Abstract
Redox reactions are ubiquitous in biological processes. Enzymes involved in redox metabolism often use cofactors in order to facilitate electron-transfer reactions. Common redox cofactors include micronutrients such as vitamins and metals. By far, while iron is the main metal cofactor, riboflavin is the most important organic cofactor. Notably, the metabolism of iron and riboflavin seem to be intrinsically related across life kingdoms. In bacteria, iron availability influences expression of riboflavin biosynthetic genes. There is documented evidence for riboflavin involvement in surpassing iron-restrictive conditions in some species. This is probably achieved through increase in iron bioavailability by reduction of extracellular iron, improvement of iron uptake pathways and boosting hemolytic activity. In some cases, riboflavin may also work as replacement of iron as enzyme cofactor. In addition, riboflavin is involved in dissimilatory iron reduction during extracellular respiration by some species. The main direct metabolic relationships between riboflavin and iron in bacterial physiology are reviewed here.
Keywords: riboflavin, iron, uptake, redox, cofactor
Introduction
Redox reactions involving electron transfers among molecules are highly required in central biological processes, from CO2 fixation and oxidative phosphorylation to protein folding and cell signaling. Many of the enzymes that catalyze biological electron transfer utilize diverse vitamins and/or metals as cofactors. Riboflavin and iron are the main cofactors, each one assisting about 17% of the cofactor-requiring enzymes (Monteverde et al., 2017). Within a cell, iron can function in a number of different forms, such as being part of heme, forming iron-sulfur clusters or as mono- and di-nuclear non-heme iron in some proteins (Caza and Kronstad, 2013). Riboflavin, also known as vitamin B2, is usually transformed into riboflavin-5′-phosphate (FMN) and flavin adenine dinucleotide (FAD), which are the main flavin cofactors.
Iron is mainly present in two oxidation states, ferrous (Fe2+) and ferric (Fe3+). The interconversion between these two states enables single electron transfers (Sheldon et al., 2016). With a few exceptions, iron is essential for bacteria. Although iron is one of the most abundant elements on earth, due to the oxidative atmosphere and basic pH the insoluble Fe3+ form is predominant, mainly forming iron oxides. Moreover, under physiological conditions, the iron concentration is some orders of magnitude lower than that needed to support bacterial growth (Sheldon et al., 2016). Thus, iron bioavailability is usually limiting. Hence, bacteria have evolved an assortment of strategies to solubilize and internalize iron and to compete for iron against a host or other microorganisms. These include the expression of siderophores, which are low molecular mass, iron-chelating molecules. Siderophores have a high affinity for ferric ion and can internalize environmental iron or hijack Fe3+ bound to mammalian host proteins such as transferrin. Bacteria may also obtain iron through direct interaction of bacterial receptors with host transferrin, heme and hemoproteins and subsequent internalization and dissociation of their coordinated iron molecules. Many species can also produce iron reductases, which may be secreted or associated with the membrane, in order to reduce Fe3+ into its more soluble Fe2+ form and uptake it through ferrous ion transport systems (Sheldon et al., 2016; Butt and Thomas, 2017; Pi and Helmann, 2017a; Pokorzynski et al., 2017; Sánchez et al., 2017).
A riboflavin molecule consists of an isoalloxazine ring with a substitution at N10 with a ribityl chain. The main biological active forms of riboflavin are its phosphorylated derivative FMN and the adenylated derivative of this, FAD. Flavins may alternate between three redox forms: oxidized, one-electron reduced or two-electron reduced. This allows them to mediate both one-electron and two-electron transfer reactions as well as electron bifurcation processes, which consists in the branching of two electrons from a single donor into two different single-electron acceptors (Abbas and Sibirny, 2011; Haase et al., 2014; Beztsinna et al., 2016; Buckel and Thauer, 2018). Most bacteria synthesize riboflavin through the riboflavin biosynthetic pathway (RBP), which starts from guanosine-5′-triphosphate and ribulose-5-phosphate. The RBP has been extensively reviewed in previous works (Fischer and Bacher, 2005, 2011; Haase et al., 2014). The bacterial enzymes forming the RBP are encoded by the ribABDEH genes and may be localized within a single operon or scattered in the genome (Vitreschak et al., 2002; García-Angulo, 2017). Also, transport systems from different families may internalize riboflavin in bacteria (Vitreschak et al., 2002; Vogl et al., 2007; Rodionov et al., 2009; Hemberger et al., 2011; Deka et al., 2013; García Angulo et al., 2013; Sun et al., 2013; Gutiérrez-Preciado et al., 2015; Jaehme and Slotboom, 2015; Rodionova et al., 2015; García-Angulo, 2017). Thus, bacteria may obtain the riboflavin by de novo biosynthesis and/or by internalization from the environment.
Both iron and riboflavin may have been present in early earth and be recruited by primordial biological systems to perform biotic functions early in evolution. In the highly reductive atmosphere of the Archean, when life began, the abundant Fe2+ could have been the main electron donor for photosynthesis. Both cofactors participate in ancient biochemical pathways such as CO2 fixation, where the electron bifurcation capacity of flavins is required. (Sousa et al., 2013, 2018; Knoll et al., 2016; Monteverde et al., 2017; Sánchez et al., 2017). Thus, organisms across kingdoms have conserved a dependence on riboflavin and iron to perform basic processes, mainly based on their redox properties. Moreover, it has been documented a metabolic crosstalk between riboflavin and iron in a number of organisms including animals, plants, yeast, and bacteria (Welkie and Miller, 1960; Buzina et al., 1979; Charoenlarp et al., 1980; Powers et al., 1988; Crossley et al., 2007; Chazarreta-Cifre et al., 2011; Hsu et al., 2011; Zhang Y. et al., 2015; Chen et al., 2017; Xin et al., 2017). In many cases, these two cofactors are involved in similar processes, sometimes with enzymes requiring both cofactors at the same time. It has also been documented the replacement of enzymes on the basis of cofactor availability and the development of an iron-riboflavin regulatory feedback. This review focuses on current research on the main metabolic interrelationships between iron and riboflavin in bacteria.
Iron-Riboflavin Regulatory Interplay in Bacteria
In bacteria, Fur is the main regulator of iron homeostasis. Fur is a transcriptional regulator that upon interaction with iron, binds to consensus DNA operators named Fur boxes close to promoters of target genes to regulate transcription. Bacterial Fur regulons include many iron uptake genes. Under low iron, iron dissociates from Fur leaving it unable to interact with Fur boxes and thus allowing the expression of such iron acquisition genes. When intracellular iron levels are sufficient, Fur binds iron and represses the genes coding for transport of iron, avoiding toxicity caused by a potential surplus. Excess iron is detrimental because of generation of reactive oxygen species through the Fenton reaction. Indirectly, Fur-iron may also positively regulate the expression of iron-storage and iron-utilizing proteins as well as iron efflux systems (Massé et al., 2007; Pi et al., 2016; Pi and Helmann, 2017a). Although initially recognized as a repressor, the Fur-iron complex is also able to directly activate genes not related to iron metabolism (Craig et al., 2011). Moreover, evidence for iron-independent Fur regulation has been obtained in some bacteria (Carpenter et al., 2009; Agriesti et al., 2014).
In many cases, the expression of RBP and riboflavin uptake genes are under the control of the FMN riboswitch (Vitreschak et al., 2002; Pedrolli et al., 2012; García Angulo et al., 2013; Pedrolli and Mack, 2014; Gutiérrez-Preciado et al., 2015; Pedrolli D.B. et al., 2015; Pedrolli D. et al., 2015; García-Angulo, 2017). This is a sequence conserved in the leader regions of messenger RNA which switches between alternative secondary structures depending on FMN binding status. Under high flavins concentrations, FMN binds the aptamer sequence within the riboswitch, promoting the formation of a secondary structure that blocks transcription, translation or both, of downstream coding sequences. When intracellular flavins concentration drop, FMN dissociates from the riboswitch, which allows genetic expression. This way, the expression of both riboflavin supply pathways, biosynthesis and uptake, is keenly linked to the intracellular flavin requirements. Nonetheless, not all bacterial RBP and riboflavin transport genes conserve a FMN riboswitch and likely, other species-specific regulatory traits may exist (García-Angulo, 2017).
It has long been known that iron physiological status influences riboflavin biosynthesis. A study on a Clostridium acetobutylicum strain used for large scale production of riboflavin, showed that the presence of iron decreased riboflavin yield, while the iron chelator bipyridine highly increased the production of the vitamin (Hickey, 1945). More recently, it was shown in this species that both iron starvation and Fur deletion highly increase the transcription of the RBP operon (Vasileva et al., 2012). Iron starvation induces the secretion of riboflavin in Methylocystis sp., a methanotrophic bacterium (Balasubramanian et al., 2010). Also, both iron deprivement and elimination of Fur increase the riboflavin uptake activity in Campylobacter jejuni (Crossley et al., 2007). Riboflavin production and transcription of the bifunctional ribBA gene are highly increased in low iron in Helicobacter pylori. In this species, a putative Fur box is found in ribBA (Worst et al., 1998). Transcriptomics approaches have also confirmed that iron and Fur negatively regulate the expression of the RBP operon in C. acetobutylicum and Caulobacter crescentus (da Silva Neto et al., 2013; Nguyen et al., 2016). In the latter, a putative Fur box is upstream of the RBP operon. In C. acetobutylicum, iron uptake genes and RBP genes are co-regulated by the carbon starvation regulator CsrA (Tan et al., 2015). These studies suggest that in some bacteria, a fall in iron supply may induce an increase in riboflavin biosynthesis and/or uptake. Nonetheless, iron effects over the expression of riboflavin provision genes are not always equal. Vibrio vulnificus is a bacterium causing wound infections and septicemia whose pathogenic potential is tightly related to the iron availability. In this pathogen, iron restriction was found to upregulate ribH, but to downregulate ribE, ribA, and ribB homologs, while elimination of Fur downregulated ribA and ribB but overexpressed ribE and ribH (Pajuelo et al., 2016). In V. cholerae, a phylogenetically related pathogen, the riboflavin regulon presents a high degree of overlap with the iron regulon (Mey et al., 2005; Sepúlveda-Cisternas et al., 2018). In this bacterium, iron regulates the expression of the RBP genes and of the ribN riboflavin importer in a riboflavin-dependent way. This regulation is gene-specific, as while iron repressed ribB in the presence of extracellular riboflavin, it induced the expression of ribD and ribN in the absence of exogenous riboflavin. Reciprocally, riboflavin repressed the expression of the tonB1 gene encoding a protein involved in the function of various iron uptake systems, but only under iron-replete conditions (Sepúlveda-Cisternas et al., 2018). Thus, iron levels may regulate the status of riboflavin provision in a gene-specific fashion and reciprocally, riboflavin exerts regulatory effects over iron acquisition genes.
Riboflavin Production as a Mean to Increase Iron Availability
Increases in riboflavin biosynthesis may help overcome iron restrictive conditions. H. pylori ribBA bifunctional gene has been shown to restore growth in low iron to a siderophore-deficient Escherichia coli strain. The mechanism through which riboflavin biosynthesis relieves iron stress in this species has not been fully elucidated. It was shown that the riboflavin production mediated by H. pylori RibBA increases extracellular iron reduction (Worst et al., 1998). Conversion of exogenous Fe3+ into Fe2+ increases iron solubility and may make it suitable for uptake through ferrous ion internalization systems. In C. jejuni, riboflavin biosynthesis also increases reduction of extracellular Fe3+ through an unknown mechanism (Crossley et al., 2007) (Figure 1A). In addition, both the bifunctional RibBA and a monofunctional RibA protein of H. pylori are able to confer hemolysis properties to E. coli. (Bereswill et al., 1998; Worst et al., 1998). Likely, hemolysis itself may be a strategy to increase iron bioavailability by releasing the iron contained in erythrocytes in an infection setting. Nonetheless, the mechanism through which riboflavin biosynthesis boosts hemolysis is not clear. Riboflavin has been shown to direct light-mediated rat and human erythrocytes hemolysis but only in the presence of other factors such as serum, oxygen, copper, azide, and aminophylline (Suzuki et al., 1982; Ali and Naseem, 2002; Ali et al., 2005). Moreover, riboflavin alone does not directly promote hemolysis in agar plates were H. pylori experiments were conducted (Bereswill et al., 1998). Thus, hemolysis may be given by augmented riboflavin biosynthesis intermediaries or increases in the expression and/or activity of a hemolysis factor (Figure 1A).
FIGURE 1.
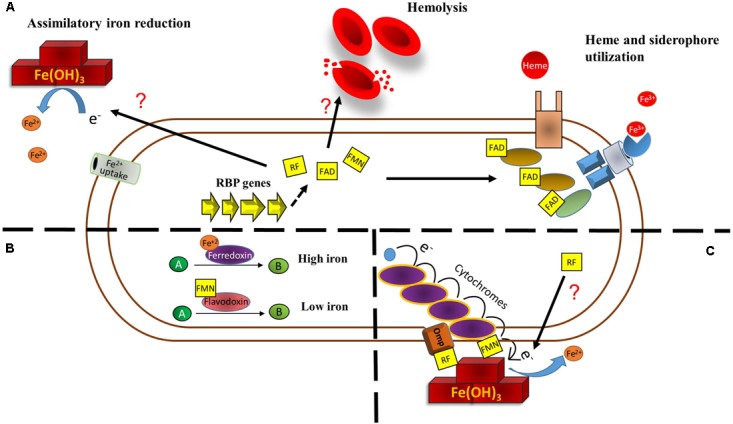
Main direct metabolic relationships among riboflavin and iron in bacteria. (A) Riboflavin may help overcome iron restriction status in three principal ways. Riboflavin increases extracellular iron reduction by a yet uncharacterized mechanism. This increases iron solubility and could make it prone to uptake by ferrous iron uptake systems. Increases in bacterial riboflavin expression produce a higher degree of hemolysis, which can increase hemoglobin availability. Also, flavins function as cofactor for enzymes involved in heme utilization and iron release from siderophores. (B) Under iron restriction, enzymes using iron as redox cofactor to catalyze a reaction may be replaced by enzymes that use flavins as cofactor. (C) Riboflavin increases dissimilatory ferric reduction. In S. oneidensis, extracellular ferric iron is used as final electron acceptor to complete the electron transport chain from quinol. This is achieved by a set of cytochromes and an outer membrane protein with the aid of flavins. In other bacteria, the mechanism has not been characterized.
Riboflavin is also required as cofactor for enzymes involved in iron acquisition. In Staphylococcus aureus, FAD functions as cofactor for IruO, a reductase of IsdG and IsdI, which are enzymes required for the degradation of heme. IruO is also directly required for iron reduction during its release from siderophores (Kobylarz et al., 2017). In addition, two more other bacterial reductases required for iron release from siderophores depend upon FAD as cofactor, YqjH in E. coli (Bamford et al., 2008) and FscN in Thermobifida fusca (Li et al., 2015). Moreover, dihydroflavin produced by the NADH:flavin oxidoreductase FerA acts as the electron donor during reductive release of Fe3+ bound to siderophore in Paracoccus denitrificans. This process is necessary for this species to grow under iron limitation (Mazoch et al., 2004; Sedlácek et al., 2009; Sedláček et al., 2016). Thus, by increasing riboflavin production, bacteria may increase the activity of some components of iron acquisition pathways (Figure 1A).
Riboflavin to Replace Iron as Cofactor
In many bacteria, iron restriction highly induces the expression of iron uptake systems and also triggers a response known as iron-sparing. This consists in the downregulation of non-essential iron-dependent enzymes in order to prioritize iron for fundamental functions. For example, iron-starving Mycobacterium tuberculosis and Bacillus subtilis downregulate aconitase and succinate dehydrogenase (Janagama et al., 2010; Smaldone et al., 2012), while Agrobacterium tumefaciens downregulates a system encoding a multicopper oxidase, cytochrome c556 and a NAD-dependent formate dehydrogenase complex, all of them needing iron as cofactor (Heindl et al., 2015). Strikingly, the iron-sparing response may also include the replacement of iron-cofactored enzymes by flavin-dependent enzymes. Early work showed that ferredoxin and flavodoxin alternate to catalyze a dehydration reaction depending on the iron status during fermentation of glutamate in Acidaminococcus fermentans (Thamer et al., 2003). In C. acetobutylicum, the expression of ferredoxin is decreased and a flavodoxin encoded by fdl1 is highly induced under iron deficiency (Vasileva et al., 2012; Nguyen et al., 2016). The fdl1 gene conserves a Fur box. In these conditions the riboflavin biosynthetic operon is also induced. In B. subtilis, the Fur-regulated flavodoxins YkuNOP are expressed under iron deprivement also likely to replace ferredoxin (Baichoo et al., 2002). Notably, in this species the YkuN and YkuP flavodoxins, which use FMN as prosthetic group, have been experimentally shown to substitute ferredoxin to transfer electrons for desaturation of membrane phospholipids (Chazarreta-Cifre et al., 2011) (Figure 1B). A recent study demonstrated that the B. subtilis Fur regulon is induced in three sequential steps, the first one including the expression of iron-uptake systems, while expression of flavodoxins putatively to replace ferredoxin is included in the second wave. The third stage is comprised by the downregulation of superfluous iron-dependent enzymes (Pi and Helmann, 2017b).
Role of Riboflavin in Dissimilatory Iron Reduction
Under anoxic conditions, some bacteria are able to reduce extracellular insoluble metals as final electron acceptors to complete the respiratory chain. In this case, riboflavin may also work as intermediary for dissimilatory iron reduction and this phenomenon has been extensively studied in Shewanella oneidensis, a facultative anaerobe model for electrochemically active microbes (Kouzuma et al., 2018). S. oneidensis is able to reduce environmental Fe3+ and other metal oxides due to a system to transport electrons from the inner membrane across the periplasm and outer membrane to the metal surface. This pathway, known as the metal reducing (Mtr) pathway, consists of the multiheme domain c type cytrochromes CymA, MtrA, MtrC, and OmcA and the outer membrane porin MtrB. The inner-membrane CymA is proposed to transfer electrons from quinol to MtrA, which is localized in the outer membrane and interacting with MtrB. The MtrAB complex conveys the electrons to the MtrC and OmcA c cytrochromes in the bacterial surface which in turn act as reductases to transfer electrons to oxides (Brutinel and Gralnick, 2012; Shi et al., 2012; Breuer et al., 2015). Although MtrC and OmcA are able to directly transfer electrons to Fe3+ oxides, secreted riboflavin and FMN highly enhance this activity (Marsili et al., 2008; von Canstein et al., 2008). In this species, flavins are secreted through the Bfe exporter and flavin-mediated reduction accounts for most (up to 75%) of extracellular electron transfer to insoluble acceptors (Kotloski and Gralnick, 2013). Although originally a mechanism where freely diffusing flavins working as electron shuttles for Fe3+ reduction was proposed, later research documented that flavins directly interact with MtrC and OmcA to transfer electrons (Okamoto et al., 2013, 2014; Hong and Pachter, 2016; Babanova et al., 2017). While riboflavin binds OmcA (Hong and Pachter, 2016), FMN binds MtrC to form a highly reductive flavocytochrome (Edwards et al., 2015) (Figure 1C). The presence of Fe3+ in the medium increases the secretion of riboflavin and FMN (Wu et al., 2013).
Extracellular riboflavin as intermediary for dissimilatory iron reduction has been reported in other bacteria species. Geothrix fermentans, an isolate of an aquifer contaminated with petroleum, uses secreted riboflavin to shuttle electrons toward environmental Fe3+ (Mehta-Kolte and Bond, 2012). Carbohydrate oxidation is increased when using riboflavin as electron transfer mediator in the presence of crystalline Fe(OH)3 as extracellular electron sink in Clostridium beijerinckii and an uncharacterized novel rhizobial solventogenic bacterium (Popovic et al., 2017). Also, riboflavin is likely involved in the reduction of extracellular Fe3+-citrate and solid phase hydrous ferric oxide by Desulfotomaculum reducens, a sulfate-reducing Gram-positive species (Dalla Vecchia et al., 2014) and in anaerobic Fe3+ reduction by an alkaliphilic bacterial consortium (Fuller et al., 2014). Nonetheless, the mechanism through which these species achieve extracellular iron reduction with the aid of flavins has not been investigated (Figure 1C). Notably, flavins may also mediate electron transfer in the opposite direction. Riboflavin and FAD increase extracellular iron oxidation of stainless steel, a process that provides the electrons required for intracellular sulfur reduction in Desulfovibrio vulgaris (Zhang P. et al., 2015).
Concluding Remarks
Many bacteria have developed a regulatory interplay on which iron levels influence the expression of riboflavin biosynthesis and uptake systems. This is probably the reflex of the fact that there is a number of physiological traits on which riboflavin and iron metabolism directly relate. First, riboflavin could help surpass eventual iron restriction conditions. Many species have complex life cycles, being able to colonize different niches with fluctuating physical conditions and nutrient availability. Thus, iron availability may change drastically. Riboflavin increases iron bioavailability and improves iron acquisition pathways. The ability of riboflavin to transfer electrons is used by some bacteria to reduce extracellular ferric oxides making it more suitable for uptake. Flavins are used as cofactor for some proteins directly involved in iron uptake, so increasing riboflavin concentration may also make the process more active. Also, some species may economize iron by replacing iron-requiring enzymes with enzymes that perform similar functions but use riboflavin as cofactor instead. This comprises a remarkable outline of the common redox properties of both molecules. Second, riboflavin participates in dissimilatory iron reduction. Riboflavin-mediated extracellular iron reduction is employed by some species to accomplish the respiratory electron chain. Altogether, this high degree of metabolic cross-talk may be reminiscent of an early recruitment of both molecules to perform related functions during biological redox reactions.
Author Contributions
IS contributed to the general draft and designed the Figure 1. JS wrote the part corresponding to Fur regulation. VG-A wrote the main text. All authors discussed the whole review and agreed with the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was funded by CONICYT/Fondecyt (Chile) Grant Number 1150818.
References
- Abbas C. A., Sibirny A. A. (2011). Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 75 321–360. 10.1128/MMBR.00030-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agriesti F., Roncarati D., Musiani F., Del Campo C., Iurlaro M., Sparla F., et al. (2014). FeON-FeOFF: the Helicobacter pylori Fur regulator commutates iron-responsive transcription by discriminative readout of opposed DNA grooves. Nucleic Acids Res. 42 3138–3151. 10.1093/nar/gkt1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I., Sakhnini N., Naseem I. (2005). Hemolysis of human red blood cells by riboflavin-Cu(II) system: enhancement by azide. Biochemistry 70 1011–1014. 10.1007/s10541-005-0217-x [DOI] [PubMed] [Google Scholar]
- Ali L., Naseem I. (2002). Hemolysis of human red blood cells by combination of riboflavin and aminophylline. Life Sci. 70 2013–2022. 10.1016/S0024-3205(01)01540-5 [DOI] [PubMed] [Google Scholar]
- Babanova S., Matanovic I., Cornejo J., Bretschger O., Nealson K., Atanassov P. (2017). Outer membrane cytochromes/flavin interactions in Shewanella spp.-A molecular perspective. Biointerphases 12:021004. 10.1116/1.4984007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N., Wang T., Ye R., Helmann J. D. (2002). Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45 1613–1629. 10.1046/j.1365-2958.2002.03113.x [DOI] [PubMed] [Google Scholar]
- Balasubramanian R., Levinson B. T., Rosenzweig A. C. (2010). Secretion of flavins by three species of methanotrophic bacteria. Appl. Environ. Microbiol. 76 7356–7358. 10.1128/AEM.00935-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford V. A., Armour M., Mitchell S. A., Cartron M., Andrews S. C., Watson K. A. (2008). Preliminary X-ray diffraction analysis of YqjH from Escherichia coli: a putative cytoplasmic ferri-siderophore reductase. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 64 792–796. 10.1107/S174430910802352X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereswill S., Fassbinder F., Völzing C., Covacci A., Haas R., Kist M. (1998). Hemolytic properties and riboflavin synthesis of Helicobacter pylori: cloning and functional characterization of the ribA gene encoding GTP-cyclohydrolase II that confers hemolytic activity to Escherichia coli. Med. Microbiol. Immunol. 186 177–187. 10.1007/s004300050062 [DOI] [PubMed] [Google Scholar]
- Beztsinna N., Solé M., Taib N., Bestel I. (2016). Bioengineered riboflavin in nanotechnology. Biomaterials 80 121–133. 10.1016/j.biomaterials.2015.11.050 [DOI] [PubMed] [Google Scholar]
- Breuer M., Rosso K. M., Blumberger J., Butt J. N. (2015). Multi-haem cytochromes in Shewanella oneidensis MR-1: structures, functions and opportunities. J. R. Soc. Interface 12:20141117. 10.1098/rsif.2014.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutinel E. D., Gralnick J. A. (2012). Shuttling happens: soluble flavin mediators of extracellular electron transfer in Shewanella. Appl. Microbiol. Biotechnol. 93 41–48. 10.1007/s00253-011-3653-0 [DOI] [PubMed] [Google Scholar]
- Buckel W., Thauer R. K. (2018). Flavin-based electron bifurcation. A new mechanism of biological energy coupling. Chem. Rev. 118 3862–3886. 10.1021/acs.chemrev.7b00707 [DOI] [PubMed] [Google Scholar]
- Butt A. T., Thomas M. S. (2017). Iron acquisition mechanisms and their role in the virulence of Burkholderia Species. Front. Cell. Infect. Microbiol. 7:460. 10.3389/fcimb.2017.00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzina R., Jusić M., Milanović N., Sapunar J., Brubacher G. (1979). The effects of riboflavin administration on iron metabolism parameters in a school-going population. Int. J. Vitam. Nutr. Res. 49 136–143. [PubMed] [Google Scholar]
- Carpenter B. M., Whitmire J. M., Merrell D. S. (2009). This is not your mother’s repressor: the complex role of fur in pathogenesis. Infect. Immun. 77 2590–2601. 10.1128/IAI.00116-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M., Kronstad J. W. (2013). Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 3:80. 10.3389/fcimb.2013.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoenlarp P., Pholpothi T., Chatpunyaporn P., Schelp F. P. (1980). The effect of riboflavin on the hematologic changes in iron supplementation of schoolchildren. Southeast Asian J. Trop. Med. Public Health 11 97–103. [PubMed] [Google Scholar]
- Chazarreta-Cifre L., Martiarena L., de Mendoza D., Altabe S. G. (2011). Role of ferredoxin and flavodoxins in Bacillus subtilis fatty acid desaturation. J. Bacteriol. 193 4043–4048. 10.1128/JB.05103-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-T., Wang Y., Yeh K.-C. (2017). Role of root exudates in metal acquisition and tolerance. Curr. Opin. Plant Biol. 39 66–72. 10.1016/j.pbi.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Craig S. A., Carpenter C. D., Mey A. R., Wyckoff E. E., Payne S. M. (2011). Positive regulation of the Vibrio cholerae porin OmpT by iron and fur. J. Bacteriol. 193 6505–6511. 10.1128/JB.05681-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley R. A., Gaskin D. J. H., Holmes K., Mulholland F., Wells J. M., Kelly D. J., et al. (2007). Riboflavin biosynthesis is associated with assimilatory ferric reduction and iron acquisition by Campylobacter jejuni. Appl. Environ. Microbiol. 73 7819–7825. 10.1128/AEM.01919-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Neto J. F., Lourenço R. F., Marques M. V. (2013). Global transcriptional response of Caulobacter crescentus to iron availability. BMC Genomics 14:549. 10.1186/1471-2164-14-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Vecchia E., Suvorova E. I., Maillard J., Bernier-Latmani R. (2014). Fe(III) reduction during pyruvate fermentation by Desulfotomaculum reducens strain MI-1. Geobiology 12 48–61. 10.1111/gbi.12067 [DOI] [PubMed] [Google Scholar]
- Deka R. K., Brautigam C. A., Biddy B. A., Liu W. Z., Norgard M. V. (2013). Evidence for an ABC-type riboflavin transporter system in pathogenic spirochetes. mBio 4:e00615-12. 10.1128/mBio.00615-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M. J., White G. F., Norman M., Tome-Fernandez A., Ainsworth E., Shi L., et al. (2015). Redox linked flavin sites in extracellular decaheme proteins involved in microbe-mineral electron transfer. Sci. Rep. 5:11677. 10.1038/srep11677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Bacher A. (2005). Biosynthesis of flavocoenzymes. Nat. Prod. Rep. 22 324–350. 10.1039/b210142b [DOI] [PubMed] [Google Scholar]
- Fischer M., Bacher A. (2011). Biosynthesis of vitamin B2: a unique way to assemble a xylene ring. Chembiochem 12 670–680. 10.1002/cbic.201000681 [DOI] [PubMed] [Google Scholar]
- Fuller S. J., McMillan D. G. G., Renz M. B., Schmidt M., Burke I. T., Stewart D. I. (2014). Extracellular electron transport-mediated Fe(III) reduction by a community of alkaliphilic bacteria that use flavins as electron shuttles. Appl. Environ. Microbiol. 80 128–137. 10.1128/AEM.02282-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Angulo V. A., Bonomi H. R., Posadas D. M., Serer M. I., Torres A. G., Zorreguieta Áet al. (2013). Identification and characterization of RibN, a novel family of riboflavin transporters from Rhizobium leguminosarum and other proteobacteria. J. Bacteriol. 195 4611–4619. 10.1128/JB.00644-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Angulo V. A. (2017). Overlapping riboflavin supply pathways in bacteria. Crit. Rev. Microbiol. 43 196–209. 10.1080/1040841X.2016.1192578 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Preciado A., Torres A. G., Merino E., Bonomi H. R., Goldbaum F. A., García-Angulo V. A. (2015). Extensive identification of bacterial riboflavin transporters and their distribution across bacterial species. PLoS One 10:e0126124. 10.1371/journal.pone.0126124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase I., Gräwert T., Illarionov B., Bacher A., Fischer M. (2014). Recent advances in riboflavin biosynthesis. Methods Mol. Biol. 1146 15–40. 10.1007/978-1-4939-0452-5_2 [DOI] [PubMed] [Google Scholar]
- Heindl J. E., Hibbing M. E., Xu J., Natarajan R., Buechlein A. M., Fuqua C. (2015). Discrete responses to limitation for iron and manganese in Agrobacterium tumefaciens: influence on attachment and biofilm formation. J. Bacteriol. 198 816–829. 10.1128/JB.00668-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger S., Pedrolli D. B., Stolz J., Vogl C., Lehmann M., Mack M. (2011). RibM from Streptomyces davawensis is a riboflavin/roseoflavin transporter and may be useful for the optimization of riboflavin production strains. BMC Biotechnol. 11:119. 10.1186/1472-6750-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey R. J. (1945). The inactivation of iron by 2,2’-bipyridine and its effect on riboflavin synthesis by Clostridium acetobutylicum. Arch. Biochem. 8 439–447. [PubMed] [Google Scholar]
- Hong G., Pachter R. (2016). Bound flavin-cytochrome model of extracellular electron transfer in Shewanella oneidensis: analysis by free energy molecular dynamics simulations. J. Phys. Chem. B 120 5617–5624. 10.1021/acs.jpcb.6b03851 [DOI] [PubMed] [Google Scholar]
- Hsu P.-C., Yang C.-Y., Lan C.-Y. (2011). Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot. Cell 10 207–225. 10.1128/EC.00158-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehme M., Slotboom D. J. (2015). Diversity of membrane transport proteins for vitamins in bacteria and archaea. Biochim. Biophys. Acta 1850 565–576. 10.1016/j.bbagen.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Janagama H. K., Senthilkumar, Bannantine J. P., Kugadas A., Jagtap P., Higgins L.,et al. (2010). Iron-sparing response of Mycobacterium avium subsp. paratuberculosis is strain dependent. BMC Microbiol. 10:268. 10.1186/1471-2180-10-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A. H., Bergmann K. D., Strauss J. V. (2016). Life: the first two billion years. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150493. 10.1098/rstb.2015.0493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobylarz M. J., Heieis G. A., Loutet S. A., Murphy M. E. P. (2017). Iron uptake oxidoreductase (IruO) uses a flavin adenine dinucleotide semiquinone intermediate for iron-siderophore reduction. ACS Chem. Biol. 12 1778–1786. 10.1021/acschembio.7b00203 [DOI] [PubMed] [Google Scholar]
- Kotloski N. J., Gralnick J. A. (2013). Flavin electron shuttles dominate extracellular electron transfer by Shewanella oneidensis. mBio 4:e00553-12. 10.1128/mBio.00553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzuma A., Ishii S., Watanabe K. (2018). Metagenomic insights into the ecology and physiology of microbes in bioelectrochemical systems. Bioresour. Technol. 255 302–307. 10.1016/j.biortech.2018.01.125 [DOI] [PubMed] [Google Scholar]
- Li K., Chen W.-H., Bruner S. D. (2015). Structure and mechanism of the siderophore-interacting protein from the fuscachelin gene cluster of Thermobifida fusca. Biochemistry 54 3989–4000. 10.1021/acs.biochem.5b00354 [DOI] [PubMed] [Google Scholar]
- Marsili E., Baron D. B., Shikhare I. D., Coursolle D., Gralnick J. A., Bond D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105 3968–3973. 10.1073/pnas.0710525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Salvail H., Desnoyers G., Arguin M. (2007). Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10 140–145. 10.1016/j.mib.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Mazoch J., Tesarík R., Sedlácek V., Kucera I., Turánek J. (2004). Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans. Eur. J. Biochem. 271 553–562. 10.1046/j.1432-1033.2003.03957.x [DOI] [PubMed] [Google Scholar]
- Mehta-Kolte M. G., Bond D. R. (2012). Geothrix fermentans secretes two different redox-active compounds to utilize electron acceptors across a wide range of redox potentials. Appl. Environ. Microbiol. 78 6987–6995. 10.1128/AEM.01460-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey A. R., Wyckoff E. E., Kanukurthy V., Fisher C. R., Payne S. M. (2005). Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect. Immun. 73 8167–8178. 10.1128/IAI.73.12.8167-8178.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteverde D. R., Gómez-Consarnau L., Suffridge C., Sañudo-Wilhelmy S. A. (2017). Life’s utilization of B vitamins on early Earth. Geobiology 15 3–18. 10.1111/gbi.12202 [DOI] [PubMed] [Google Scholar]
- Nguyen N.-P.-T., Linder S., Flitsch S. K., Schiel-Bengelsdorf B., Dürre P., Soucaille P. (2016). Cap0037, a novel global regulator of Clostridium acetobutylicum metabolism. mBio 7:e01218-16. 10.1128/mBio.01218-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto A., Hashimoto K., Nealson K. H., Nakamura R. (2013). Rate enhancement of bacterial extracellular electron transport involves bound flavin semiquinones. Proc. Natl. Acad. Sci. U.S.A. 110 7856–7861. 10.1073/pnas.1220823110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto A., Kalathil S., Deng X., Hashimoto K., Nakamura R., Nealson K. H. (2014). Cell-secreted flavins bound to membrane cytochromes dictate electron transfer reactions to surfaces with diverse charge and pH. Sci. Rep. 4:5628. 10.1038/srep05628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajuelo D., Hernández-Cabanyero C., Sanjuan E., Lee C.-T., Silva-Hernández F. X., Hor L.-I., et al. (2016). Iron and Fur in the life cycle of the zoonotic pathogen Vibrio vulnificus. Environ. Microbiol. 18 4005–4022. 10.1111/1462-2920.13424 [DOI] [PubMed] [Google Scholar]
- Pedrolli D., Langer S., Hobl B., Schwarz J., Hashimoto M., Mack M. (2015). The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J. 282 3230–3242. 10.1111/febs.13226 [DOI] [PubMed] [Google Scholar]
- Pedrolli D. B., Kühm C., Sévin D. C., Vockenhuber M. P., Sauer U., Suess B., et al. (2015). A dual control mechanism synchronizes riboflavin and sulphur metabolism in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 112 14054–14059. 10.1073/pnas.1515024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrolli D. B., Mack M. (2014). Bacterial flavin mononucleotide riboswitches as targets for flavin analogs. Methods Mol. Biol. 1103 165–176. 10.1007/978-1-62703-730-3_13 [DOI] [PubMed] [Google Scholar]
- Pedrolli D. B., Matern A., Wang J., Ester M., Siedler K., Breaker R., et al. (2012). A highly specialized flavin mononucleotide riboswitch responds differently to similar ligands and confers roseoflavin resistance to Streptomyces davawensis. Nucleic Acids Res. 40 8662–8673. 10.1093/nar/gks616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H., Helmann J. D. (2017a). Ferrous iron efflux systems in bacteria. Metallomics 9 840–851. 10.1039/c7mt00112f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H., Helmann J. D. (2017b). Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 114 12785–12790. 10.1073/pnas.1713008114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H., Patel S. J., Argüello J. M., Helmann J. D. (2016). The Listeria monocytogenes Fur-regulated virulence protein FrvA is an Fe(II) efflux P1B4-type ATPase. Mol. Microbiol. 100 1066–1079. 10.1111/mmi.13368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorzynski N. D., Thompson C. C., Carabeo R. A. (2017). Ironing out the unconventional mechanisms of iron acquisition and gene regulation in chlamydia. Front. Cell. Infect. Microbiol. 7:394. 10.3389/fcimb.2017.00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic J., Ye X., Haluska A., Finneran K. T. (2017). Ferric iron and extracellular electron shuttling increase xylose utilization and butanol production during fermentation with multiple solventogenic bacteria. Appl. Microbiol. Biotechnol. 101 8053–8061. 10.1007/s00253-017-8533-9 [DOI] [PubMed] [Google Scholar]
- Powers H. J., Wright A. J., Fairweather-Tait S. J. (1988). The effect of riboflavin deficiency in rats on the absorption and distribution of iron. Br. J. Nutr. 59 381–387. 10.1079/BJN19880047 [DOI] [PubMed] [Google Scholar]
- Rodionov D. A., Hebbeln P., Eudes A., ter Beek J., Rodionova I. A., Erkens G. B., et al. (2009). A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 191 42–51. 10.1128/JB.01208-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionova I. A., Li X., Plymale A. E., Motamedchaboki K., Konopka A. E., Romine M. F., et al. (2015). Genomic distribution of B-vitamin auxotrophy and uptake transporters in environmental bacteria from the Chloroflexi phylum. Environ. Microbiol. Rep. 7 204–210. 10.1111/1758-2229.12227 [DOI] [PubMed] [Google Scholar]
- Sánchez M., Sabio L., Gálvez N., Capdevila M., Dominguez-Vera J. M. (2017). Iron chemistry at the service of life. IUBMB Life 69 382–388. 10.1002/iub.1602 [DOI] [PubMed] [Google Scholar]
- Sedláček V., Klumpler T., Marek J., Kučera I. (2016). Biochemical properties and crystal structure of the flavin reductase FerA from Paracoccus denitrificans. Microbiol. Res. 18 9–22. 10.1016/j.micres.2016.04.006 [DOI] [PubMed] [Google Scholar]
- Sedlácek V., van Spanning R. J. M., Kucera I. (2009). Ferric reductase A is essential for effective iron acquisition in Paracoccus denitrificans. Microbiol. Read. Engl. 155 1294–1301. 10.1099/mic.0.022715-0 [DOI] [PubMed] [Google Scholar]
- Sepúlveda-Cisternas I., Lozano Aguirre L., Fuentes Flores A., Vásquez Solis de Ovando I., García-Angulo V. A. (2018). Transcriptomics reveals a cross-modulatory effect between riboflavin and iron and outlines responses to riboflavin biosynthesis and uptake in Vibrio cholerae. Sci. Rep. 8:3149. 10.1038/s41598-018-21302-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon J. R., Laakso H. A., Heinrichs D. E. (2016). Iron acquisition strategies of bacterial pathogens. Microbiol. Spectr. 4:VMBF-0010-2015. 10.1128/microbiolspec.VMBF-0010-2015 [DOI] [PubMed] [Google Scholar]
- Shi L., Rosso K. M., Clarke T. A., Richardson D. J., Zachara J. M., Fredrickson J. K. (2012). Molecular underpinnings of Fe(III) oxide reduction by Shewanella oneidensis MR-1. Front. Microbiol. 3:50. 10.3389/fmicb.2012.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaldone G. T., Revelles O., Gaballa A., Sauer U., Antelmann H., Helmann J. D. (2012). A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. J. Bacteriol. 194 2594–2605. 10.1128/JB.05990-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa F. L., Preiner M., Martin W. F. (2018). Native metals, electron bifurcation, and CO2 reduction in early biochemical evolution. Curr. Opin. Microbiol. 43 77–83. 10.1016/j.mib.2017.12.010 [DOI] [PubMed] [Google Scholar]
- Sousa F. L., Thiergart T., Landan G., Nelson-Sathi S., Pereira I. A. C., Allen J. F., et al. (2013). Early bioenergetic evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20130088. 10.1098/rstb.2013.0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun E. I., Leyn S. A., Kazanov M. D., Saier M. H., Novichkov P. S., Rodionov D. A. (2013). Comparative genomics of metabolic capacities of regulons controlled by cis-regulatory RNA motifs in bacteria. BMC Genomics 14:597. 10.1186/1471-2164-14-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Miura T., Ogiso T. (1982). Riboflavin photosensitized hemolysis of rat erythrocytes in the presence of serum. J. Pharmacobiodyn. 5 568–575. 10.1248/bpb1978.5.568 [DOI] [PubMed] [Google Scholar]
- Tan Y., Liu Z.-Y., Liu Z., Zheng H.-J., Li F.-L. (2015). Comparative transcriptome analysis between csrA-disruption Clostridium acetobutylicum and its parent strain. Mol. Biosyst. 11 1434–1442. 10.1039/c4mb00600c [DOI] [PubMed] [Google Scholar]
- Thamer W., Cirpus I., Hans M., Pierik A. J., Selmer T., Bill E., et al. (2003). A two [4Fe-4S]-cluster-containing ferredoxin as an alternative electron donor for 2-hydroxyglutaryl-CoA dehydratase from Acidaminococcus fermentans. Arch. Microbiol. 179 197–204. 10.1007/s00203-003-0517-8 [DOI] [PubMed] [Google Scholar]
- Vasileva D., Janssen H., Hönicke D., Ehrenreich A., Bahl H. (2012). Effect of iron limitation and fur gene inactivation on the transcriptional profile of the strict anaerobe Clostridium acetobutylicum. Microbiol. Read. Engl. 158 1918–1929. 10.1099/mic.0.056978-0 [DOI] [PubMed] [Google Scholar]
- Vitreschak A. G., Rodionov D. A., Mironov A. A., Gelfand M. S. (2002). Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30 3141–3151. 10.1093/nar/gkf433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl C., Grill S., Schilling O., Stülke J., Mack M., Stolz J. (2007). Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J. Bacteriol. 189 7367–7375. 10.1128/JB.00590-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Canstein H., Ogawa J., Shimizu S., Lloyd J. R. (2008). Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74 615–623. 10.1128/AEM.01387-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welkie G. W., Miller G. W. (1960). Iron nutrition of Nicotiana tabacum L. in relation to riboflavin, riboflavin-5-phosphate, and flavin adenine dinucleotide content. Plant Physiol. 35 516–520. 10.1104/pp.35.4.516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worst D. J., Gerrits M. M., Vandenbroucke-Grauls C. M., Kusters J. G. (1998). Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J. Bacteriol. 180 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Cheng Y.-Y., Li B.-B., Li W.-W., Li D.-B., Yu H.-Q. (2013). Electron acceptor dependence of electron shuttle secretion and extracellular electron transfer by Shewanella oneidensis MR-1. Bioresour. Technol. 136 711–714. 10.1016/j.biortech.2013.02.072 [DOI] [PubMed] [Google Scholar]
- Xin Z., Pu L., Gao W., Wang Y., Wei J., Shi T., et al. (2017). Riboflavin deficiency induces a significant change in proteomic profiles in HepG2 cells. Sci. Rep. 7:45861. 10.1038/srep45861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Xu D., Li Y., Yang K., Gu T. (2015). Electron mediators accelerate the microbiologically influenced corrosion of 304 stainless steel by the Desulfovibrio vulgaris biofilm. Bioelectrochemistry 101 14–21. 10.1016/j.bioelechem.2014.06.010 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Dai Z., Krivoruchko A., Chen Y., Siewers V., Nielsen J. (2015). Functional pyruvate formate lyase pathway expressed with two different electron donors in Saccharomyces cerevisiae at aerobic growth. FEMS Yeast Res. 15:fov024. 10.1093/femsyr/fov024 [DOI] [PubMed] [Google Scholar]


