Abstract
The dysbiosis of the oral microbiome is associated with both localized and systemic diseases. Modulating the resident microbial communities by the dietary consumption of probiotics has become an appealing means to promote host health by either restoring host-microbe balance or preventing dysbiosis. Most probiotics strategies target the intestinal microbiome, but little is known about their impact on the oral microbiome. We analyzed here the saliva microbiome from 21 volunteers, longitudinally collected before, during, and after consumption of a commercial probiotic and a standard yoghurt using 16S amplicon sequencing. The alpha diversity of the saliva microbiome had a statistically significant increase (P-value = 0.0011) in one of the groups that consumed the probiotic. The overall structure of the microbiome was however not significantly impacted by the probiotic, although oligotyping analysis revealed that both Streptococci and Lactobacilli present in the probiotic product persisted in the saliva microbiome. In contrast, non-probiotic yoghurt consumption had a lesser impact on the overall diversity and Lactobacillus and Streptococcus persistence. Our results suggest that consumption of commercial probiotics in healthy subjects increase the overall diversity of the oral cavity microbiome in the short term, but such dietary interventions are not able to substantially modify the structure of the microbiome.
Introduction
The role of the human microbiome in human health has been investigated extensively, highlighting its importance in immune system regulation1,2, nutrient absorption3, and weight balance4. While the majority of these studies have focused on the gut microbiome, the oral microbiome has also received attention5,6, particularly in association with the initiation and development of common oral diseases such as dental caries7–9.
The potential to modify the microbiome by introducing live microbial strains that are presumed to be beneficial to health, so called probiotics, is an active area of research. In this context, most studies have investigated the impact of probiotic bacterial species on the gut microbiome. Such bacteria usually belong to the Bifidobacterium, Lactobacillus and Streptococcus genera10,11, and their potential beneficial effects have been investigated both in mouse models12–14 and human trials15–17. While several products are commercially available for the modulation of the human gut microbiome18,19, only a few have been specifically proposed for the oral cavity20.
We previously described the short-term changes induced by a commercial probiotic product in the taxonomic composition of the saliva microbiome21. This study suggested that the overall diversity of the microbiome increased with the consumption of a probiotic product, but because of the limited sample size and the cross-sectional rather than longitudinal study design, additional evidence is required. In this study, we further investigate the impact of a probiotic and standard yoghurt intake in terms of their effect on the composition and structure of the oral microbiome and evaluate the persistence of the probiotic strains.
Results
The saliva microbiome after consumption of a probiotic product
In this study we collected longitudinal saliva samples from 21 healthy volunteers, divided into three groups of seven volunteers (Fig. 1A). Each of these three group was asked to follow a probiotic-free diet for at least two weeks, after which the first set of saliva samples were collected to define the baseline microbiome configuration (T1). The following day (T2), the first and the third group of volunteers consumed the probiotic product (at breakfast and lunch respectively) consisting of a commercial fermented milk drink (“Coop Probiotico Bianco” [COOP Italia s.c. Bologna (IT)], 100 gr) containing S. thermophilus, L. delbrueckii subsp. Bulgaricus and L. paracasei strains, while the second group consumed the same amount of a non-probiotic yoghurt (“Mila Bianco” [Mila latte montagna, Alto Adige (IT)], 100 gr). On the last day of sampling (T3), the first group reverted back to the normal diet while the second and the third groups ate the probiotic product.
Figure 1.
Study design, composition and diversity of the saliva microbiome. (A) Study design comprising three groups of seven healthy volunteers each. The baseline is defined at T1, without any probiotic or non-probiotic yoghurt ingestion. Group 1 was designed to evaluate the influence of a single probiotic intake, while Group 2 was designed to assess the differences between probiotic and non-probiotic yoghurt consumption. The last group was used to investigate a repeated probiotic intake (after lunch at both T2 and T3). All samples were collected after lunch, while yoghurt or probiotic ingestion occurred at breakfast (~5 hours before sample collection) or at lunch (~1 hour before sample collection) as reported in the figure. (B) Taxonomic profiling at genus level for all groups and timepoints. (C) Principal Coordinates Analysis (PCoA) plot of OTU-based beta diversity (weighted Unifrac) with samples colored by group. (D) Quantitative taxonomic composition of the probiotic fermented milk drink (“Probiotic”) and the non-probiotic commercial yoghurt (“Yoghurt”) as derived by OTU analysis and (E) by oligotyping analysis.
The saliva microbiome was investigated by 16S rRNA gene sequencing producing a total of 4.6 million reads (39,3 k ± 14,2 k reads per sample, Supplementary Table 2). We also sequenced the probiotic and non-probiotic products to assess their composition (56,3 k and 12,0 k reads respectively). By applying the QIIME pipeline22 to the dataset (see Methods), we generated a total of 4503 operational taxonomic units (OTUs) clustered at 97% identity with an open-reference OTU picking strategy.
The three most abundant phyla in the saliva samples were Proteobacteria, Firmicutes and Bacteroidetes, with average relative abundances of 36.4%, 28.7%, and 18%, respectively. Fusobacteria (10.5%) and Actinobacteria (5.5%) were less abundant, but still present in all samples (minimum abundance of 0.2% and 0.9% respectively) (Supplementary Figure 1A). At the genus level, Prevotella, Neisseria, Haemophilus, and Streptococcus were the dominant genera (Fig. 1B), which is consistent with previous studies of the healthy oral microbiome6,23. The three sets of biological replicates we performed showed a very low technical variability (Supplementary Figure 1B).
The complexity of the saliva microbial communities in subjects who ate the probiotic drink tended to be higher than for those who ate the standard yoghurt (Supplementary Figure 2A), confirming our previous finding21 although the result was not statistically significant (P-value 0.081). The samples associated with repeated probiotic intake were characterized by a higher alpha diversity than standard diet and single probiotic intake ones (Supplementary Figure 2C) but, again, this was not supported by statistical significance.
Samples did not show a strong grouping by probiotic intake when displaying their inter-sample beta diversity with ordination plots (Fig. 1C), which was expected based on our previous observations21. Instead samples tended to cluster by subject, according to both standard OTU analysis and subspecies level oligotyping analysis24 (Supplementary Figure 3) of the four most abundant genera (Streptococcus, Haemophilus, Prevotella, and Neisseria), confirming the presence of a personal microbial signature in the saliva25,26 (Supplementary Figures 4 and 5). We also profiled both the probiotic fermented milk drink and the non-probiotic yoghurt (Fig. 1D) finding that, in both communities, Streptococcus and Lactobacillus were the most dominant genera as these encompass the bacteria responsible for fermentation of the products. In the probiotic product, Streptococcus was enriched (76%) compared to the non-probiotic (53%), while Lactobacillus was characterized by an opposite trend (20% in probiotic, 41% in non-probiotic yoghurt). This is compatible with the known composition of the probiotic, enriched in S. thermophilus, L. delbrueckii subsp. bulgaricus and L. paracasei strains. Both the probiotic and non-probiotic product shared the most abundant Streptococcus oligotype and the four most abundant Lactobacillus oligotypes (Fig. 1E).
Evidence of increased complexity of the oral microbiome after short-term probiotic consumption
The longitudinal analysis of the alpha diversity in the three groups (Fig. 2 and Supplementary Figure 6) revealed that probiotic intake is significantly associated with an increase in the microbial community diversity for Group 1 (P-value of 0.0011 between T1 and T2). The non-probiotic yoghurt consumption group also showed an increased alpha diversity but to a lesser extent than its probiotic counterpart (P-value of 0.0204 between T1 and T2 in Group 2). Group 3 also consumed the probiotic, but the observed increase in average alpha diversity was not significant. Probiotic consumption is thus associated with a higher, albeit limited, enrichment in the microbial complexity of the saliva, compared to the non-probiotic yoghurt and standard diet.
Figure 2.
Longitudinal alpha diversity (PD-WT metric) across groups normalized to the baseline (T1). (* for P-values <0.05).
We further investigated the differences in microbiome composition across time-points and study groups. Ordination analysis showed no apparent clustering by group, suggesting that the overall structure of the saliva microbiome is not affected by short-term probiotic intake (Fig. 1C). Indeed, no statistically relevant difference was found in beta diversity analysis across timepoints and groups (Supplementary Figure 7). Moreover, no significant variations were observed when comparing intra- and inter-samples distances at different timepoints (Supplementary Figure 8). Although the probiotic consumption does not change the microbiome composition to a large extent, the large overall variability could potentially hide the small-scale changes driven by probiotic intake but biomarkers detection analysis with multiple hypothesis testing correction27 resulted in no statistically significant differentially abundant features.
Overall, despite an increase in microbiome complexity after probiotic intake, no strong variation in the taxonomic structure of the saliva microbiome was observed.
Persistence of the probiotic strains in the saliva microbiome
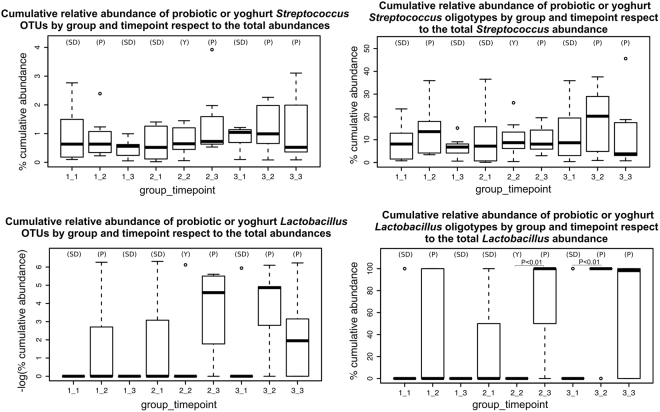
We then investigated whether bacteria present in the probiotic product (S. thermophilus, L. delbrueckii subsp. bulgaricus and L. paracasei) were detectable in the saliva samples to evaluate the presence and persistence of the probiotic strains in the saliva. As a genus, Streptococcus was present in all subjects, and even OTU analysis cannot reach the resolution needed to track the probiotic strain. Nevertheless, finer resolution oligotyping showed a higher abundance of the probiotic Streptococcus oligotypes in subjects who ate the probiotic with respect to non-probiotic oligotypes of the same genus (37.1% average increase after probiotic consumption in Group 1 and 28.7% average increase in Group 3, P-values: 0.37 and 0.51, respectively) (Fig. 3). When the subjects reverted to the standard diet also the probiotic-specific Streptococcus oligotypes returned to a condition similar to the baseline, confirming the short time persistence of oral probiotic organisms (P-value: 0.17, 51.6% average decrease in the only group with standard diet following the probiotic consumption timepoint, Group 1). The same oligotyping analysis performed for probiotic Lactobacillus highlighted a stronger effect compared to Streptococcus, probably because the former is more likely to be confounded with other Streptococci commonly present in the human saliva. The probiotic Lactobacillus OTUs and oligotypes were indeed found enriched (P-values OTUs for the three study groups: 0.21, 0.12 and 0.22; P-values oligotypes: 0.27, 0.008 and 0.004) only in groups which consumed the fermented milk product (Lactobacillus oligotypes relative abundances increased on average 66.7% and 83.3% after probiotic consumption at breakfast in Group 1 and at lunch in Group 3, respectively), with the only exception of the first time-point of the second group (associated with standard diet), in which the Lactobacillus oligotypes were present at relatively high abundance (28.6%). In contrast, no significant enrichment was found associated with the non-probiotic yoghurt consumed at breakfast (Fig. 3). In general, the probiotic-associated Lactobacillus OTUs and oligotypes showed a longer term persistence in the oral microbiome than the Lactobacillus OTUs and oligotypes associated with the non-probiotic yoghurt intake.
Figure 3.
Relative abundances of Streptococcus and Lactobacillus OTUs and oligotypes across timepoints and study groups. (SD) shows the standard diet, (Y) the non-probiotic yoghurt and (P) the probiotic intake.
Discussion
In this work we extended our previous analysis21 to verify the hypothesis that the intake of commercially available probiotic products has a directly effect on the diversity and composition of the saliva microbiome, at least at short timescales. Our longitudinal study design showed that the diversity of the saliva microbiome is increased after consumption of the probiotic product. The overall taxonomic and abundance distribution of bacterial genera is however minimally influenced by probiotic intake even though we verified that the bacteria present in the probiotic product persist to a certain extent in the saliva microbiome. In particular, the impact of probiotic Streptococci is limited in that it only slightly modifies the overall abundance of this already resident genus. Stronger support for short-term persistence was found for Lactobacillus, probably because this organism is not common in the oral microbiome. Altogether, our study confirms that the oral microbiome can potentially be modulated by commercial probiotic products, but that this modulation is limited in both the breadth of the change and in its temporal persistence. Additional studies are required to verify whether continued (e.g. daily) intake of probiotics for longer periods can have a larger and more long-lasting impact on the structure of the oral microbial communities.
Methods
Cohort and Study design
Saliva samples were collected from 21 age-matched healthy volunteers (10 males, 11 females, avg. age 23 s.d. 1,23) that were sampled at three different time-points, following the protocol in Fig. 1A and as described below. We collected 21 samples for each time-point and, for the first time-point, three samples were collected in duplicate as controls for a total of 66 samples. All participants gave their informed consent, and this study was approved by the ethical committee of the University of Trento (Reference number 2015-014); all experiments were performed in accordance with the relevant guidelines and regulations for human subjects research. Each volunteer completed a questionnaire about oral care habits (including frequency of teeth brushing, use of dental floss or mouthwash). Current smokers and individuals using antibiotic treatments in the last six months were excluded from the study. Volunteers were instructed to self-collect the saliva samples and were asked not to brush their teeth, use dental floss or mouthwash before sample collection. They were also requested to avoid intimate kissing. Self-collection was performed under supervision and directly in the laboratory. Volunteers were randomly assigned into three groups of seven subjects (Fig. 1A). For all the three groups, no administration of probiotic or non-probiotic yoghurt was perfomed for the first time-point in order to define the baseline. The second collection after the probiotic administration for the first and the third group and the non-probiotic yoghurt for the second group. At the third sampling, the first group returned to the standard diet, the second switched to the probiotic and the third group repeated the probiotic intake. The probiotic was a commercial fermented milk drink (“Coop Probiotico Bianco” [COOP Italia s.c. Bologna (IT)], 100 g) enriched in S. thermophilus, L. delbrueckii subsp. bulgaricus, L. paracasei strains and supplemented with vitamin B6 and vitamin D, conserved at +4 °C until consumption. The same amount of non-probiotic commercial yoghurt (“Mila Bianco” [Mila latte montagna, Alto Adige (IT)], 100 g) was administered as a control.
Sample collection and DNA extraction
Samples, including technical replicates, were collected through a passive drooling procedure in sterile 50 ml screw top tubes (approximately 3 ml of saliva per sample). About 30 minutes before collection, all volunteers performed a mouth rinse with drinking water. Samples were vortexed before 2 ml of each was centrifuged at 10,000 × g for 10 minutes to collected the bacterial pellets. Total DNA was extracted using the PowerSoil DNA Isolation Kit (MoBio), according to manufacturer’s instructions. The same extraction procedure was also performed on 2 ml of the probiotic drink, the non-probiotic yoghurt and pure water as a negative control. DNA was eluted in 100 μl molecular-grade water and the DNA purity, yield, and concentration was assessed with the NanoDrop ND-2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). DNA was stored at −20 °C until used.
16S rRNA hypervariable region V4 amplification library preparation and sequencing
The V4 hypervariability region of 16 rRNA gene was amplified using Illumina adapted primer 515F (GTGYCAGCMGCCGCGGTAA), and Illumina adapted barcoded 806R primers (GGACTACNVGGGTWTCTAAT) as described previously28,29. The complete list of sample-specific barcodes is included as supplementary material (Supplementary Table 1). Each sample was PCR amplified in duplicate 25 µl reactions each comprising: 10 µl 5 PRIME HotMasterMix (Invitrogen; Life Technologies Europe BV, Monza, Italy) 0.5 µl each of forward and reverse primers (final concentration of 200 nM), 1 µl template DNA (10 ng/µl) and PCR grade water up to the final volume of 25 µl. The thermal cycling conditions were: 94 °C 3 minutes initial denaturation, 35 cycles of 94 °C (45 s), 50 °C (60 s), 72 °C (90 s) followed by a final extension of 10 minutes at 72 °C.
The duplicate amplicon libraries were pooled, visualized and extracted from a 1.5% agarose gel using the Wizard SV Gen and PCR Clean-Up System (Promega Italia s.r.l., Milano, Italy), as described by the manufacturer. The libraries were further cleaned using Agencourt AMPure Xp purification (Beckman Coulter s.r.l, Milan, Italy), following manufacturer’s instructions, and quantified using the Qubit Fluorometric Quantitation system (ThermoFisher Scientific-Invitrogen; Life Technolo-gies Europe BV, Monza, Italy). Finally, equal molarities of amplicons were pooled and sequencing was performed with Illumina MiSeq Reagent Kit v2 (300 cycles) as described in29.
Data analysis
The Illumina MiSeq sequencing run produced 4.6 million paired-end reads after merging with fastq-join30 using default parameters. Joined reads had an average length of 250 bp. We obtained on average 39.3 thousands reads for each sample. Average phred quality score per read was 37. QIIME22 was used to demultiplex and trim low quality reads, reducing the initial reads to 3.38 million (26.5% reduction on average). Trimming was performed with 25 as minimum acceptable quality and 75% as minimum acceptable read length after trimming. FastQC31 was used to analyze overall quality of the raw data. The median sequence length of joined reads after pre-processing was 253 bp. Operational Taxonomic Units (OTU) were built using 5 as minimum OTU cardinality using UCLUST32. The few OTUs present also in the negative controls were discarded. RDP33 was then used to perform the taxonomic profiling, using Greengenes 13.8 dataset as reference sequences34. QIIME was also used to measure alpha diversity and beta diversity. All ordination plots are based on PD-whole tree metric, if not otherwise specified. Biomarker detection was performed using LEfSe27. Oligotyping24 analysis was performed by extracting reads assigned by QIIME to each genus (Streptococcus, Lactobacillus, Haemophilus, Neisseria, Prevotella) and using 10 entropy components for Streptococcus and Lactobacillus, 5 for Neisseria, 9 for Haemophilus and 15 for Prevotella; minimum substantive abundance filter threshold was set to 50 reads in all cases.
Ethics approval and consent to participate
This study was approved by the ethical committee of the University of Trento (Reference number: 2015-014) and all participants gave their signed informed consent prior to enrollment.
Consent for publication
Not applicable because no individuals person’s data are reported in any form.
Data availability
Raw sequences were deposited in the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/sra) and are available with accession numbers SRR2093962 to SRR2094047.
Electronic supplementary material
Acknowledgements
We thank Malgorzata Kos and Rossella Tomazzolli for technical support, Olivier Jousson and Alessandro Quattrone for supporting the initiative. This work was supported by the University of Trento CIBIO teaching fund and partially supported by the European Union FP7 Marie-Curie grant (PCIG13-618833), MIUR grant FIR RBFR13EWWI, Fondazione Caritro grant Rif.Int.2013.0239, and Terme di Comano grant to NS.
Author Contributions
E.D., A.T., M.A.D. and N.S. conceived the study and designed the experiments, P.F., G.C., HTM-CMB-2015, A.T., V.D.S. and R.B. performed the experiments, E.D., P.F., and N.S. analysed the data, P.F., E.D., A.T. and N.S. wrote and revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Erik Dassi and Pamela Ferretti contributed equally to this work.
A comprehensive list of consortium members appears at the end of the paper
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28491-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nicola Segata, Email: nicola.segata@unitn.it.
HTM-CMB-2015:
Alessandra Speccher, Alice Migazzi, Bartolomeo Bosco, Bodike Rajashekar, Calogero Zarbo, Claudio Ballabio, Daniele Rossetto, Eleonora Maino, Eloina Corradi, Federica Costa, Francesca Precazzini, Harun Or Rashid, Manuel Nicolussi, Mattia Bolzan, Michele Demozzi, Michele Olivieri, Nicole Zordan, Renato Pedron, Serena Manara, Setareh Rezvan, Sindhu Narasimha Naik, Solmaz Khaghani, Stefania Masella, Thomas Perli, and Virginia Pierini
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews Immunology. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krajmalnik-Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutrition in clinical practice: official publication of the American Society for Parenteral and Enteral Nutrition. 2012;27(2):201–214. doi: 10.1177/0884533611436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiBaise JK, Frank DN, Mathur R. Impact of the Gut Microbiota on the Development of Obesity: Current Concepts. The American Journal of Gastroenterology Supplements. 2012;1(1):22–27. doi: 10.1038/ajgsup.2012.5. [DOI] [Google Scholar]
- 5.Human Microbiome Project C: Structure, function and diversity of the healthy human microbiome. Nature486(7402), 207–214 (2012) [DOI] [PMC free article] [PubMed]
- 6.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome biology. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Kolltveit KM, Tronstad L, Olsen I. Systemic diseases caused by oral infection. Clin Microbiol Rev. 2000;13(4):547–558. doi: 10.1128/CMR.13.4.547-558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wade WG. The oral microbiome in health and disease. Pharmacological research. 2013;69(1):137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Lopez A, Camelo-Castillo A, Ferrer MD, Simon-Soro A, Mira A. Health-Associated Niche Inhabitants as Oral Probiotics: The Case of Streptococcus dentisani. Front Microbiol. 2017;8:379. doi: 10.3389/fmicb.2017.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wescombe PA, Heng NC, Burton JP, Chilcott CN, Tagg JR. Streptococcal bacteriocins and the case for Streptococcus salivarius as model oral probiotics. Future microbiology. 2009;4(7):819–835. doi: 10.2217/fmb.09.61. [DOI] [PubMed] [Google Scholar]
- 11.McMahon SB, Morrison JF. Factors that determine the excitability of parasympathetic reflexes to the cat bladder. The Journal of physiology. 1982;322:35–43. doi: 10.1113/jphysiol.1982.sp014020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugahara H, et al. Probiotic Bifidobacterium longum alters gut luminal metabolism through modification of the gut microbial community. Scientific reports. 2015;5:13548. doi: 10.1038/srep13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vieira AT, et al. Oral treatment with Bifidobacterium longum 51A reduced inflammation in a murine experimental model of gout. Beneficial microbes. 2015;6(6):799–806. doi: 10.3920/BM2015.0015. [DOI] [PubMed] [Google Scholar]
- 14.Shoaib A, Dachang W, Xin Y. Determining the role of a probiotic in the restoration of intestinal microbial balance by molecular and cultural techniques. Genetics and molecular research: GMR. 2015;14(1):1526–1537. doi: 10.4238/2015.February.20.8. [DOI] [PubMed] [Google Scholar]
- 15.Simon MC, et al. Intake of Lactobacillus reuteri improves incretin and insulin secretion in glucose-tolerant humans: a proof of concept. Diabetes care. 2015;38(10):1827–1834. doi: 10.2337/dc14-2690. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, et al. 454 pyrosequencing reveals changes in the faecal microbiota of adults consuming Lactobacillus casei Zhang. FEMS microbiology ecology. 2014;88(3):612–622. doi: 10.1111/1574-6941.12328. [DOI] [PubMed] [Google Scholar]
- 17.Plaza-Diaz J, et al. Pyrosequencing analysis reveals changes in intestinal microbiota of healthy adults who received a daily dose of immunomodulatory probiotic strains. Nutrients. 2015;7(6):3999–4015. doi: 10.3390/nu7063999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fooks LJ, Gibson GR. Probiotics as modulators of the gut flora. The British journal of nutrition. 2002;88(Suppl 1):S39–49. doi: 10.1079/BJN2002628. [DOI] [PubMed] [Google Scholar]
- 19.Arora T, Singh S, Sharma RK. Probiotics: Interaction with gut microbiome and antiobesity potential. Nutrition (Burbank, Los Angeles County, Calif) 2013;29(4):591–596. doi: 10.1016/j.nut.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Nase L, et al. Effect of long-term consumption of a probiotic bacterium, Lactobacillus rhamnosus GG, in milk on dental caries and caries risk in children. Caries research. 2001;35(6):412–420. doi: 10.1159/000047484. [DOI] [PubMed] [Google Scholar]
- 21.Dassi E, et al. Enhanced microbial diversity in the saliva microbiome induced by short-term probiotic intake revealed by 16S rRNA sequencing on the IonTorrent PGM platform. Journal of biotechnology. 2014;190:30–39. doi: 10.1016/j.jbiotec.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewhirst FE, et al. The human oral microbiome. Journal of bacteriology. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(28):E2875–2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall MW, et al. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes. 2017;3:2. doi: 10.1038/s41522-016-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donati C, et al. Uncovering oral Neisseria tropism and persistence using metagenomic sequencing. Nature microbiology. 2016;1(7):16070. doi: 10.1038/nmicrobiol.2016.70. [DOI] [PubMed] [Google Scholar]
- 27.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ea-utils: Command-line tools for processing biological sequencing data https://github.com/ExpressionAnalysis/ea-utils.
- 31.FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 32.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences were deposited in the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/sra) and are available with accession numbers SRR2093962 to SRR2094047.