Abstract
Multidrug efflux pumps confer resistance to their bacterial hosts by pumping out a diverse range of compounds, including most antibiotics. Being more familiar with the details of functional dynamics and conformations of these types of pumps could help in discovering approaches to stop them functioning properly. Computational approaches, particularly conventional molecular dynamics simulations followed by diverse post simulation analysis, are powerful methods that help researchers by opening a new window to study phenomena that are not detectable in as much detail in vitro or in vivo as they are in silico. In this study, accelerated molecular dynamics simulations were applied to study the dynamics of AcrB efflux pump transporters in interaction with PAβN and tetracycline as an inhibitor and a substrate, respectively, to compare the differences in the dynamics and consequently the mechanism of action of the pump. The different dynamics for PAβN -bound form of AcrB compared to the TET-bound form is likely to affect the rotating mechanism typically observed for AcrB transporter. This shows the dynamics of the active AcrB transporter is different in a substrate-bound state compared to an inhibitor-bound state. This advances our knowledge and helps to unravel the mechanism of tripartite efflux pumps.
Subject terms: Computational models, Antimicrobial resistance
Introduction
Overexpression of the resistance nodulation cell division (RND) type of tripartite efflux pumps in the Gram-negative pathogens is a major component of multidrug resistance (MDR)1. These types of efflux pumps recognize a diverse range of compounds and pump them out from the bacteria cell via proton-motive force (PMF) secondary transporters like AcrB, part of AcrAB-TolC efflux pump, present in Klebsiella pneumoniae and Escherichia coli2–8.
The substrate specificity and selectivity of AcrAB-TolC efflux pump depends on the homotrimer structure of AcrB, with threefold asymmetric conformation, in which each monomer adopts a different conformation, access, binding and extrusion5,9–14. AcrB of K. pneumoniae, that has similar sequence to the solved crystal structure of AcrB of E. coli, pumps out the compounds employing a rotating mechanism that is allosterically coupled, in which each monomer successively adopts one of the three conformations10,11,14–20. Recent computational works have further shed lights on the relationship between functional rotation and substrate transport21–23. Structures with bound drugs revealed two discrete multisite binding pockets separated by a switch loop, with the distal pocket in the binding (tight) state and a proximal pocket in the structure24,25 (Fig. 1). The proximal and distal binding pockets that are involved in forming the multibinding site of AcrB, as the substrate-selective part of the pump, play key roles in its specificity and binding to substrates2,24–32. The G-loop controls the access of substrates to the distal pocket by forming a boundary between the proximal and distal binding pockets2. Under the G-loop, there is a narrow channel that connects proximal and distal pockets to each other (Fig. 1). The pockets are enriched in aromatic, polar and charged amino acid residues that form favourable interactions with the transported substrates. The microenvironment of the distal binding pocket within AcrB has been studied in detail33,34. Since the distal binding pocket includes many hydrophobic, polar and charged residues, this microenvironment mediates extrusion of a wide range of compounds by AcrB4,35,36. Based on the multisite-drug-oscillation hypothesis by Yamaguchi8, the spacious multidrug-binding pocket may have numerous binding sites even for a single substrate, suggesting that substrates may move between binding sites during transport. This hypothesis could explain the apparently broad substrate specificity of cell membrane exporters and their highly efficient ejection of drugs from the bacteria cells.
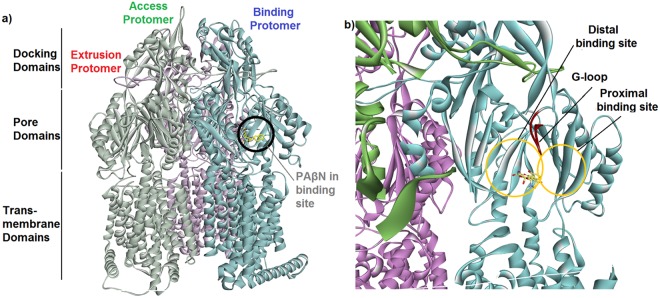
Figure 1.
(a) The proposed homotrimer model structure of the AcrB efflux pump transporter in complex with PAβN. It represents the structure that was obtained after carrying out the homology modelling, minimization and equilibration. The full-space complete homotrimer structure of AcrB has been shown on the right side of the picture, and each subunit has been represented by different colours; The binding site in binding monomers determined by SMINA molecular docking somewhere close to the distal pocket has been represented by a black circle (b) Tetracycline in the multi-binding sites within the binding protomer of the AcrB transporter.
Substrates that are taken up from the entrance ports, in particular the cleft, of an efflux pump could be transported through dual multidrug-binding pockets by a peristaltic mechanism at the substrate translocation channel37 through both proximal and distal pockets, and could be potentially extruded from the top exit port. Actually, the asymmetric trimer is supportive of a rotating access mechanism for drug binding and release (Fig. S1).
Designing effective inhibitors to stop these types of pumps from effluxing out antibiotics requires an understanding of the functional dynamics and molecular mechanisms of conformational changes of this type of protein. There are only a few solved structures for AcrB of E. coli in complex with different compounds10,20,25,38 and no structure for AcrB of K. pneumoniae.
Conventional molecular dynamics (cMD) simulations enable simulations on the order of tens to hundreds of nanoseconds; however, longer simulations are required to monitor biological processes that typically occur on longer time scales of up to milliseconds or more39. Recently developed advanced sampling techniques such as accelerated molecular dynamics (aMD) simulations allows access to these longer time scale molecular events beyond those obtainable with cMD40,41. aMD reduces the height of local barriers which allows faster calculations and time efficient simulation of biomacromolecules42. It simplifies the sampling by requiring only the evolution of a single copy of the system and does not require any previous knowledge of the shape of the potential energy profile39.
aMD simulations allow determination of time-dependent protein conformational changes which enables sampling the conformational space more efficiently than conventional molecular dynamics simulations43–48 and has been fully integrated into commonly used software such as Amber39,49. Advanced sampling techniques employed by aMD extend the time scale of MD simulations that converges the correct canonical probability distribution and enables rapid sampling of the conformational space42.
Nikaido et al. in their recent works2,50,51 studied a truncated model of AcrB transporter from E. coli in interaction with different ligands by cMD simulations. This showed the important role of the G- loop and the deep hydrophobic groove of the distal site, indicating that the binding mode of compounds to the transporter alters the efflux of other substrates. Similarly, Fischer and Kandt studied the conformational changes in the porter domain of AcrB from E. Coli conventional molecular dynamics52. In this study, we use the sampling power of aMD simulations to reveal the time-dependent conformational changes in the homology-modelled multidrug efflux pump AcrB from K. pneumoniae (SI), in interaction with Phenylalanyl-arginine-β-naphthylamide (PAβN), an inhibitor and modulator of AcrB7,53, and tetracycline (TET), a substrate for the transporter (Fig. 2) after generating the starting complexes by molecular docking. Principal component analysis (PCA) has been used to investigate the correlations among various important regions of the complexes in the course of aMD simulations and to distinguish between different conformational states.
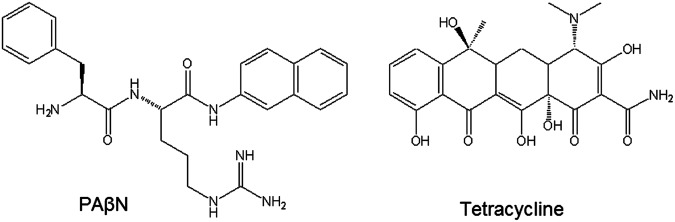
Figure 2.
Chemical structures of the compounds, PAβN and Tetracycline, used in this study.
Results and Discussion
Structures of the complexes
Figure 1 shows the generated structure and important parts of AcrB transporter for Klebsiella pneumoniae by homology modelling using Swiss Model webserver with sequence identity 91.6% against the template, crystal structure of AcrB of Escherichia coli (PDB id code 4DX5). The trimer of the AcrB protein was obtained as the final model from homology modelling in a 3D PDB structure format. The template that was used for the homology modelling was a monomer structure and, therefore, the generated model was also a monomer structure. The assembly procedure was performed using the Accelrys discovery studio. The accuracy and validity of the generated AcrB model was examined in detail, and was shown in the different panels of Fig. S2. According to the SMINA molecular docking results, the location for the binding of PAβN to the protein structure was identified in the multisite binding pocket within the binding protomer of the transporter (Fig. 3). The docked complex of PAβN-AcrB was comparable to the previous structures determined for ligand-bound AcrB by Nakashima et al.24. Only one PAβN was bound to the binding monomer of homotrimer, and it was bound only to the multi-binding site. Also, other favorable docked poses showed that PAβN could bind to the access monomer of the AcrB homotrimer, which suggests that PAβN could be forced to pass through the path, during the dynamic of the efflux process, by a transient conformational change from the access form to the binding form, and PAβN would move to the gate of the distal pocket in the binding state. There were strong hydrophobic interactions between PAβN and Phe616 of AcrB which is located at the tip of the hairpin-like G-loop and forms a partition between the proximal binding pocket and distal binding pocket at the top of the channel between the two pockets. GOLD molecular docking of PAβN and TET to the located binding site by SMINA also showed that the affinity of PAβN to the AcrB transporter (binding energy -39.9 kcal/mol and score 37.3) is much more favourable over TET (binding energy -25.6 kcal/mol and score 22.2). Phe-cluster residues, including Phe136, Phe178, Phe616, Phe627 and Phe665, provided effective Pi interactions between the ligand and the transporter. These strong interactions resulted in higher score and favourable docking energy.
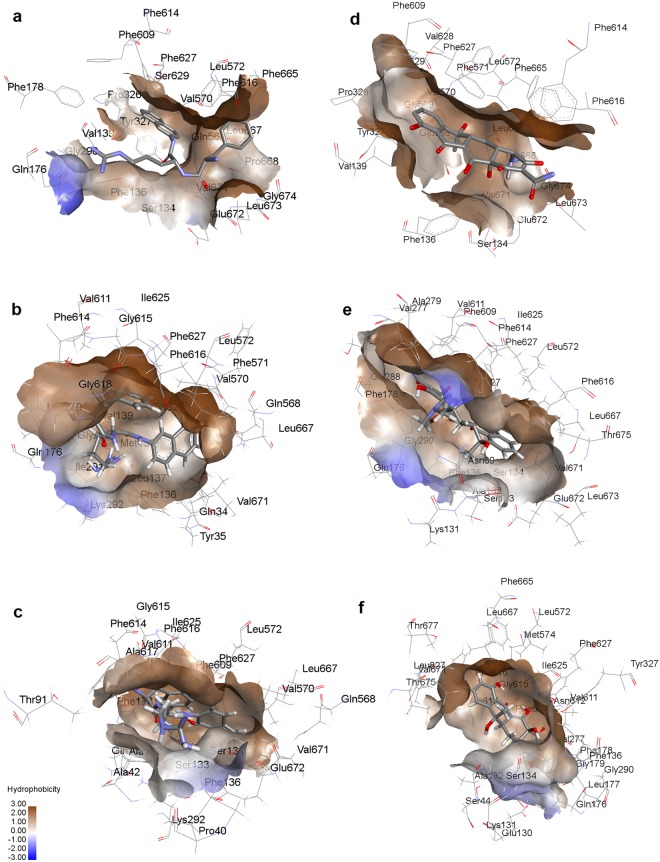
Figure 3.
3D structures of PAβN (right column) and TET (left column) in the multi binding site of AcrB; (a & d) after GOLD molecular docking, (b & e) average structure after 100 ns cMD and (c & f) average structure after 200 ns aMD.
Important regions of AcrB involved in interaction with ligands
Table 1 shows the residues of AcrB that interact with the ligand during the course of the simulation. They have been marked by sequence alignment of AcrB K. pneumoniae to the solved-structure of AcrB from E. coli9,51. The matched amino acids in the alignment of AcrB sequences from E.coli and K. pneumoniae occupy the same locations in their corresponding PDB structures.
Table 1.
The key residues, forming different important regions of AcrB involved in interaction with ligands or functional dynamics of the pump.
| Region | K. pneumoniae AcrB Lining residues |
|---|---|
| Proximal binding site | S79, T91, S134, S135, K292, L572, M574, Q576, F616, T623, M661, F663, F665, N666, L667, L673, T675, D680, R716, N718, E825 |
| Distal binding site | S46, Q89, S128, E130, S134, F136, Q176, L177, F178, S180, E273, N274, D276, I277, G290, Y327, L572, F609, V611, F614, F616, R619, F627 |
| Cleft | F663, F665, L667, R716, L827 |
| G-loop/Phe-loop | G615, F616, A617, G618 |
| G-loop tip | Phe616 |
| Postulate gate | Q124, Y757 |
Numbering is according to the amino acids’ positions in the K. pneumonia.
Comparative binding of inhibitor and substrate
Molecular docking showed that PAβN was bound to the narrow channel under the G-loop. The naphthylamine moiety bonded to the bottom area of the distal binding site in interaction with Phe627. On the other side of the molecule, the arginine group was in the distal binding site in interaction with Gln176, with the phenylalanine group in interaction with Leu667 of proximal binding site (Figs 3a and S3a). Tetracycline was in interaction with Ser134, a shared residue between the proximal and distal site, at the bottom of the narrow channel of the multi-binding site and mostly toward the distal pocket (Figs 3d and S3d).
The average structure extracted from cMD simulations showed that the naphthylamine moiety, phenylalanine and arginine groups of PAβN were bound to Phe136, Ph614 and Lys292 respectively, all in the distal binding site (Figs 3b and S3b). It appeared that PAβN stayed in the hydrophobic site of the distal pocket. In contrast, tetracycline was found in interaction with Phe614, Tyr327 of the distal binding site and Phe616 of G-loop between the distal and proximal sites (Figs 3e and S3e), which means the antibiotic failed to stay in the hydrophobic pocket.
The monitoring of the interaction energy between the ligands and the amino acid residues of multi-binding site in AcrB, during the course of cMD by extracting the structures every 10 ns, showed that the stability of PAβN-AcrB complex is higher compared to tetracycline-AcrB complex., There are at least three responsive amino acid Phe136, Phe616 and Phe627 with favorable interaction energy that could be observed in all 10 ns panels, whereas there is just one common responsive residue Phe614 during the cMD in the case of tetracycline (Table S1).
At the end of the simulation, the results in the average structure extracted from aMD simulations indicated that the naphthylamine moiety of PAβN was still in interaction with Tyr327 and the phenylalanine moiety was in contact with Phe616 and Phe178 of the hydrophobic groove, in the distal binding site (Figs 3c and S3c). Tetracycline, which was bound to a residue of the distal pocket, Phe614, was mostly in interaction with the residues of G-loop, Gly615, Phe616 and Ala617, as well as a residue from the proximal site, Leu667 (Fig. 3f and S3f). This data suggests that PAβN could effectively interact with the hydrophobic groove of the distal pocket, as expected of a potential efflux pump inhibitor, which, by peripherally binding to the hydrophobic trap, could occlude the passageway and consequently interfere with the binding of the other compounds2,50,51. On the other hand, tetracycline formed weaker interactions with the hydrophobic pocket during the course of the simulation, which would eventually result in efflux through the transporter. This results in a good agreement with what Kinana and coworkers showed regarding the effects of various amino acids of binding pocket in substrate binding54. The monitoring of the interaction energies between the ligands and the key amino acid residues of multi-binding site in AcrB, during the course of aMD by extracting the average structures (Table S2), showed that the stability of PAβN-AcrB complex is higher compared to TET-AcrB complex. There were three amino acids that formed favourable interactions with PAβN compared to a single amino acid that interacted with Tetracycline during the course of the simulation (Table S3 & S4).
Monitoring the distances between the ligands and some of the key residues of the multi-binding site of AcrB (Fig. S4) showed that, in the case of PAβN, the distances reach a steady state very soon via tight binding of the ligand to the deep hydrophobic groove, but for tetracycline they are fluctuating and variable, which means the ligand formed transient interactions moving around the multi-binding site. These results are in a good agreement with the multisite-drug-oscillation hypothesis Yamaguchi and co-workers for AcrB8. According to this hypothesis, the substrate is oscillating and may be just occluded in the distal pocket without a specific binding site, as we observed that tetracycline oscillates in the AcrB multibinding site in the spacious drug binding pocket. Also, it can explain the difference in the drug specificity in a certain exporter like AcrB, where tetracycline is exported and PAβN is not exported.
Binding free energy
cMD simulations followed by calculation of the relative binding free energy of the inhibitor and substrate. Table 2 summarizes the calculated values of the different energy contributions to the relative binding free energy. The calculated binding free energy of tetracycline −11 kcal/mol is notably weaker than PAβN with −28 kcal/mol. The GB approach that resulted in a slightly lower binding energy for both substrate and inhibitor still suggests a more favorable complex in PAβN-bound state. It implies PAβN formed a stable complex that could inhibit the transporter, and that tetracycline is highly likely to end up being pumped out due to the weaker interaction.
Table 2.
Calculated energy contributions to form the AcrB–PAβN and AcrB–TET complexes (kcal/mol) and inhibition constants (Kd in Molar) with standard errors of the mean (in parentheses) after cMD.
| Energy distributions | AcrB-PAβN | AcrB-TET |
|---|---|---|
| ΔEele | −16.7 (2.1) | −20.1 (2.2) |
| ΔEvdw | −49.2 (2.0) | −41.6 (1.1) |
| ΔEsol | 37.8 (3.3) | 49.7 (3.5) |
| ΔGPB | −28.1 (2.9) | −11.9 (1.9) |
| ΔGGB | −35.9 (2.3) | −16.3 (2.3) |
| −TΔS | 18.7 | 18.9 |
| ΔGbind | −9.4 (0.7) | 6.9 (0.5) |
| Kd* | 1.4 × 10−7 | 1.2 × 105 |
| Kd (Bulk)** | 8.4 × 1017 | 7.3 × 1029 |
*Kd obtained by using ΔG = RT ln Kd formula.
**Calculated by considering Avogadro’s number.
The entropy contribution in ligand binding was calculated to determine the absolute binding free energy and dissociation constant for the systems. The calculated values clearly show that the tendency of tetracycline is to be dissociated from the complex, but that PAβN is likely to remain as a complex with AcrB owing to its favourable free energy of binding. Also, comparison of the value of Kd in interaction of PAβN with AcrB (1.4 × 10−7) and AdeB (4.9 × 10−6) in our recent study55 showed more binding strength and affinity of the inhibitor to the AcrB transporter over AdeB. This suggests that PAβN is an effective inhibitor of AcrB and explains why it does not inhibit the AdeB efflux pump.
Principal component analysis
After running aMD simulations, PCA was performed to characterize the conformational transitions in the AcrB transporter, modulated by different ligands, with different roles as the potent inhibitor and substrate. The dendrogram data (Fig. S5) appeared to fall into six and seven clusters for PAβN- and TET- complexes, respectively, and the PCA plots are coloured to show this. By considering six and seven clusters in PAβN- and TET- complexes respectively, it can be seen that AcrB in complex with inhibitor has different dynamics and functions compared to the AcrB in complex with the substrate.
Figures 4–6 show conformer plots that display the relationships between different conformers, highlighting the major differences between structures and enabling the interpretation and characterization of multiple inter-conformer relationships. They have been generated in three different panels for diverse protomers in AcrB; Access (loose)/Binding (tight)/Extrusion (open) subunits. PCA highlights that PAβN- and TET-bound conformations are not only different in the binding protomer (Fig. 5) but also that they are completely diverse in their access and extrusion conformations (Figs 4 and 6). In addition, substrate-bound structures are different from inhibitor-bound ones, in general, as they possess diverse conformer graphs with diverse conformational clustering.
Figure 4.
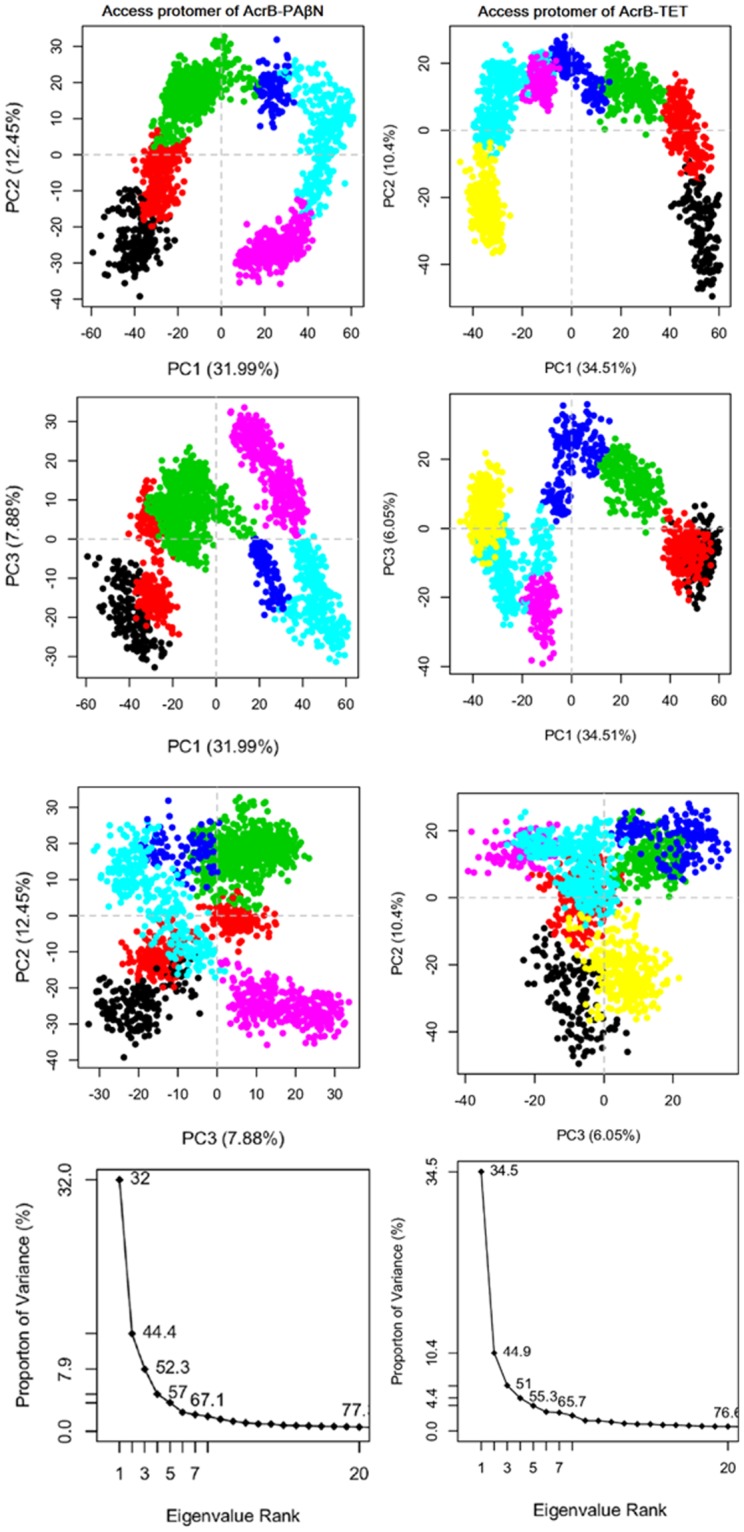
Row (1–3) Conformer plot of PCA data colored by cluster after calculation of cluster groups in the access protomer of AcrB-PAβN (left panels) and AcrB-TET (right panels); Row (4). The rank ordering of the eigenvalues of the covariance matrix. Eigenvalue spectrum: Results obtained from diagonalization of the atomic displacement correlation matrix of Cα atom coordinates from the first snapshot structures. Inset shows histograms for the projection of the distribution of structures onto the first six principal components.
Figure 6.
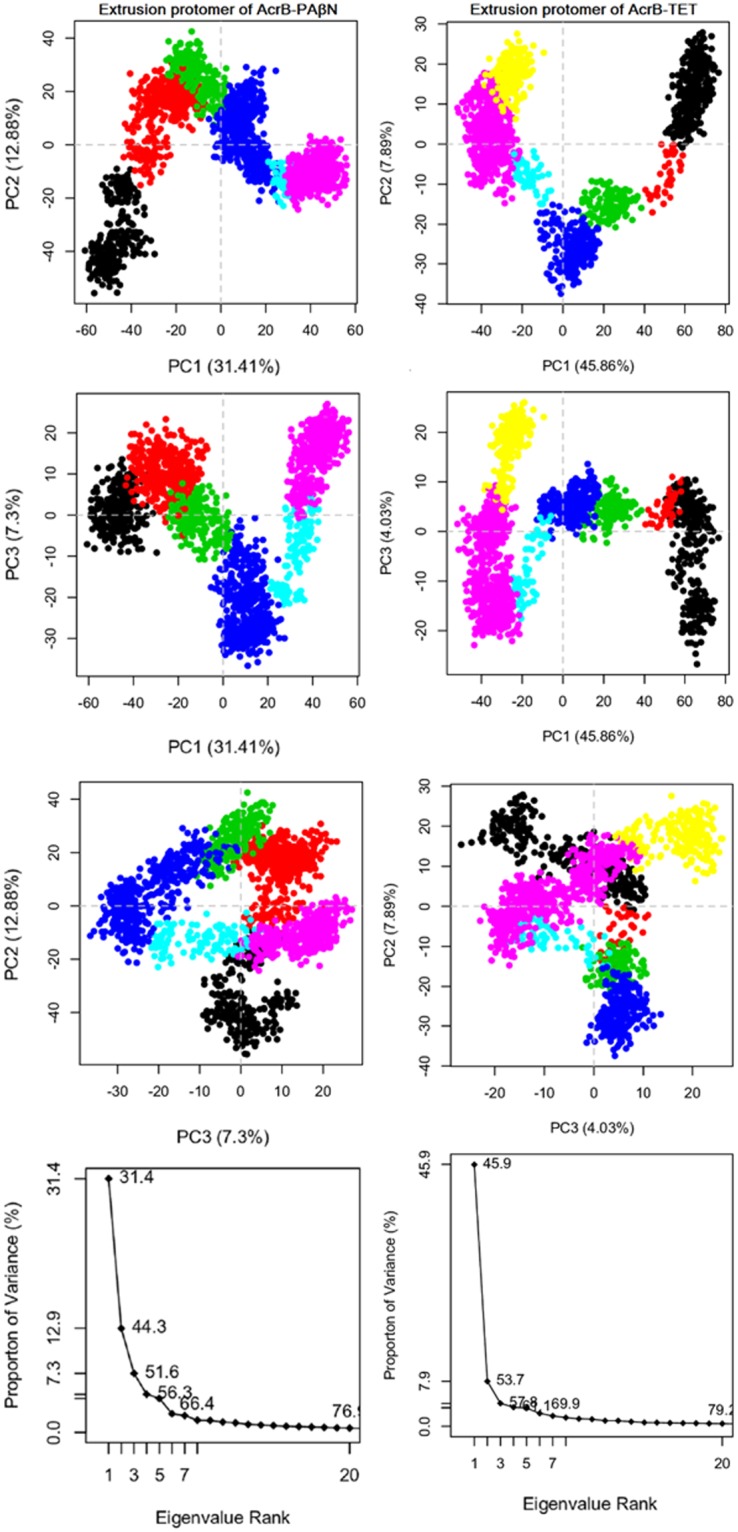
Row (1–3) Conformer plot of PCA data colored by cluster after calculation of cluster groups in extrusion protomer of AcrB-PAβN (left panels) and AcrB-TET (right panels); Row (4) The rank ordering of the eigenvalues of the covariance matrix. Eigenvalue spectrum: Results obtained from diagonalization of the atomic displacement correlation matrix of Cα atom coordinates from the first snapshot structures. Inset shows histograms for the projection of the distribution of structures onto the first six principal components.
Figure 5.
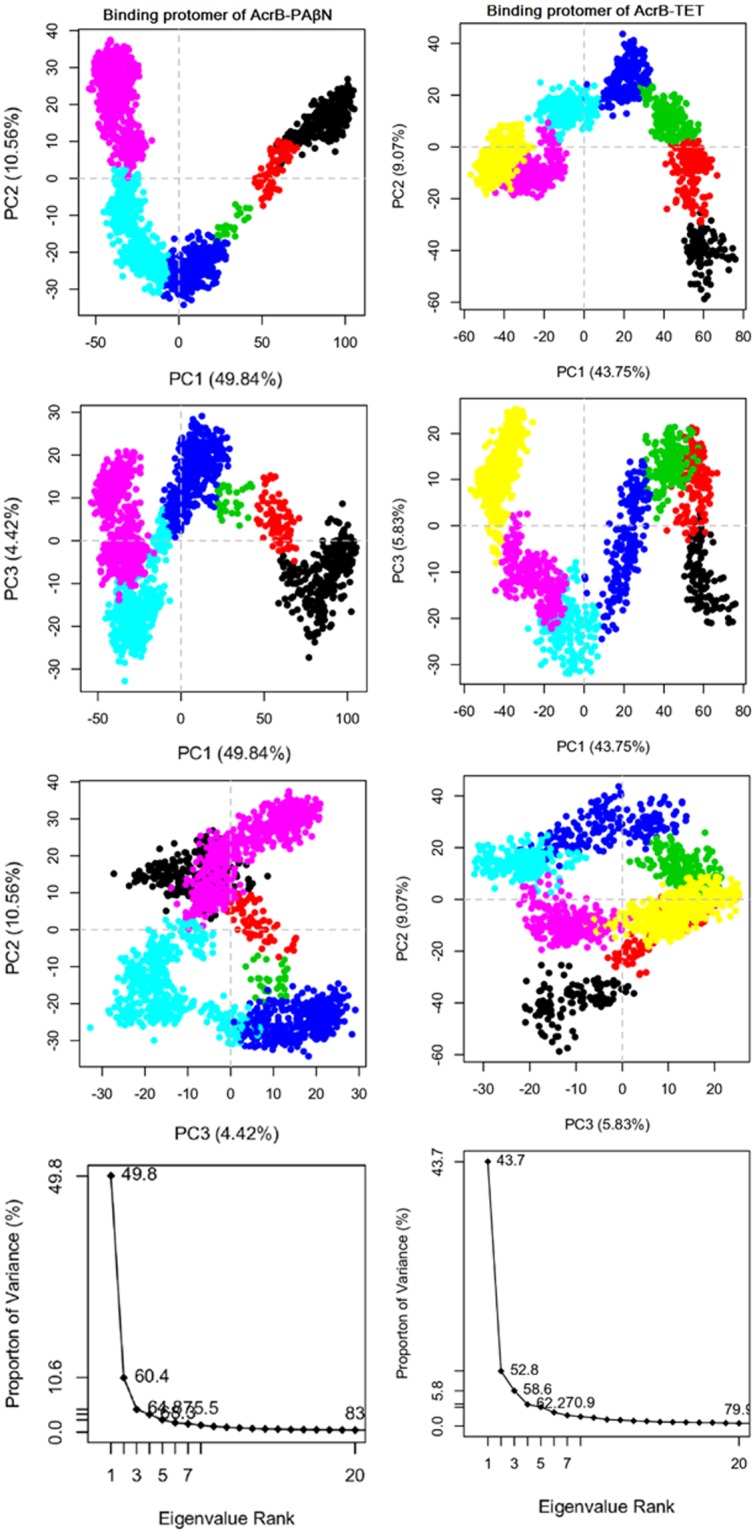
Row (1–3) Conformer plot of PCA data colored by cluster after calculation of cluster groups in binding protomer of AcrB-PAβN (left panels) and AcrB-TET (right panels); Row (4) The rank ordering of the eigenvalues of the covariance matrix. Eigenvalue spectrum: Results obtained from diagonalization of the atomic displacement correlation matrix of Cα atom coordinates from the first snapshot structures. Inset shows histograms for the projection of the distribution of structures onto the first six principal components.
The last row in the conformer graphs of Figs 4–6 is the eigenvalue spectrum obtained from the principal component analysis of the computationally determined conformers. The magnitude of each eigenvalue is expressed as the proportion of the total variance (mean-square fluctuation) captured by the corresponding eigenvector. Labels on each point indicate the cumulative sum of variance accounted for by a particular eigenvector and its preceding eigenvectors.
The fluctuations captured by the first principal component were visualized as a trajectory for each complex. They were generated as movies in PyMOL visualizer software for PAβN-bound (Movie S1) and TET-bound (Movie S2) transporters. The movies show a different motion in AcrB transporter in interaction with different compounds applied in this study. There is another twist in the transmembrane domain of PAβN-bound transporter compared to TET-bound transporter, so the observed motion in AcrB-TET complex is not present in AcrB- PAβN complex. Also, movies S3 and S4 were generated to show the dynamics of the AcrB pump in interaction with PAβN and TET respectively using extracted snapshots during the aMD simulations by VMD and movies were created by VideoMach software.
Discussion
We examined the binding of PAβN and tetracycline to the multi binding pocket of the binding protomer of AcrB, at first by blind docking with the program Autodock SMINA and then using GOLD56 flexible molecular docking in the SMINA-located site. The study was followed by conventional molecular dynamics simulations combined by accelerated molecular dynamics, for a whole model of AcrB transporter used to study the dynamics of all the three protomers of the pump.
The accurate computational observation and direct assessment of functionally important protein dynamics and conformations, without any timescale limitations, were performed using an accelerated molecular dynamics simulation approach, by simulating the transition between minima of different potential energy. By this approach, we have gained insight to the comparative function of AcrB in interaction with its substrate or inhibitor. The results show that the motion of the transporter was induced by the ligand in the interaction with the multi-binding site. The induced motion is ligand-dependent, which means it is different for the two bound compounds. The different dynamics for PAβN-bound form of AcrB compared to the Tet-bound form is likely to affect the rotating mechanism typically observed for AcrB transporter. This type of motion dependency has implications for understanding the ligand binding mechanisms in the AcrB tripartite efflux pump.
In agreement with Nikaido et al., whose work showed that the binding position of the naphthylamine moiety of PAβN is the reason why it produces efflux inhibition50,51, the results of this study showed a strong interaction between naphthylamine moiety of PAβN and the hydrophobic trap of distal binding site. Nikaido et al. also suggested that the largest fraction of the binding energy comes from the trap and PAβN interacts strongly with the trap51. Similarly, binding free energy calculations showed a very favorable free energy of binding for PAβN in interaction with AcrB. In addition, monitoring of the interaction energy between this inhibitor and key residues of binding site indicate that Ala132, Ala617, Ser134, Phe627, Phe136 and Phe616 in AcrB-PAβN complex with the most favorable energy of interaction mostly belong to the hydrophobic trap.
Nikaido et al. computationally showed that PAβN as an inhibitor of AcrB from E. coli tends to move out the distal pocket at least partially, getting into contact with G-loop, and is thought to control the access of substrates to distal pocket2. In another study they showed that the mode of binding of PAβN to AcrB affects the efflux of other compounds50. In addition, they experimentally and computationally showed that PAβN inhibited the efflux of other drugs by binding to the hydrophobic trap of the distal binding pocket, and also by interfering with the binding of other drug substrates to the upper part of the binding pocket. Our results also showed that PAβN occludes the channel by sticking to the hydrophobic binding pocket and interaction with other residues within the multi-binding site. Occlusion of the binding protomer affects the regular dynamics and allosteric switching motions of three protomer of the transporter. Consequently, normal rotatingmotion which leads to the extrusion of the compound could be corrupted. Abnormality in the motion of the transporter causes dramatic changes to the pump’s compound extrusion ability. The new dominant dynamics induced by the interaction of the inhibitor with the transporter, by blocking the binding monomer, corrupt the switching of the functional dynamics and prevent the transporter from extruding the compounds. It seems that the efflux pump works by a ligand-dependent dynamics approach that fully affects the functional features of the transporter. Another study on AcrB of E. coli showed that the assembly of the RND-type efflux systems is dynamically regulated in response to external stimuli57.
This study showed AcrB could adopt different dynamics when it interacts with its substrate Tetracycline compared to the inhibitor PAβN-bound form. The simulation data suggests the correlations among the distal and proximal pockets of the multi-binding site, Phe cluster and cleft during the inhibitor binding event could result in the inhibition of the rotating mechanism and prevent the exporter from extruding any other substrates, leading to the inhibition of the tripartite pump. The information obtained from the study will contribute to the design of new, effective and selective efflux pump inhibitors that could play key roles in interrupting the rotating mechanism and allosteric conformational changes that could reverse antimicrobial resistance2.
Methods
cMD and aMD simulations were performed using the program AMBER 1258. MD simulations started from the docked structures, obtained from blind docking by AutoDock SMINA59, which followed by flexible docking using the GOLD56 program. MD simulations of efflux pump complexes have been carried out without the lipid bilayer due to the size of the efflux pump systems. Vargiu2 and Kinana50 showed that the results for systems with and without the lipid bilayer are essentially the same and equally valid. However, the transporter was not truncated and the whole trimeric protein structure was used in this study. In this study, the power of GPU acceleration combined with the sampling power of aMD raising minima, were used to study conformational transitions that occur in AcrB in interaction with substrate and inhibitor, separately. Also, the CUDA implementation of PMEMD was used to carry out the simulations on GPUs.
Generating the structures
The structure of the Klebsiella pneumoniae AcrB transporter was generated by homology modelling using Swiss Model webserver, by applying Uniprot code Q93K40 as the amino acid sequence. The template was AcrB of Escherichia coli, with PDB ID code 4DX5 and resolution 1.9 Å. The sequence identity between the target and template was 91.6%. The accuracy and validity of the generated AcrB model was examined by Phyre260 and the MPI Bioinformatics toolkit61. Phenylalanyl-arginine-β-naphthylamide (PAβN) and tetracycline (TET) PDB structures were generated by Chem3D 15.0. All the receptor and ligand structures were minimized by the SYBYL program.
Molecular docking
The starting structures for running simulations were obtained by molecular docking of the ligands to the AcrB transporter. The orientation of the ligands within the multi-binding site of the binding protomer was taken from docking calculations performed with the AutoDock SMINA package59 through blind molecular docking, to find the cavity for ligands with the best affinity among all the probable ones. All the parameters were kept at their default values for running SMINA.
Then, an evaluated flexible molecular docking was performed using GOLD56 molecular docking into the SMINA-located binding site. The Genetic algorithm (GA) is used in GOLD ligand docking to examine the ligand conformational flexibility, along with the partial flexibility of the protein. The maximum number of runs was set to 20 for each compound, with the default parameters (100 population size, 5 for the number of islands, 100,000 number of operations and 2 for the niche size). Default cut off values of 2.5 Å (dH-X) for hydrogen bonds and 4.0 Å for van-der-Waals distance were used. When the solutions attained RMSD values within 1.5 Å, GA docking was terminated.
Molecular dynamics (MD) simulations
In this stage, the method section includes (i) preparatory energy minimizations, heating the system, cMD equilibration, (ii) building a simulation environment suitable for aMD simulations.
cMD simulations were performed using the AMBER 12 package program58. A time step of 2 fs was used. Periodic boundary conditions were used, and electrostatic interactions were treated using the particle-mesh Ewald method, with a real-space cutoff of 12 Å and a grid spacing of 1 Å per grid point in each dimension. The van der Waals interactions were modeled with a Lennard–Jones potential, using a 12 Å cutoff. The simulations were performed in NPT ensemble, which means the temperature and pressure conditions were constant, and the temperature was kept at 300 K by applying the Langevin thermostat to all heavy atoms, with the Langevin damping constant set to 5 ps-1. The pressure was kept at 1 atm using the isotropic position scaling protocol used in AMBER.
The minimization was performed in two phases, and each phase was performed in two stages. In the first phase, ions and all water molecules were minimized for 2500 cycles of steepest descent followed by 2500 cycles of conjugate gradient minimization. Afterward, the entirety of the systems were minimized for a total of 10000 cycles without restraint, wherein 5500 cycles of steepest descent were followed by 4500 cycles of conjugate gradient minimization. After minimizations, the systems were heated for 500 ps while the temperature was raised from 0 to 300 K, and then equilibration was performed without a restraint for 1000 ps while the temperature was kept at 300 K. Sampling of reasonable configurations was conducted by running 100 ns simulations for each system.
Using the relaxed starting structure, obtained from cMD, the necessary information was calculated to set the necessary aMD parameters. Calculations of the added values for performing aMD simulations were shown at the end of the SI file as a note. From cMD run, we obtained the average total potential energy of −54,6820 and −59,5545 kcal/mol, and average dihedral energy of 32671 and 32605 kcal/mol for AcrB-PAβN and AcrB-TET systems, respectively. Using this information, and considering AcrB has 3093 residues and that the two mentioned complex systems have 196809 and 211425 atoms, respectively, the aMD parameters were calculated; Average total potential energy threshold (EthreshP) were obtained as −515,331 and −561,717 kcal/mol; Inverse strength boost factor for the total potential energy (alphaP) were obtained 31,489 and 33,828 kcal/mol; Average dihedral energy threshold (EthreshD) were obtained 45,047 and 44,961 kcal/mol; Inverse strength boost factor for the dihedral energy (alphaD) were obtained 2,475 and 2,471 kcal/mol, for AcrB-PAβN and AcrB-TET systems respectively. Then, the full 200 ns aMD simulations were run for each system.
Postprocessing analyses: MM-PBSA/MM_GBSA calculation
The free energy of binding of the compounds to AcrB was evaluated by means of the molecular mechanic energies combined with the Poisson–Boltzmann and generalized Born and surface area continuum solvation (MM/PBSA/MM/GBSA) postprocessing methods62,63. In these method, the binding free energy of each compound is evaluated as
| 1 |
with Gcom, Grec, and Glig being the absolute free energies of complex, receptor, and ligand, respectively, averaged over the equilibrium trajectory of the complex (single-trajectory approach). According to these schemes, the free-energy difference can be decomposed as
| 2 |
where ΔEMM is the difference in the molecular mechanics energy, ΔGsolv is the solvation-free energy, and ΔSconf is the solute conformational entropy. The first two terms were calculated with the following equations:
| 3 |
| 4 |
EMM includes the molecular mechanics energy contributed by the bonded (Ebond, Eangle, and Etorsion) and nonbonded (Evdw and Eele, calculated with no cutoff) terms of the force field. ΔGsolv is the solvation-free energy, which can be modeled as the sum of an electrostatic contribution (ΔGsolv,p, evaluated using the MMGBSA or MM-PBSA approach) and a nonpolar one (ΔGsolv,np = γΔSA + b, proportional to the difference in solvent-exposed surface area, ΔSA).
In this approach for the fully equilibrated structures (Fig. S6), 20 snapshots were collected from the last 200 ps of cMD simulations of complex systems for post processing analysis of free energy calculation. The RMSD of the ligands (Fig S7) with respect to the last conformation of the corresponding ligands sampled during the MD were calculated which showed the ligands in complex with the systems reached steady state at the end of the simulation The ΔGPB term was calculated by solving the finite-difference Poisson-Boltzmann equation using the internal PBSA program. The SCALE value was set to 2. The Parse radii were employed for all atoms64. The solvent probe radius was set at 1.4 Å (with the radii in the prmtop files). MM-PBSA running was performed with the pbsa module (PROC = 2). The value of the exterior dielectric constant was set at 80, and the solute dielectric constant was set at 165. The nonpolar contribution was determined on the basis of the solvent accessible surface area (SASA) using the LCPO method66 and CAVITY-OFFSET set at 0.00.
Finally, the conformational entropy contribution, to estimate the absolute binding free energy, was calculated through normal-mode analysis using the ‘nmode’ module of AMBER.
Postprocessing analyses: Analysis of protein conformational change using principal component analysis (PCA)
The original trajectory files produced by aMD were significantly large, and we can’t include them to Bio3d package installed in R program due to space limitations, so the trajectories in each aMD simulations were down sampled with an interval of 100. The points represented in the conformer plots were computationally clustered and colored by the cluster. This was performed by creating a distance matrix of the principal components of interest.
PCA reduces the dimensionality of large data sets by calculating a covariance matrix and its eigenvectors. Vectors with the highest eigenvalues become the most significant principal components. When principal components are plotted against each other, similar structures cluster. Each cluster then theoretically shows a different protein conformational state. To avoid sample noise from random fluctuations48,67, following aMD simulations of the AcrB transporter, the PCA was calculated only for Cα atoms. Then, each protomer was selected in a separate PCA analysis, which was a good discriminator of conformations.
With the Bio3D package installed in R, the plot command has been overloaded to create a default PCA plot with four graphs. Three are the z-scores of the first three principal components plotted against each other in two dimensions. The last is a scree plot showing how much of the variance of the data set is captured by each principal component.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
We thank the King’s College High Performance Computing (HPC) system staffs, at Faculty of Natural and Mathematical Sciences (NMS/Ada), particularly Dr. Andreas Biternas, HPC and Linux Lead. We also thank BBSRC (Sparking Impact Award) for providing funding for the early part of the work.
Author Contributions
S.J., K.M.R. and J.M.S. conceived the study, S.J., K.M.R. and J.M.S. designed the experiments, S.J. and K.M.R. conducted the experiments. All authors contributed to analysis and interpretation of the data. S.J. and K.M.R. wrote the manuscript and all authors edited the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28531-6.
References
- 1.Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev. 2015;28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol. 2011;77:1–60. doi: 10.1002/9780470920541.ch1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma D, et al. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padilla E, et al. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2010;54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sjuts H, et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc Natl Acad Sci USA. 2016;113:3509–3514. doi: 10.1073/pnas.1602472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi A, Nakashima R, Sakurai K. Structural basis of RND-type multidrug exporters. Front Microbiol. 2015;6:327. doi: 10.3389/fmicb.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 10.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 11.Du D, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 13.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeger MA, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 15.Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol. 2009;191:1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takatsuka Y, Nikaido H. Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J Bacteriol. 2007;189:8677–8684. doi: 10.1128/JB.01127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz R, Vargiu AV, Collu F, Kleinekathofer U, Ruggerone P. Functional rotation of the transporter AcrB: insights into drug extrusion from simulations. PLoS Comput Biol. 2010;6:e1000806. doi: 10.1371/journal.pcbi.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao XQ, Kenzaki H, Murakami S, Takada S. Drug export and allosteric coupling in a multidrug transporter revealed by molecular simulations. Nat Commun. 2010;1:117. doi: 10.1038/ncomms1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tornroth-Horsefield S, et al. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure. 2007;15:1663–1673. doi: 10.1016/j.str.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Wang, Z. et al. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife6, 10.7554/eLife.24905 (2017). [DOI] [PMC free article] [PubMed]
- 21.Zuo Z, Weng J, Wang W. Insights into the Inhibitory Mechanism of D13-9001 to the Multidrug Transporter AcrB through Molecular Dynamics Simulations. J Phys Chem B. 2016;120:2145–2154. doi: 10.1021/acs.jpcb.5b11942. [DOI] [PubMed] [Google Scholar]
- 22.Zuo Z, Wang B, Weng J, Wang W. Stepwise substrate translocation mechanism revealed by free energy calculations of doxorubicin in the multidrug transporter AcrB. Sci Rep. 2015;5:13905. doi: 10.1038/srep13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vargiu AV, et al. Water-mediated interactions enable smooth substrate transport in a bacterial efflux pump. Biochim Biophys Acta. 2018;1862:836–845. doi: 10.1016/j.bbagen.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 25.Eicher T, et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci USA. 2012;109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohnert JA, et al. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol. 2008;190:8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohnert JA, Schuster S, Fahnrich E, Trittler R, Kern WV. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF) J Antimicrob Chemother. 2007;59:1216–1222. doi: 10.1093/jac/dkl426. [DOI] [PubMed] [Google Scholar]
- 28.Bohnert JA, Karamian B, Nikaido H. Optimized Nile Red efflux assay of AcrAB-TolC multidrug efflux system shows competition between substrates. Antimicrob Agents Chemother. 2010;54:3770–3775. doi: 10.1128/AAC.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wehmeier C, Schuster S, Fahnrich E, Kern WV, Bohnert JA. Site-directed mutagenesis reveals amino acid residues in the Escherichia coli RND efflux pump AcrB that confer macrolide resistance. Antimicrob Agents Chemother. 2009;53:329–330. doi: 10.1128/AAC.00921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vargiu AV, et al. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: explanation and rationale by molecular dynamics simulations. J Am Chem Soc. 2011;133:10704–10707. doi: 10.1021/ja202666x. [DOI] [PubMed] [Google Scholar]
- 31.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Husain F, Nikaido H. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol Microbiol. 2010;78:320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai T, et al. Functionality mapping on internal surfaces of multidrug transporter AcrB based on molecular theory of solvation: implications for drug efflux pathway. J Phys Chem B. 2011;115:8288–8295. doi: 10.1021/jp2015758. [DOI] [PubMed] [Google Scholar]
- 34.Ramaswamy VK, Vargiu AV, Malloci G, Dreier J, Ruggerone P. Molecular Rationale behind the Differential Substrate Specificity of Bacterial RND Multi-Drug Transporters. Sci Rep. 2017;7:8075. doi: 10.1038/s41598-017-08747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukagoshi N, Aono R. Entry into and release of solvents by Escherichia coli in an organic-aqueous two-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol. 2000;182:4803–4810. doi: 10.1128/JB.182.17.4803-4810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 38.Oswald C, Tam HK, Pos KM. Transport of lipophilic carboxylates is mediated by transmembrane helix 2 in multidrug transporter AcrB. Nat Commun. 2016;7:13819. doi: 10.1038/ncomms13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce LC, Salomon-Ferrer R. Augusto, F. d. O. C., McCammon, J. A. & Walker, R. C. Routine Access to Millisecond Time Scale Events with Accelerated Molecular Dynamics. J Chem Theory Comput. 2012;8:2997–3002. doi: 10.1021/ct300284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voter AF. Hyperdynamics: Accelerated Molecular Dynamics of Infrequent Events. Phys. Rev. Lett. 1997;78:3908. doi: 10.1103/PhysRevLett.78.3908. [DOI] [Google Scholar]
- 41.Voter AF. A method for accelerating the molecular dynamics simulation of infrequent events. The Journal of Chemical Physics. 1997;106:4665. doi: 10.1063/1.473503. [DOI] [Google Scholar]
- 42.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 43.Bucher D, Grant BJ, Markwick PR & McCammon, M. Accessing a hidden conformation of the maltose binding protein using accelerated molecular dynamics. PLoS Comput Biol. 7 (2011). [DOI] [PMC free article] [PubMed]
- 44.Grant BJ, Gorfe AA, McCammon JA. Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLoS Comput Biol. 2009;5:e1000325. doi: 10.1371/journal.pcbi.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mucksch C, Urbassek HM. Enhancing protein adsorption simulations by using accelerated molecular dynamics. PLoS One. 2014;8:e64883. doi: 10.1371/journal.pone.0064883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Oliveira CA, Grant BJ, Zhou M, McCammon JA. Large-scale conformational changes of Trypanosoma cruzi proline racemase predicted by accelerated molecular dynamics simulation. PLoS Comput Biol. 2011;7:e1002178. doi: 10.1371/journal.pcbi.1002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmon L, et al. Multi-timescale conformational dynamics of the SH3 domain of CD2-associated protein using NMR spectroscopy and accelerated molecular dynamics. Angew Chem Int Ed Engl. 2012;51:6103–6106. doi: 10.1002/anie.201202026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas JR, Gedeon PC, Grant BJ, Madura JD. LeuT conformational sampling utilizing accelerated molecular dynamics and principal component analysis. Biophys J. 2012;103:L1–3. doi: 10.1016/j.bpj.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinana AD, Vargiu AV, May T, Nikaido H. Aminoacyl beta-naphthylamides as substrates and modulators of AcrB multidrug efflux pump. Proc Natl Acad Sci USA. 2016;113:1405–1410. doi: 10.1073/pnas.1525143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother. 2014;58:6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer N, Kandt C. Porter domain opening and closing motions in the multi-drug efflux transporter AcrB. Biochim Biophys Acta. 2013;1828:632–641. doi: 10.1016/j.bbamem.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 53.Saw HT, Webber MA, Mushtaq S, Woodford N, Piddock LJ. Inactivation or inhibition of AcrAB-TolC increases resistance of carbapenemase-producing Enterobacteriaceae to carbapenems. Journal of Antimicrobial Chemotherapy. 2016;71:1510–1519. doi: 10.1093/jac/dkw028. [DOI] [PubMed] [Google Scholar]
- 54.Kinana AD, Vargiu AV, Nikaido H. Effect of site-directed mutations in multidrug efflux pump AcrB examined by quantitative efflux assays. Biochem Biophys Res Commun. 2016;480:552–557. doi: 10.1016/j.bbrc.2016.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamshidi S, Sutton JM, Rahman KM. Computational Study Reveals the Molecular Mechanism of the Interaction between the Efflux Inhibitor PAβN and the AdeB Transporter from Acinetobacter baumannii. ACS Omega. 2017;2:3002–3016. doi: 10.1021/acsomega.7b00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nissink JW, et al. A new test set for validating predictions of protein-ligand interaction. Proteins. 2002;49:457–471. doi: 10.1002/prot.10232. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto K, et al. Substrate-dependent dynamics of the multidrug efflux transporter AcrB of Escherichia coli. Sci Rep. 2016;6:21909. doi: 10.1038/srep21909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Case, D. A. et al. AMBER 12. University of California, San Francisco (2012).
- 59.Koes DR, Baumgartner MP, Camacho CJ. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J Chem Inf Model. 2013;53:1893–1904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alva V, Nam SZ, Soding J, Lupas AN. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016;44:W410–415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang C, Greene D, Xiao L, Qi R, Luo R. Recent Developments and Applications of the MMPBSA Method. Front Mol Biosci. 2017;4:87. doi: 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sitkoff D, Sharp KA, Honig B. Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models. J. Phys. Chem. 1994;98:1978–1988. doi: 10.1021/j100058a043. [DOI] [Google Scholar]
- 65.Wang W, Kollman PA. Free energy calculations on dimer stability of the HIV protease using molecular dynamics and a continuum solvent model. J Mol Biol. 2000;303:567–582. doi: 10.1006/jmbi.2000.4057. [DOI] [PubMed] [Google Scholar]
- 66.Weiser J, Shenkin PS, Still WC. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO) J. Comput. Chem. 1999;20:217–230. doi: 10.1002/(SICI)1096-987X(19990130)20:2<217::AID-JCC4>3.0.CO;2-A. [DOI] [Google Scholar]
- 67.Gedeon PC, Thomas JR, Madura JD. Accelerated molecular dynamics and protein conformational change: a theoretical and practical guide using a membrane embedded model neurotransmitter transporter. Methods Mol Biol. 2015;1215:253–287. doi: 10.1007/978-1-4939-1465-4_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).