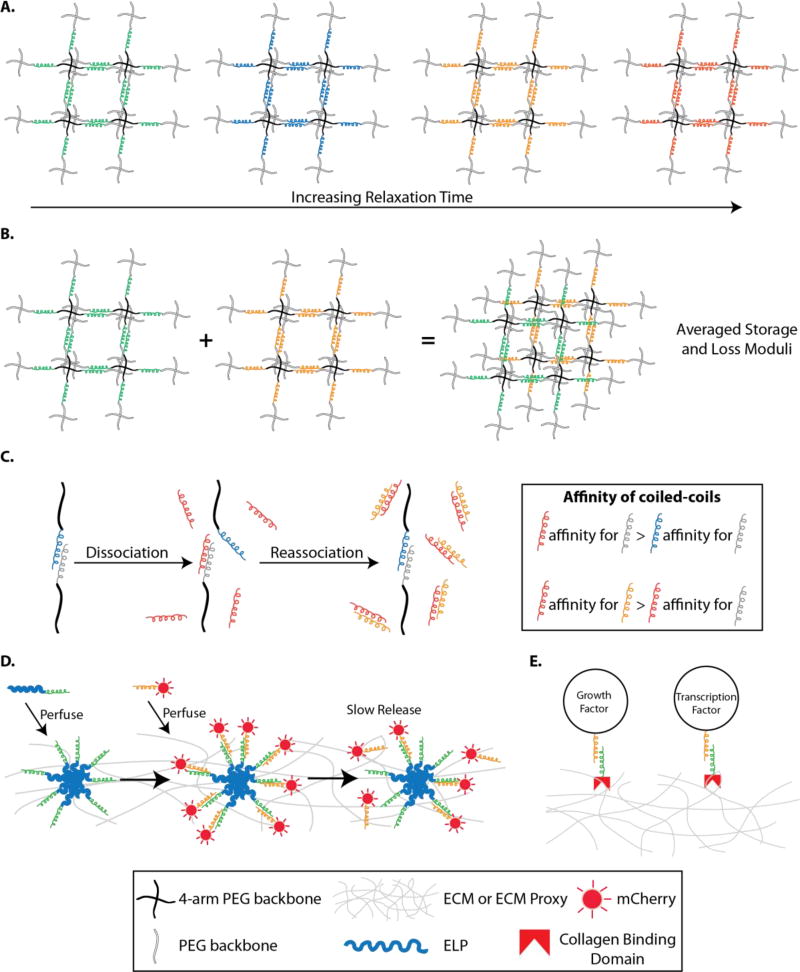
Figure 3. Coiled-coils.
The primary structure of coiled coils can be modified to provide an unlimited number of these domains/modules to be used for structurally programming the ECM. A. Mixing PEG backbones (grey) conjugated with different coiled-coiled units (green, blue, yellow, red, and grey coils), the storage and loss moduli can be custom made to achieve desired ECM. B. The dynamic behavior of hydrogels linked via coiled-coil domains can be controlled by programming coiled-coil primary amino acid structures. C. A coiled-coil polymer system is capable of dissociation and re-association in response to the introduction of other coiled-coil domains. These networks can be disrupted through the introduction of a non-PEGylated coiled-coil domain with a higher affinity for one of the PEG-coil domains (boxed coiled-coils), thereby displacing the other PEG-coil binding partner from participating in crosslinking. D. When basic leucine zipper coils (ZR) are fused to ELP (thick blue line) and mCherry (red sunshine) and mixed together, coacervates are formed to obtain slow, controlled release of mCherry-ZE. E. The combination of collagen binding domains (red broken squares) and coiled coils provides a means to decorate the ECM with growth factors.
The ability to use parts from orthogonal systems to induce changes in behavior is central to synthetic biology. Along these lines, a coiled-coil polymer system was developed that dissociates and re-associates in response to the introduction of coiled-coil domains from various polymers [227]. In this example, four different polymers, each composed of a PEG backbone with coiled-coil domains conjugated at the termini, were constructed to create various PEG-coil species. By mixing different PEG-coil domains together, it was shown that molecular networks could be dissociated with the introduction of a coiled-coil lacking PEG, which has a higher affinity than the PEG-coil domains This resulted in displacing the other PEG-coil binding partner from participating in crosslinking (Fig. 3C). Importantly, these molecular networks could be re-formed through reintroducing a coiled-coil domain lacking PEG. This flexible and modular approach to designing hydrogels could be useful in the context of tissue engineering to create injectable and durable hydrogels. The switching behavior of such networks has the potential to control the tethering/dissociation of ligands or growth factors to the material network with the addition of other coiled-coil domains. Additionally, it is well within the reach to imagine that these principles could be applied to recombinant proteins, and perhaps, with a genetic program controlling the temporal expression of these different materials to augment mechanical and chemical properties of the matrix over time.
A second type of coiled-coil being used to tailor biomaterials is the leucine zipper coiled coil (ZR). Leucine zipper proteins are a group of transcriptional regulators that dimerize to form a DNA binding domain. It is believed that this dimerization results from the hydrophobic association of the a-helices of the two leucine zipper monomers into a coiled coil [228]. Recently, the self-assembly of two fusion proteins, one containing an elastin like polypeptide (ELP) with a leucine zipper coiled-coil, and the other containing the mCherry fluorescent protein with a leucine zipper coiled-coil was observed to create a hydrogel (Fig. 3D) [228]. Performing temperature transitions to induce the formation of an ELP hydrogel showed to drastically alter the diffusion coefficient compared to the ELP hydrogel without the leucine zipper coiled-coils, creating a slow and controlled release of the mCherry coiled-coil. Therefore, coiled-coil domains can be used to program the release of small molecules like genetic inducers to control encapsulated cells containing genetic circuits, or growth factors for guiding stem cell fate decisions for tissue engineering applications.
Similarly, the fibronectin-derived collagen binding domain (CBD) as well as coiled-coil domains were combined for the purpose of non-covalently immobilizing the growth factors, basic fibroblast growth factor (bFGF) and endothelial growth factor (EGF), to a hydrogel [229]. In this study, the CBD was genetically fused the C-terminal of the Kcoil coiled-coil, as well as bFGF and EGF, both with an Ecoil domain (Kcoil’s binding partner) (Fig. 3E). This novel approach to functionalize the extracellular environment has demonstrated that growth factors can be tethered to an ECM for influencing cell behavior.
3.4 Covalent Modules
3.4.1 SpyCatcher-SpyTag
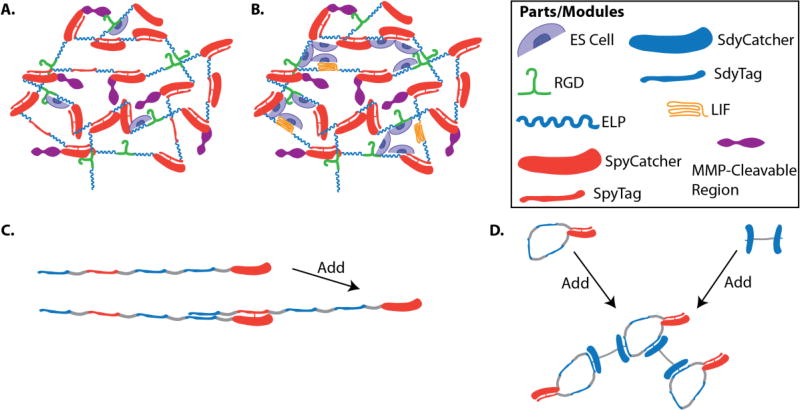
The SpyTag-SpyCatcher system is a protein coupling tool, harvested from S. pyogenes, is used for irreversible peptide-protein ligation that uses peptide-protein partners to covalently lock them together [230]. This system was derived from the second immunoglobulin-like collagen adhesion domain (CnaB2) of the S. pyogenes fibronectin binding protein FbaB, which naturally forms an isopeptide bond [231]. To create this modular tool the CnaB2 was split into two pieces, a short 13 amino acid SpyTag and a larger 138 amino acid SpyCatcher that spontaneously forms an isopeptide bond that imparts significant stability to the molecule (Fig. 4). The SpyCatcher-SpyTag system has been applied to the culture of stem cells through the incorporation of bioactive agents into recombinant hydrogels [205]. To create a custom made ECM that includes the SpyCatcher-SpyTag system, SpyCatcher and SpyTag modules can be encoded into the DNA sequence of recombinant proteins that form hydrogels (Fig. 4A). Moreover, designing biomaterials using these modules coupled with functional domains can produce materials with conjugated biomolecules to enhance cell function. For example, it was shown that conjugating Leukemia Inhibitory Factor (LIF), a cytokine that maintains embryonic stem cell pluripotency, to a hydrogel dramatically improved the number of undifferentiated embryonic stem cells compared to gels without the conjugated LIF (Fig. 4B) [205]. Altogether, the SpyCatcher-SpyTag is a modular system can be used for a variety of applications to produce novel protein architectures, including joining functionalized polypeptide strands to proteins (Fig. 4C) at specific times during cell culture [230], provide an environment to improve viability of cells [232], enzyme stability [233], and vaccine optimization [234].
Figure 4. Spy Systems.
Elastin like polypeptides ELPs (blue thick lines) harboring two SpyCatcher domains (red thick domain), an RGD region (green), and an MMP-cleavable sequence (purple) can be mixed with A. ELPs containing SpyTags (thinner red line) either flanking the ELP or placed internally, or B. with ELPs containing two terminal SpyTags along with LIF (yellow), a cytokine that aides in maintaining ES cell pluripotency. C. The SpyCatcher/SpyTag can be used to assemble functionalized peptides. D. SdyCatcher/SdyTag enables 3D architectures can be formed when cyclic SpyCatcher/SpyTag and cyclic SdyCatcher/SdyTag modules are mixed together.
3.4.2 SdyTag-SdyTag
Similar domains for protein-peptide covalent cross-linking have been found in S. dysgalactiae called SdyCatcher and SdyTag [235]. To demonstrate how these novel modules can be used to functionalize biomaterials, it was shown that SpyCatcher:SpyTag-EGFP-SdyTag:SdyCatcher structures could be assembled. In order to achieve higher-order 3D structures, the cross-linking of circularized SpyCatcher/SpyTag and SdyCatcher-SdyCatcher proteins was performed (Fig. 4D). It is possible that these novel 3D ECM structures can be translated by cells to self-assemble post-translationally that offers an interesting new direction for cells to create their own custom-made ECM to control their own cell fate.
Altogether, the Spy systems have garnered much attention, and are being utilized in the modular assembly of antibodies [236, 237], virus-like particles [238], and microcapsules [239]. While the Spy and Spy-like systems have shown impressive results in bacteria, their applications in higher organisms is only just beginning. For example, it was recently demonstrated that the SpyCatcher-SpyTag system is functional in C. elegans, human embryonic kidney (HEK) cells, and in primary neuronal cultures for selectively labeling membrane-localized proteins [240]. We forecast the increased utilization of these components by tissue engineering technologies to come.
CONCLUSIONS
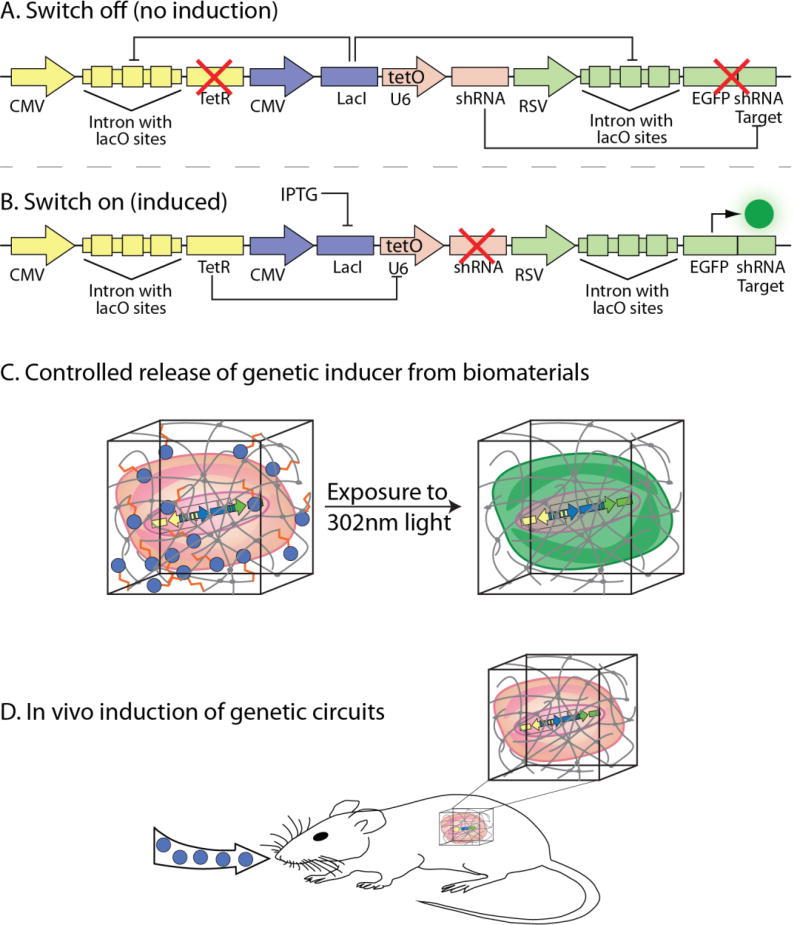
Studies have shown that cell fate decisions are strongly influenced by both intrinsic and extrinsic factors. Therefore, engineering approaches that couple synthetic biology and biomaterials should improve our ability to control cell behavior. This can be accomplished by taking a bottom-up approach to assemble functional genetic modules to control intrinsic gene expression patterns, and similarly, assemble functional extracellular matrix domains/modules to create custom-made microenvironments for directing cell behavior. It has previously been shown that interfacing synthetic biology with biomaterials enables the creation of genetically interactive biomaterials. In these studies, inducer molecules for the genetic circuit, LTRi_EGFP (Fig. 5A and B) [3], was attached to the biomaterial scaffold with various triggers of release [241, 242], allowing for spatial and temporal control of gene expression in vitro (Fig. 5C). Similar studies have shown that biomaterials can be used to implant cells containing the LTRi_EGFP genetic circuit at various locations in mice and gene expression could be controlled by adding various concentrations of the inducer to their drinking water (Fig. 5D) [241].
Figure 5. Interfacing synthetic biology and biomaterials.
A. LTRi_EGFP genetic circuit in the absence of inducer in is the off state and does not express Enhanced Green Fluorescence Protein (EGFP). B. When inducer is added, this flips the genetic switch on and EGFP is expressed. C. Genetically interactive biomaterials have been modified with genetic inducer molecules (blue circles) attached to the scaffold via photolabile bonds (orange lines). In the absence of 302nm of light, the inducer molecules remain attached to the biomaterial and do not activate EGFP expression. Upon exposure to 302nm of light, the photolabile bond breaks, releasing the inducer molecules, which activate EGFP expression. D. Cells harboring LTRi_EGFP were encapsulated into biomaterials and implanted into mice showed EGFP expression only when the genetic inducer was added to the drinking water.
Looking ahead, taking a bottom-up approach to design and construct gene networks to regulate intrinsic gene expression profiles, in addition to building complex biomaterial scaffolds with functional domains/modules to construct structurally programmable ECMs for controlling the extrinsic signals that cells are exposed to has the potential to revolutionize our ability to guide the growth and behavior of cells that will ultimately impact basic research, pharmaceutics, and medicine.
Acknowledgments
We gratefully acknowledge the funding from the University of Utah startup funds, a University of Utah SEED grant [10045314], the National Science Foundation CAREER program [CBET-1554017], the Office of Naval Research Young Investigator Program [N00014-16-1-3012], and the National Institutes of Health Trailblazer Award [1R21EB025413-01].
References
- 1.Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proc Natl Acad Sci U S A. 2012;109(15):5850–5855. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang AL, Wolf JJ, Smolke CD. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr Opin Biotechnol. 2012;23(5):679–688. doi: 10.1016/j.copbio.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130(2):363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald M, et al. Adoption of the Q Transcriptional System for Regulating Gene Expression in Stem Cells. ACS Synth Biol. 2017 doi: 10.1021/acssynbio.7b00149. [DOI] [PubMed] [Google Scholar]
- 5.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 6.Green AA, et al. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159(4):925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horner M, Weber W. Molecular switches in animal cells. FEBS Lett. 2012;586(15):2084–2096. doi: 10.1016/j.febslet.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Kramer BP, et al. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004;22(7):867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 9.Mandal M, et al. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113(5):577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 10.Polstein LR, et al. An Engineered Optogenetic Switch for Spatiotemporal Control of Gene Expression, Cell Differentiation, and Tissue Morphogenesis. ACS Synth Biol. 2017;6(11):2003–2013. doi: 10.1021/acssynbio.7b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidl SR, et al. Refactoring and optimization of light-switchable Escherichia coli two-component systems. ACS Synth Biol. 2014;3(11):820–831. doi: 10.1021/sb500273n. [DOI] [PubMed] [Google Scholar]
- 12.Weber W, Fussenegger M. Molecular diversity--the toolbox for synthetic gene switches and networks. Curr Opin Chem Biol. 2011;15(3):414–420. doi: 10.1016/j.cbpa.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Aulehla A, Pourquie O. Oscillating signaling pathways during embryonic development. Curr Opin Cell Biol. 2008;20(6):632–637. doi: 10.1016/j.ceb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Danino T, et al. A synchronized quorum of genetic clocks. Nature. 2010;463(7279):326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403(6767):335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 16.Fung E, et al. A synthetic gene-metabolic oscillator. Nature. 2005;435(7038):118–122. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
- 17.Judd EM, Laub MT, McAdams HH. Toggles and oscillators: new genetic circuit designs. Bioessays. 2000;22(6):507–509. doi: 10.1002/(SICI)1521-1878(200006)22:6<507::AID-BIES3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Stricker J, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456(7221):516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tigges M, et al. A tunable synthetic mammalian oscillator. Nature. 2009;457(7227):309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 20.Friedland AE, et al. Synthetic gene networks that count. Science. 2009;324(5931):1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber W, et al. A synthetic time-delay circuit in mammalian cells and mice. Proc Natl Acad Sci U S A. 2007;104(8):2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnet J, et al. Amplifying genetic logic gates. Science. 2013;340(6132):599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 23.Hasty J, et al. Noise-based switches and amplifiers for gene expression. Proc Natl Acad Sci U S A. 2000;97(5):2075–2080. doi: 10.1073/pnas.040411297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guet CC, et al. Combinatorial synthesis of genetic networks. Science. 2002;296(5572):1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 25.Mukherji S, van Oudenaarden A. Synthetic biology: understanding biological design from synthetic circuits. Nat Rev Genet. 2009;10(12):859–871. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schukur L, Fussenegger M. Engineering of synthetic gene circuits for (re-)balancing physiological processes in chronic diseases. Wiley Interdiscip Rev Syst Biol Med. 2016;8(5):402–422. doi: 10.1002/wsbm.1345. [DOI] [PubMed] [Google Scholar]
- 27.Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31(5):448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 28.Auslander D, et al. A synthetic multifunctional mammalian pH sensor and CO2 transgene-control device. Mol Cell. 2014;55(3):397–408. doi: 10.1016/j.molcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Bowsher CG, Swain PS. Environmental sensing, information transfer, and cellular decision-making. Curr Opin Biotechnol. 2014;28C:149–155. doi: 10.1016/j.copbio.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Brenner M, Cho JH, Wong WW. Synthetic biology: Sensing with modular receptors. Nat Chem Biol. 2017;13(2):131–132. doi: 10.1038/nchembio.2290. [DOI] [PubMed] [Google Scholar]
- 31.Daringer NM, et al. Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth Biol. 2014;3(12):892–902. doi: 10.1021/sb400128g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng J, et al. A general strategy to construct small molecule biosensors in eukaryotes. Elife. 2015:4. doi: 10.7554/eLife.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia JR, et al. Microbial nar-GFP cell sensors reveal oxygen limitations in highly agitated and aerated laboratory-scale fermentors. Microb Cell Fact. 2009;8:6. doi: 10.1186/1475-2859-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Looger LL, et al. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423(6936):185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 35.Saeidi N, et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz KA, et al. Rewiring human cellular input-output using modular extracellular sensors. Nat Chem Biol. 2017;13(2):202–209. doi: 10.1038/nchembio.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slomovic S, Collins JJ. DNA sense-and-respond protein modules for mammalian cells. Nat Methods. 2015;12(11):1085–1090. doi: 10.1038/nmeth.3585. [DOI] [PubMed] [Google Scholar]
- 38.Smole A, et al. A Synthetic Mammalian Therapeutic Gene Circuit for Sensing and Suppressing Inflammation. Mol Ther. 2017;25(1):102–119. doi: 10.1016/j.ymthe.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu F, Menn DJ, Wang X. Quorum-sensing crosstalk-driven synthetic circuits: from unimodality to trimodality. Chem Biol. 2014;21(12):1629–1638. doi: 10.1016/j.chembiol.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youk H, Lim WA. Secreting and sensing the same molecule allows cells to achieve versatile social behaviors. Science. 2014;343(6171):1242782. doi: 10.1126/science.1242782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auslander S, et al. Programmable single-cell mammalian biocomputers. Nature. 2012;487(7405):123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 42.Callura JM, et al. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc Natl Acad Sci U S A. 2010;107(36):15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27(5):465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gitzinger M, et al. The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res. 2012;40(5):e37. doi: 10.1093/nar/gkr1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guido NJ, et al. A bottom-up approach to gene regulation. Nature. 2006;439(7078):856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 46.Isaacs FJ, et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22(7):841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 47.Kaern M, Blake WJ, Collins JJ. The engineering of gene regulatory networks. Annu Rev Biomed Eng. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- 48.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11(5):367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150(3):647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Litcofsky KD, et al. Iterative plug-and-play methodology for constructing and modifying synthetic gene networks. Nat Methods. 2012;9(11):1077–1080. doi: 10.1038/nmeth.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber W, et al. A synthetic mammalian electro-genetic transcription circuit. Nucleic Acids Res. 2009;37(4):e33. doi: 10.1093/nar/gkp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paddon CJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496(7446):528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 53.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 54.Danino T, et al. Programmable probiotics for detection of cancer in urine. Sci Transl Med. 2015;7(289):289ra284. doi: 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Folcher M, Fussenegger M. Synthetic biology advancing clinical applications. Curr Opin Chem Biol. 2012;16(3–4):345–354. doi: 10.1016/j.cbpa.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10(11):785–794. doi: 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Higashikuni Y, Chen WC, Lu TK. Advancing therapeutic applications of synthetic gene circuits. Curr Opin Biotechnol. 2017;47:133–141. doi: 10.1016/j.copbio.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 58.Kim T, et al. A synthetic erectile optogenetic stimulator enabling blue-light-inducible penile erection. Angew Chem Int Ed Engl. 2015;54(20):5933–5938. doi: 10.1002/anie.201412204. [DOI] [PubMed] [Google Scholar]
- 59.Kojima R, Aubel D, Fussenegger M. Toward a world of theranostic medication: Programming biological sentinel systems for therapeutic intervention. Adv Drug Deliv Rev. 2016;105(Pt A):66–76. doi: 10.1016/j.addr.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 60.Nissim L, Bar-Ziv RH. A tunable dual-promoter integrator for targeting of cancer cells. Mol Syst Biol. 2010;6:444. doi: 10.1038/msb.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossger K, Charpin-El-Hamri G, Fussenegger M. A closed-loop synthetic gene circuit for the treatment of diet-induced obesity in mice. Nat Commun. 2013;4:2825. doi: 10.1038/ncomms3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie Z, et al. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333(6047):1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 63.Ye H, Aubel D, Fussenegger M. Synthetic mammalian gene circuits for biomedical applications. Curr Opin Chem Biol. 2013;17(6):910–917. doi: 10.1016/j.cbpa.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Ye H, et al. Pharmaceutically controlled designer circuit for the treatment of the metabolic syndrome. Proc Natl Acad Sci U S A. 2013;110(1):141–146. doi: 10.1073/pnas.1216801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye H, et al. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332(6037):1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 66.Ye H, Fussenegger M. Synthetic therapeutic gene circuits in mammalian cells. FEBS Lett. 2014;588(15):2537–2544. doi: 10.1016/j.febslet.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 67.Auslander S, Fussenegger M. Engineering Gene Circuits for Mammalian Cell-Based Applications. Cold Spring Harb Perspect Biol. 2016;8(7) doi: 10.1101/cshperspect.a023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deans TL. Parallel Networks: Synthetic Biology and Artificial Intelligence. ACM Journal on Emerging Technologies in computing systems (JETC) 2014;11(3) [Google Scholar]
- 69.Baylin SB, et al. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10(7):687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 70.Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 71.van ‘t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 72.Caddeo S, Boffito M, Sartori S. Tissue Engineering Approaches in the Design of Healthy and Pathological In Vitro Tissue Models. Front Bioeng Biotechnol. 2017;5:40. doi: 10.3389/fbioe.2017.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 74.van den Broek LJ, et al. Human hypertrophic and keloid scar models: principles, limitations and future challenges from a tissue engineering perspective. Exp Dermatol. 2014;23(6):382–386. doi: 10.1111/exd.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anesiadis N, et al. Analysis and design of a genetic circuit for dynamic metabolic engineering. ACS Synth Biol. 2013;2(8):442–452. doi: 10.1021/sb300129j. [DOI] [PubMed] [Google Scholar]
- 76.Auslander S, Wieland M, Fussenegger M. Smart medication through combination of synthetic biology and cell microencapsulation. Metab Eng. 2012;14(3):252–260. doi: 10.1016/j.ymben.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 77.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292(5518):929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 78.Jeong H, et al. The large-scale organization of metabolic networks. Nature. 2000;407(6804):651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 79.Park JH, Lee SY. Towards systems metabolic engineering of microorganisms for amino acid production. Curr Opin Biotechnol. 2008;19(5):454–460. doi: 10.1016/j.copbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 80.Teixeira AP, Fussenegger M. Synthetic biology-inspired therapies for metabolic diseases. Curr Opin Biotechnol. 2017;47:59–66. doi: 10.1016/j.copbio.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Bloom RJ, Winkler SM, Smolke CD. Synthetic feedback control using an RNAi-based gene-regulatory device. J Biol Eng. 2015;9:5. doi: 10.1186/s13036-015-0002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andrianantoandro E, et al. Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100073. 0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Basu S, et al. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci U S A. 2004;101(17):6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen AY, et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat Mater. 2014;13(5):515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol. 2013;2(10):604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miyamoto T, et al. Synthesizing biomolecule-based Boolean logic gates. ACS Synth Biol. 2013;2(2):72–82. doi: 10.1021/sb3001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Purcell O, Lu TK. Synthetic analog and digital circuits for cellular computation and memory. Curr Opin Biotechnol. 2014;29C:146–155. doi: 10.1016/j.copbio.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roquet N, Lu TK. Digital and analog gene circuits for biotechnology. Biotechnol J. 2014;9(5):597–608. doi: 10.1002/biot.201300258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roquet N, et al. Synthetic recombinase-based state machines in living cells. Science. 2016;353(6297):aad8559. doi: 10.1126/science.aad8559. [DOI] [PubMed] [Google Scholar]
- 90.Yang L, et al. Permanent genetic memory with >1-byte capacity. Nat Methods. 2014;11(12):1261–1266. doi: 10.1038/nmeth.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chan G, Mooney DJ. New materials for tissue engineering: towards greater control over the biological response. Trends Biotechnol. 2008;26(7):382–392. doi: 10.1016/j.tibtech.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 92.Deans TL, Elisseeff JH. Mimicking extracellular matrix to direct stem cell differentiation. World Stem Cell Report, Genetics Policy Institute; 2009. [Google Scholar]
- 93.Deans TL, Elisseeff JH. Stem cells in musculoskeletal engineered tissue. Curr Opin Biotechnol. 2009;20(5):537–544. doi: 10.1016/j.copbio.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 94.Deans TL, Elisseeff JH. The life of a cell: probing the complex relationships with the world. Cell Stem Cell. 2010;6(6):499–501. doi: 10.1016/j.stem.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 95.Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9(1):37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- 96.Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9(1):11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 97.Watt FM, Hogan BL. Out of Eden: stem cells and their niches. Science. 2000;287(5457):1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 98.Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453(7193):306–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
- 99.Aamodt JM, Grainger DW. Extracellular matrix-based biomaterial scaffolds and the host response. Biomaterials. 2016;86:68–82. doi: 10.1016/j.biomaterials.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009;5(1):1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 101.Beachley VZ, et al. Tissue matrix arrays for high-throughput screening and systems analysis of cell function. Nat Methods. 2015;12(12):1197–1204. doi: 10.1038/nmeth.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen FM, Liu X. Advancing biomaterials of human origin for tissue engineering. Prog Polym Sci. 2016;53:86–168. doi: 10.1016/j.progpolymsci.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen XD, et al. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. J Bone Miner Res. 2007;22(12):1943–1956. doi: 10.1359/jbmr.070725. [DOI] [PubMed] [Google Scholar]
- 104.Coburn JM, et al. Bioinspired nanofibers support chondrogenesis for articular cartilage repair. Proc Natl Acad Sci U S A. 2012;109(25):10012–10017. doi: 10.1073/pnas.1121605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32(12):3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 107.Gilbert TW, Sellaro TL, Badylak SF. Decellularization of tissues and organs. Biomaterials. 2006;27(19):3675–3683. doi: 10.1016/j.biomaterials.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 108.Griffith LG. Emerging design principles in biomaterials and scaffolds for tissue engineering. Ann N Y Acad Sci. 2002;961:83–95. doi: 10.1111/j.1749-6632.2002.tb03056.x. [DOI] [PubMed] [Google Scholar]
- 109.Griffith LG, Naughton G. Tissue engineering--current challenges and expanding opportunities. Science. 2002;295(5557):1009–1014. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 110.Hillel AT, et al. Photoactivated composite biomaterial for soft tissue restoration in rodents and in humans. Sci Transl Med. 2011;3(93):93ra67. doi: 10.1126/scitranslmed.3002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parmar PA, et al. Collagen-mimetic peptide-modifiable hydrogels for articular cartilage regeneration. Biomaterials. 2015;54:213–225. doi: 10.1016/j.biomaterials.2015.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Porzionato A, et al. Decellularized Human Skeletal Muscle as Biologic Scaffold for Reconstructive Surgery. Int J Mol Sci. 2015;16(7):14808–14831. doi: 10.3390/ijms160714808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sachlos E, Czernuszka JT. Making tissue engineering scaffolds work. Review: the application of solid freeform fabrication technology to the production of tissue engineering scaffolds. Eur Cell Mater. 2003;5:29–39. doi: 10.22203/ecm.v005a03. discussion 39–40. [DOI] [PubMed] [Google Scholar]
- 114.Bashor CJ, et al. Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annu Rev Biophys. 2010;39:515–537. doi: 10.1146/annurev.biophys.050708.133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Benenson Y. Biomolecular computing systems: principles, progress and potential. Nat Rev Genet. 2012;13(7):455–468. doi: 10.1038/nrg3197. [DOI] [PubMed] [Google Scholar]
- 116.Brophy JA, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014;11(5):508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ho P, Chen YY. Mammalian synthetic biology in the age of genome editing and personalized medicine. Curr Opin Chem Biol. 2017;40:57–64. doi: 10.1016/j.cbpa.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Karlsson M, Weber W. Therapeutic synthetic gene networks. Curr Opin Biotechnol. 2012;23(5):703–711. doi: 10.1016/j.copbio.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 119.Mathur M, Xiang JS, Smolke CD. Mammalian synthetic biology for studying the cell. J Cell Biol. 2017;216(1):73–82. doi: 10.1083/jcb.201611002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Breithaupt H. The engineer’s approach to biology. EMBO Rep. 2006;7(1):21–23. doi: 10.1038/sj.embor.7400607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chin JW. Programming and engineering biological networks. Curr Opin Struct Biol. 2006;16(4):551–556. doi: 10.1016/j.sbi.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 122.Slusarczyk AL, Lin A, Weiss R. Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet. 2012;13(6):406–420. doi: 10.1038/nrg3227. [DOI] [PubMed] [Google Scholar]
- 123.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 124.Liu HS, et al. Lac/Tet dual-inducible system functions in mammalian cell lines. Biotechniques. 1998;24(4):624–628. 630–622. doi: 10.2144/98244st03. [DOI] [PubMed] [Google Scholar]
- 125.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25(6):1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bertram R, Hillen W. The application of Tet repressor in prokaryotic gene regulation and expression. Microb Biotechnol. 2008;1(1):2–16. doi: 10.1111/j.1751-7915.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Deuschle U, Meyer WK, Thiesen HJ. Tetracycline-reversible silencing of eukaryotic promoters. Mol Cell Biol. 1995;15(4):1907–1914. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aubel D, Fussenegger M. Mammalian synthetic biology--from tools to therapies. Bioessays. 2010;32(4):332–345. doi: 10.1002/bies.200900149. [DOI] [PubMed] [Google Scholar]
- 129.Auslander S, Auslander D, Fussenegger M. Synthetic Biology-The Synthesis of Biology. Angew Chem Int Ed Engl. 2017;56(23):6396–6419. doi: 10.1002/anie.201609229. [DOI] [PubMed] [Google Scholar]
- 130.Auslander S, Fussenegger M. From gene switches to mammalian designer cells: present and future prospects. Trends Biotechnol. 2013;31(3):155–168. doi: 10.1016/j.tibtech.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 131.Bacchus W, Aubel D, Fussenegger M. Biomedically relevant circuit-design strategies in mammalian synthetic biology. Mol Syst Biol. 2013;9:691. doi: 10.1038/msb.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Black JB, Perez-Pinera P, Gersbach CA. Mammalian Synthetic Biology: Engineering Biological Systems. Annu Rev Biomed Eng. 2017;19:249–277. doi: 10.1146/annurev-bioeng-071516-044649. [DOI] [PubMed] [Google Scholar]
- 133.Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12(5):381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 134.Collins J. Synthetic Biology: Bits and pieces come to life. Nature. 2012;483(7387):S8–10. doi: 10.1038/483S8a. [DOI] [PubMed] [Google Scholar]
- 135.Collins JJ, et al. Synthetic biology: How best to build a cell. Nature. 2014;509(7499):155–157. doi: 10.1038/509155a. [DOI] [PubMed] [Google Scholar]
- 136.Deans TL, Grainger DW, Fussenegger M. Synthetic Biology: Innovative approaches for pharmaceutics and drug delivery. Adv Drug Deliv Rev. 2016;105(Pt A):1–2. doi: 10.1016/j.addr.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 137.Dobrin A, Saxena P, Fussenegger M. Synthetic biology: applying biological circuits beyond novel therapies. Integr Biol (Camb) 2015 doi: 10.1039/c5ib00263j. [DOI] [PubMed] [Google Scholar]
- 138.Greber D, Fussenegger M. Mammalian synthetic biology: engineering of sophisticated gene networks. J Biotechnol. 2007;130(4):329–345. doi: 10.1016/j.jbiotec.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 139.Haellman V, Fussenegger M. Synthetic Biology-Toward Therapeutic Solutions. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 140.Kobayashi H, et al. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101(22):8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lienert F, et al. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat Rev Mol Cell Biol. 2014;15(2):95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat Biotechnol. 2009;27(12):1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.MacDonald IC, Deans TL. Tools and applications in synthetic biology. Adv Drug Deliv Rev. 2016;105(Pt A):20–34. doi: 10.1016/j.addr.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 144.Ruder WC, Lu T, Collins JJ. Synthetic biology moving into the clinic. Science. 2011;333(6047):1248–1252. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 145.Way JC, et al. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014;157(1):151–161. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 146.Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nat Rev Genet. 2012;13(1):21–35. doi: 10.1038/nrg3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guye P, et al. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Saxena P, et al. A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat Commun. 2016;7:11247. doi: 10.1038/ncomms11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gordley RM, Gersbach CA, Barbas CF., 3rd Synthesis of programmable integrases. Proc Natl Acad Sci U S A. 2009;106(13):5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Daniel R, et al. Synthetic analog computation in living cells. Nature. 2013;497(7451):619–623. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- 151.Ajo-Franklin CM, et al. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21(18):2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Burrill DR, et al. Synthetic memory circuits for tracking human cell fate. Genes Dev. 2012;26(13):1486–1497. doi: 10.1101/gad.189035.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Burrill DR, Silver PA. Making cellular memories. Cell. 2010;140(1):13–18. doi: 10.1016/j.cell.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Inniss MC, Silver PA. Building synthetic memory. Curr Biol. 2013;23(17):R812–816. doi: 10.1016/j.cub.2013.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kotula JW, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci U S A. 2014;111(13):4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Weinberg BH, et al. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat Biotechnol. 2017;35(5):453–462. doi: 10.1038/nbt.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xie M, Haellman V, Fussenegger M. Synthetic biology-application-oriented cell engineering. Curr Opin Biotechnol. 2016;40:139–148. doi: 10.1016/j.copbio.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 159.Dow LE, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33(4):390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fu Y, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kalhor R, Mali P, Church GM. Rapidly evolving homing CRISPR barcodes. Nat Methods. 2017;14(2):195–200. doi: 10.1038/nmeth.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zalatan JG, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160(1–2):339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dai WJ, et al. CRISPR-Cas9 for in vivo Gene Therapy: Promise and Hurdles. Mol Ther Nucleic Acids. 2016;5:e349. doi: 10.1038/mtna.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Maeder ML, Gersbach CA. Genome-editing Technologies for Gene and Cell Therapy. Mol Ther. 2016 doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ousterout DG, et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Perez-Pinera P, Ousterout DG, Gersbach CA. Advances in targeted genome editing. Curr Opin Chem Biol. 2012;16(3–4):268–277. doi: 10.1016/j.cbpa.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Savic N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 172.Long C, et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345(6201):1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li HL, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4(1):143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Duportet X, et al. A platform for rapid prototyping of synthetic gene networks in mammalian cells. Nucleic Acids Res. 2014;42(21):13440–13451. doi: 10.1093/nar/gku1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Inniss MC, et al. A novel Bxb1 integrase RMCE system for high fidelity site-specific integration of mAb expression cassette in CHO Cells. Biotechnol Bioeng. 2017;114(8):1837–1846. doi: 10.1002/bit.26268. [DOI] [PubMed] [Google Scholar]
- 176.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 177.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 178.Bloom RJ, Winkler SM, Smolke CD. A quantitative framework for the forward design of synthetic miRNA circuits. Nat Methods. 2014;11(11):1147–1153. doi: 10.1038/nmeth.3100. [DOI] [PubMed] [Google Scholar]
- 179.Pitner RA, Scarpelli AH, Leonard JN. Regulation of Bacterial Gene Expression by Protease-Alleviated Spatial Sequestration (PASS) ACS Synth Biol. 2015;4(9):966–974. doi: 10.1021/sb500302y. [DOI] [PubMed] [Google Scholar]
- 180.Hartfield RM, et al. Multiplexing Engineered Receptors for Multiparametric Evaluation of Environmental Ligands. ACS Synth Biol. 2017;6(11):2042–2055. doi: 10.1021/acssynbio.6b00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Irvine DJ. A Receptor for All Occasions. Cell. 2016;164(4):599–600. doi: 10.1016/j.cell.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 182.Morsut L, et al. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell. 2016;164(4):780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Roybal KT, et al. Precision Tumor Recognition by T Cells With Combinatorial Antigen-Sensing Circuits. Cell. 2016;164(4):770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vonderheide RH, June CH. Engineering T cells for cancer: our synthetic future. Immunol Rev. 2014;257(1):7–13. doi: 10.1111/imr.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Engler AJ, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 186.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7(9):733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guilak F, et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5(1):17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Legant WR, et al. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods. 2010;7(12):969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.McBeath R, et al. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 190.Trappmann B, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11(7):642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 191.Yang C, et al. Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014;13(6):645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Crowder SW, et al. Material Cues as Potent Regulators of Epigenetics and Stem Cell Function. Cell Stem Cell. 2016;18(1):39–52. doi: 10.1016/j.stem.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. 2014;13(6):547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol. 2017;18(12):728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Watt FM, Huck WT. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol. 2013;14(8):467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 196.Madl CM, et al. Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat Mater. 2017;16(12):1233–1242. doi: 10.1038/nmat5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Huber MC, et al. Introducing a combinatorial DNA-toolbox platform constituting defined protein-based biohybrid-materials. Biomaterials. 2014;35(31):8767–8779. doi: 10.1016/j.biomaterials.2014.06.048. [DOI] [PubMed] [Google Scholar]
- 198.Winnacker M. Recent advances in the synthesis of functional materials by engineered and recombinant living cells. Soft Matter. 2017;13(38):6672–6677. doi: 10.1039/c7sm01000a. [DOI] [PubMed] [Google Scholar]
- 199.Abdeen AA, Saha K. Manufacturing Cell Therapies Using Engineered Biomaterials. Trends Biotechnol. 2017;35(10):971–982. doi: 10.1016/j.tibtech.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hiew VV, Simat SFB, Teoh PL. The Advancement of Biomaterials in Regulating Stem Cell Fate. Stem Cell Rev. 2018;14(1):43–57. doi: 10.1007/s12015-017-9764-y. [DOI] [PubMed] [Google Scholar]
- 201.Liu Z, et al. Looking into the Future: Toward Advanced 3D Biomaterials for Stem-Cell-Based Regenerative Medicine. Adv Mater. 2018 doi: 10.1002/adma.201705388. [DOI] [PubMed] [Google Scholar]
- 202.Nuttelman CR, Tripodi MC, Anseth KS. Synthetic hydrogel niches that promote hMSC viability. Matrix Biol. 2005;24(3):208–218. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 203.Parisi-Amon A, et al. Protein-Nanoparticle Hydrogels That Self-assemble in Response to Peptide-Based Molecular Recognition. ACS Biomater. Sci. Eng. 2017;3(5):750–756. doi: 10.1021/acsbiomaterials.6b00286. [DOI] [PubMed] [Google Scholar]
- 204.Sawkins MJ, et al. Hydrogels derived from demineralized and decellularized bone extracellular matrix. Acta Biomater. 2013;9(8):7865–7873. doi: 10.1016/j.actbio.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sun F, et al. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proc Natl Acad Sci U S A. 2014;111(31):11269–11274. doi: 10.1073/pnas.1401291111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103(4):655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chilkoti A, Christensen T, MacKay JA. Stimulus responsive elastin biopolymers: Applications in medicine and biotechnology. Curr Opin Chem Biol. 2006;10(6):652–657. doi: 10.1016/j.cbpa.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.MacEwan SR, Chilkoti A. Elastin-like polypeptides: biomedical applications of tunable biopolymers. Biopolymers. 2010;94(1):60–77. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 209.MacEwan SR, Chilkoti A. Applications of elastin-like polypeptides in drug delivery. J Control Release. 2014;190:314–330. doi: 10.1016/j.jconrel.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Nettles DL, Chilkoti A, Setton LA. Applications of elastin-like polypeptides in tissue engineering. Adv Drug Deliv Rev. 2010;62(15):1479–1485. doi: 10.1016/j.addr.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Roberts S, Dzuricky M, Chilkoti A. Elastin-like polypeptides as models of intrinsically disordered proteins. FEBS Lett. 2015;589(19 Pt A):2477–2486. doi: 10.1016/j.febslet.2015.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Stayton PS, et al. Control of protein-ligand recognition using a stimuli-responsive polymer. Nature. 1995;378(6556):472–474. doi: 10.1038/378472a0. [DOI] [PubMed] [Google Scholar]
- 213.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 214.Burdick JA, Anseth KS. Photoencapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials. 2002;23(22):4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 215.Liu JC, Heilshorn SC, Tirrell DA. Comparative cell response to artificial extracellular matrix proteins containing the RGD and CS5 cell-binding domains. Biomacromolecules. 2004;5(2):497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 216.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238(4826):491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 217.Salinas CN, Anseth KS. The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials. 2008;29(15):2370–2377. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24(24):4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 219.Morrison CJ, et al. Matrix metalloproteinase proteomics: substrates, targets, and therapy. Curr Opin Cell Biol. 2009;21(5):645–653. doi: 10.1016/j.ceb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 220.Parmar PA, et al. Enhanced articular cartilage by human mesenchymal stem cells in enzymatically mediated transiently RGDS-functionalized collagen-mimetic hydrogels. Acta Biomater. 2017;51:75–88. doi: 10.1016/j.actbio.2017.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513(1):30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 222.Wong Po Foo CT, et al. Two-component protein-engineered physical hydrogels for cell encapsulation. Proc Natl Acad Sci U S A. 2009;106(52):22067–22072. doi: 10.1073/pnas.0904851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21(10):375–382. [PubMed] [Google Scholar]
- 224.McAlinden A, et al. Alpha-helical coiled-coil oligomerization domains are almost ubiquitous in the collagen superfamily. J Biol Chem. 2003;278(43):42200–42207. doi: 10.1074/jbc.M302429200. [DOI] [PubMed] [Google Scholar]
- 225.Armony G, et al. Cross-linking reveals laminin coiled-coil architecture. Proc Natl Acad Sci U S A. 2016;113(47):13384–13389. doi: 10.1073/pnas.1608424113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dooling LJ, Tirrell DA. Engineering the Dynamic Properties of Protein Networks through Sequence Variation. ACS Cent Sci. 2016;2(11):812–819. doi: 10.1021/acscentsci.6b00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Danmark S, Aronsson C, Aili D. Tailoring Supramolecular Peptide-Poly(ethylene glycol) Hydrogels by Coiled Coil Self-Assembly and Self-Sorting. Biomacromolecules. 2016;17(6):2260–2267. doi: 10.1021/acs.biomac.6b00528. [DOI] [PubMed] [Google Scholar]
- 228.Tropsha A, et al. Do interhelical side chain-backbone hydrogen bonds participate in formation of leucine zipper coiled coils? Proc Natl Acad Sci U S A. 1991;88(21):9488–9492. doi: 10.1073/pnas.88.21.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Addi C, et al. A highly versatile adaptor protein for the tethering of growth factors to gelatin-based biomaterials. Acta Biomater. 2017;50:198–206. doi: 10.1016/j.actbio.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 230.Reddington SC, Howarth M. Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher. Curr Opin Chem Biol. 2015;29:94–99. doi: 10.1016/j.cbpa.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 231.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 2012;109(12):E690–697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Gao X, et al. Engineering Protein Hydrogels Using SpyCatcher-SpyTag Chemistry. Biomacromolecules. 2016;17(9):2812–2819. doi: 10.1021/acs.biomac.6b00566. [DOI] [PubMed] [Google Scholar]
- 233.Schoene C, et al. SpyTag/SpyCatcher cyclization confers resilience to boiling on a mesophilic enzyme. Angew Chem Int Ed Engl. 2014;53(24):6101–6104. doi: 10.1002/anie.201402519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Liu Z, et al. A novel method for synthetic vaccine construction based on protein assembly. Sci Rep. 2014;4:7266. doi: 10.1038/srep07266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tan LL, Hoon SS, Wong FT. Kinetic Controlled Tag-Catcher Interactions for Directed Covalent Protein Assembly. PLoS One. 2016;11(10):e0165074. doi: 10.1371/journal.pone.0165074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Alam MK, et al. Synthetic Modular Antibody Construction by Using the SpyTag/SpyCatcher Protein-Ligase System. Chembiochem. 2017;18(22):2217–2221. doi: 10.1002/cbic.201700411. [DOI] [PubMed] [Google Scholar]
- 237.Yumura K, et al. Use of SpyTag/SpyCatcher to construct bispecific antibodies that target two epitopes of a single antigen. J Biochem. 2017;162(3):203–210. doi: 10.1093/jb/mvx023. [DOI] [PubMed] [Google Scholar]
- 238.Brune KD, et al. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci Rep. 2016;6:19234. doi: 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Schloss AC, et al. Fabrication of Modularly Functionalizable Microcapsules Using Protein-Based Technologies. ACS Biomater. Sci. Eng. 2016;2(11):1856–1861. doi: 10.1021/acsbiomaterials.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bedbrook CN, et al. Genetically Encoded Spy Peptide Fusion System to Detect Plasma Membrane-Localized Proteins In Vivo. Chem Biol. 2015;22(8):1108–1121. doi: 10.1016/j.chembiol.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Deans TL, et al. Regulating synthetic gene networks in 3D materials. Proc Natl Acad Sci U S A. 2012;109(38):15217–15222. doi: 10.1073/pnas.1204705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Singh A, Deans TL, Elisseeff JH. Photomodulation of Cellular Gene Expression in Hydrogels. Acs Macro Letters. 2013;2(3):269–272. doi: 10.1021/mz300591m. [DOI] [PubMed] [Google Scholar]