Abstract
Background and Objectives
Previous studies have demonstrated the metabolism of tibolone through sulfation, with the cytosolic sulfotransferase (SULT) SULT2A1 as the major responsible enzyme. The current study aimed to investigate how SULT2A1 genetic polymorphisms may affect the dehydroepiandrosterone (DHEA)- and tibolone-sulfating activity of SULT2A1.
Methods
Site-directed mutagenesis was employed to generate cDNAs encoding ten different SULT2A1 allozymes. Recombinant SULT2A1 allozymes were expressed in BL21 E. coli cells, and purified using glutathionine-Sepharose affinity chromatography. An established sulfotransferase assay was used to analyze DHEA- and tibolone-sulfating activity of the purified SULT2A1 allozymes.
Results
The nine human SULT2A1 allozymes plus the wild-type SULT2A1 were found to display differential sulfating activity toward DHEA and tibolone. Kinetic analysis revealed that different SULT2A1 allozymes exhibited differential substrate affinity and catalytic efficiency toward the two substrates tested.
Conclusion
The results obtained provided useful information concerning the differential metabolism of tibolone through sulfation in individuals with different SULT2A1 genotypes.
1. Introduction
Tibolone, a synthetic steroid hormone drug, is commonly used in hormone replacement therapy in postmenopausal women as well as for the treatment of endometriosis [1–8]. The metabolism of tibolone in humans has been shown to involve both Phase I and Phase II metabolic reactions, with sulfation being the most important one in the latter category [9]. Using recombinant human cytosolic sulfotransferases (SULTs), it has been demonstrated that tibolone may be subjected to SULT-mediated sulfation, with SULT2A1 being the major responsible enzyme [10]. In addition to its therapeutic role, there are adverse effects associated with the use of tibolone, with studies showing correlations with breast, kidney, liver and vascular disorders [11]. Further investigation into the metabolism of tibolone, in particular sulfation, may provide insightful information concerning reasons underlying these health risks.
The cytosolic sulfotransferases (SULTs) are a major group of Phase II enzymes involved in the metabolism of key endogenous compounds such as catecholamine neurotransmitters and thyroid/steroid hormones, as well as a variety of xenobiotics including drugs [12–15]. SULT enzymes catalyze the transfer of the sulfonate group from the donor compound, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to the hydroxyl or amine group of substrate compounds [16]. The addition of the sulfonate group may inactivate and/or increase the water-solubility of the substrate compounds, thereby facilitate their excretion from the body [12–15]. In humans, there are thirteen SULTs that display distinct substrate specificity and/or tissue distribution [17]. As mentioned above, sulfation has been shown to be the most important Phase II reaction in the metabolism of tibolone in vivo [9], and that SULT2A1 has been identified as a major enzyme mediating the sulfation of tibolone [10]. While the major enzyme capable of sulfating tibolone has been identified, a lesser understood aspect is how genetic polymorphisms of the responsible enzyme, SULT2A1, may affect the differential metabolism of tibolone in different individuals. Like with many other genes, single nucleotide polymorphisms (SNPs) of SULT genes have been reported [15, 18, 19]. An earlier study sequencing DNA samples from African-American and Caucasian-American subjects revealed three non-synonymous coding SNPs (cSNPs) for the SULT2A1 gene [20]. Interestingly, the corresponding allozymes expressed in COS-7 cells displayed differential sulfating activity toward dehydroepiandrosterone (DHEA), an endogenous substrate [12]. These findings, therefore, raised an interesting issue whether SULT2A1 genetic polymorphisms may have an impact on the metabolism of tibolone through sulfation, thereby influencing the efficacy of the drug in different individuals.
In this study, we report the generation, expression, and purification of ten SULT2A1 allozymes. The differential sulfating activities of the purified SULT2A1 allozymes toward tibolone and DHEA were examined and their kinetic constants determined.
2. Materials and Methods
2.1. Materials
Dehydroepiandrosterone (DHEA), tibolone, dithiothreitol (DTT), dimethylsulfoxide (DMSO), 3-(N-morpholino) propanesulfonic acid (MOPS), and isopropyl-1-thio-β-D-galactopyranoside (IPTG) were products of Sigma Chemical Company (St. Louis, MO USA). Carrier-free sodium [35S]sulfate was a product of Perkin-Elmer (Waltham, MA USA). Ecolume scintillation cocktail was purchased from MP Biomedical (Solon, OH USA). Cellulose thin-layer chromatography (TLC) plates were from EMD chemicals (Gibbstown, NJ USA). Recombinant human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase was prepared as previously described [21]. EX Taq DNA polymerase was a product of Takara Bio (Mountain View, CA USA). Protein molecular weight markers were from New England Biolabs, Inc. (Ipswich, MA USA). Oligonucleotide primers were synthesized by MWG Biotech (Louisville, KY USA). All other reagents were of the highest grades commercially available.
2.2. Preparation of SULT2A1 allozymes
The QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA USA) was used for the generation of cDNAs encoding SULT2A1 allozymes. Briefly, wild-type SULT2A1 cDNA packaged in pGEX-2TK prokaryotic expression vector was used as the template in conjunction with specific mutagenic primers (see Table 1 for the mutagenic primers used). The amplification conditions were 12 cycles of 30 s at 95°C, 1 min at 55°C, and 6 min at 68°C. The “mutated” SULT2A1 sequences were verified by nucleotide sequencing [22]. pGEX-2TK vector harboring individual mutated SULT2A1 sequence was transformed into competent XL1-Blue E. coli cells. The transformed cells, grown to A600 nm = ~0.5 in 1 liter of Luria broth (LB) medium supplemented with 100 μg/ml ampicillin and induced with 0.1 mM IPTG overnight at room temperature, were collected by centrifugation and homogenized in 20 ml of an ice-cold lysis buffer (10 mM Tris-HCl, pH 8.0, 150 mm NaCl, and 1 mM EDTA) using an Aminco French press. The crude homogenate thus prepared was subjected to centrifugation at 10,000 × g for 30 min at 4°C. The supernatant collected was fractionated using 0.5 ml of glutathione-Sepharose, and the bound fusion protein was treated with 2 ml of a thrombin digestion buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2.5 mM CaCl2) containing 5 units/ml bovine thrombin. Following a 1-h incubation at room temperature with constant agitation, the preparation was subjected to centrifugation. The recombinant enzyme present in the supernatant collected was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to the enzymatic characterization as described below.
Table 1.
Primer sets used in the site-directed mutagenesis of the cDNA encoding human human SULT2A1
| SULT2A1 Allozyme | Amino Acid Substitution | Mutagenic Primer Set |
|---|---|---|
| SULT2A1*2 | Met57Thr | 5′-gagattctctgcctgacgcactccaagggggat-3′ 5′-atccccttggagtgcgtcaggcagagaatctc-3′ |
| SULT2A1*3 | Thr90Ser | 5′-acagcactcagtgaatcggagagtccacgttta-3′ 5′-taaacgtggactctccgattcactgagtgctgt-3′ |
| SULT2A1*4 | Leu159Val | 5′-tgtcaaggaaccgtggtatatggtcatggttt-3′ 5′-aaacatgacccatataccacagttccttgaca-3′ |
| SULT2A1*5 | Glu186Val | 5′-ttactgagttatgaggtgctgaaacaggacaca 3′ 5′-tgtgtcctgtttcagcacctcataactcagtaa-3′ |
| SULT2A1*6 | Met57Thr/Glu186Val | *2 + *5 |
| SULT2A1*7 | Ala63Pro | 5′-ccaagggggatcccaagtggatc-3′ 5′-gatcacttgggatcccccttgg-3′ |
| SULT2A1*8 | Lys227Glu | 5′-tgaaagaaaacgagatgtccaat-3′ 5′-attggacatctcgttttctttca-3′ |
| SULT2A1*9 | Ala261Thr | 5′-acttcacagtgacccaagctgaa-3′ 5′-ttcagcttgggtcactgtgaagt-3′ |
| SULT2A1*10 | Ala63Pro/Ala261Thr | *7 + *9 |
SULT, cytosolic sulfotransferase.
2.3. Enzymatic assay
The sulfating activity of the recombinant human SULT2A1 allozymes was determined using PAP[35S] as the sulfonate donor. The reaction mixture for the standard enzymatic assay, prepared in a final volume of 20 μl, contained, 50 mM MOPS at pH 7.0, 14 μM of PAP[35S], 1 mM DTT, and 10 μM substrate (DHEA or tibolone). Controls with water or DMSO replacing substrate were also included. The reaction was started by the addition of the enzyme, allowed to continue at 37°C for 10 minutes, and terminated by placing the thin-walled tube containing the reaction mixture on a heating block at 100°C for 2 minute. Heated reaction mixture was subjected to centrifugation to pellet down the precipitates formed. Afterwards, 2 μl of the reaction mixture was spotted on a cellulose TLC plate and the spotted TLC plate was subjected to TLC analysis using a solvent system containing n-butanol, isopropanol, 88% formic acid and H2O in a ratio of 3:1:1:1 (by volume) [23]. Upon completion of TLC, the TLC plate was air-dried and autoradiographed by using an X-ray film. The radioactive spot corresponding to the sulfated product was located and cut out and eluted in 0.5 ml water in a glass vial. 4.5 ml of Ecolume scintillation liquid was added to each vial, mixed thoroughly, and the radioactivity therein was counted by using a liquid scintillation counter. The cpm count obtained was used to calculate the specific activity in the unit of nmol of sulfated product/minute/mg enzyme. For the kinetic studies on the sulfation of DHEA and tibolone, different concentrations (1 μM, 2 μM, 5 μM, 10 μM, and 20 μM) of these substrate compounds and 50 mM MOPS buffer at pH 7.0 were used. The specific activity data were analyzed based on the Michaelis-Menten equation to calculate the kinetic constants. GraphPad Prisim6 software was used in data analysis.
2.4. Miscellaneous methods
The sulfonate donor, PAP [35S], was synthesized from ATP and carrier-free [35S]sulfate using the bifunctional human ATP sulfurylase/APS kinase [21]. The synthesized PAP[35S] was adjusted to the desired concentration and specific activity by the addition of nonradioactive (cold) PAPS. SDS-PAGE was performed on a 12% polyacrylamide gel using the method of Laemmli [24]. Protein determination was based on the method of Bradford [25] with bovine serum albumin as a standard.
3. Results
3.1. Preparation of human SULT2A1 allozymes
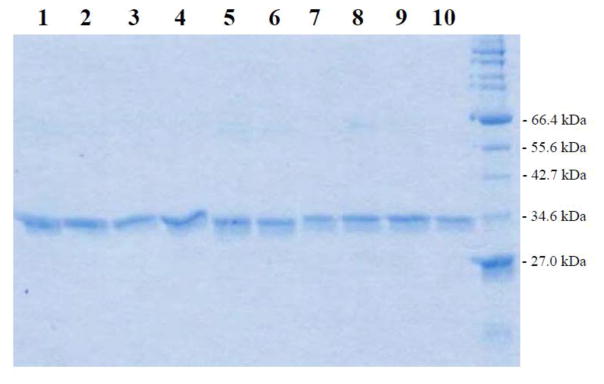
Based on the procedure described in the “Materials and Methods” section, cDNAs encoding different SULT2A1 allozymes packaged in pGEX-2TK prokaryotic expression vector were individually transformed into BL 21 E. coli host cells for expressing the recombinant enzymes. As shown in Figure 1, the recombinant SULT2A1 allozymes fractionated from the homogenates of transformed E. coli cells using glutathione-Sepharose and cleaved off the bound fusion proteins by thrombin digestion appeared to be highly homogeneous upon SDS-polyacrylamide gel electrophoresis, with apparent molecular weights similar to predicted molecular weight (33,780) of SULT2A1.
Figure 1.
SDS gel electrophoretic pattern of the purified human SULT2A1 allozymes. SDS-PAGE was performed on a 12% gel, followed by Coomassie blue staining. Samples analyzed in lanes 1 through 1 correspond to SULT2A1 allozymes 1 through 10. Positions of protein molecular weight markers co-electrophoresed are marked on the right.
3.2. Sulfation of DHEA and tibolone by human SULT2A1 allozymes
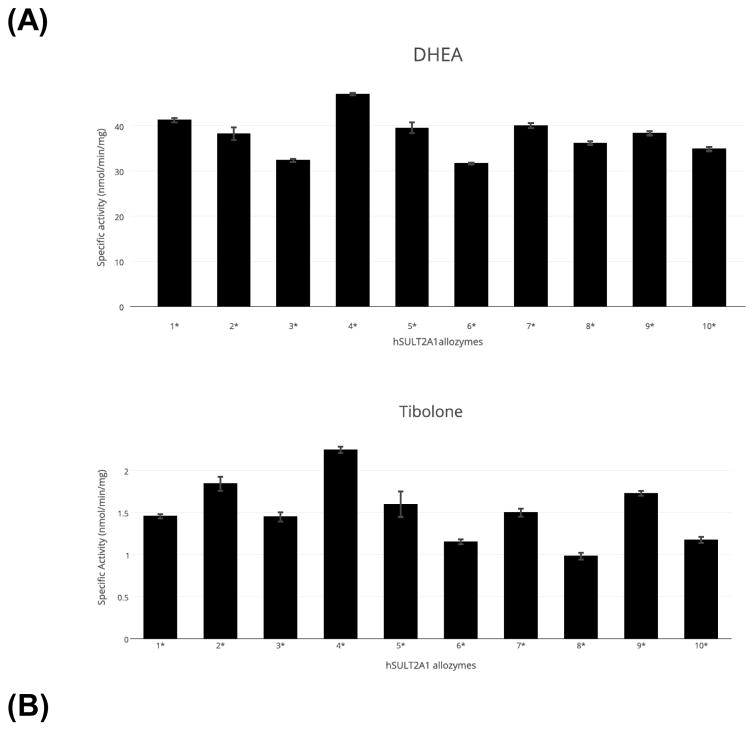
Purified SULT2A1 allozymes were assayed for sulfating activity using DHEA and tibolone as substrates. As shown in Figure 2A, the ten allozymes exhibited specific activities ranging 31.64 – 47.08 nmol/min/mg with DHEA as the substrate. With tibolone as the substrate, the allozymes exhibited specific activities ranging 0.98 – 2.25 nmol/min/mg. It is noted that both substrates were tested at a concentration of 10 μM. While the variation in DHEA-sulfating activity among the ten SULT2A1 allozymes was not as dramatic as that previously reported [20], the differences are significant and may have functional implications in vivo. The variation in tibolone-sulfating activity, on the other hand, was considerably bigger (close to 2.3 fold) among the ten SULT2A1 allozymes (Figure 2B).
Figure 2.
Specific activities of the sulfation of (A) DHEA and (B) tibolone by human SULT2A1 allozymes. Concentration of DHEA or tibolone used in the enzymatic assays was 10 μM. Data shown represent mean ± standard deviation derived from four independent experiments.
3.3. Kinetics of the sulfation of DHEA and tibolone by human SULT2A1 allozymes
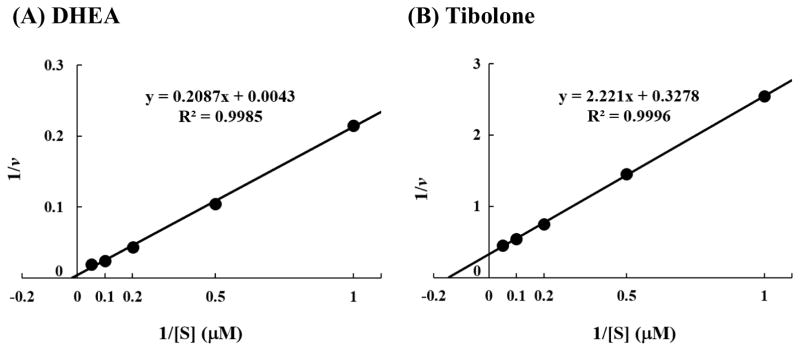
Enzymatic assays were carried out using varying concentrations of DHEA or tibolone. Data obtained were used to generate Lineweaver-Burke double-reciprocal plots in order to calculate the Km, Vmax, and Vmax/Km for each of the ten SULT2A1 allozymes in catalyzing the sulfation of DHEA or tibolone. Figure 3 shows the Lineweaver-Burke double reciprocal plots of the sulfation of, respectively, DHEA and tibolone by the wild-type SULT2A1 (SULT2A1*1). The values of Km, Vmax, and Vmax/Km determined for the ten SULT2A1 allozymes are compiled in Table 2. With DHEA as the substrate, the Km values determined for the ten SULT2A1 allozymes spanned a range of 8.71 – 11.91 μM, while the Vmax values spanned a range of 61.41 – 93.64 nmol/min/mg enzyme. Calculated Vmax/Km determined for the ten allozymes varied in the range of 6.39 – 8.46, which in general correlate reasonably well with the specific activity data shown in 2A. With tibolone as the substrate, the Km values determined for the ten SULT2A1 allozymes spanned a range of 8.69 – 15.98 μM, while the Vmax values spanned a range of 2.33 – 5.16 nmol/min/mg enzyme. Calculated Vmax/Km determined for the ten allozymes varied in the range of 0.18 – 0.47, which also correlate reasonably well with the specific activity data shown in Figure 2B.
Figure 3.
Lineweaver-Burke double-reciprocal plot of the sulfation of (A) DHEA and (B) tibolone by wild-type human SULT2A1 (SULT2A1*1).
Table 2.
Kinetic constants of the sulfation of DHEA and tibolone by human SULT2A1 allozymes
| DHEA as Substrate | |||
|---|---|---|---|
| Vmax (nmol/min/mg) | Km (μM) | Vmax/Km | |
| SULT2A1*1 | 83.2 ± 4.6 | 10.9 ± 1.3 | 7.63 |
| SULT2A1*2 | 75.5 ± 4.6 | 9.96 ± 1.4 | 7.58 |
| SULT2A1*3 | 76.8 ± 3.7 | 11.9 ± 1.2 | 6.45 |
| SULT2A1*4 | 93.6 ± 6.7 | 11.1 ± 1.7 | 8.46 |
| SULT2A1*5 | 84.6 ± 5.3 | 11.8 ± 1.5 | 7.14 |
| SULT2A1*6 | 61.4 ± 3.6 | 9.6 ± 3.6 | 6.37 |
| SULT2A1*7 | 81.6 ± 5.0 | 11.1 ± 1.4 | 7.38 |
| SULT2A1*8 | 73.5 ± 3.4 | 10.8 ± 1.1 | 6.79 |
| SULT2A1*9 | 73.3 ± 5.2 | 10.0 ± 1.5 | 7.34 |
| SULT2A1*10 | 62.8 ± 5.3 | 8.7 ± 1.4 | 7.22 |
| Tibolone as Substrate | |||
| SULT2A1*1 | 3.6 ± 0.2 | 13.0 ± 1.7 | 0.28 |
| SULT2A1*2 | 3.6 ± 0.1 | 8.7 ± 0.8 | 0.41 |
| SULT2A1*3 | 2.8 ± 0.2 | 8.9 ± 1.4 | 0.32 |
| SULT2A1*4 | 5.2 ± 0.3 | 10.9 ± 1.5 | 0.47 |
| SULT2A1*5 | 3.4 ± 0.2 | 10.1 ± 1.4 | 0.34 |
| SULT2A1*6 | 2.9 ± 0.2 | 14.3 ± 2.0 | 0.20 |
| SULT2A1*7 | 3.7 ± 0.4 | 12.6 ± 2.3 | 0.29 |
| SULT2A1*8 | 2.8 ± 0.3 | 16.0 ± 3.0 | 0.18 |
| SULT2A1*9 | 3.8 ± 0.4 | 11.0 ± 2.2 | 0.34 |
| SULT2A1*10 | 2.3 ± 0.2 | 8.9 ± 1.6 | 0.26 |
Data shown represent mean ± standard deviation calculated based on the results obtained from four independent kinetic experiments.
SULT, cytosolic sulfotransferase.
4. Discussion
The current study was designed to clarify how genetic polymorphisms may impact the sulfation of tibolone in different individuals with different SULT2A1 genotypes. Evidence indicating individual variation in the enzymatic activity of SULT2A1, previously named the DHEA sulfotransferase, was first reported in 1993 [26]. Of the 94 human hepatic tissue samples analyzed, a 4.6-fold variation in DHEA-sulfating activity was observed. Moreover, the frequency distribution of DHEA-sulfating activity among the samples tested appeared to be bimodal, with approximately 25% in a high activity subgroup [26]. These findings provided the first indication of underlying genetic variations that may influence the sulfating activity of DHEA. In a later study sequencing DNA samples from 120 human subjects, three non-synonymous coding SNPs for the SULT2A1 gene were detected [20]. The corresponding SULT2A1 allozymes, expressed in COS-1 cells, displayed differential DHEA-sulfating activity. These data showed clearly a genotype-phenotype relationship for SULT2A1. It is thus important to clarify further how genetic polymorphisms of SULT2A1 may influence the metabolism of not only the endogenous compounds such as DHEA, but also drugs including tibolone, through sulfation. By searching the NCBI databases, we have identified ten non-synonymous coding SNPs for SULT2A1 (cf. Table 1). To find out how different amino acid changes may affect the sulfating activity of the SULT2A1 protein products, our approach was to prepare recombinant SULT2A1 allozymes for enzymatic characterization with DHEA and tibolone as substrates.
Previous studies have revealed DHEA as a major endogenous substrate for SULT2A1 [27]. For tibolone, SULT2A1 was reported to be the major enzyme responsible for its sulfation [9]. In view of the genetic polymorphisms of SULT2A1, it is an interesting question whether different SULT2A1 allozymes may display differential sulfating activities toward not only the endogenous DHEA, but also drugs such as tibolone. Data from the enzymatic assays showed indeed significant differences in both DHEA- and tibolone-sulfating activities among the ten SULT2A1 allozymes analyzed. To examine further the effects of genetic polymorphisms on the sulfation of DHEA and tibolone by SULT2A1, kinetic experiments were performed on each of the ten SULT2A1 allozymes. Kinetic constants (Km, Vmax, and Vmax/Km) obtained indicated that the ten SULT2A1 allozymes displayed differential affinities for DHEA or tibolone, as well as intrinsic catalytic activities toward the two substrates. Collectively, the results derived from the above-mentioned studies provide useful information concerning the enzymatic mechanism underlying the differential sulfation of DHEA and tibolone by different SULT2A1 allozymes. It thus appears that SULT2A1 polymorphisms may have implications in not only the metabolism of key endogenous compounds such as DHEA, but also drugs like tibolone that may be metabolized through sulfation by SULT2A1.
5. Conclusions
The current study was designed to clarify how SULT2A1 genetic polymorphisms may affect the DHEA- and tibolone-sulfating activities of SULT2A1 allozymes. Results obtained showed indeed differential DHEA- and tibolone-sulfating activities among the ten SULT2A1 allozymes analyzed. Kinetic analysis revealed further that different SULT2A1 allozymes exhibited differential substrate affinity and catalytic efficiency toward the two substrates tested. Additional studies are warranted in order to clarify how SULT2A1 polymorphisms may impact on the expression and stability of SULT2A1 allozymes and thus the metabolism and pharmacokinetics of tibolone in vivo.
Key Points.
Human SULT2A1 allozymes displayed differential sulfating activities toward DHEA and tibolone.
Kinetic analysis revealed that different SULT2A1 allozymes exhibited differential substrate affinity and catalytic efficiency toward DHEA and tibolone.
Acknowledgments
Funding: This study was supported in part by a grant from National Institutes of Health (Grant # R03HD071146).
Footnotes
Compliance with Ethical Standards: This work complies with all ethical standards.
Conflict of Interest: All authors declare that they have no conflict of interest.
References
- 1.Moore RA. Livial: a review of clinical studies. Br J Obstet Gynaecol. 1999;106(Suppl 19):1–21. [PubMed] [Google Scholar]
- 2.Swegle JM, Kelly MW. Tibolone: a unique version of hormone replacement therapy. Ann Pharmacother. 2004;38:874–881. doi: 10.1345/aph.1D462. [DOI] [PubMed] [Google Scholar]
- 3.Kenemans P, Speroff L. Tibolone: clinical recommendations and practical guidelines. A report of the International Tibolone Consensus Group. Maturitas. 2005;51:21–28. doi: 10.1016/j.maturitas.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Lazovic G, Radivojevic U, Marinkovic J. Tibolone: the way to beat many a postmenopausal ailments. Expert Opin Pharmacother. 2008;9:1039–1047. doi: 10.1517/14656566.9.6.1039. [DOI] [PubMed] [Google Scholar]
- 5.Biglia N, Maffei S, Lello S, Nappi RE. Tibolone in postmenopausal women: a review based on recent randomised controlled clinical trials. Gynecol Endocrinol. 2010;26:804–814. doi: 10.3109/09513590.2010.495437. [DOI] [PubMed] [Google Scholar]
- 6.Lundstrom E, Christow A, Kersemaekers W, Svane G, Azavedo E, Söderqvist G, Mol-Arts M, Barkfeldt J, von Schoultz B. Effects of tibolone and continuous combined hormone replacement therapy on mammographic breast density. Am J Obstet Gynecol. 2002;186:717–722. doi: 10.1067/mob.2002.121896. [DOI] [PubMed] [Google Scholar]
- 7.Cummings SR, Ettinger B, Delmas PD, Kenemans P, Stathopoulos V, Verweij P, Mol-Arts M, Kloosterboer L, Mosca L, et al. The effects of tibolone in older postmenopausal women. N Engl J Med. 2008;359:697–708. doi: 10.1056/NEJMoa0800743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Archer DF, Stathopoulos V, Helmond F. Endometrial effects of tibolone. J Clin Endocrinol Metab. 2007;92:911–918. doi: 10.1210/jc.2006-2207. [DOI] [PubMed] [Google Scholar]
- 9.Vos RM, Krebbers SF, Verhoeven CH, Delbressine LP. The in vivo human metabolism of tibolone. Drug Metab Dispos. 2002;30:106–112. doi: 10.1124/dmd.30.2.106. [DOI] [PubMed] [Google Scholar]
- 10.Falany J, Macrina N, Falany C. Sulfation of tibolone and tibolone metabolites by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 2004;88:383–391. doi: 10.1016/j.jsbmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Zayas-Jaime FJ, Ornelas-Aguirre JM, Perez-Napoles DE. Reasons for the Abandonment of hormone replacement therapy with tibolone in menopausal women. Ginecol Obstet Mex. 2013;81:593–601. [PubMed] [Google Scholar]
- 12.Falany CN, Roth JA. Properties of human cytosolic sulfotransferases involved in drug metabolism. In: Jeffery EH, editor. Human Drug Metabolism; from Molecular Biology to Man. CRC Press; Boca Raton: 1994. pp. 101–115. [Google Scholar]
- 13.Mulder GM, Jakoby WB. Sulfation. In: Mulder GJ, editor. Conjugation Reactions in Drug Metabolism. Taylor and Francis; London: 1990. pp. 107–161. [Google Scholar]
- 14.Weinshilboum RM, Otterness DM. Sulfotransferase enzymes. In: Kaufman FC, editor. Conjugation-Deconjugation Reactions in Drug Metabolism and Toxicity. Springer-Verlag; Berlin: 1994. pp. 45–78. [Google Scholar]
- 15.Glatt H, Engelke CE, Pabel U, Teubner W, Jones AL, Coughtrie MW, Andrae U, Falany CN, Meinl W. Sulfotransferases: genetics and role in toxicology. Toxicol Lett. 2000;112–113:341–348. doi: 10.1016/s0378-4274(99)00214-3. [DOI] [PubMed] [Google Scholar]
- 16.Lipmann F. Biological sulfate activation and transfer. Science. 1958;128:575–580. doi: 10.1126/science.128.3324.575. [DOI] [PubMed] [Google Scholar]
- 17.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 18.Glatt H, Engelke CE, Pabel U, Teubner W, Jones AL, Coughtrie MW, Andrae U, Falany CN, Meinl W. Sulfotransferases: genetics and role in toxicology. Toxicol Lett. 2000;112–113:341–348. doi: 10.1016/s0378-4274(99)00214-3. [DOI] [PubMed] [Google Scholar]
- 18.Lindsay J, Wang LL, Li Y, Zhou SF. Structure, function and polymorphism of human cytosolic sulfotransferases. Curr Drug Metab. 2008;9:99–105. doi: 10.2174/138920008783571819. [DOI] [PubMed] [Google Scholar]
- 19.Daniels J, Kadlubar S. Sulfotransferase genetic variation: from cancer risk to treatment response. Drug Metab Rev. 2013;45:415–422. doi: 10.3109/03602532.2013.835621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomae BA, Eckloff BW, Freimuth RR, Wieben ED, Weinshilboum RM. Human sulfotransferase SULT2A1 pharmacogenetics: genotype-to-phenotype studies. Pharmacogenomics J. 2002;2:48–56. doi: 10.1038/sj.tpj.6500089. [DOI] [PubMed] [Google Scholar]
- 21.Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, Takayanagi K, Natori Y, Liu MC. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. Biosci Biotechnol Biochem. 1998;62:1037–1040. doi: 10.1271/bbb.62.1037. [DOI] [PubMed] [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu MC, Lipmann F. Decrease of tyrosine-O-sulfate-containing proteins found in rat fibroblasts infected with Rous sarcoma virus or Fujinami sarcoma virus. Proc Natl Acad Sci USA. 1984;81:3695–3698. doi: 10.1073/pnas.81.12.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Aksoy IA, Sochorová V, Weinshilboum RM. Human liver dehydroepiandrosterone sulfotransferase: nature and extent of individual variation. Clin Pharmacol Ther. 1993;54:498–506. doi: 10.1038/clpt.1993.181. [DOI] [PubMed] [Google Scholar]
- 27.Comer KA, Falany JL, Falany CN. Cloning and expression of human liver dehydroepiandrosterone sulphotransferase. Biochem J. 1993;289:233–240. doi: 10.1042/bj2890233. [DOI] [PMC free article] [PubMed] [Google Scholar]