Abstract
Due to its substantial nutritional value, quinoa (Chenopodium quinoa Willd.) is currently attracting worldwide attention. Quinoa is characterized by a high adaptability to various environmental conditions. This is the first report on the phytochemical and genetic evaluation of quinoa germplasms introduced to Egypt, and the results could be used to implement propagation techniques in the future. For phytochemical characterization, 41 traits, including primary and secondary metabolites, antioxidant molecules, sugars, organic acids and fatty acids, were evaluated. At the same time, 4 RAPD and 7 ISSR markers were used for genetic analysis. UPGMA analysis of RAPD and ISSR polymorphic markers, their combined dataset and phytochemical traits were used to evaluate genetic relationships among genotypes. The quinoa genotypes displayed reasonable variation in the studied phytochemical traits. The results of the genetic analysis confirmed that RAPD and ISSR markers could be used to distinguish effectively quinoa genotypes. The phytochemical and genetic characterization reported herein will be a promising guide for breeding seed quality in quinoa.
Electronic supplementary material
The online version of this article (10.1007/s12298-018-0541-4) contains supplementary material, which is available to authorized users.
Keywords: Quinoa, Phytochemical traits, Molecular markers, Genetic analysis
Introduction
Quinoa (Chenopodium quinoa Willd.) is an annual broadleaf plant belonging to the Amaranthaceae family. Quinoa is an allotetraploid (2n = 4x = 36, with a base chromosome number of x = 9) and exhibits disomic inheritance for most qualitative traits (Risi and Galwey 1984). Quinoa seed is covered with a perianth, whose color is the same as that of the stem (Ruales and Nair 1993). Quinoa is considered a pseudocereal and could be used as an alternative to cereals.
Due to their high protein content composed of an incredible mixture of essential amino acids, quinoa seeds have potential nutritive value (Valencia-Chamorro 2003). Moreover, their relatively high content of beneficial fatty acids and tocopherols could make quinoa seeds a potentially valuable candidate as a new oil crop (Vidueiros et al. 2015). Quinoa seeds are characterized as having a high starch content; however, quinoa seeds cannot be used solely for bread making, as they lack gluten.
Interest in quinoa has increased in recent decades because of the enormous adaptability of the species to different agro-ecological conditions as well as its potential contribution in the fight against hunger and malnutrition (Zurita-Silva et al. 2014). Its capability of contributing to global food security was documented in 2013 (International Year of Quinoa). Quinoa cultivation has expanded beyond its traditional range to more than 70 countries during the past decade (FAO 2012).
Quinoa seeds are considered a good source of fiber, vitamins and minerals as well as health-benefiting phytochemicals (Varli and Sanlier 2016). Compared with whole cereal seeds, quinoa seeds, in addition to their dietary value, contain high amounts of antioxidant-phenolic compounds (Repo-Carrasco-Valencia et al. 2010). Quinoa seeds are also characterized by high contents of isoflavones, which oppose vessel contraction and reduce arterial resistance in humans (Vega-Galvez et al. 2010). Saponins in quinoa have physiological roles, as they act as insecticides, bactericides and fungicides (Vega-Galvez et al. 2010).
Molecular markers are powerful tools for the detection of genetic variability in various species and are important for germplasm conservation and cultivar identification. Furthermore, molecular markers allow the development of enhanced breeding programs, including marker-assisted selection and marker-aided backcross breeding (Maughan et al. 2004). The first DNA-based markers in quinoa were reported by Fairbanks et al. (1993), who used rapid amplified polymorphic DNA (RAPD) techniques. These DNA markers have enabled the revealing of genetic diversity between quinoa and other Chenopodiaceae species (Rana et al. 2010). Inter-simple sequence repeat (ISSR) markers are simpler to use than simple sequence repeat (SSR) markers, which are frequently used for genotyping quinoa. ISSR markers are inexpensive and are easily scored manually, and prior knowledge of the target sequences flanking the repeat regions is not necessary, which is not true for SSR markers (Ana-Cruz et al. 2017). In a recent study, EST-SSR markers were used to estimate the genetic polymorphism among four quinoa accessions (Zhang et al. 2016). Additionally, development of novel markers through whole-genome re-sequencing, like InDel (insertion/deletion) would provide knowledge on genomic variation, population structure, and genetic diversity in quinoa (Zhang et al. 2017).
Despite its agronomic potential, quinoa is still an underutilized crop, and relatively few active breeding programs exist (Zurita-Silva et al. 2014). In addition, no attempts have been made to characterize or evaluate the phytochemicals and genetic structure of the germplasm introduced to Egypt. The lack of characterization and evaluation studies could limit selection and improvement as well as cultivation expansion. To assess the genetic structure of the present genotypes, PCR-based RAPD and ISSR-DNA fingerprinting constitute one of the best tools. The phytochemical, nutritional and genetic characterization reported in the present study will be of particular value in ongoing efforts to both accelerate the improvement of quinoa and develop core collections that can be used by traditional breeding programs in Egypt.
Materials and methods
Plant material
Seeds of five genotypes of quinoa (Chenopodium quinoa Willd.) were obtained from the Egyptian Desert Research Center (DRC). The provided genotypes are known as “kvl-sra2, kvl-sra3, regalona, Q37 and Q52”. The origin of the first two genotypes is Denmark, while the origin of last three genotypes is Chile. The five genotypes were planted at Ras Sudr Experimental Farm Station of DRC, South Sinai, Egypt under saline soil and water of Ras Sudr conditions. The plantation period was from mid of November 2016 to April 2017. At full maturity stage, seeds were harvested and air-dried, healthy and uniform seeds from each genotype were selected for phytochemical and molecular analysis. Three biological replicates per each genotype were ground to a fine powder and sieved through 2.0-mm sieve for extraction.
Carbon, hydrogen and nitrogen (CHN) analysis
The CHN content of the seed powders was analyzed using an Elementar vario MACRO cube analyzer (Hanau, Germany); the combustion tube temperature was 1150 °C, the reduction tube temperature was 850 °C, and the carrier gas was ultrapure helium. The C and H in the combustion gases were measured by infrared detectors, and the N was measured by a thermal conductivity detector (the CHN data are summarized in supplementary data Table S1).
Phytochemical characterization
The total protein (TP) content was assessed by quantifying the total N and converting it to protein via multiplication by a factor of 6.25. Ninhydrin assays described by Lee and Takahashi (1966) were then used to determine the total free amino acids (TFAs). Total lipids (TLs) were extracted and quantified in accordance with a modified protocol reported by Bligh and Dyer (1959).
Alkaloids were extracted and determined in accordance with the method of Harborne (1973), and the total phenolic content was determined by the method of Saeedeh and Asna (2007). Saponins were estimated in accordance with the methods of Hiai et al. (1975), and total tannins were evaluated using the European Community (2000) reference method. All quantified phytochemicals were expressed as milligrams per gram of dry weight (d wt).
Estimation of antioxidants
The technique reported by Chang et al. (2002) was used to estimate total flavonoids using quercetin as a standard flavonoid. The ß-carotene and lycopene contents of the seed extracts were determined by the method of Nagata and Yamashita (1992).
Free sugar and organic acid HPLC analyses
Free sugar and organic acid extractions were performed in accordance with the methods of Silva et al. (2013). A definite mass and 50 ml of 0.01 N H2SO4 were mixed and stirred (300 rpm) for 30 min. The extracts were then filtered, evaporated to dryness (40 °C) and redissolved in 1 ml of 0.01 N H2SO4, followed by filtration using a 0.22-μm membrane. Sugar and organic acid analyses were assessed by reversed-phase HPLC using a Shimadzu 10AVP HPLC system (Shimadzu Corp., Japan) equipped with RID-10A refractive index detector. A 20-µl sample was injected onto an Aminex HPX-87H column (BioRad, CA, USA) and then eluted isocratically using 5 mM degassed and filtered H2SO4 for 25 min at a flow rate of 0.6 ml min−1 and a column temperature of 65 °C. The free sugars and organic acids were quantified by comparing their absorbance with those of external standards.
GC–MS of fatty acids
The extracted oils were trans-esterified according to Freedman and Butterfield (1986) by refluxing the oils with 50 ml of methanol and 10 wt% sulfuric acid for 24 h at 85 °C. The mixture was then washed thrice to remove all methanol, glycerol and acid catalysts. The chloroform was then removed using a rotary evaporator, and the produced fatty acids methyl esters (FAMEs) were dissolved in 10 ml of ethyl acetate. The fatty acid profile of the FAMEs was determined by GC–MS using an Agilent 7890A gas chromatograph equipped with a capillary column (60 m × 0.250 mm internal diameter) coated with DB-23 ([50%-cyanopropyl]-methylpolysiloxane) for the stationary phase (0.25 μm film thickness); He was used for the mobile phase (flow rate: 1.2 ml min−1). The instrument was coupled to an Agilent 5975C inert MSD with a triple axis detector (Agilent Technologies, CA, USA). One microliter of FAME solution was loaded onto the column, which was preheated to 150 °C. This temperature was held for 5 min, after which it was increased to 250 °C at a rate of 4 °C min−1 and then held for 2 min.
Genetic characterization
Extraction and purification of genomic DNA
One hundred micrograms of seed was ground to a powder in liquid N, after which the powder was transferred to prewarmed cetyltrimethyl ammonium bromide extraction buffer and incubated for 30 min at 65 °C. The total genomic DNA was isolated in accordance with the methods of Clarke (2009), although some modifications, including the addition of polyvinylpyrrolidone and mercaptoethanol, were performed. The DNA concentration and purity were determined by a Nanodrop ND-1000 spectrophotometer. The 260/280 absorption ratio ranged from 1.7 to 1.8, which indicates DNA purity. For genetic characterization, 4 RAPD and 7 ISSR primers were used (the details of the selected primers are listed in supplementary data Table S2).
RAPD-PCR
RAPD fingerprinting was performed using the indicated primers (1.0 μl) in a 20-μl reaction volume containing genomic DNA (100 ng), sterilized double-distilled H2O (7 μl) and Dream Taq Green PCR Master Mix #k 1081 (Thermo Fisher Scientific, UK) (10 μl). PCR amplification was performed as follows: initial denaturation at 94 °C for 5 min; 40 cycles of 94 °C for 1 min, 37 °C for 30 s and 72 °C for 2 min; and a final extension for 8 min at 72 °C.
ISSR-PCR
Seven ISSR primers were used as generic primers in the PCR amplification of the ISSR regions. The amplification mixture was the same as that described for the RAPD technique. The thermocycling parameters were as follows: an initial denaturation step for at 94 °C for 5 min; 40 cycles of 94 °C for 1 min, 48–52 °C for 30 s (depending on the primer), and 72 °C for 1 min; and a final extension cycle at 72 °C for 8 min.
Agarose gel electrophoresis
After the completion of PCR, 10 μl of the PCR-amplified products was electrophoresed using Tris–borate ethylenediaminetetraacetic acid buffer in a submarine gel apparatus at 100 V for 1 h. The gels were visualized under UV light and imaged using a gel documentation system (CFW-1312 M; Bio-Rad). All reactions were performed at least twice, and only stable products were scored. Thermo Scientific Gene-Ruler DNA ladders that were 1 kb or 100 bp in length (100 ng μl−1) were used as molecular size standards.
Data analysis
Each RAPD and ISSR-PCR amplification product was considered a dominant allele for a given locus. The presence or absence of an amplified fragment was scored as 1 or 0, respectively. The data were analyzed using NTSYS-pc 2.1 software. Dendrograms were constructed from the similarity matrix data of the RAPD and ISSR cluster analyses, their combined dataset and 41 phytochemical traits by the use of the unweighted pair group method with arithmetic mean (UPGMA).
Polymorphic information content (PIC) values were computed by using the formula of Roldan-Ruiz et al. (2000): PICi = 2fi (1 − fi), where PICi is the polymorphic information content “i”, fi is the frequency of the amplified allele (band present), and (1 − fi) is the frequency of the null allele (band absent). PIC estimates range from 0 (monomorphic) to 1 (highly polymorphic). The resolving power (Rp) of each primer was calculated as described by Prevost and Wilkinson (1999): Rp = ΣIb, where Ib is the band informativeness, which is calculated from the formula: 1 − [2 × (0.5 − p)], and p is the proportion of genotypes composing the band. The marker index (MI) was calculated in accordance with the methods of Powell et al. (1996) as: MI = PIC × number of polymorphic loci. Shannon’s diversity index (Shannon and Weaver 1949) was calculated by the formula H′ = − Σpi ln pi.
Statistical analysis
The data were analyzed by one-way ANOVA using Costat software. Significant differences among genotypes were calculated by Duncan’s multiple range test at 0.05%. Pearson’s correlation coefficients between phytochemical traits were used to calculate the level and type of association between each pair of traits using SPSS software.
Results and discussion
Phytochemical characterization
The primary and secondary metabolites and antioxidant composition of the five quinoa genotypes are shown in Table 1. TP accounted for highly significant variation among genotypes, as it ranged from 120.3 mg g−1 d wt (kvl-sra3) to 190.3 mg g−1 d wt (kvl-sra2). These values are similar to those reported by Bhargava et al. (2007) (125.5–210.2 mg g−1 d wt) but superior to those reported by Gonzalez et al. (2012) (91.5–155.3 mg g−1 d wt).
Table 1.
Primary metabolites, secondary metabolites (mg g−1 d wt) and antioxidant molecules (µg g−1 d wt) of five quinoa genotypes
| Genotype | Primary metabolites (mg g−1 d wt) | Secondary metabolites (mg g−1 d wt) | Antioxidants (µg g−1 d wt) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total proteins | Amino acids | Total lipids | Alkaloids | Phenolics | Saponins | Tannins | Flavonoids | β-carotene | Lycopene | |
| kvl-sra2 | 190.3 ± 1.3A | 9.0 ± 0.1B | 80.4 ± 1.1A | 4.18 ± 0.3A | 1.63 ± 0.3A | 2.76 ± 0.2B | 0.31 ± 0.1A | 272.3 ± 11.7B | 10.69 ± 1.03A | 1.98 ± 0.54AB |
| kvl-sra3 | 120.3 ± 2.2D | 8.9 ± 0.1B | 78.0 ± 4.3A | 4.24 ± 0.5A | 0.90 ± 0.1B | 2.98 ± 0.3B | 0.29 ± 0.0A | 199.7 ± 7.0C | 10.50 ± 1.56A | 2.02 ± 0.18AB |
| regalona | 173.4 ± 1.3B | 9.0 ± 0.1B | 78.3 ± 1.9A | 4.82 ± 0.4A | 0.66 ± 0.3B | 4.12 ± 0.3A | 0.23 ± 0.4C | 260.7 ± 18.7B | 12.93 ± 1.85A | 1.48 ± 0.12B |
| Q37 | 147.5 ± 3.5C | 8.6 ± 0.1C | 62.0 ± 1.7C | 4.85 ± 0.3A | 1.18 ± 0.3B | 3.27 ± 0.2B | 0.25 ± 0.2BC | 288.8 ± 7.0A | 12.02 ± 1.21A | 1.92 ± 0.29AB |
| Q52 | 149.7 ± 1.3C | 9.4 ± 0.0A | 69.3 ± 3.3B | 4.82 ± 0.3A | 2.02 ± 0.1A | 3.31 ± 0.1B | 0.28 ± 0.0AB | 127.1 ± 7.0D | 12.65 ± 1.32A | 2.40 ± 0.22A |
| P | ** | ** | ** | NS | ** | ** | ** | ** | NS | NS |
| LSD | 2.73 | 0.16 | 4.92 | 0.48 | 0.44 | 0.46 | 0.03 | 14.45 | 2.59 | 0.56 |
NS Non Significant
Means within the same column followed by different letters are significantly different (P < 0.05)
** Significant at 0.01 level
The TFAs did not significantly differ among genotypes. The lowest TFA content occurred in Q37 (8.6 mg g−1 d wt), and the highest content occurred in Q52 (9.4 mg g−1 d wt). Gonzalez et al. (2012) reported that all quinoa genotypes are rich in essential amino acids. This fact supports the need for quinoa in the human diet to treat malnutrition and improve general health.
At the same time, the TL content highly significantly varied among genotypes. The highest quinoa seed lipid content (80.4 mg g−1 d wt) occurred in kvl-sra2; the lowest (62.0 mg g−1 d wt) in Q37. The lipid contents reported in this study are within the range proposed by Koziol (1992), who reported a quinoa lipid content of 52.0–97.0 mg g−1 d wt. Compared with many gluten-free foodstuffs containing relatively high amounts of lipids but lower mineral and vitamin contents, quinoa is an outstanding gluten-free grain with high vitamin and mineral contents, making it a potentially fundamental aspect of any hygienic, gluten-free diet (Gordillo-Bastidas et al. 2016).
Highly significant differences (P < 0.05) were recorded for the investigated secondary metabolites (phenolics, saponins and tannins) among all the genotypes; however the genotypic variation did not significantly affect alkaloid concentrations. Alkaloid concentrations ranged from 4.18 mg g−1 d wt in genotype kvl-sra 2 to 4.85 mg g−1 d wt in genotype Q37. No data currently exist about the alkaloid contents of quinoa; such data could be intensely studied in the future.
The lowest phenolic content (0.66 mg g−1 d wt) was recorded for regalona; however, the highest content (2.02 mg g−1 d wt) was recorded for Q52. These values are in line with those of Miranda et al. (2010), who recorded values ranging from 1.59 to 2.84 mg g−1 d wt in Chilean quinoa. Furthermore, Tang and Tsao (2017) stated that ferulic acid-4-glucoside was the principal free phenolic compound, while vanillic and ferulic acids were the major conjugated phenolics. Such a high content of phenolics in quinoa seeds confirms the strong antioxidant and anticancer potential of seed extracts (Gawlik-Dziki et al. 2013).
In contrast to the phenolic content, regalona exhibited the highest saponin content (4.12 mg g−1 d wt); at the same time, the lowest saponin content (2.76 mg g−1 d wt) occurred in kvl-sra2. Vega-Galvez et al. (2010) stated that the pericarp, which can be detached before consumption to increase the palatability of quinoa seed, is rich in saponins. Saponin content is essential to differentiate between bitter and sweet quinoa genotypes. Knowledge about saponin content in different genotypes and its biosynthesis regulation may aid the further development of sweet cultivars (Fiallos-Jurado et al. 2016). In light of the above data, kvl-sra2 is the most ideal genotype for human consumption.
The content of tannins was lower than that of the other secondary metabolites in the five quinoa genotypes. The tannin content, which is an anti-nutritional factor, varied from 0.23 mg g−1 d wt in regalona to 0.31 mg g−1 d wt in kvl-sra2. Nevertheless, Chauhan et al. (1992) reported higher tannin contents in quinoa seeds (0.53%) and affirmed that detaching the seed pericarp reduced the tannin contents to 0.28%. The low tannin content in quinoa increases its nutritional value, palatability and digestibility.
The differences in antioxidant molecules among quinoa genotypes were highly significant for flavonoids but were not significant for β-carotene or lycopene. The highest flavonoid content (288.8 µg g−1 d wt) was recorded in Q37, whereas the lowest content (127.1 µg g−1 d wt) was recorded in Q52. These flavonoid values are considerably higher than those obtained by Carciochi et al. (2014) (110.6 µg g−1 d wt). Those authors reported that quinoa seeds are composed of 0.37% flavonoids, which increased fourfold 3 d after germination.
The mean values of β-carotene among quinoa genotypes varied between 10.5 µg g−1 d wt (kvl-sra3) and 12.65 µg g−1 d wt (Q52). Until now, no published data were available concerning the β-carotene content of quinoa seeds. However, the β-carotene content in quinoa leaves has been estimated and ranged from 4.3 to 19.5 μg g−1 fresh weight (f wt) (Sharma et al. 2012). In addition to the antioxidant capability of β-carotene, its presence in quinoa seeds makes them a dietary supplement in the case of vitamin A deficiency.
The Q52 genotype had the highest lycopene content (2.40 µg g−1 d wt), and the regalona had the lowest content (1.48 µg g−1 d wt). Lycopene exhibits many health benefits. In addition to its potent antioxidative potential, lycopene can prevent carcinogenesis and atherogenesis by stabilizing biomolecules such as lipids, proteins and DNA (Agarwal and Rao 1998). Importantly, compared with the other genotypes, Q52 had higher β-carotene and lycopene contents but a lower flavonoid content.
Organic acid and sugar contents
The results of the HPLC analysis of organic acids in the quinoa genotypes revealed four compounds (oxalate, tartrate, succinate and lactate), whose contents significantly varied (Table 2; Fig. S1). The highest organic acid contents were recorded in genotypes Q52 and kvl-sra3 (56.35 and 55.02 µg g−1 d wt, respectively); the regalona and Q37 genotypes had medium organic acid contents (41.19 and 38.99 µg g−1 d wt, respectively), and the kvl-sra2 genotype had the lowest content (18.37 µg g−1 d wt) of organic acid. Succinate was the predominant organic acid in all genotypes except kvl-sra2. Tartrate was the second most predominant organic acid, although this was not the case in the regalona and Q37 genotypes. Oxalate and lactate were common in all genotypes, although the content of each was low. Organic acids increase the therapeutic and nutritional value of quinoa and in addition to increasing mineral absorption, organic acids regulate pepsin activity by reducing gastric pH (Lückstädt and Mello 2011).
Table 2.
Organic acids and free sugars composition (µg g−1 d wt) of five quinoa genotypes
| RT (min) | Compound | Genotype | ANOVA | |||||
|---|---|---|---|---|---|---|---|---|
| kvl-sra2 | kvl-sra3 | regalona | Q37 | Q52 | P | LSD | ||
| Organic acids (µg g−1 d wt) | ||||||||
| 6.33 | Oxalic | 6.82 ± 0.02B | 4.85 ± 0.01C | 8.88 ± 0.02A | 2.26 ± 0.01D | 1.96 ± 0.01E | ** | 0.02 |
| 8.02 | Tartaric | 11.21 ± 0.10B | 11.87 ± 0.17B | 0.00 ± 0.00C | 0.00 ± 0.00C | 16.52 ± 1.04A | ** | 0.86 |
| 11.00 | Succinic | 0.00 ± 0.00E | 37.98 ± 0.28A | 32.01 ± 0.06D | 36.43 ± 0.10C | 37.51 ± 0.20B | ** | 0.29 |
| 12.50 | Lactic | 0.34 ± 0.00B | 0.32 ± 0.00C | 0.30 ± 0.01D | 0.30 ± 0.02D | 0.36 ± 0.01A | ** | 0.02 |
| Total | 18.37 | 55.02 | 41.19 | 38.99 | 56.35 | |||
| Sugar (mg g−1 d wt) | ||||||||
| 7.14 | Mannitol | 0.71 ± 0.02C | 0.50 ± 0.02E | 0.64 ± 0.01D | 0.82 ± 0.00B | 1.18 ± 0.03A | ** | 0.03 |
| 8.78 | Glucose | 4.90 ± 0.10C | 4.93 ± 0.03C | 4.88 ± 0.01C | 5.85 ± 0.05A | 5.40 ± 0.20B | ** | 0.19 |
| 9.52 | Cellobiose | 1.46 ± 0.00A | 1.26 ± 0.01B | 1.03 ± 0.03C | 1.38 ± 0.02AB | 1.44 ± 0.16AB | ** | 0.13 |
| 10.38 | Arabinose | 3.96 ± 0.32A | 0.00 ± 0.00B | 0.00 ± 0.00B | 0.00 ± 0.00B | 0.00 ± 0.00B | ** | 0.26 |
| 14.64 | Galactose | 0.88 ± 0.01D | 1.08 ± 0.03A | 0.94 ± 0.01B | 0.92 ± 0.01C | 0.78 ± 0.01C | ** | 0.03 |
| 15.90 | Maltose | 0.23 ± 0.01D | 0.25 ± 0.01C | 0.28 ± 0.01B | 0.11 ± 0.02E | 0.35 ± 0.01A | ** | 0.02 |
| Total | 12.14 | 8.02 | 7.77 | 9.08 | 9.15 | |||
RT retention time (min)
Means within the same column followed by different letters are significantly different (P < 0.05)
** Significant at 0.01 level
The results of the HPLC analysis of quinoa seeds revealed numerous free sugars: mannitol, glucose, cellobiose, arabinose, galactose and maltose (Table 2; Fig. S1). Among these sugars, glucose exhibited the highest concentration, followed by cellobiose, galactose, mannitol and maltose. Their levels differed significantly among genotypes. The highest contents of glucose, cellobiose, galactose and mannitol were observed in Q37, kvl-sra2, kvl-sra3 and Q52, respectively. However, the highest arabinose content was characteristic of kvl-sra2. On the other hand, maltose was the least abundant sugar (0.11–0.35 mg g−1 d wt). The highest total sugar content occurred in kvl-sra2 (12.14 mg g−1 d wt), but the lowest total sugar content occurred in regalona (7.77 mg g−1 d wt). The sugar composition of quinoa seeds has been shown to differ among various studies (Gonzalez et al. 1989, Abugoch 2009).
GC–MS of fatty acids
The fatty acid composition of quinoa genotypes is listed in Table 3 and Fig. S2. The most commonly abundant fatty acids were α-linolenic (C18:3), palmitic (C16:0) and linoleic acid (C18:2). Linoleic acid occurred as the 9,12-cis isoform in the kvl-sra2, kvl-sra3 and regalona genotypes and as the 9-cis, 11-trans isoform in the Q37 and Q52 genotypes. Generally, linoleic acid accounted for more than half of the total fatty acids (53.8–58.1%). Unsaturated fatty acids (UFAs) accounted for the highest content of total fatty acids (86.3–86.8%); the most common UFAs were oleic (18.4–24.1%), α-linolenic (5.6–6.1%) and erucic acid (1.1–2.0%). Moreover, the various isoforms of the UFAs varied among the studied genotypes.
Table 3.
Fatty acids profile of five quinoa genotypes
| No | RT | FAMEs (chemical name) | Chemical formula | No of C atoms | kvl-sra2 | kvl-sra3 | regalona | Q37 | Q52 |
|---|---|---|---|---|---|---|---|---|---|
| Area % | |||||||||
| 1 | 12.074 | Methyl tetradecanoate | C14H28O2 | C14:0 | 0.4 | 0.3 | 0.3 | 0.3 | 0.3 |
| 2 | 12.464 | Methyl octanoate | C8H16O2 | C8:0 | 0.5 | ND | ND | ND | ND |
| 3 | 14.336 | Hexadecanoic acid, methyl ester | C16H32O2 | C16:0 | 10.6 | 10.8 | 10.9 | 10.6 | 10.8 |
| 4 | 14.910 | n-Decanoic acid | C10H20O2 | C10:0 | 0.9 | ND | ND | ND | ND |
| 5 | 15.192 | Hexadecanoic acid, 14-methyl-, methyl ester | C16H32O2 | C16:0 | ND | 0.2 | 0.2 | 0.2 | 0.2 |
| 6 | 16.709 | Hexacosanoic acid, methyl ester | C16H32O2 | C16:0 | ND | 0.2 | 0.2 | 0.3 | ND |
| 7 | 17.100 | 13,16-Octadecadiynoic acid, methyl ester | C18H32O2 | C18:2 | ND | 0.2 | 0.2 | ND | ND |
| 8 | 17.759 | Methyl stearate | C18H36O2 | C18:0 | 0.8 | 0.6 | 0.7 | 0.7 | 0.9 |
| 9 | 18.486 | 9-Octadecenoic acid (Z)-, methyl ester | C18H34O2 | C18:1 | 24.1 | 18.4 | 18.4 | 23.1 | 22.7 |
| 10 | 19.913 | 9,12-Octadecadienoic acid (Z,Z)-, methyl ester | C18H32O2 | C18:2 | 53.8 | 58.1 | 58.1 | ND | ND |
| 11 | 20.034 | Methyl 9-cis,11-trans-octadecadienoate | C18H32O2 | C18:2 | ND | ND | ND | 54.5 | 55.3 |
| 12 | 21.381 | 9,12,15-Octadecatrienoic acid, methyl ester, (Z,Z,Z)- | C18H30O2 | C18:3 | 6.0 | 6.1 | 6.1 | 5.6 | 5.9 |
| 13 | 24.066 | Eicosanoic acid, methyl ester | C20H40O2 | C20:0 | ND | 0.4 | 0.4 | 0.5 | 0.5 |
| 14 | 24.815 | Methyl 13-eicosenoate | C20H38O2 | C20:1 | 1.8 | 1.3 | 1.3 | ND | ND |
| 15 | 25.006 | Methyl 9-eicosenoate | C20H38O2 | C20:1 | ND | ND | ND | 1.4 | 1.4 |
| 16 | 35.789 | Docosanoic acid, methyl ester | C22H44O2 | C22:0 | ND | 1.1 | 1.0 | 0.8 | 0.8 |
| 17 | 36.588 | Methyl 11-docosenoate | C22H42O2 | C22:1 | 1.1 | 2.0 | 2.0 | 1.9 | 1.3 |
| 18 | 38.624 | Cyclopropanetetradecanoic acid, 2-octyl-, methyl ester | C25H48O2 | C25:1 | ND | 0.2 | 0.2 | ND | ND |
| Total unsaturated FAMEs (%) | 86.8 | 86.4 | 86.3 | 86.6 | 86.5 | ||||
| Total saturated FAMEs (%) | 13.2 | 13.6 | 13.7 | 13.4 | 13.5 | ||||
RT retention time (min), ND not detected
The major saturated fatty acids (SFAs) were palmitic (10.6–10.9%), stearic (0.6–0.9%), and myristic acid (0.3–0.4%). Caprylic acid (C8:0) and capric acid (C10:0) were characteristic of kvl-sra2. However, arachidic acid (C20:0) and behenic (C22:0) acid were recorded in all quinoa genotypes except kvl-sra2. The results showed that the percentage of SFAs did not vary significantly (13.2–13.7%) among genotypes and that the type of SFAs was most characteristic for the genotypes.
The common UFAs obtained herein were also reported by Ruales and Nair (1993) [linoleic acid (52.3%), oleic acid (24.1%) and linolenic acid (3.8%)]. In addition, Miranda et al. (2012) reported 10% palmitic acid, 30% oleic acid, 57% linoleic acid and 12% α-linolenic acid.
Correlation between phytochemical traits
In light of the nutritional traits of quinoa seeds, 300 correlation coefficients were evaluated (Table 4). Fifty-five of those traits exhibited highly significant values (P < 0.01) with an r ≥ 0.62, and 52 of those traits exhibited significant associations (P < 0.05) with an r ≥ 0.36. The C content was strongly correlated with N, TP, oxalate and arabinose (0.91, 0.91, 0.62, and 0.85, respectively) and inversely associated with succinate and SFAs (− 0.89 and − 0.46, respectively), and the H content was strongly associated galactose (0.76). At the same time, N was positively correlated with TP and arabinose (1.00 and 0.71, respectively) and negatively correlated with succinate (− 0.79). In addition, the TP content was positively associated with arabinose (0.71) but negatively associated with succinate (− 0.79). By the same pattern, TFAs was strongly correlated with tartrate, lactose and maltose (0.68, 0.79 and 0.89, respectively) but inversely correlated with flavonoids (− 0.72).
Table 4.
Pearson’s correlation coefficients among nutritious traits of quinoa genotypes
| C | H | N | TPs | FAA | TLs | Alk | Phen | Sap | Tan | Flav | β-Car | Lyco | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | 1 | ||||||||||||
| H | − 0.25 | 1 | |||||||||||
| N | 0.91** | − 0.46* | 1 | ||||||||||
| TPs | 0.91** | − 0.46* | 1.00** | 1 | |||||||||
| FAA | 0.41 | − 0.15 | 0.20 | 0.20 | 1 | ||||||||
| TLs | 0.56* | 0.47* | 0.32 | 0.32 | 0.25 | 1 | |||||||
| Alk | − 0.27 | − 0.48* | − 0.02 | − 0.02 | 0.10 | − 0.51* | 1 | ||||||
| Phen | 0.31 | − 0.44* | 0.18 | 0.18 | 0.48* | − 0.21 | − 0.07 | 1 | |||||
| Sap | − 0.11 | − 0.02 | 0.13 | 0.13 | 0.09 | − 0.13 | 0.59* | − 0.42 | 1 | ||||
| Tan | 0.29 | 0.24 | − 0.03 | − 0.03 | 0.19 | 0.34 | − 0.58* | 0.50* | − 0.77** | 1 | |||
| Flav | 0.20 | − 0.13 | 0.45* | 0.45* | − 0.72** | 0.02 | − 0.07 | − 0.52* | 0.06 | − 0.28 | 1 | ||
| β-Car | − 0.08 | − 0.37 | 0.10 | 0.10 | 0.17 | − 0.36 | 0.23 | 0.12 | 0.54* | − 0.54* | − 0.12 | 1 | |
| Lyco | − 0.09 | − 0.18 | − 0.27 | − 0.27 | 0.44* | − 0.23 | 0.01 | 0.41 | − 0.45* | 0.32 | − 0.53* | − 0.29 | 1 |
| Oxal | 0.62** | 0.26 | 0.60** | 0.60** | − 0.01 | 0.79** | − 0.28 | − 0.44 | 0.25 | − 0.09 | 0.46* | − 0.04 | − 0.64** |
| Tart | 0.18 | 0.17 | − 0.19 | − 0.19 | 0.68** | 0.24 | − 0.37 | 0.66** | − 0.54* | 0.73** | − 0.78** | − 0.24 | 0.63** |
| Succ | − 0.89** | 0.17 | − 0.79** | − 0.79** | − 0.09 | − 0.50* | 0.49* | − 0.25 | 0.41 | − 0.46* | − 0.42 | 0.29 | 0.07 |
| Lact | 0.37 | − 0.15 | 0.09 | 0.09 | 0.79** | 0.11 | − 0.16 | 0.79** | − 0.36 | 0.61** | − 0.70** | − 0.05 | 0.62** |
| Mann | 0.04 | − 0.64** | 0.08 | 0.08 | 0.57* | − 0.54* | 0.46* | 0.76** | 0.03 | 0.01 | − 0.55* | 0.36 | 0.50* |
| Gluc | − 0.53* | − 0.55* | − 0.28 | − 0.28 | − 0.30 | − 0.95** | 0.50* | 0.28 | − 0.03 | − 0.26 | 0.03 | 0.23 | 0.26 |
| Cello | 0.16 | − 0.36 | 0.03 | 0.03 | 0.17 | − 0.33 | − 0.19 | 0.79** | − 0.73** | 0.61** | − 0.20 | − 0.26 | 0.63** |
| Arab | 0.85** | − 0.15 | 0.71** | 0.71** | 0.09 | 0.47* | − 0.54* | 0.33 | − 0.53* | 0.55* | 0.36 | − 0.35 | 0.03 |
| Galc | − 0.43 | 0.76** | − 0.50* | − 0.50* | − 0.61** | 0.32 | − 0.36 | − 0.74** | − 0.06 | 0.00 | 0.29 | − 0.37 | − 0.32 |
| Malt | 0.26 | 0.15 | 0.03 | 0.03 | 0.89** | 0.42 | 0.03 | 0.29 | 0.20 | 0.18 | − 0.78** | 0.19 | 0.20 |
| USFs | 0.46* | − 0.47* | 0.41 | 0.41 | − 0.02 | − 0.12 | − 0.28 | 0.54* | − 0.62** | 0.43 | 0.27 | − 0.19 | 0.19 |
| SFAs | 0− .46* | 0.44 | − 0.41 | − 0.41 | 0.00 | 0.07 | 0.28 | − 0.50* | 0.55* | − 0.43 | − 0.26 | 0.24 | − 0.27 |
| Oxal | Tart | Succ | Lact | Mann | Gluc | Cello | Arab | Galc | Malt | USFs | SFAs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | ||||||||||||
| H | ||||||||||||
| N | ||||||||||||
| TPs | ||||||||||||
| FAA | ||||||||||||
| TLs | ||||||||||||
| Alk | ||||||||||||
| Phen | ||||||||||||
| Sap | ||||||||||||
| Tan | ||||||||||||
| Flav | ||||||||||||
| β-Car | ||||||||||||
| Lyco | ||||||||||||
| Oxal | 1 | |||||||||||
| Tart | − 0.29 | 1 | ||||||||||
| Succ | − 0.53* | − 0.14 | 1 | |||||||||
| Lact | − 0.31 | 0.90** | − 0.25 | 1 | ||||||||
| Mann | − 0.62** | 0.37 | 0.17 | 0.61** | 1 | |||||||
| Gluc | − 0.81** | − 0.23 | 0.43 | − 0.10 | 0.56* | 1 | ||||||
| Cello | − 0.58* | 0.58* | − 0.35 | 0.63** | 0.54* | 0.43 | 1 | |||||
| Arab | 0.43 | 0.24 | − 0.99** | 0.33 | − 0.13 | − 0.38 | 0.46* | 1 | ||||
| Galc | 0.27 | − 0.26 | 0.21 | − 0.58* | − 0.90** | − 0.35 | − 0.48* | − 0.21 | 1 | |||
| Malt | 0.14 | 0.62** | 0.10 | 0.62** | 0.35 | − 0.49* | − 0.14 | − 0.11 | − 0.33 | 1 | ||
| USFs | − 0.09 | 0.22 | − 0.68** | 0.30 | 0.18 | 0.19 | 0.74** | 0.74** | − 0.37 | − 0.34 | 1 | |
| SFAs | 0.14 | − 0.22 | 0.68** | − 0.42 | − 0.18 | − 0.17 | − 0.72** | − 0.74** | 0.37 | 0.35 | − 0.85** | 1 |
* Significant at 0.05 level; ** Significant at 0.01 level
The TLs in quinoa seeds were linearly correlated with oxalate (0.79) but negatively correlated with glucose (− 0.95). Phenolics were positively correlated with tartrate, lactose, mannose and cellobiose (0.66, 0.79, 0.76 and 0.79, respectively) but negatively correlated with galactose (− 0.74). Nevertheless, saponins were negatively correlated with tannins, cellobiose and UFAs (− 0.77, − 0.73 and − 0.62, respectively). Tannins were positively correlated with tartrate (0.73), and flavonoids were inversely correlated with tartrate, lactose and maltose (− 0.78, − 0.70 and − 0.78, respectively). Lycopene was strongly correlated with tartrate (0.63), lactose (0.62) and cellobiose (0.63) but was inversely correlated with oxalate (− 0.64).
Oxalate was negatively correlated with mannose and glucose (− 0.62 and − 0.81, respectively). Tartrate was strongly correlated with lactose (0.90) and maltose (0.62). Succinate was positively correlated with SFAs (0.68) but negatively correlated with arabinose and UFAs (− 0.99 and − 0.68, respectively). At the same time, lactose was strongly correlated with cellobiose (0.63) and maltose (0.62), and mannose was inversely correlated with galactose (− 0.90). Cellobiose was strongly correlated with UFAs (0.74) but inversely correlated with SFAs (− 0.72). Arabinose showed correlations having r values of 0.74 and − 0.74 with UFAs and SFAs, respectively. Finally, UFAs were inversely correlated (− 0.85) with SFAs.
Genetic characterization
The patterns of amplification products obtained from the RAPD and ISSR primers were reproducible and stable and thus suitable for genotypic analysis (Fig. S3). A summary of the analysis of the data derived from both marker types is listed in Table 5.
Table 5.
Summary of genetic analysis of five quinoa genotypes using RAPD and ISSR markers
| Primer | Range of band size (bp) | Total Bands | Polymorphic bands | % of polymorphism | PIC | (MI) | Rp | H′ |
|---|---|---|---|---|---|---|---|---|
| RAPD | ||||||||
| OPC-2 | 1677–342 | 15 | 3 | 20.00 | 0.064 | 0.192 | 27.6 | 2.67 |
| OPG-2 | 2795–272 | 9 | 3 | 33.33 | 0.142 | 0.426 | 16.0 | 1.64 |
| OPG-19 | 1110–235 | 8 | 3 | 37.50 | 0.120 | 0.360 | 13.6 | 2.02 |
| OPO-10 | 1906–301 | 17 | 11 | 64.71 | 0.273 | 3.003 | 26.8 | 2.81 |
| Total | 49 | 26 | 53.06 | |||||
| Mean | 12.25 | 5.00 | 38.89 | 0.150 | 1.000 | 21.00 | 2.29 | |
| ISSR | ||||||||
| UBC 842 | 900–139 | 15 | 1 | 6.67 | 0.032 | 0.032 | 29.2 | 2.70 |
| UBC 845 | 900–258 | 26 | 26 | 100.00 | 0.400 | 10.400 | 17.6 | 3.16 |
| UBC 855 | 1040–228 | 8 | 6 | 75.00 | 0.340 | 2.040 | 9.6 | 1.98 |
| UBC 856 | 928–192 | 13 | 1 | 7.69 | 0.037 | 0.037 | 25.2 | 2.56 |
| UBC 868 | 1111–372 | 7 | 1 | 14.29 | 0.064 | 0.064 | 13.6 | 1.94 |
| UBC 889 | 791–261 | 12 | 2 | 16.67 | 0.067 | 0.134 | 21.6 | 2.44 |
| Total | 85 | 37 | 43.53 | |||||
| Mean | 12.14 | 5.29 | 31.47 | 0.13 | 1.820 | 17.83 | 2.31 | |
PIC polymorphic information content, MI marker index, Rp resolving power, H′ Shannon’s diversity index
RAPD analysis
Four RAPD primers were used to investigate the genetic structure and pattern of genetic variation among the five quinoa genotypes. The primers revealed a total of 49 loci, which varied in size from 235 to 2795 bp, that were 53.06% polymorphic and had an average of 12.25 loci primer−1. The polymorphism percentage ranged from 20 to 64.71%. The highest number of polymorphic loci obtained with primer OPO-10 was 11; their band size ranged from 301 to 1906 bp. The major PIC average was 0.273 (OPO-10); the minor, 0.064 (OPC-2). The maximal Rp values were 27.6 and 26.8 for OPC-2 and OPO-10, respectively, whereas the OPG-19 primer exhibited the lowest Rp value (13.6). The highest MI value (3.003) was recorded for OPO-10; the lowest (0.192) for OPC-2. Shannon’s diversity index average was 2.29; the lowest value (1.64) was recorded for OPG-2, while the highest value (2.81) was recorded for OPO-10. Among the studied RAPD primers, OPO-10 was the most informative.
The ability of the RAPD technique to characterize genotypic structure and measure genetic diversity among quinoa genotypes has been reported in few studies. The first investigation was performed by Fairbanks et al. (1993), who characterized 16 quinoa accessions using RAPD markers. Del Castillo et al. (2007) used 10 RAPD markers for the genetic characterization of 87 Bolivian quinoa accessions. Ruas et al. (1999) used 33 RAPD primers to study the genetic relationship among 19 accessions of six species of Chenopodium. In addition, the genetic diversity and relationships among 55 accessions belonging to 14 species of Chenopodium was estimated by Rana et al. (2010) using RAPD techniques.
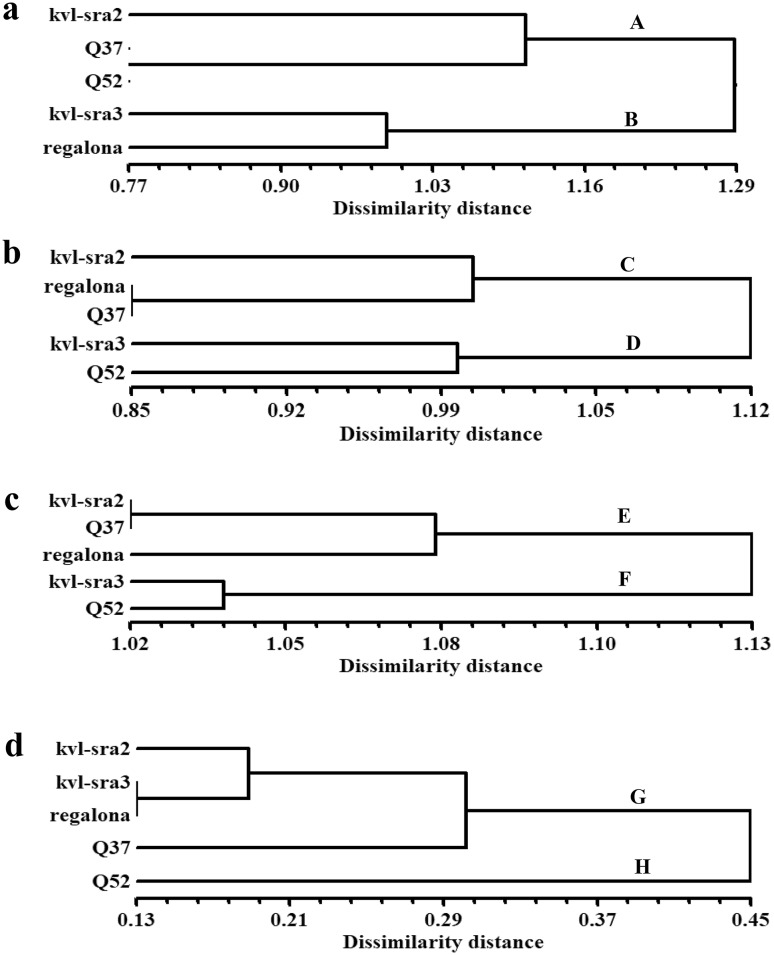
A dendrogram constructed from UPGMA cluster analysis of the RAPD markers is shown in Fig. 1a. The five genotypes separated into two main clusters at a dissimilarity distance of 1.29. One of the clusters (A) included three genotypes (kvl-sra2, Q37 and Q52), while the other (B) grouped the genotypes kvl-sra3 and regalona at a dissimilarity distance of approximately 0.98. In the first cluster, the Q37 and Q52 genotypes were delimited together at a dissimilarity distance of 0.77 but were separated from kvl-sra2 by a distance of approximately 1.08.
Fig. 1.
Dendrograms presenting genetic relationship among five quinoa genotypes revealed by UPGMA cluster analysis based on different marker types. a RAPD, b ISSR, c combined dataset of RAPD and ISSR and d phytochemical traits
ISSR analysis
The seven ISSR primers used identified 85 total loci across the five genotypes; 37 loci were polymorphic, accounting for 43.53% of the polymorphism. The number of amplified loci varied from four (UBC-817) to 26 (UBC-845); their sizes ranged from 139 to 1111 bp, and there was an average of 12.14 loci primer−1. Only one primer (UBC-845) exhibited 100% polymorphism; in contrast, no polymorphism was recorded for primer UBC-817. The overall PIC values ranged from 0.00 (UBC-817) to 0.40 (UBC-845). In addition, the average Rp was 17.83; the highest value (29.2) was recorded for UBC-842, and the lowest value (8.0) was recorded for UBC-817. The maximal MI value (10.4) was recorded for UBC-845, while no Rp was detected for UBC-817. Shannon’s diversity index averaged 2.31; the lowest value (1.39) was recorded for UBC-817, and the highest value (3.16) was recorded for UBC-845. The results of the ISSR data analysis showed that UBC-845 was the most informative primer. Studies evaluating the genetic diversity at inter and/or intraspecific levels have reported the efficiency of ISSR techniques, as has occurred for Lemna spp. (El-Kholy et al. 2015) and rice cultivars (Alam et al. 2016); these reports are in addition to those at the individual level, as reported for quinoa (Ana-Cruz et al. 2017).
The UPGMA analysis of the ISSR primers was used to generate a dendrogram based on the dissimilarity matrix (Fig. 1b). The dendrogram separated the five genotypes into two main clusters (C and D) at a dissimilarity distance of 1.12. The first cluster (C) was divided into two branches at a dissimilarity distance of approximately 1.00. The first branch comprised kvl-sra2, while the second branch grouped regalona and Q37 at a dissimilarity distance of 0.85. On the other hand, the second cluster (D) delimited kvl-sra2 and Q52 together at a dissimilarity distance of 1.00.
Our results with respect to the ISSR markers revealed moderate genetic variability among the studied genotypes. This finding is in accordance with the results of Ana-Cruz et al. (2017), and this level of genetic variability could be attributed to the reproduction processes of the species and seed exchange between farmers. Microsatellite markers (SSR) had been used efficiently in genetic characterization within quinoa germplasm from Ecuador, Colombia, Peru, Bolivia, Chile and Argentina (Fuentes et al. 2012).
With respect to polymorphism, RAPD markers were superior to ISSR markers; the RAPD markers detected 53.06% polymorphism, while the ISSR markers detected only 43.53%. Furthermore, the average numbers of polymorphic loci primer−1 and average PIC and Rp values were slightly higher for the RAPD markers than for the ISSR ones. However, the ISSR markers were more effective than were RAPD markers regarding MI and H′.
The genetic dissimilarity distances among the studied genotypes were calculated based on the data derived from the RAPD and ISSR makers, their combination and the phytochemicals. The results of the RAPD marker analysis showed that the genetic dissimilarity values ranged from 0.77 to 0.129, while the results of the ISSR marker analysis showed that the values ranged from 0.85 to 1.12. The combined data of both markers revealed a dissimilarity distance that ranged from 1.02 to 1.13. The above results are in agreement with those reported for Mangifera indica (Gajera et al. 2011). Finally, a dissimilarity distance of 0.13–0.45 was recorded on the basis of phytochemical traits.
The comparison between the RAPD- and ISSR-derived dendrograms showed differences in the clustering patterns of genotypes within clusters. These differences could have occurred because each marker targets different sequences throughout the genome. Therefore, both markers are valuable for genetic characterization and diversity analysis in quinoa. The combined dataset of both markers consisted of 134 loci, which were used to generate a dendrogram based on the UPGMA analysis (Fig. 1c). The delimitation of the five genotypes was similar to that produced by the ISSR data analysis albeit with slight differences in dissimilarity distances between genotypes. The UPGMA-based dendrogram showed two main clusters (E and F) that exhibited a dissimilarity distance ranging from 1.02 to 1.13.
In the main dataset, to evaluate the phenetic relationships among the studied cultivars, NTSYS-pc software was used to analyze 41 phytochemicals traits. The generated dendrogram separated the five genotypes into a main cluster (G) and one branch (H) at a dissimilarity distance of 0.45 (Fig. 1d). The cluster (G) was subdivided at a dissimilarity distance of approximately 0.3 into a subcluster containing three genotypes (kvl-sra2, kvl-sra3 and regalona) and a branch comprising the Q37 genotype. On the other hand, the Q52 genotype did not group with any of the genotypes and was delimited alone at a dissimilarity distance of 0.45.
Compared with the dendrograms generated by RAPD and ISSR markers or their combined data analysis, the dendrogram based on phytochemical properties showed a completely different topology. This finding indicates that the genotypic structure of the studied cultivars based on phytochemical traits is unclear, as is the case with the genotypic traits, and is in complete agreement with the results of Vidueiros et al. (2015). This result could be explained by environmental and genotype-environment interactions that influence the phenotypic (phytochemical) characters. Vidueiros et al. (2015) studied the nutritional characterization, diversity and interrelationships among the nutritional traits of 21 quinoa accessions; their study was based on twelve nutritive characters for the nutritional characterization and genetic diversity evaluation of Northwest Argentina genotypes. However, in the present investigation, 41 phytochemical and nutritive characters were used to evaluate five genotypes introduced to Egypt. The results of the interrelationships among traits could be used in breeding programs aiming to improve quinoa seed quality.
Conclusion
The present study provides knowledge about the phytochemical traits and genetic structure of five quinoa genotypes introduced to Egypt. The phytochemical properties considerably varied among genotypes. Nevertheless, unlike the molecular data, the phytochemical traits did not show a clear genotypic structure. The phytochemical, nutritional and genetic characterization results reported in the present study will be promising for guiding the breeding of quinoa seed quality in Egypt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors declare no competing interests. This work was supported by Tanta and Kafrelsheikh Universities. Authors would express their deep thanks for Dr. Christopher J. Chuck, Bath University, UK for technical support during HPLC and GC–MS analysis and interpretation of data.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12298-018-0541-4) contains supplementary material, which is available to authorized users.
References
- Abugoch JLE. Quinoa (Chenopodium quinoa Willd.): composition, chemistry, nutritional and functional properties. Adv Food Nutr Res. 2009;58:1–31. doi: 10.1016/S1043-4526(09)58001-1. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Rao AV. Tomato lycopene and low density lipoprotein oxidation: a human dietary intervention study. Lipids. 1998;33:981–984. doi: 10.1007/s11745-998-0295-6. [DOI] [PubMed] [Google Scholar]
- Alam SM, Siddika S, Haque E. Genetic diversity of some upland and lowland rice cultivars in Bangladesh using RAPD, ISSR and SSR markers. Nucleus. 2016;59:15–23. doi: 10.1007/s13237-015-0148-x. [DOI] [Google Scholar]
- Ana-Cruz MC, Helena ME, Yacenia MC. Molecular characterization of Chenopodium quinoa Willd. using inter-simple sequence repeat (ISSR) markers. Afr J Biotechnol. 2017;16:483–489. [Google Scholar]
- Bhargava A, Shukla S, Ohri D. Genetic variability and interrelationship among various morphological and quality traits in quinoa (Chenopodium quinoa Willd.) Field Crops Res. 2007;101:104–116. doi: 10.1016/j.fcr.2006.10.001. [DOI] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;3:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- Carciochi RA, Manrique GD, Dimitrov K. Changes in phenolic composition and antioxidant activity during germination of quinoa seeds (Chenopodium quinoa Willd.) Int Food Res J. 2014;21:767–773. [Google Scholar]
- Chang C, Yang M, Wen H, Chern J. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J Food Drug Anal. 2002;10:178–182. [Google Scholar]
- Chauhan GS, Eskin NAM, Tkachuk R. Nutrients and antinutrients in quinoa seed. Cereal Chem. 1992;69:85–88. [Google Scholar]
- Clarke JD. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5177. [DOI] [PubMed] [Google Scholar]
- Del Castillo C, Winkel T, Mahy G, Bizoux JP. Genetic structure of quinoa (Chenopodium quinoa Willd) from the Bolivian Altiplano as revealed by RAPD markers. Gen Res Crop Evol. 2007;54:897–905. doi: 10.1007/s10722-006-9151-z. [DOI] [Google Scholar]
- El-Kholy AS, Youssef MS, Eid EM. Genetic diversity of Lemna gibba L. and L. minor L. populations in Nile Delta based on biochemical and ISSR markers. Egypt J Exp Biol Bot. 2015;11:11–19. [Google Scholar]
- European Community Reference method of tannins dosages. J Offic des Commun Euro L. 2000;197:18–20. [Google Scholar]
- Fairbanks D, Waldrigues A, Ruas CF, Maughan PJ, Robison LR, Andersen WR, Riede CR, Pauley CS, Caetano LG, Arantes OM, Fungaro MHP, Vidotto MC, Jankevicius SE. Efficient characterization of biological diversity using field DNA extraction and random amplified polymorphic DNA markers. Revis Bras Gen. 1993;16:11–22. [Google Scholar]
- Fiallos-Jurado J, Pollier J, Moses T, Arendt P, Barriga-Medina N, Morillo E, Arahana V, Torres M, Goossens A, Leon-Reyes A. Saponin determination, expression analysis and functional characterization of saponin biosynthetic genes in Chenopodium quinoa leaves. Plant Sci. 2016;250:188–197. doi: 10.1016/j.plantsci.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Food Agriculture Organization of United Nations (FAO) (2012) International year of quinoa IYQ-2013. http://www.rlc.fao.org/en/about-fao/iyq-2012/. Accessed 11 Mar 2013
- Freedman B, Butterfield RPE. Transesterification kinetics of soybean oil. J Am Oil Chem Soc. 1986;63:1375–1380. doi: 10.1007/BF02679606. [DOI] [Google Scholar]
- Fuentes F, Bazile D, Bhargava A, Martinez EA. Implications of farmers’ seed exchanges for on-farm conservation of quinoa, as revealed by its genetic diversity in Chile. J Agric Sci. 2012;150(6):702–716. doi: 10.1017/S0021859612000056. [DOI] [Google Scholar]
- Gajera HP, Tomar RS, Patel SV, Viradia RR, Golakiya BA. Comparison of RAPD and ISSR markers for genetic diversity analysis among different endangered Mangifera indica genotypes of Indian Gir forest region. J Plant Biochem Biotechnol. 2011;20:217–223. doi: 10.1007/s13562-011-0049-2. [DOI] [Google Scholar]
- Gawlik-Dziki U, Swieca M, Sulkowski M, Dziki D, Baraniak B, Czy J. Antioxidant and anticancer activities of Chenopodium quinoa leaves extracts—in vitro study. Food Chem Toxicol. 2013;57:154–160. doi: 10.1016/j.fct.2013.03.023. [DOI] [PubMed] [Google Scholar]
- Gonzalez JA, Roldan A, Gallardo M, Escudero T, Prado FE. Quantitative determinations of chemical compounds with nutritional value from Inca crops: Chenopodium quinoa (‘quinoa’) Plant Foods Hum Nutr. 1989;39:331–337. doi: 10.1007/BF01092070. [DOI] [PubMed] [Google Scholar]
- Gonzalez JA, Konishi Y, Bruno M, Valoya M, Prado FE. Interrelationships among seed yield, total protein and amino acid composition of ten quinoa (Chenopodium quinoa) cultivars from two different agroecological regions. J Sci Food Agric. 2012;92:1222–1229. doi: 10.1002/jsfa.4686. [DOI] [PubMed] [Google Scholar]
- Gordillo-Bastidas E, Díaz-Rizzolo DA, Roura E, Massanés T, Gomis R. Quinoa (Chenopodium quinoa Willd), from nutritional value to potential health benefits: an integrative review. J Nutr Food Sci. 2016;6:1–10. [Google Scholar]
- Harborne JB. Phytochemical methods. London: Chapman and Hall; 1973. pp. 49–188. [Google Scholar]
- Hiai S, Oura H, Hamanake H, Odakea Y. A colour reaction of penaxadiol with vanillin and sulfuric acid. Planta Med. 1975;28:131–138. doi: 10.1055/s-0028-1097841. [DOI] [PubMed] [Google Scholar]
- Koziol M. Chemical composition and nutritional evaluation of quinoa (Chenopodium quinoa Willd) J Food Compost Anal. 1992;5:35–68. doi: 10.1016/0889-1575(92)90006-6. [DOI] [Google Scholar]
- Lee YP, Takahashi T. An improved colorimetric determination of amino acids with the use of ninhydrin. Anal Biochem. 1966;14:71–73. doi: 10.1016/0003-2697(66)90057-1. [DOI] [Google Scholar]
- Lückstädt C, Mello S. The use of organic acids in animal nutrition, with special focus on dietary potassium diformate under European and Austral-Asian conditions. Recent Adv Anim Nutr Aust. 2011;18:123–130. [Google Scholar]
- Maughan PJ, Bonifacio A, Jellen E, Stevens M, Coleman C, Ricks M, Mason S, Jarvis D, Gardunia B, Fairbanks D. A genetic linkage map of quinoa (Chenopodium quinoa) based on AFLP, RAPD and SSR markers. Theor Appl Genet. 2004;109:1188–1189. doi: 10.1007/s00122-004-1730-9. [DOI] [PubMed] [Google Scholar]
- Miranda M, Vega-Galvez A, Lopez J, Parada G, Sanders M, Aranda M, Uribe E, Scala KD. Impact of air-drying temperature on nutritional properties, total phenolic content and antioxidant capacity of quinoa seeds (Chenopodium quinoa Willd.) Ind Crops Prod. 2010;32:258–263. doi: 10.1016/j.indcrop.2010.04.019. [DOI] [Google Scholar]
- Miranda M, Vega-Galvez A, Martinez E, Lopez J, Rodriguez MJ, Henriquez K, Fuentes F. Genetic diversity and comparison of physicochemical and nutritional characteristics of six quinoa (Chenopodium quinoa Willd.) genotypes cultivated in Chile. Ciênc Tecnol Aliment. 2012;32:835–843. doi: 10.1590/S0101-20612012005000114. [DOI] [Google Scholar]
- Nagata M, Yamashita I. Simple method for simultaneous determination of chlorophyll and carotenoids in tomato fruit. Nippon Shokuhin Kogyo Gakkaish. 1992;39:925–928. doi: 10.3136/nskkk1962.39.925. [DOI] [Google Scholar]
- Powell WM, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed. 1996;2:225–238. doi: 10.1007/BF00564200. [DOI] [Google Scholar]
- Prevost A, Wilkinson MJ. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet. 1999;98:107–112. doi: 10.1007/s001220051046. [DOI] [Google Scholar]
- Rana TS, Narzary D, Ohri D. Genetic diversity and relationships among some wild and cultivated species of Chenopodium L. (Amaranthaceae) using RAPD and DAMD methods. Curr Sci. 2010;98:840–846. [Google Scholar]
- Repo-Carrasco-Valencia R, Hellstrom JK, Pihlava JM, Mattila PH. Flavonoids and other phenolic compounds in Andean indigenous grains: Quinoa (Chenopodium quinoa), kañiwa (Chenopodium pallidicaule) and kiwicha (Amaranthus caudatus) Food Chem. 2010;120:128–133. doi: 10.1016/j.foodchem.2009.09.087. [DOI] [Google Scholar]
- Risi J, Galwey NW. The Chenopodium grains of the Andes: Inca crops for modern agriculture. Adv Appl Biol. 1984;10:145–216. [Google Scholar]
- Roldan-Ruiz I, Calsyn E, Gilliland TJ, Coll R, Van Eijk MJT, De Loose M. Estimating genetic conformity between related ryegrass (Lolium) varieties. 2. AFLP characterization. Mol Breed. 2000;6:593–602. doi: 10.1023/A:1011398124933. [DOI] [Google Scholar]
- Ruales J, Nair BM. Content of fat, vitamins and minerals in quinoa (Chenopodium quinoa, Willd) seeds. Food Chem. 1993;48:131–136. doi: 10.1016/0308-8146(93)90047-J. [DOI] [Google Scholar]
- Ruas P, Bonifacio A, Ruas C, Fairbanks D, Andersen W. Genetic relationship among 19 accessions of six species Chenopodium L., by random amplified polymorphic DNA fragments (RAPD) Euphytica. 1999;105:25–32. doi: 10.1023/A:1003480414735. [DOI] [Google Scholar]
- Saeedeh A, Asna U. Antioxidant properties of various solvent extracts of mulberry (Morus indica L.) leaves. Food Chem. 2007;102:1233–1240. doi: 10.1016/j.foodchem.2006.07.013. [DOI] [Google Scholar]
- Shannon CE, Weaver W. The mathematical theory of communication. Urbana: University of Illinois Press; 1949. [Google Scholar]
- Sharma KD, Bindal G, Rathour R, Rana JC. ß-carotene and mineral content of different Chenopodium species and the effect of cooking on micronutrient retention. Int J Food Sci Nutr. 2012;63:290–295. doi: 10.3109/09637486.2011.624493. [DOI] [PubMed] [Google Scholar]
- Silva TMS, Santos FP, Evangelista-Rodrigues A, Silva EMS, Silva GS, Novais JS, Santos FAR, Camara CA. Phenolic compounds, melissopalynological, physicochemical analysis and antioxidant activity of jandaira (Melipona subnitida) honey. J Food Compost Anal. 2013;29:10–18. doi: 10.1016/j.jfca.2012.08.010. [DOI] [Google Scholar]
- Tang Y, Tsao R. Phytochemicals in quinoa and amaranth grains and their antioxidant, anti-inflammatory, and potential health beneficial effects: a review. Mol Nutr Food Res. 2017;61:1–16. doi: 10.1002/mnfr.201600767. [DOI] [PubMed] [Google Scholar]
- Valencia-Chamorro SA. Quinoa. In: Trugo L, Finglas PM, Caballero B, editors. Encyclopedia of food science and nutrition. Amsterdam: Academic Press; 2003. pp. 4895–4902. [Google Scholar]
- Varli SN, Sanlier N. Nutritional and health benefits of quinoa (Chenopodium quinoa Willd.) J Cereal Sci. 2016;69:371–376. doi: 10.1016/j.jcs.2016.05.004. [DOI] [Google Scholar]
- Vega-Galvez A, Miranda M, Vergara J, Uribe E, Puente L, Martínez EA. Nutrition facts and functional potential of quinoa (Chenopodium quinoa Willd.), an ancient Andean grain: a review. J Sci Food Agric. 2010;90:2541–2547. doi: 10.1002/jsfa.4158. [DOI] [PubMed] [Google Scholar]
- Vidueiros SM, Curti RN, Dyner LM, Binaghi MJ, Peterson G, Bertero HD, Pallaro AN. Diversity and interrelationships in nutritional traits in cultivated quinoa (Chenopodium quinoa Willd.) from Northwest Argentina. J Cereal Sci. 2015;62:87–93. doi: 10.1016/j.jcs.2015.01.001. [DOI] [Google Scholar]
- Zhang TF, Qi WC, MF G ZX, Li T, Zhao H. Exploration and transferability evaluation of EST-SSRs in quinoa. Acta Agron Sin. 2016;42(4):492–500. doi: 10.3724/SP.J.1006.2016.00492. [DOI] [Google Scholar]
- Zhang T, Gu M, Liu Y, Lv Y, Zhou L, Lu H, Liang S, Bao H, Zhao H. Development of novel InDel markers and genetic diversity in Chenopodium quinoa through whole-genome re-sequencing. BMC Genom. 2017;18(1):685–700. doi: 10.1186/s12864-017-4093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Silva A, Fuentes F, Zamora P, Jacobsen SE, Schwember AR. Breeding quinoa (Chenopodium quinoa Willd.): potential and perspectives. Mol Breed. 2014;34:13–30. doi: 10.1007/s11032-014-0023-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.