Graphical abstract
Keywords: Rhizobia, Genista microcephala, Argyrolobium uniflorum, SDS-PAGE, Gammaproteobacteria, Heavy metal tolerance
Abstract
A phenotypic characterization of thirteen root nodule bacteria recovered from wild legumes (Genista microcephala and Argyrolobium uniflorum) growing in arid eco-climate zones (Northeastern Algeria) was conducted using analysis of sixty-six phenotypic traits (carbohydrate and nitrogen assimilation, vitamin requirements, growth temperature, salinity/pH tolerance and enzyme production). Furthermore, SDS-PAGE profiles of total cell protein, antibiotic susceptibility and heavy metal resistance were performed. The results showed that the isolates can grow at pH 4 to 10, salt concentration (0–5%) and temperature up to 45 °C. The rhizobia associated with Genista microcephala and Argyrolobium uniflorum were able to produce different hydrolytic enzymes including cellulose, pectinase and urease, with remarkable tolerance to toxic metals such as zinc, lead, copper, and mercury. Numerical analysis of the phenotypic characteristics revealed that the rhizobial isolates formed four main distinct groups showing high levels of similarity with Gammaproteobacteria. The salt tolerant and heavy metals resistance patterns found among the indigenous rhizobial strains are reflecting the environmental stresses pressure and make the strains good candidates for plant successful inoculation in arid areas.
Introduction
Arid lands represent nearly 85% of the total area in Algeria. They are characterized by high temperature, erratic rainfall, low relative humidity and productive soil, and large seasonal and annual variations [1]. The naturally growing leguminous plants living in such regions are subject to severe environmental conditions, leading to a disturbance of plant–microbe symbioses, which are a critical ecological factor in helping further plant cultivation in degraded lands [2], [3].
Root nodule bacteria, collectively called rhizobia, are soil bacteria that can establish a nitrogen-fixing symbiosis with various naturally growing trees and herbs, in cultivated and non-cultivated lands, including members of the leguminous plants Leguminosae native to arid regions [4]. Generally, rhizobia species have been classified into six genera, all belonging to α-proteobacteria, the fast and moderately fast-growing genera Rhizobium, Allorhizobium, Mesorhizobium and Ensifer (formerly Sinorhizobium); the slow-growing genus Bradyrhizobium; and the genus Azorhizobium [5], [6], which so far comprise approximately 100 defined species [7]. However, other non-classical rhizobia have been reported belonging to the β-proteobacteria [8], and γ-proteobacteria [9] although their nodulating ability was not clearly demonstrated.
The nitrogen-fixing leguminous plants are key components of the natural succession in arid Mediterranean ecosystems, upon establishing rhizobial and mycorrhizal symbioses, which constitute a fundamental source of nitrogen input to the ecosystem [10]. These symbioses increase soil fertility and quality and enhance the establishment of key plant species [3]. Compared with the nitrogen-fixing heterotrophs and associative bacteria, rhizobia-legume symbioses represent the major mechanism of biological nitrogen fixation in arid lands [10]. Therefore, their potential environmental and biotechnological applications have received much interest [11], [12].
Significant bioclimatic belts in different regions of the Mediterranean Basin give rise to very diverse forms of vegetation and present an extraordinary wealth of over 500 endemic pastoral species [13]. In Algeria, pastoral and forage systems are remarkably diverse, and the endemism is important in Fabaceae and Poaceae [14], [15]. The tribe Genisteae (Family Fabaceae) contains approximately 140 shrubby plant species [16], mainly distributed in the Mediterranean region. Genista microcephala (Coss. & Durieu) and Argyrolobium uniflorum ((Decne.) Jaub. & Spach) are endemic shrubs from North Africa. They are common in eastern Algeria [17] and colonize the forests, rocky hills and low mountains. The two wild legumes are important forage and/or pasture plants playing a fundamental role in the process of restoring the ecological balance of their environment.
Endosymbiotic bacteria from G. microcephala and A. uniflorum growing in an arid ecoclimate zone from Tunisia have been described [18], [19]. However, no data about bacteria able to nodulate G. microcephala and A. uniflorum from Algeria are available. Rhizobial strains isolated from these plants are associated with different species of rhizobia, predominantly of the genera Rhizobium, Sinorhizobium Phyllobacterium and Ensifer [18], [19], [20], [21]. Considering the major ecological role of G. microcephala and A. uniflorum in Algerian arid zones, the present work aimed to characterize root symbiotic nitrogen-fixing bacteria using numerical taxonomy of phenotypic characteristics such as protein profile and antibiotic and heavy metal resistance.
Material and methods
Sampling zone and plant material
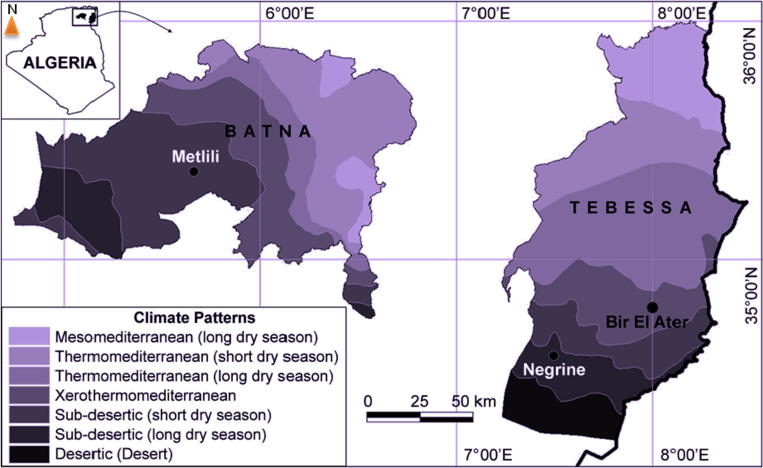
Two wild plant and endemic legume species belonging to the tribe Genisteae were collected in February 2017 (Northeastern Algeria). (i) Argyrolobium uniflorum (Decne.) Jaub. & Spach was collected from two sites, Bir El-Ater (xero-thermo-Mediterranean climate) (coordinate 34°43′24″N, 8°2′33″E, Tebessa) and Negrine (subdesert area) (coordinate 34°29′24″N, 7°33′1″E, Tebessa), and (ii) Genista microcephala Coss. & Durieu was sampled from a subdesert zone (Metlili 35°15′49″N, 5°39′8″E) located near the province of Batna (Fig. 1). The climate of these regions is thermo-Mediterranean, i.e., Mediterranean semiarid, with dry hot summers (maximum temperature recorded in July = 35 °C, precipitation = 10 mm) and relatively cold winters (minimum temperature in January = 1.7 °C with precipitation = 27 mm).
Fig. 1.
Geographic location and climate patterns map of the sampled sites (Solid circles) in northeastern Algeria.
Nodule collection and storage
Nodules were harvested from healthy green plants according to Vincent [22] and Beck et al. [23]. Only red-pink and large nodules indicating the presence of active leghemoglobin and nitrogen fixation were selected. For short conservation and immediate use, nodules were stored at 4 °C and desiccated under CaCl2 for a long period of storage.
Bacterial isolation
Isolation of indigenous nitrogen-fixing bacteria from nodules was determined according to the method described by Somasegaran and Hoben [24]. Briefly, conserved nodules were rehydrated in sterile distilled water for 24 h at 4 °C and then for one hour at room temperature. The rehydrated nodules were surface-sterilized by immersing in 95% ethanol for 5 to 10 s and 0.1% mercuric chloride solution for 2 min. Then, nodules were rinsed ten times and were kept for one hour in sterile water. In aseptic conditions, nodules were individually crushed with sterile water; then, aliquots of 100 μL were separately spread on Congo red-Yeast Mannitol Agar (CR-YMA) (g/L) (yeast extract, 0.5; mannitol, 10.0; K2HPO4, 0.5; MgSO4 7H2O, 0.20; NaCl 0.10, Congo red, 0.025; agar, 15) and glucose peptone agar (GPA) (g/L) (peptic digest, 20; dextrose, 10; NaCl, 5; agar, 15) with bromocresol purple (0.04 g/L). Plates were incubated at 30 °C for 3 to 6 days, and single colonies were picked and surface-streaked several times until purification. Pure cultures were maintained on YMA slants at 4 °C or in 25% glycerol at −80 °C.
Nodulation tests and symbiotic efficiency
The ability of bacterial isolates to infect their original host was determined using the jar nodulation test of Leonard [22]. Seeds of A. uniflorum and G. microcephala were sterilized for ten seconds in 95% ethanol and three minutes in 0.1% HgCl2. Then, they were scarified using concentrated sulfuric acid for six minutes and rinsed 10 times with sterile water. For imbibing, seeds were kept in the last water rinse for two hours. After seed germination on Tryptone Yeast Agar (TYA) (g/L) (Casein hydrolysate, 6; yeast extract, 3; agar 15), three plants per jar were inoculated with 1 mL of an original bacterial isolate suspension (DO = 0.1, approximately 106 cell/mL).
Phenotypic characterization of isolates
Pure isolates were characterized on the basis of their microscopic, morphological and biochemical characteristics using standard methods. For comparison, five reference strains were used in the study (Table 1). The reference strains were originally isolated from wild legumes growing in arid environments. Phenotypic characteristics were determined on YMA, CR-YMA, and BC-GPA [22]. The 3-ketolactose test and calcium glycerophosphate precipitation were conducted as described [25], [26], [27]. Growth on 10% litmus milk was also used to differentiate between contaminants and rhizobial shapes that have rapid growth. Growth temperature at (4 °C, 20 °C, 28 °C, 37 °C, 45 °C and 50 °C), salt tolerance (0.5%, 1%, 2%, 3%, 5% and 10%) and pH range of growth (pH 3.5 to 10) (at intervals of 0.5) were assessed on yeast mannitol broth. The growth results were recorded by measuring the optical density (OD) at 600 nm after 24 h incubation at 28 °C.
Table 1.
Reference strains included in this study.
| Code | Host species | Strain | References |
|---|---|---|---|
| A6 | H. coronarium | Rhizobium sullae sp. nov. RHA6 | Benguedouar et al. [28] |
| Hca1 | H. carnosum | Pseudomonas sp. KD | Benhizia et al. [9] |
| Hp7 | H. pallidum | Enterobacter kobei | |
| Hs1 | H. spinosissimum | Pseudomonas sp. NZ096 | |
| HnA | H. naudinianum | Panotoea agglomerans | Torrche et al. [29] |
Carbohydrate assimilation and utilization of nitrogen sources
Carbohydrate assimilation screening was carried out using nine substrates as a sole carbon source (1% w/v: arabinose, fructose, glucose, lactose, maltose, raffinose, sorbitol, sucrose and xylose) on modified YMB where yeast extract was replaced by NH4Cl at 0.1% (w/v) and mannitol by one of the tested carbohydrates [30]. Nitrogen assimilation was determined on defined medium 8 as described by Vincent [22], where sodium glutamate was replaced by one of the following amino acids at 0.1%: alanine, arginine, asparagine, cysteine, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine. The determination of vitamin needs was conducted on BIII medium (g/L) (mannitol, 10; sodium glutamate, 1.1; K2HPO4, 0.23; MgSO4 7H2O, 0.1; trace element stock 1.0 mL) [31], using the following vitamins at 0.1% (Riboflavin, p-aminobenzoic acid, nicotinic acid, biotin, thiamine-HCl, Ca-pantothenate and pyridoxine). The results were recorded by measuring the optical density at 600 nm after 24 h incubation at 28 °C. Specific enzymes (cellulase, nitrate reductase, pectinase, tryptophan deaminase, tryptophanase, and urease) were determined according to the methods described by Joffin and Leyral [32].
Antibiotic susceptibility and heavy metal resistance
The agar dilution method on TYA was used to determine the intrinsic antibiotic resistance and heavy metal tolerance among the bacterial isolates. The following antibiotics (spectinomycin, erythromycin, rifampicin, gentamycin, streptomycin, kanamycin, and chloramphenicol) were used at different concentrations ranging from 0.5 to 5000 μg/mL. In addition, the heavy metals (HgCl2, ZnCl2, CuCl2, Pb(CH3COO)2 and SbO3) were supplemented at final concentrations of 0.5 to 6000 μg/mL. Then, 10 μL of the bacterial suspensions (2 × 108 c.f.u./mL) was inoculated on the surface of each plate and incubated at 30 °C up to 7 days. The isolates were considered resistant when visible growth occurred.
SDS-PAGE of whole cell proteins
Symbiotic isolates and reference strains were grown at 28 °C for 48 h on TY broth and SDS-PAGE of whole cell proteins was carried out on 12.52% (w/v) gradient polyacrylamide gels as described by Lammeli [33]. Gels were loaded with approximately 50 μg of protein preparation per lane and run at 40 mA with 120 V starting voltage for 4 h. After migration, bands were visualized by 0.01% Coomassie Brilliant Blue R250 stain for one night with gentle stirring.
Data analysis
Phenotypic characteristics and normalized densitometric traces of the protein electrophoretic patterns were clustered using agglomerative hierarchical clustering (AHC) [34]. The results of the phenotypic characterization were converted into a binary dataset, which was used to estimate the simple matching similarity coefficient of each strain pair and to generate a similarity matrix. All data analysis was performed using the statistical software XLStat version 2014 (www.xlstat.com).
Results and discussion
Phenotypic characterization
A total of thirteen rhizobium isolates were selected. Authentication of the isolated bacteria as root nodule bacteria is based upon the aptitude to nodulate their native host legumes. All thirteen isolates were rod shaped, Gram-negative, non-spore forming and fast-growing bacteria (visible growth within 2 days) with acidification of the YMA-BTB. Only two isolates (A and E) showed 3-ketoglucosidase activity following oxidation of C3-glucosyl saccharides. Neither browning or formation of precipitate were observed after 72 h of growth on agar mannitol and calcium glycerophosphate (Table 2). All selected isolates showed slow growth on litmus milk medium associated with a proteolytic activity. Similar findings were reported with rhizobia isolated from Hedysarum coronarium and Medicago ciliaris showing slow growth rates and failure of 3-ketoglucosidase formation [35], [36], [37].
Table 2.
Enzymatic activities and distinctive tests among rhizobia isolates and references strains. +, growth or positive reaction; −, no growth or negative reaction. TDA, tryptophan deaminase.
| Isolates | TDA | Nit | Urease | Cellulase | Tryptophanase | Pectinase | Calcium glycerophosphate | 3-ceto lactose | Litmus milk | |
|---|---|---|---|---|---|---|---|---|---|---|
| Reference strains | Hs1 | + | + | + | − | − | + | − | − | − |
| Hp7 | + | + | + | − | − | + | − | − | − | |
| Hca1 | − | + | + | − | − | + | − | − | − | |
| A6 | + | + | + | + | − | + | − | − | + | |
| HnA | + | + | + | − | − | + | − | − | − | |
| Argyrolobium uniflorum | N1 | − | + | + | + | − | + | − | − | + |
| N2 | + | + | + | + | − | + | − | − | + | |
| YN12 | + | + | + | + | − | + | − | − | + | |
| AN123′ | − | + | + | + | − | + | − | − | + | |
| AN11′ | + | + | + | + | − | + | − | − | + | |
| AB2′ | − | + | + | + | − | + | − | − | + | |
| B3 | + | + | + | + | − | + | − | − | + | |
| Genista microcephala | D | + | + | + | + | − | + | − | − | + |
| E | + | + | + | + | − | + | − | + | + | |
| K | + | + | + | + | − | + | − | − | + | |
| F | + | + | + | + | − | + | − | − | + | |
| A | + | + | + | + | − | + | − | + | + | |
| M | + | + | + | + | − | + | − | − | + | |
Physiological characterization
Physiological and metabolic properties of the isolates are presented in Table 3. The measurements of the optical density indicated clear differences in carbohydrate assimilation of isolates according to their symbiotic partner. Isolates from A. uniflorum showed maximum growth in media containing glucose, maltose, arabinose, fructose and sucrose. Conversely, G. microcephala isolates presented low growth rates using carbohydrates as a sole carbon source. No or low growth was recorded on maltose, raffinose, arabinose and sucrose. Similarly, fast-growing isolates were found predominantly in root nodule bacteria associated with indigenous legumes in Eastern Algeria [9], [36]. Howieson and McInnes [37] reported that most legumes in the Mediterranean area appear to be nodulated by fast-growing bacteria. The fast-growing rhizobia were considered acidifying bacteria [38], [39], whereas slow-growing rhizobia were more limited in their ability to use diverse carbon sources. Therefore, the fast-growing isolates may be attributed to the soil types within the respective collection regions, as well as variation in the indigenous legume flora. However, inability of isolates from G. microcephala to grow on sucrose or lactose may indicate the lack of a disaccharide uptake system [40].
Table 3.
Phenotypic characteristics of rhizobia isolates. +, growth or positive reaction; −; no growth or negative reaction.
| Characteristics |
A. uniforlum |
G. microcephala |
Reference strains |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AB2′ | B3 | AN11′ | AN123′ | N2 | N1 | YN12 | D | E | K | F | A | M | A6 | HS1 | HnA | HCa1 | HP7 | ||
| Temperature | −4 °C | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + | + |
| 28 °C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| 45 °C | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − | + | − | + | |
| 50 °C | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | |
| pH | 3.5−10 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NaCl % | 0.5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| 5 | + | + | + | + | + | + | + | − | − | + | + | + | + | + | + | + | + | + | |
| 10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Carbon source | Glucose | + | + | + | + | + | + | + | + | − | + | − | − | − | + | + | + | + | + |
| Maltose | + | + | + | + | + | + | + | − | − | − | − | − | − | + | + | + | + | + | |
| Raffinose | + | + | + | − | + | − | + | − | − | − | − | + | − | + | + | + | + | + | |
| Xylose | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | |
| Arabinose | + | + | + | + | + | + | + | − | − | + | − | − | − | + | + | + | + | + | |
| Fructose | + | + | + | + | + | + | + | + | + | + | − | + | − | + | + | + | + | + | |
| Lactose | + | + | − | − | + | + | + | + | − | + | − | − | − | + | + | + | + | + | |
| Sorbitol | + | + | + | − | − | + | − | − | + | + | − | − | − | + | + | + | + | + | |
| Sucrose | + | + | + | + | + | + | + | − | − | + | − | − | − | + | + | + | + | + | |
| Vitamins | PAA | + | + | + | + | − | − | + | − | − | − | − | + | + | − | + | − | − | − |
| Biotin | + | + | + | + | + | + | + | − | − | − | − | + | − | − | + | − | + | + | |
| Perydoxine | + | + | + | − | + | + | + | − | − | − | − | + | + | + | + | + | + | + | |
| Thiamine | + | + | + | + | + | + | + | − | + | − | + | + | + | + | + | − | − | + | |
| Riboflavin | + | + | + | + | + | + | + | + | − | − | − | + | − | − | + | − | − | + | |
| Panthotenate | + | + | + | + | + | + | + | − | − | − | − | − | + | − | + | − | − | − | |
| Nicotinic Ac | + | + | + | + | + | + | + | − | − | − | − | + | + | − | + | − | − | − | |
| Amino acids | Valine | − | + | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + |
| Tyrosine | + | + | + | + | + | − | + | − | − | − | − | + | − | + | + | + | + | + | |
| Leucine | − | − | − | + | − | + | − | + | + | + | + | + | + | + | + | + | + | + | |
| Proline | + | + | + | + | + | − | + | + | + | − | − | + | − | + | − | + | + | + | |
| Threonine | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | − | − | |
| Isoleucine | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | |
| Phenylalanine | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Tryptophan | − | + | + | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | |
| Lysine | − | − | + | − | − | − | − | − | − | − | + | − | + | − | + | − | − | − | |
| Glycine | + | − | + | + | − | + | − | + | − | − | − | + | + | + | + | + | + | + | |
| Serine | − | − | + | − | − | − | + | − | − | − | − | − | + | + | + | + | + | + | |
| Histidine | + | + | − | − | − | + | + | + | + | + | + | + | − | − | + | − | + | + | |
| Arginine | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | |
| Methionine | − | − | − | − | − | − | − | − | + | − | − | − | + | + | + | + | + | + | |
| Alanine | + | + | − | − | − | − | + | + | + | + | + | + | + | − | − | + | − | − | |
| Asparagine | − | − | + | − | − | − | − | − | − | − | − | − | − | + | + | − | − | + | |
| Cysteine | + | − | − | − | − | − | − | − | + | + | − | + | + | + | + | + | − | + | |
| Glutamine | − | − | − | + | − | − | − | + | + | + | − | − | − | − | + | − | − | + | |
The isolates showed maximum growth using leucine and proline as nitrogen sources. Furthermore, the results indicated that asparagine does not promote the growth of all isolates.
The following isolates AN123, N1, and AB2′ do not possess tryptophan deaminase, and no indole formation was noted among the isolates, whereas the other tested bacteria produce indole-3-acetic acid and tryptophan deaminase. All the rhizobial isolates showed pectinolytic and cellulolytic activities, while no cellulolytic activity was detected in Gammaproteobacteria Hs1, HnA, Hp7, and HcA1 used as reference strains. Previously, production of hydrolases has been reported among rhizobia [37]. Moreover, cellulolytic activity was observed in all microsymbionts belonging to Rhizobium and Bradyrhizobium [37] but not for Hedysarum associated bacteria [9].
Werner et al. [41] reported that vitamin requirements for rhizobia were highly variable (e.g.) cell growth was stimulated by biotin for Bradyrhizobium and thiamine for Rhizobium, while the presence of biotin, thiamine, and riboflavin limit the growth of Sinorhizobium meliloti [42].
NaCl tolerance, pH effect, and growth temperature
All the tested bacteria presented a broad spectrum of pH tolerance, as they were able to grow in acidic and alkaline pH values ranging from pH 3.5 to pH 10. All selected bacteria but two isolates (D and E), were more tolerant to salt up to 5% NaCl (w/v) (Table 3). It was reported that salinity inhibits nitrogen fixation by increasing the resistance to oxygen diffusion in the nodules with consequent inhibition of nitrogenase activity [43], [44]. Similarly, salinity tolerance up to 800 mM NaCl was noted among rhizobia isolated from Medicago ciliaris and Medicago polymorpha collected in the Sebkha of Misserghine (Northwestern Algeria) [45]. Furthermore, thermotolerance variability was noted among rhizobia isolates, which were able to grow up to 45° C. Heat stress and temperature adaptation in rhizobia has been widely studied [46], showing that root nodule bacteria are mesophilic, and can grow at temperatures ranging from 28 °C to 37 °C [44].
Resistance to antibiotics and heavy metals
The use of high quality, effective rhizobia on agriculture has contributed substantially to the economy of farming systems through the biological nitrogen fixation in the rhizosphere. However, the rhizosphere comprises large populations of antibiotic-producing microorganisms, which affect susceptible rhizobia [47]. Thus, antibiotic resistance is an extremely valuable and positive selection marker to select symbiotically effective bacteria.
The antibiotic susceptibility patterns of the selected isolates are presented in Table 4. The results show that all isolates were resistant to spectinomycin, erythromycin, and gentamycin. However, they were more susceptible to kanamycin, chloramphenicol, streptomycin, and rifampicin. Several researchers have reported antibiotic/rhizobia interactions and it has been noted that fast-growing bacteria are more sensitive to antibiotics than slow-growing rhizobia [48], [49].
Table 4.
Antibiotic susceptibility and heavy metal tolerance of rhizobia isolates. (Spect: spectynomycin; Gent: gentamicin; Kan: kanamycin; CHL: chloramphenicol; Strep: streptomycin; Rif: rifampicin; Ery; erythromycin).
| Origin | Strains | MIC (µg/mL) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spec | Gent | Kan | CHL | Strep | Rif | Ery | SbO3 | ZnCl2 | CuCl2 | HgCl2 | Pb(CH3COO)2 | ||
| Reference strains | Hs1 | >5000 | 300 | 20 | 100 | 500 | 200 | >5000 | >6000 | 2750 | 1500 | 250 | 2250 |
| Hp7 | >5000 | 300 | 300 | 100 | 600 | 50 | >5000 | >6000 | 2500 | 1500 | 250 | 2250 | |
| Hca1 | >5000 | 400 | 20 | 100 | 400 | 100 | >5000 | >6000 | 2250 | 1500 | 250 | 2250 | |
| A6 | >5000 | 1250 | 600 | 500 | 1250 | 200 | >5000 | >6000 | 2750 | 1500 | 500 | 2250 | |
| HnA | >5000 | 100 | 20 | 300 | 400 | 50 | >5000 | >6000 | 2750 | 1500 | 750 | 2250 | |
| Argyrolobium Uniflorum | N1 | >5000 | 800 | 300 | 400 | 600 | 150 | >5000 | >10000 | 2100 | 1600 | 750 | 1700 |
| N2 | >5000 | 800 | 300 | 400 | 600 | 50 | >5000 | >10000 | 1600 | 1550 | 600 | 1700 | |
| YN12 | >5000 | 1000 | 300 | 400 | 600 | 150 | >5000 | >10000 | 1600 | 1500 | 500 | 1700 | |
| AN123′ | >5000 | 1000 | 600 | 400 | 600 | 150 | >5000 | >10000 | 1800 | 1550 | 800 | 1700 | |
| AN11′ | >5000 | 1200 | 600 | 400 | 600 | 50 | >5000 | >10000 | 2100 | 1600 | 800 | 1700 | |
| AB2′ | >5000 | 1250 | 300 | 400 | 600 | 150 | >5000 | >10000 | 2100 | 1200 | 600 | 1700 | |
| B3 | >5000 | 800 | 600 | 400 | 1000 | 50 | >5000 | >10000 | 1800 | 1600 | 300 | 1700 | |
| Genista microcephala | D | >5000 | 1250 | 600 | 400 | 600 | 200 | >5000 | >10000 | 1800 | 1500 | 800 | 1700 |
| E | >5000 | 800 | 600 | 400 | 1000 | 200 | >5000 | >10000 | 1600 | 1050 | 300 | 1700 | |
| K | >5000 | 700 | 300 | 400 | 600 | 200 | >5000 | >10000 | 1600 | 1500 | 500 | 1700 | |
| F | >5000 | 300 | 300 | 300 | 600 | 150 | >5000 | >10000 | 1600 | 1500 | 800 | 1700 | |
| A | >5000 | 1200 | 600 | 400 | 600 | 150 | >5000 | >10000 | 1800 | 1500 | 1000 | 1700 | |
| M | >5000 | 800 | 600 | 400 | 600 | 150 | >5000 | >10000 | 2100 | 1000 | 500 | 1700 | |
In addition, the tested strains showed higher MIC values for antimony up to 10 mg/mL and less resistance to mercury (Table 4). Isolates (N1, AN11′, and B3) presented maximum lead and copper tolerance of 1.7 mg/mL and 1.6 mg/mL, respectively (Table 4). Zinc resistance was reported at (2.1 mg/mL) for the isolates (N1, AN11′, AB2′, and M). The pattern of metal tolerance was in the order Sb > Zn > Pb > Cu > Hg. In soil, the bacterial population would have been exposed to heavy metals that allow the ability to grow and survive at high toxic metal concentrations [48]. The results of such pressure as well as other environmental conditions, such as temperature, salinity, and pH, can contribute to the selection of metal tolerance among different rhizobia species indicating their ability to survive in contaminated soils as described elsewhere [50]. This result is consistent with the literature showing that the Rhizobium group was resistant to high concentrations of arsenate, zinc, copper, and even mercury [51].
Numerical analysis of phenotypic traits
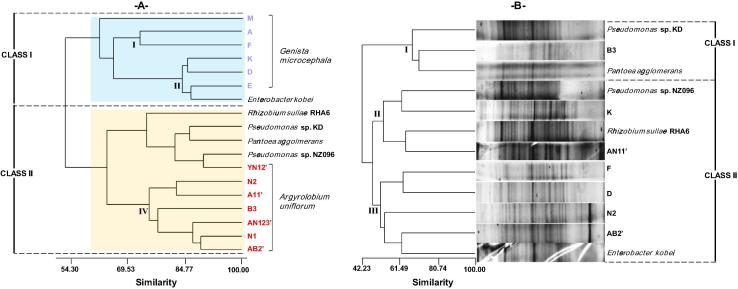
In this study, thirteen rhizobia isolates were characterized, and 66 phenotypic traits were included for numerical analysis. Agglomerative hierarchical clustering showed that below the boundary level of 62% average similarity, the tested isolates can be grouped into two class and four clusters (Fig. 2A). Class I grouped all isolates recovered from G. microcephala (Metlili) in which Cluster I was composed of two isolates (A and F) at 74.22% similarity; Cluster II was represented by three isolates (K, D, and E) together with the reference strain Enterobacter kobei. Class II compiled the six bacterial isolates (AB2, N1′, B3, N2, AN11 and YN12′) originating from nodules of A. uniflorum collected in Bir El-Ater and Negrine.
Fig.2.
Dendrogram showing the phenotypic relationships (A) and normalized sodium dodecyl sulfate-polyacrylamide gel electrophoresis patterns of the rhizobia strains nodulating Genista microcephala and Argyrolobium uniflorum (B).
Analysis of protein profiles
As shown in Fig. 2. protein analysis showed that at 45.61% similarity, the isolates formed three distinct classes with reference strains. Cluster I grouped one isolate (B3) from A. uniflorum (Bir El-Ater) and two reference strains (Pseudomonas sp. KD and Pantoea agglomerans). The two isolates (K and AN11′) from G. microcephala and A. uniflorum were clustered with Pseudomonas sp. NZ096 and Rhizobium sullae at 62.61% and 64.35% similarity, respectively (Cluster II). Cluster III classified the isolate (AB2′) from A. uniflorum (Bir El-Ater) and Enterobacter kobei at 62.61%. The comparison of the total protein profiles obtained by electrophoresis in the presence of SDS can be highly standardized for grouping a number of strains [52] and the strains with identical protein gel electropherograms may constitute a very homogeneous cluster with most likely high internal molecular homologies. In addition, several studies have revealed a great similarity between the content of protein and DNA/DNA hybridization [53]. However, recently, Benguedouar et al. [28] have showed a limited use of SDS-PAGE for rhizobia identification at the species level.
Since the biological resources of Algerian arid regions are little known [54], recently, work has been intended to characterize rhizobia in nodulating endemic legumes using both phenotypic and molecular approaches. Merrabt et al. [45] have studied symbiosis in saline soil regions of two legumes Medicago ciliaris and Medicago polymorpha and rhizobial strains belonging to Rhizobium, Sinorhizobium, Phyllobacterium, and Agrobacterium were characterized using partial sequencing of the 16S rRNA gene. Similarly, a genetic diversity study was conducted among rhizobia isolates from annual Medicago spp. (Medicago arabica, Medicago polymorpha, Medicago minima and Medicago orbicularis) located in semi-arid zones [55]. Riah et al. [56] have characterized Rhizobium isolates from lentil (Lens culinaris), and pea (Pisum sativum) plants growing in two eco-climatic zones (sub-humid and semi-arid) using PCR-restriction fragment length polymorphism (RFLP) of the 16S–23SrRNA intergenic region (IGS), and the nodD-F symbiotic region. Indeed, Torche et al. [29] have investigated rhizobia from root nodules of two wild legume species Hedysarum naudinianum and H. perrauderianum using both culture-dependent methods and 16S amplicon cloning which revealed, in both plants, the presence of a Mesorhizobium sp. Furthermore, Bradyrhizobium characterization was reported from root nodules of Cytisus villous [57].
Conclusions
In general, phenotypic studies showed a large physiological and biochemical diversity of selected isolates, exhibiting the basic characteristics of rhizobia and displaying high levels of similarity with Gammaproteobacteria. In addition, the isolates showed variable tolerance to different stress factors (temperature, pH, salinity, antibiotics and heavy metals), which allowed for the selection of good candidates for future research. In fact, they are multipurpose bacteria with very interesting characteristics, which offers these legumes important ecological advantages and may improve symbiotic characteristics for others. Further molecular characterization of bacterial isolates from G. microcephala and A. uniflorum using conventional methods should be performed for further examination of diversity.
Conflict of interest
The authors declare that they have no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Zahran HH. Legumes-microbes interactions under stressed environments. In: Saghir Khan M, Musarrat J, Zaidi A, editors. Microbs for legumes improvement; 2010. p. 353–387.
- 2.Dommergues Y., Dohoux E., Dien H. Les arbres fixateurs d’azote caractéristique fondamentales et rôle dans l’aménagement des écosystèmes méditerranéens et tropicaux. Edition Espaces. 1998;34:15–16. [Google Scholar]
- 3.Requena N., Perez-Solis E., Azcon-Aguilar C., Jeffries P., Barea J.M. Management of indigenous plant–microbe symbioses aids restoration of desertified ecosystems. Appl Environ Microbiol. 2001;67:495–498. doi: 10.1128/AEM.67.2.495-498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandamme P., Goris J., Chen W.M., de Vos P., Willems A. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov. nodulate the roots of tropical legumes. Syst Appl Microbiol. 2002;25:507–512. doi: 10.1078/07232020260517634. [DOI] [PubMed] [Google Scholar]
- 5.Nzoué A., Miché L., Klonowska A., Laguerre G., De Lajudie P., Moulin L. Multilocus sequence analysis of Bradyrhizobia isolated from Aeschynomenespecies in Senegal. Syst Appl Microbiol. 2009;32:400–412. doi: 10.1016/j.syapm.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Pongslip N. Bentham Science Publishers; 2012. Phenotypic and genotypic diversity of rhizobia. [Google Scholar]
- 7.Gyaneshwar P., Hirsch A.M., Moulin I., Chen W.M., Elliott G.N., Bontemps C. Legume-nodulating betaproteobacteria, diversity, host range and future prospects. Mol Plant Microb Interact. 2011;24:1276–1288. doi: 10.1094/MPMI-06-11-0172. [DOI] [PubMed] [Google Scholar]
- 8.Moulin L., Munive A., Dreyfus B., Boivin-Masson C. Nodulation of legumes by members of the beta-subclass of prote, obacteria. Nature. 2001;411:948–950. doi: 10.1038/35082070. [DOI] [PubMed] [Google Scholar]
- 9.Benhizia Y., Benhizia H., Benguedouar A., Muresu R., Giacomini A., Squartini A. Gamma proteobacteria can nodulate legumes of the genus Hedysarum. Syst Appl Microbiol. 2004;27:462–468. doi: 10.1078/0723202041438527. [DOI] [PubMed] [Google Scholar]
- 10.Zahran H.H. Rhizobia from wild legumes: diversity, taxonomy, ecology, nitrogen fixation and biotechnology. J Biotechnol. 2001;91:143–153. doi: 10.1016/s0168-1656(01)00342-x. [DOI] [PubMed] [Google Scholar]
- 11.Kalita M., Stepkowski T., Lotock B., Malek W. Phylogeny of nodulation genes and symbiotic properties of Genista tinctoria bradyrhizobia. Arch Microbiol. 2006;186:87–97. doi: 10.1007/s00203-006-0124-6. [DOI] [PubMed] [Google Scholar]
- 12.Cardinale M., Bonnì M.L., Marsala S., Puglia A.M., Quatrini P. Diversity of rhizobia nodulating wild shrubs of Sicily and some neighbouring islands. Arch Microbiol. 2008;190:461–470. doi: 10.1007/s00203-008-0394-2. [DOI] [PubMed] [Google Scholar]
- 13.Abdelguerfi A, Abdelguerfi-Laouar M. Les ressources génétiques d'intérêt fourrager et-ou pastoral: diversité, collecte et valorisation au niveau méditerranéen. In: Ferchichi A, Ferchichi A, editors. Réhabilitation des pâturages et des parcours en milieux méditerranéens. Cahiers Options Méditerranéennesm, vol. 62. Zaragoza: CIHEAM; 2004. p. 29–41.
- 14.Tani K.C., Le Bourgeois T., Munoz F. Aspects floristiques de la flore des champs du domaine phytogéographique oranais (Nord-Ouest algérien) et persistance d’espèces rares et endémiques. Flora Mediterranea. 2009:5–22. [Google Scholar]
- 15.Bensizerara D., Menasria T., Melouka M., Cheriet L., Chenchouni H. Antimicrobial activity of xerophytic plant (Cotula cinerea Delile) extracts against some pathogenic bacteria and fungi. Jordan J Biol Sci. 2013;6:266–271. [Google Scholar]
- 16.Duran A., Dural H. Genista vuralii (Fabaceae), a new species from Turkey. Ann Botanici Fennici. 2003;40:113–116. [Google Scholar]
- 17.Maire R. Flore de l’Afrique du Nord; Lechevalier, Paris: 1987. Leguminosae; p. 16. [Google Scholar]
- 18.Zakhia F., Jeder H., Domergue O., Willems A., Cleyet-Marel J.C., Gillis M. Characterisation of wild legume nodulating bacteria (LNB) in the infra-arid zone of Tunisia. Syst Appl Microbiol. 2004;27:380–395. doi: 10.1078/0723-2020-00273. [DOI] [PubMed] [Google Scholar]
- 19.Mahdhi M., De Lajudie P., Mars M. Phylogenetic and symbiotic characterization of rhizobialbacteria nodulating Argyrolobium uniflorum in Tunisian arid soils. Can J Microbiol. 2008;54:209–217. doi: 10.1139/w07-131. [DOI] [PubMed] [Google Scholar]
- 20.Merabet C., Martens M., Mahdhi M., Zakhia F., Sy A., Le Roux C. Multilocus sequence analysis of root nodule isolates from Lotus arabicus (Senegal), Lotus creticus, Argyrolobium uniflorum and Medicago sativa (Tunisia) and description of Ensifer numidicus sp. nov. and Ensifer garamanticus sp. nov. Int J Syst Evol Microbiol. 2010;60:664–674. doi: 10.1099/ijs.0.012088-0. [DOI] [PubMed] [Google Scholar]
- 21.Mahdhi M., Nzoue A., Gueye F., Merabet C., de Lajudie P., Mars M. Phenotypic and genotypic diversity of Genista saharae microsymbionts from the infra-arid region of Tunisia. Lett Appl Microbiol. 2007;45:604–609. doi: 10.1111/j.1472-765X.2007.02233.x. [DOI] [PubMed] [Google Scholar]
- 22.Vincent J.M. Blackwell Scientific Publication Ltd.; Oxford, United Kingdom: 1970. The manual for the principal study of root nodule bacteria. [Google Scholar]
- 23.Beck DP, Materon LA, Afandi F. Pratical Rhizobium - Legume Technology Manual. ICARDA Syria; 1993.
- 24.Somasegaran P., Hobenh J. Springler verlage; New York: 1994. Handbook for Rhizobia. [Google Scholar]
- 25.Hofer A.V.A. Characterization of Bacterium radiobacter (Beijerinck and Van Delden) J Bacteriol. 1941;41:193–224. doi: 10.1128/jb.41.2.193-224.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernaerts J.E., De Ley J. A biochemical test for crown gall bacteria. Nature. 1963;199:406–407. [Google Scholar]
- 27.Jordan D.C. Familly III. Rhizobiaceae. In: Krieg N.R., Holt J.G., editors. vol. 1. The Williams & Wilkins. Co.; Baltimore: 1984. pp. 234–245. (Bergey’s manual of systemetic bacteriology). [Google Scholar]
- 28.Benguedouar A., Corich V., Giacomini A., Squartini A., Nuti M. Characterization of symbiotic bacteria from the Mediterranean legume crop Hedysarum coronarium (sulla) by multilocus enzyme electrophoresis. Agricoltura Mediterranea. 1997;127:173–177. [Google Scholar]
- 29.Torche A., Benhizia H., Rosselli R., Romoli O., Zanardo M., Baldan E. Characterization of bacteria associated with nodules of two endemic legumes of Algeria, Hedysarum naudinianum and H. perrauderianum. Ann Microbiol. 2014 10.1007/s13213-013-0745-3. [Google Scholar]
- 30.Vandamme P., Pot B., Gillis M., De Vos P., Kerster S.K., Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dazzo F.B. Leguminous root nodules. In: Burns R., Slater J., editors. Experimental microbial ecology. Blackwell Scientific Publication; Oxford: 1982. pp. 431–446. [Google Scholar]
- 32.Joffin JN, Leyral G. Microbiologie technique. Dictionnaire des techniques. Tome I. Canopé - CRDP de Bordeaux. France; 2001.
- 33.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bactériophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Sneath P.H.A., Sokal R.R. San Francisco W.H. Freeman & Co; 1973. Numerical taxonomy: the principles and practice of numerical classification; p. 573. [Google Scholar]
- 35.Cheriet D., Ouartasi A., Chekireb D., Babaarbi S. Phenotypic and symbiotic characterization of rhizobia isolated from Medicago ciliaris L. growing in Zerizer from Algeria. Afr. J Microbiol Res. 2014;8:1763–1778. [Google Scholar]
- 36.Struffi P., Corich V., Giacomini A., Benguedouar A., Squartini A., Cassella S. Metabolic properties, stress tolerance and macromolecular profiles of rhizobia nodulating Hedysarum coronarium. J Appl Microbiol. 1998;48:81–89. doi: 10.1046/j.1365-2672.1997.00318.x. [DOI] [PubMed] [Google Scholar]
- 37.Howieson JG, McInnes A. The legume-rhizobia symbiosis. Does it vary for the tropics relative to the Mediterranean basin? In: Gomide JA, Matto WRS, da Silva SC, editors. Proceedings of the XIX international grasslands congress, Brazil. Brazil: Brazilian Society of Animal Husbandry; 2001. p. 585–590.
- 38.Castro S., Carrera I., Martinez-Drets G. Methods to evaluate nodulation competitiveness between Sinorhizobiummeliloti strains using melanin production as a marker. J Microbiol Methods. 2000;41:173–177. doi: 10.1016/s0167-7012(00)00151-2. [DOI] [PubMed] [Google Scholar]
- 39.Safronova V.I., Piluzza G., Belimov A.A., Bullitta S. Phenotypic and genotypicanalysis of rhizobia isolated from pasture legumes native of Sardinia and Asinara Island. Antonie Van Leeuwenhoek. 2004;2:115–127. doi: 10.1023/B:ANTO.0000020278.58236.77. [DOI] [PubMed] [Google Scholar]
- 40.Marsudi N.D.S., Glenn A.R., Dilworth M.J. Identification and characterization of fast- and slow-growing root nodule bacteria from southwestern Australian soils able to nodulate Acacia saligna. Soil Biol Biochem. 1999;31 1229 123. [Google Scholar]
- 41.Werner D. Philips-University Marburg Germany; Edition Chapman and Hall: 1992. Symbioses of plants and microbes. [Google Scholar]
- 42.Karunakaran R., Ebert K., Harvey S., Leonard M.E., Ramachandran V., Poole P. Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. vicia strain 3841. J Bacteriol. 2006;188:6661–6668. doi: 10.1128/JB.00641-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Igual L., Vel Azquez E., Mateos P.F., Rodrequez-Barruecol C., Cerventes E., Martinez Molina E. Cellulase isoenzyme profiles in Frankia strains belonging to different cross-inoculation groups. Plant Soil. 2001;229:35–39. [Google Scholar]
- 44.Zahran H.H. Rhizobium Legume symbiosis and nitrogen fixation under sever conditions and in an arid climate. Microbiol Mol BiolRev. 1999;63:968–989. doi: 10.1128/mmbr.63.4.968-989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merabet C., Bekki A., Benrabah N., BabaAhmed Bey M., Bouchentouf I., Ameziane H. Distribution of Medicago spieces and their microsymbionts in a saline region of Algeria. Arid Land Res Manage. 2006;20:1–13. [Google Scholar]
- 46.Naamala J., Jaiswal S.K., Dakora F.D. Antibiotics resistance in Rhizobium: type, process, mechanism and benefit for agriculture. Curr Microbiol. 2016:1–13. doi: 10.1007/s00284-016-1005-0. [DOI] [PubMed] [Google Scholar]
- 47.Lira M.D., Lima A.S.T., Arruda J.R.F., Smith D.L. Effect of root temperature on nodule development of bean, lentil and pea. Soil Biol Biochem. 2005;37:235–239. [Google Scholar]
- 48.Maatallah J., Berraho E., Sanjuan J., Lluch C. Phenotypic characterization of rhizobia isolated from chickpea (Cicerarietinum) growing in Moroccan soils. Agronomie. 2002;22:321–329. [Google Scholar]
- 49.Margesin R., Płaza G.A., Kasenbacher S. Characterization of bacterial communities at heavy-metal-contaminated sites. Chemosphere. 2011;82:1583–1588. doi: 10.1016/j.chemosphere.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 50.Alikhani A.H., Yakhchali B. Potential use of Iranian rhizobial strains as plant growth-promoting rhizobacteria (PGPR) and effects of selected strains on growth characteristics of wheat, corn and alfalfa. Desert. 2010;14:27–35. [Google Scholar]
- 51.Zerhari K., Aurag J., Khbaya B., Kharchaf D., Filali-Maltouf A. Phenotypic characteristics of rhizobia isolates nodulating Acacia species in the arid and Saharan regions of Morocco. Lett Appl Microbiol. 2000;30:351–357. doi: 10.1046/j.1472-765x.2000.00730.x. [DOI] [PubMed] [Google Scholar]
- 52.Carrasco J.A., Armario P., Pajuelo E., Burgos A., Caviedes M.A., Lopez R. Isolation and characterization of symbiotically effective Rhizobium resistant to arsenic and heavy metals after the toxic spill at the Aznalcollar pyrite mine. Soil Biol Biochem. 2005;37:1131–1140. [Google Scholar]
- 53.Keresters K., Pot B., Denettinek D., Torek H., Vancanneyt M., Auterin L. Plenum Press; New York: 1994. Identification and typing of bacteria by protein electrophoresis. Bacterial diversity and systematics; pp. 51–66. [Google Scholar]
- 54.Menasria T., Aguilera M., Hacene H., Benammar L., Ayachi A., Bachir A. Diversity and bioprospecting of extremely halophilic Archaea isolated from Algerian arid and semi-arid wetland ecosystems for halophilic-active hydrolytic enzymes. Microbiol Res. 2018;207:289–298. doi: 10.1016/j.micres.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 55.Sebbane N., Sahnoune M., Zakhia F., Willems A., Benallaoua S., De La Judie P. Phenotypical andgenotypical characteristics of root-nodulating bacteria isolated from annual Medicago spp. in soummam Valley (Algeria) Lett Appl Microbiol. 2006;42:235–241. doi: 10.1111/j.1472-765X.2005.01846.x. [DOI] [PubMed] [Google Scholar]
- 56.Riah N., BénaG Djekoun A, Heulin K., de Lajudie P., Laguerre G. Genotypic and symbiotic diversity of Rhizobium populations associated with cultivated lentil and pea in sub-humid and semi-arid regions of Eastern Algeria. Syst Appl Microbiol. 2014;37:368–375. doi: 10.1016/j.syapm.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 57.Ahnia H., Boulila F., Boulilan A., Boucheffa K., Duran D., Bourebaba Y. Cytisus villosus from Northeastern Algeria is nodulated by genetically diverse Bradyrhizobium strains. Antonie van Leeuwenhoek. 2014;105:1121–1129. doi: 10.1007/s10482-014-0173-9. [DOI] [PubMed] [Google Scholar]