Abstract
The method of delivery of CRISPR-Cas9 into target cells is a strong determinant of efficacy and specificity in genome editing. Even though high efficiency of Cas9 delivery is necessary for optimal editing, its long-term and high levels of expression correlate with increased off-target activity. We developed vesicles (VEsiCas) carrying CRISPR-SpCas9 ribonucleoprotein complexes (RNPs) that are efficiently delivered into target cells through the fusogenic glycoprotein of the vesicular stomatitis virus (VSV-G). A crucial step for VEsiCas production is the synthesis of the single guide RNA (sgRNA) mediated by the T7 RNA polymerase in the cytoplasm of producing cells as opposed to canonical U6-driven Pol III nuclear transcription. In VEsiCas, the absence of DNA encoding SpCas9 and sgRNA allows rapid clearance of the nuclease components in target cells, which correlates with reduced genome-wide off-target cleavages. Compared with SpCas9 RNPs electroporation, which is currently the method of choice to obtain transient SpCas9 activity, VEsiCas deliver the nuclease with higher efficiency and lower toxicity. We show that a wide variety of cells can be edited through VEsiCas, including a variety of transformed cells, induced pluripotent stem cells (iPSCs), and cardiomyocytes, in vivo. VEsiCas is a traceless CRISPR-Cas9 delivery tool for efficient and safe genome editing that represents a further advancement toward the therapeutic use of the CRISPR-Cas9 technology.
Keywords: CRISPR-Cas9, VEsiCas, RNPs, genome editing, VLPs, nuclease, ribonucleoprotein complex, viral-like particles, SpCas9, GUIDE-seq
Introduction
CRISPR-Cas9-mediated genome editing technologies have tremendous potential for both basic and clinical applications.1, 2, 3, 4, 5, 6, 7, 8, 9, 10 In CRISPR-Cas9 applications, cleavage specificity and delivery efficiency strongly determine the outcome of genome editing.11 High expression levels of Cas9 for long periods result in high numbers of non-specific cleavages.12, 13 So, by affecting the levels and kinetics of expression, the method of delivery is a key determinant of editing efficiency and specificity. In fact, successful genome editing can be obtained through DNA-free delivery of CRISPR-SpCas9 ribonucleoprotein complexes (RNPs), which allows transient expression of the RNA-guided nuclease.14, 15, 16 However, the advantages offered by RNP delivery via physical (electroporation)14 or chemical (lipid- and polymer-based reagents)16, 17 methods are counterbalanced by strong limitations of their in vivo use.18 Opposed to these methods, viral vectors, including those of retroviral origin, are widely used for efficient delivery of Cas9 and sgRNA genes both in vitro and in vivo.8, 19, 20 Nevertheless, these delivery tools are generally not ideal for transient therapeutic approaches because of long-term transgene expression and potential risks for insertional mutagenesis.13, 21 Among the non-integrating vectors, which, in principle, should prevent mutagenic integration,22, 23, 24 those derived from adeno-associated viruses (AAVs) are suitable only for the expression of small transgenes (no greater than ∼4 kb) and are thus not fully compatible with the CRISPR-Cas9 technology. To circumvent the genotoxicity generated by retroviral vectors while preserving viral delivery efficiency, an integration-defective lentiviral vector (IDLV) vector carrying a single guide RNA (sgRNA) transgene and packaged with the SpCas9 protein has been developed.25 A major improvement in the completely traceless delivery of exogenous protein cargos into cells is potentially offered by the development of virus-like particles (VLPs).26 The viral origin of VLPs assures the efficient transduction of target cells even though no viral genomes is carried by the particles, thus allowing rapid clearance of the shuttled protein and RNAs. VLPs have been mainly used in the past decade for vaccination purposes27 or for the delivery of exogenous proteins.26, 28 To minimize the viral elements, protein cargo delivery can be obtained with vesicles made exclusively with the envelope glycoprotein of the vesicular stomatitis virus (VSV-G).29
Here we developed efficient and traceless delivery of SpCas9 and sgRNAs consisting of vesicles, named VEsiCas, that carry SpCas9 RNPs and are decorated with the fusogenic VSV-G. To obtain complete incorporation of sgRNAs into VEsiCas, we exploited the T7 RNA polymerase transcription system, which provides high rates of cytoplasmic sgRNA transcription in producing cells. VEsiCas were efficiently used in this study for genome editing applications with a variety of cell types in culture as well as in vivo after injection into the cardiac muscle of a mouse model. Finally, we tested genome editing procedures where simultaneous targeting of more than one locus is required, such as for genomic deletions or for Cas9-nickase applications, demonstrating the plasticity of VEsiCas for more complex strategies of genetic surgery.
Results
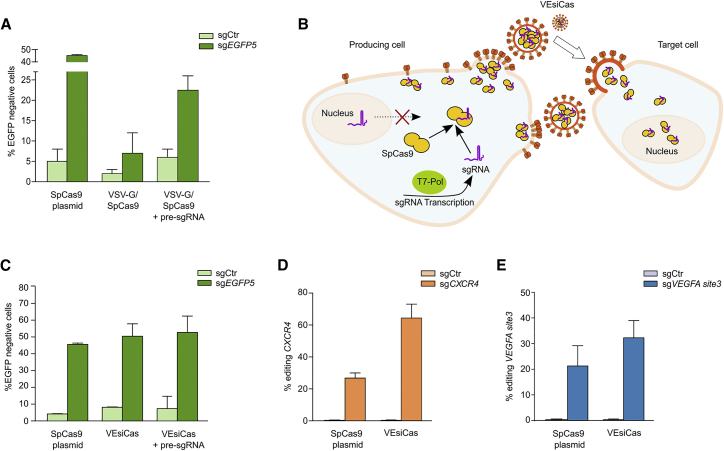
Design and Development of VEsiCas
VSV-G-induced vesicles have been reported to mediate protein transfer in the absence of additional viral components.29 We tested whether VSV-G vesicles could be adapted to DNA-free delivery of CRISPR-Cas9 RNPs. SpCas9 and an sgRNA toward the EGFP coding sequence (sgEGFP5) were expressed together with VSV-G in HEK293T cells, and the derived conditioned clarified medium was applied to a fluorescent reporter cell line, HEK293-EGFP. To evaluate the efficacy of EGFP editing mediated by the VSV-G vesicle treatments, a comparative analysis was performed with SpCas9-sgRNA plasmid transfections, used as a benchmark, similar to previous reports.14, 15, 16, 30 The expression of EGFP was poorly altered under these conditions, indicating inefficient genome editing, whereas efficient editing was observed by transfecting SpCas9 together with the sgRNA (Figure 1A). The limited editing activity was not explained by the lack of delivered protein because both vesicles and target cells were positive for SpCas9 (Figure S1A). These results prompted us to evaluate whether the limiting factor for vesicle activity could be insufficient incorporation of sgRNA. To this aim, the HEK293-EGFP target cells were transfected with plasmids expressing sgEGFP5 before treatment with VSV-G vesicles purified from HEK293T cells expressing SpCas9-sgRNA. Under these experimental conditions, we obtained editing levels that were closer to those observed in cells transfected with SpCas9 and the sgRNA (Figure 1A, compare the sixth and second columns of the graph). These results clearly suggested that the limited editing observed with the SpCas9/VSV-G preparations was due to inefficient delivery of the sgRNA. We speculated that poor sgRNA delivery could be due to inefficient formation of SpCas9-sgRNA RNPs during vesicle production. In particular, the RNA polymerase III (Pol III)-synthesized sgRNAs in the nuclei may be poorly coupled with cytoplasmic SpCas9 to form RNPs at cell periphery, close to the nascent VSV-G vesicles. To test this hypothesis, we employed a T7 RNA polymerase-driven transcription system31, 32 that catalyzes RNA synthesis in the cytoplasm (schematized in Figure 1B). The sgRNAs were cloned downstream of the T7 promoter, and the 5′ hepatitis delta virus (HDV) ribozyme was introduced between the sgRNA coding sequence and the T7 RNA polymerase terminator to induce the formation of mature sgRNAs with unmodified 3′ constant regions.33 The VSV-G-enveloped SpCas9 vesicles were produced in cell lines stably expressing the T7 RNA polymerase and resistant to toxicity induced by high levels of uncapped 5′-triphosphate cytoplasmic RNA generated by this transcriptional system32, 34, 35 (Figure S1B). The derived VSV-G-enveloped SpCas9 Vesicles, VEsiCas, produced in BSR-T7/5 cells expressing sgEGFP5, were verified for SpCas9 incorporation (Figure S1C), observing that SpCas9 was about 1.5%–2% of the total protein content of VEsiCas (Figure S1D). VEsiCas produced in BSR-T7/5 cells induced at least 50% loss of EGFP fluorescence in HEK293-EGFP cells, very similarly to knockouts observed with SpCas9-sgRNA plasmid transfection or VEsiCas treatments of cells pre-transfected with sgEGFP5 (Figure 1C). To test VEsiCas properties in gene substitution experiments, non-fluorescent cells stably transfected with a single copy of the EGFP Y66S variant were treated with VEsiCas together with a donor DNA carrying a truncated wild-type EGFP sequence corresponding to the Y66S mutated region. The production of EGFP fluorescent cells indicated proper repair of the EGFP gene by homology-directed repair (HDR), demonstrating the efficacy of VEsiCas in knockin applications (Figure S2). VEsiCas were then tested toward two genomic loci, CXCR4 and VEGFA, commonly used as benchmarks in genome-editing experiments;11 at these loci, VEsiCas induced similar percentages of insertions or deletions (indels) as those obtained with SpCas9-sgRNA plasmid transfection (Figures 1D and 1E). These results were obtained with multiple rounds (three times) of VEsiCas spinoculation, with single spinoculation treatments producing cumulative editing effects (Figure S3).
Figure 1.
Design and Genome Editing Activity of VEsiCas
(A) EGFP disruption assay with VSV-G/SpCas9 vesicles produced in HEK293T cells. Shown are percentages of EGFP knockout HEK293-EGFP cells generated by transfection of SpCas9 (SpCas9 plasmid) together with EGFP targeting (sgEGFP5) or control (sgCtr) sgRNA or transduction with VSV-G/SpCas9 vesicles carrying U6-transcribed sgRNA. Where indicated, HEK293-EGFP cells were pre-transfected with sgEGFP5 or sgCtr (+ pre-sgRNA) prior to VSV-G/SpCas9 vesicle treatment. Data are presented as mean ± SEM for n = 2 independent experiments. (B) Schematic of VEsiCas production in BSR-T7/5 cells. T7 RNA polymerase, expressed in the cytosol, regulates cytosolic sgRNA expression by means of the T7 promoter. Vesicles decorated with VSV-G, expressed by BSR-T7/5 producer cells, bud incorporating SpCas9 complexed with sgRNA to form VEsiCas. In target cells, VEsiCas release active SpCas9-sgRNA complexes that enter the nuclei through two nuclear localization sequences introduced in SpCas9. (C) Genome activity of VEsiCas produced in BSR-T7/5 on HEK293-EGFP cells. Shown are percentages of non-fluorescent HEK293-EGFP cells following transfection of SpCas9 (SpCas9 plasmid) together with sgRNAs (sgEGFP5 or sgCtr) or treatment with VEsiCas carrying sgRNAs (sgEGFP5 or sgCtr) either with or without pre-transfection with sgRNAs, as indicated. Data are presented as mean ± SEM for n = 2 independent experiments. (D and E) VEsiCas-mediated editing of the CXCR4 (D) and VEGFA site3 (E) genomic loci. Percentages of indel formation in HEK293T cells were measured through TIDE analysis following transfection of SpCas9 (SpCas9 plasmid) together with sgRNAs (sgCXCR4, sgVEGFA site3, or sgCtr) or after three sequential treatments with VEsiCas carrying sgRNAs (sgCXCR4, sgVEGFA site3, or sgCtr). Data are presented as mean ± SEM for n = 2 independent experiments.
SpCas9 pseudotransduction was also tested using lentivirus-based VLPs (lenti-VLPs). The HIV-1 Gag domain or a reduced portion of it (MinimalGag) were reported to generate VLPs, described to efficiently transfer protein cargos to recipient cells.36 SpCas9 fused to Gag or MinimalGag was verified as functionally active in genome editing activity against the EGFP locus (Figures S4A–S4C). The two SpCas9-Gag chimeras were used to obtain lenti-VLPs in BSR-T7/5 cells, which produced high percentages of indels in the EGFP, CXCR4, and VEGFA loci (Figures S4D–S4G). However, because no dramatic improvements in genome editing efficacy were obtained with lenti-VLPs, VEsiCas carrying exclusively the VSV-G viral element were used hereafter.
Overall, our data clearly show that VEsiCas efficiently deliver SpCas9-sgRNA RNPs free from encoding DNA or additional elements of viral origin. A key factor to obtain highly efficient genome editing particles was the relocation of sgRNA expression from the nucleus to the cytoplasm of producing cells, which was obtained with cytoplasmic T7 RNA polymerase in appropriate permissive cells.
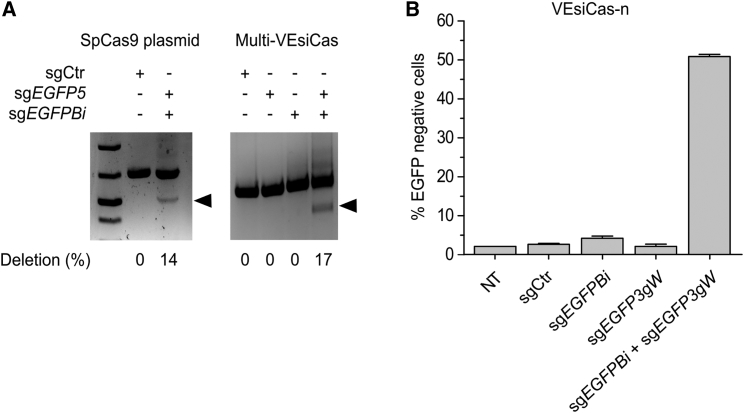
Multiplexed VEsiCas
Because genome editing applications such as targeted genomic deletions may require the simultaneous delivery of more than one sgRNA, we evaluated the possibility to incorporate multiple guides into VEsiCas (Multi-VEsiCas). Multi-VEsiCas were produced in BSR-T7/5 cells expressing two T7-driven EGFP-targeting sgRNAs (sgEGFP5 and sgEGFPBi). Incubation of HEK293-EGFP reporter cells with Multi-VEsiCas carrying both sgRNAs generated the expected deletion in the EGFP locus with ∼17% efficiency, which was similar to the one obtained with transient transfection of plasmids encoding SpCas9 and the corresponding sgRNAs (∼14%) (Figure 2A). Conversely, VEsiCas carrying each individual sgRNA (either sgEGFP5 or sgEGFPBi) were not able to produce any detectable deletion into the target locus (Figure 2A). The flexibility of the VEsiCas delivery platform was further demonstrated by incorporating the SpCas9 nickase (SpCas9-n, D10A mutant)37 to generate VEsiCas-n carrying two closely positioned sgRNAs targeting the EGFP coding sequence. VEsiCas-n were prepared either with individual sgRNAs (sgEGFPBi or sgEGFP3gW) or with the sgRNAs in combination (sgEGFPBi and sgEGFP3gW). A single treatment of HEK293-EGFP cells with VEsiCas-n carrying both sgRNAs produced a robust decrease in the number of fluorescent cells (50%), whereas VEsiCas-n delivering individual sgRNAs did not downregulate EGFP expression, as expected (Figure 2B). These data indicate that more than one sgRNA can be delivered simultaneously by VEsiCas, demonstrating their applicability to generate deletions and to deliver SpCas9 nickase.
Figure 2.
Genome Editing by Multiplexed VEsiCas
(A) Gene deletion using VEsiCas. Shown is gel electrophoresis analysis of the EGFP locus deletion obtained in HEK293-EGFP cells through Multi-VEsiCas (right) treatment or by transfection with SpCas9 together with specific sgRNAs (sgEGFP5 and sgEGFPBi) (left). The amount of deletion (percent) was quantified by densitometry. Arrowheads indicate the expected band corresponding to PCR amplification of the deleted EGFP locus. (B) Activity of VEsiCas delivering SpCas9 nickase. Shown are percentages of EGFP knockout cells following incubation with VEsiCas-n, carrying SpCas9 nickase loaded with sgRNA toward two sites in the EGFP locus (sgEGFP3gW and sgEGFPBi) or with sgCtr. NT, not treated. Data are presented as mean ± SEM for n = 2 independent experiments.
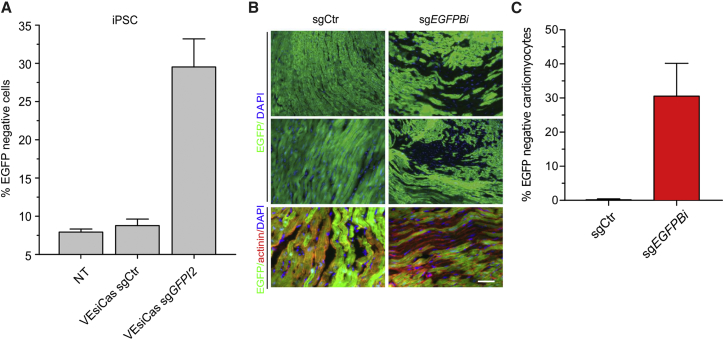
VEsiCas Editing Efficiency in Cells and In Vivo
Increasing amounts of VEsiCas resulted into a proportional increase in editing activity (Figure S3). Moreover, side-by-side comparison with electroporation protocols, which are often used to deliver Cas9 into cells for genome editing, revealed that VEsiCas are more efficient. In fact, compared with electroporation, a lesser amount of SpCas9 delivered by VEsiCas was required to obtain similar percentages of EGFP knockout cells (Figure S5A). Moreover, viability assays using the same experimental conditions as in Figure S5A revealed that, conversely to electroporation, VEsiCas induced minimal cell toxicity (Figure S5B). The efficacy of VEsiCas was tested in different cell lines, including adherent cells (HeLa), suspension cells (J-Lat-A1), and human induced pluripotent stem cells (iPSCs) expressing EGFP, showing a robust decrease in the number of fluorescent cells in all tested models (Figure 3A; Figure S6). Lastly, VEsiCas were tested in the cardiac muscle of EGFP transgenic mice. VEsiCas were injected into the hearts of 5-day-old mice, which were analyzed for levels of EGFP fluorescence 10 days after treatment. The cardiac tissue from treated animals was stained using antibodies against α-actinin to specifically evaluate EGFP fluorescence in cardiomyocytes. As shown in Figure 3B, large areas of non-fluorescent cardiomyocytes could be observed close to the VEsiCas injection site. The efficacy of VEsiCas in vivo editing was quantified by measuring the percentage of EGFP-negative cardiomyocytes in at least three sections obtained from 5 mice derived from either control (VEsicas sgRNA Ctr) or sgEGFPBi VEsiCas groups. The data reported in Figure 3C show that, compared with the control, sgEGFPBi VEsiCas-treated mice showed around 30% more EGFP-negative cardiomyocytes.
Figure 3.
Genome Editing Activity in Human iPSCs and in the Heart of an EGFP Mouse Model
(A) VEsiCas activity in induced pluripotent stem cells (iPSCs). Shown is the percentage of non-fluorescent iPSCs stably expressing EGFP after treatment with VEsiCas carrying either sgCtr or sgGFPI2. Data are presented as mean ± SEM for n = 2 independent experiments. (B) VEsiCas-mediated gene disruption in the cardiac muscle of EGFP mice. Shown are fluorescence microscopy images of cardiac tissue sections from EGFP transgenic mice 10 days after intra-cardiac injection of VEsiCas carrying sgCtr or sgEGFPBi. DAPI was used as a nuclear counterstain. Immunostaining for α-actinin was performed to identify cardiomyocytes (bottom). A representative sample of n = 5 experiments is shown. Scale bar, 100 μm. (C) Quantification of VEsiCas EGFP knockout in cardiac tissue. The bar graph shows the percentage of non-fluorescent cardiomyocytes following VEsiCas treatments (mean ± SEM of n = 5 independent experiments).
Overall, these data show that VEsiCas are efficient tools to deliver SpCas9 RNPs for genome editing in culture cells as well as in vivo.
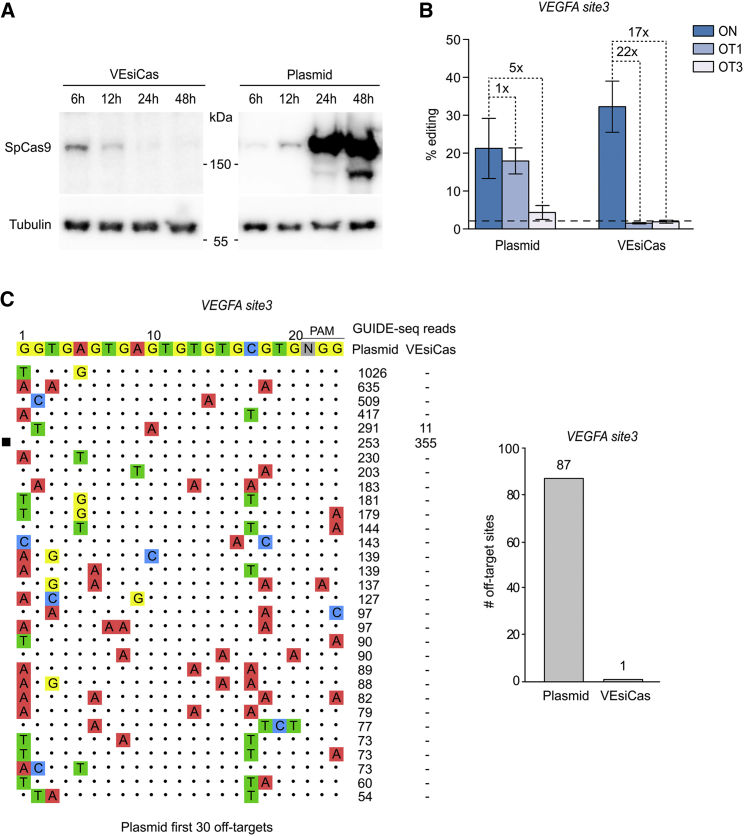
Limited Off-Target Activity by SpCas9 Delivered through VEsiCas
The off-target activity produced by Cas9 is still one of the main limitations for its therapeutic use. The transient expression of the nuclease in target cells has been shown to limit non-specific cleavages.13, 14 To address this point, the kinetics of SpCas9 intracellular levels delivered through VEsiCas in comparison with the amounts of nuclease expressed by transfected plasmid were examined. SpCas9 from VEsiCas was detected in target cells 6 hr after transduction and gradually disappeared within the following 18 hr (Figure 4A). Conversely, the nuclease intracellular levels produced by an expression plasmid were clearly detected 12 hr after transfection and increasingly accumulated thereafter (Figure 4A). To evaluate the extent of the off-target activity generated by SpCas9 delivered through VEsiCas or by transfection, indel formation was measured at two previously validated off-target sites associated with editing of the VEGFA locus (VEGFA OT1 and OT3).13 Tracking of indels by decomposition (TIDE) analysis38 revealed that SpCas9 delivered through VEsiCas produced higher levels of on-target activity than transfected SpCas9 while causing background levels of indels at both off-target sites (Figure 4B). Strikingly, compared with the on-target site, VEsiCas produced at least 22- and 17-fold less indels at OT1 and OT3, respectively, whereas transfected SpCas9 cleaved OT1 similarly to the on-target site, and OT3 was edited just 5 times less. A deeper off-target analysis of the VEGFA locus was performed through genome-wide unbiased identification of double strand breaks (DSBs) enabled by sequencing (GUIDE-seq),39 a genome-wide approach (Figure 4C). This analysis revealed that SpCas9 expressed from a transfected plasmid cleaved a total number of 87 off-target sites, five of them with a higher number of GUIDE-seq reads (1,026, 635, 509, 417, and 291, respectively) than the on-target site (253 reads). Conversely, GUIDE-seq analysis in VEsiCas-treated cells detected exclusively a single off-target site, with hundreds less GUIDE-seq reads than on-target (355 reads on-target and 11 reads off-target) and a 37 times improved on/off-target ratio compared with plasmid transfection.
Figure 4.
Analysis of On/Off-Target Activity Generated by VEsiCas on the VEGFA Locus
(A) Traceless delivery of SpCas9 through VEsiCas. Shown is a time course analysis of SpCas9 intracellular levels by western blot analysis in HEK293T cells transduced with VEsiCas or transfected with a plasmid expressing SpCas9. The reported time points correspond to the time of analysis following treatments (transduction or transfection). The western blot is representative of n = 2 independent experiments. (B) Targeted comparison of genome editing specificity using VEsiCas or a plasmid expressing SpCas9 in combination with sgVEGFA site3. Shown are percentages of indel formation measured by TIDE in the VEGFA site3 locus and in two sgVEGFA site3 previously validated off-target sites (OT1 and OT3) in HEK293T cells. The dashed bars represent the TIDE background. (C) Genome-wide specificity of VEsiCas targeting the VEGFA site3 locus. Shown is GUIDE-seq analysis for the sgRNA targeting VEGFA site3 locus in HEK293T cells transfected with SpCas9 or treated with VEsiCas. The black square indicates on-target sites. DNA from three biological replicates was mixed before library preparation. Right: the total number of off-targets identified for the sgVEGFA site3 with SpCas9 transfection or VEsiCas treatments.
In conclusion, the off-target analysis performed on the VEGFA locus, a gold standard for the evaluation of SpCas9 specificity,11 revealed that VEsiCas produced a more specific genomic modification, which correlates with the rapid clearance of the nuclease from the target cells.
Discussion
The CRISPR-Cas9 technology is a powerful tool for genome editing. Its translation into clinical use strongly depends on further improvements in specificity (complete abrogation of off-target activity) and delivery. These are tightly interdependent aspects of genome editing because the amount and time of Cas9 expression in recipient cells strongly correlate with the frequencies of off-target cleavages.13, 14, 40 Moreover, persistence of Cas9 expression may generate adverse immune responses toward the modified cells in vivo, further reinforcing the demand for a highly controllable method of delivery.41 Transient expression of Cas9 has been obtained through physical and chemical delivery methods,14, 16, which are not ideal for in vivo applications.18 Premiere tools for in vivo gene delivery are viral vectors, which, however, have serious limitations deriving from their potential risk of insertional mutagenesis13, 21 and permanent transgene expression that may increase the number of SpCas9 off-target cleavages.42 Genotoxicity associated with viral integration can be partially circumvented by using non-integrating viral vectors such as those derived from AAVs).24, 43 Because of size limitations, SpCas9 orthologs, such as the one from Staphylococcus aureus,44 or split-protein strategies (split-Cas9) have generally been employed with AAV vectors.45 However, the separation of CRISPR elements introduces high complexity into the system, and occasional AAV integrations have been reported.43 Similarly, unpredictable insertional mutagenesis reported with other non-integrating vectors, such as IDLVs, can be enhanced by nuclease activity beyond the background level.46, 47 Attempts to combine the advantages offered by viral delivery together with transient SpCas9 expression were recently addressed by using a self-limiting CRISPR-Cas9 lentiviral vector13 and by engineering lentiviral particles containing pre-packaged SpCas9 together with a non-integrating lentiviral vector expressing the sgRNAs.25 A further step toward the exploitation of viral delivery properties in a DNA-free context is the development of VLPs,26, 28 which have mainly been used in the past decade for vaccination approaches27 and for the delivery of exogenous proteins.26, 28 In this study, we explored the use of lentivirus-based VLPs for the delivery of SpCas9 fused to HIV-1 Gag in the absence of a viral transfer genome. To further abandon viral elements for the production of vesicles, we exploited the fusogenic properties of VSV-G alone, which is sufficient for the delivery of protein cargos.29 We discovered that VSV-G vesicles, in combination with SpCas9, were sufficiently loaded with SpCas9 protein. However, vesicles produced under standard experimental conditions using nuclear U6-based transcription for sgRNA synthesis showed poor genome-editing activity, suggesting non-sufficient amounts of incorporated sgRNA. This limitation was circumvented by favoring SpCas9 protein assembly with the sgRNA in producing cells through cytoplasmic sgRNA synthesis driven by the T7 RNA polymerase. To this end, we employed cells resistant to cytotoxicity induced by high levels of uncapped 5′-triphosphate cytoplasmic RNA, BSR-T7/5.32
A remarkable advantage offered by the VEsiCas approach is the transient nature of delivered SpCas9. As demonstrated in this study, the rapid clearance of SpCas9, which decreased as soon as 12 hr after VEsiCas treatment, strongly lowered the off-target activity associated with genome editing, as opposed to high levels of non-specific cleavages generated by plasmid transfection. Our data also prove that VEsiCas are more efficient and less toxic in delivering SpCas9 RNPs than electroporation. In fact, our system requires less SpCas9 protein, offering a clear advantage in preventing potential adverse effects of immune responses against the edited cells.41
Eventually, VEsiCas can be readily adapted to more complex genome-editing approaches, such as the use of Cas9 nickase, requiring incorporation of sgRNAs pairs; their use with other emerging RNA-guided nucleases can also be envisioned.48
Overall, the efficient and traceless delivery of CRISPR-Cas9 through the VEsiCas approach represents a further advancement toward safer in vivo genome editing.
Materials and Methods
Plasmids and Oligonucleotides
The sgRNAs for the initial experiments on VSV-G vesicle production were transcribed from a U6 promoter-driven cassette derived from pX330 (a gift from Feng Zhang, Addgene 42230)1 and cloned into the pUC19 plasmid.13 sgRNA oligos were cloned using a double BbsI site inserted before the sgRNA constant portion according to a previously published cloning strategy.1 The Gag-SpCas9 plasmid was obtained through the fusion of the Gag coding sequence with 3×FLAG-SpCas9 encoded by the pX-SpCas9 vector.13 pX-SpCas9 was obtained by removal of an NdeI fragment including the sgRNA expression cassette from pX330 and was used to express SpCas9. MinimalGag-Cas9 was assembled in pCDNA3 by subcloning the SpCas9 coding sequence from pX-SpCas9 and MinimalGag from the Δ-Zwt-p2b plasmid,36 a generous gift from H.G. Gottlinger. Afterward, an additional version of both constructs was obtained by removing, through site-directed mutagenesis, a methionine in the linker peptide derived from pX-Cas9 that led to unfused SpCas9 production (Figure S4).
For VLP and VEsiCas production in BSR-T7/5 cells, sgRNAs were transcribed from a pVAX-T7-sgRNA expression plasmid having a T7 promoter-driven cassette cloned into the pVAX plasmid at the NdeI site. sgRNA oligos were cloned in pVAX-T7-sgRNA using a double BsmBI site inserted before the sgRNA constant portion (a list of oligonucleotides used to clone sgRNAs is available in Table S1). pVAX-T7-sgRNA included a 5′ HDV ribozyme33 designed to cleave the 3′ end of the sgRNA containing the T7 terminator and the ribozyme itself. The Cas9-nickase construct was obtained by inserting the D10A mutation in the SpCas9 coding sequence. The T7 RNA polymerase coding sequence was amplified from the genome of BSR-T7/5 cells and cloned in place of EGFP into the pEGFP-N1 plasmid using the HindIII/XbaI sites to obtain the pKANA-T7-RNA-Pol plasmid. Plasmids were verified by Sanger sequencing. Information regarding relevant plasmid DNA sequences produced for this manuscript can be found in the Supplemental Information.
Cell Cultures and Transfection
BHK21-derived producing cells stably expressing T7 polymerase (BSR-T7/5) were a kind gift from Karl Conzelmann (Ludwig-Maximilians-Universität, Munich, Germany).31 To select cells that retain the T7 RNA polymerase construct, the medium was supplemented with 1 mg/mL G418 (Gibco-Life Technologies). HEK293T cells were obtained from the ATCC. HEK293-EGFP cells were generated by stable transfection of pEGFP-IRES-Puro and selected with 1 μg/mL of puromycin.13 BHK-21 (ATCC CCL-10), Vero (ATCC-CCL-81), HeLa (ATCC-CCL-2), and all cell lines described above were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, 10 U/mL penicillin, and 10 μg/mL streptomycin. J-Lat-A1 are Jurkat cells that have been latently transduced by an HIV-1 vector encoding EGFP.49 J-Lat-A1 cells were cultured in RPMI medium supplemented with 10% FBS and penicillin and streptomycin (pen/strep) antibiotics. EGFP expression was induced with 10 nM TPA (12-O-tetradecanoylphorbol-13-acetate) treatment for 24 hr. To obtain HeLa-EGFP, HeLa cells were transfected with the pEGFP-C1 plasmid (Clontech Laboratories) using FuGENE HD transfection reagent (Promega). After selection in culture medium with 400 μg/mL G418 (Life Technologies) for approximately 10 days, cells expressing high levels of EGFP were enriched by fluorescence-activated cell sorting (FACS) and propagated as a polyclonal cell population. Stock cultures of HeLa-EGFP cells (∼95% EGFP-positive cells) were maintained in culture medium supplemented with 200 μg/mL G418. Transgenic human iPSCs constitutively expressing GFP were derived from a commercial human episomal iPSC line (Gibco, Thermo Fisher Scientific) originally derived from CD34+ cord blood using an EBNA-based episomal system. Human iPSC clones stably expressing copGFP under control of the ubiquitous cytomegalovirus promoter were generated by infection with the pGZ-CMV-copGFP lentiviral vector (System Biosciences). Zeocin-based clone selection was started 72 hr after infection for 7 days. Resistant colonies were manually picked, expanded clonally, and characterized for their pluripotency competence. Human iPSC lines were grown on feeder-free Geltrex-coated dishes and cultured in StemMACS iPS-Brew XF medium (Miltenyi Biotec). All cell lines were verified to be mycoplasma-free (PlasmoTest, InvivoGen).
Transfection experiments were performed in 12–24 multi-well plates with 250–1,000 ng of each plasmid using the TransIT-LT1 (Mirus) reagent according to manufacturer’s instructions. Cells were collected 2–4 days after transfection or as described.
Gene substitution experiments through HDR of the EGFP-Y66S gene were performed using previously described cells and donor DNA.13 Briefly 293-iY66S cells (Flp-In T-REx system, Life Technologies) were generated by Flp-mediated recombination of the pcDNA5-FRT-TO-EGFP-Y66S plasmid in cells carrying a single genomic FLP recombinase target (FRT) site and stably expressing the tetracycline repressor. 293-iY66S cells were cultured in selective medium containing 15 μg/mL blasticidin and 100 μg/mL hygromycin (Life Technologies). 293-iY66S cells were transfected in 24 multi-wells with 1 μg of pcDNA5-FRT-TO-rEGFP(1-T203K-stop) donor plasmid together with either 500 ng of pcDNA3.1 or 250 ng of pX-Cas9 and 250 ng of pUC19 plasmid encoding sgRNAEGFPBi or control sgRNA (sgCtr) using TransIT-LT1 (Mirus Bio) according to manufacturer’s instructions. 16 hr after transfection with the donor plasmid, cells were washed with new medium and treated with VEsiCas (1 μg Cas9). Cells were collected and analyzed 4 days after treatment. The expression of EGFP was induced by treatment with 100 ng/mL doxycycline (Cayman Chemical) for 20 hr before fluorescence measurement.
VSV-G/SpCas9 Vesicles, Lenti-VLPs, and VEsiCas Production
For VSV-G/Cas9 vesicle production, a 100-mm dish of confluent HEK293T cells was transfected with 15 μg of pxCas9, Gag-SpCas9, or pCDNA3-MinGag-Cas; 15 μg of the desired pUC-U6-sgRNA; and 3 μg of VSV-G plasmids using the polyethyleneimine (PEI) method.50 Subsequent productions were performed in BSR-T7/5 cells using the conditions reported above, except for the pVAX-T7-sgRNA plasmid, which substituted pUC-U6-sgRNA for RNA guide expression. For Multi-VEsiCas deletion experiments, 7.5 μg of each pVAX-T7-sgRNA (sgEGFP5 and sgEGFPBi) targeting EGFP was used for particle production. For VEsiCas-n production, 15 μg of Cas9-Nickase plasmid and 7.5 μg of each pVAX-T7-sgEGFPBi and pVAX-T7-sgEGFP3gW were used. After 12 hr of incubation, the medium was replaced with fresh complete DMEM, and 48 hr later, the supernatant was collected, centrifuged at 400 × g for 5 min, and filtered through a 0.22 μm polyethersulfone (PES) filter. VLPs and VEsiCas were then concentrated and purified through a 20% sucrose cushion by ultracentrifugation for 2 hr at 150,000 × g (4°C) and suspended in suitable volumes of complete medium (DMEM, RPMI, or StemMACS iPS-Brew XF medium, according to the target cells) or 1× PBS for mouse injections and stored at −80°C. The amount of SpCas9 or Gag-SpCas9 and MinGag-SpCas9 chimeras produced in the VSV-G vesicles was evaluated through western blot analysis using purified SpCas9 as a standard. Unless indicated, for each single transduction experiment we used vesicles containing about 1 μg of SpCas9. The reference recombinant SpCas9 protein was produced in bacteria (see below) according to Gagnon et al.51 and quantified through Coomassie staining. The efficiency of SpCas9 incorporation into VEsiCas was obtained through a western blot ratio of incorporated SpCas9 protein on the total amount of proteins quantified in VEsiCas by Bradford assay (Bio-Rad).
Delivery in Target Cells
The day before transduction, 1 × 105 HEK293T, HEK293-EGFP, HeLa-EGFP, or J-LAT-A1 cells were seeded in a 24-well plate. VEsiCas and VLPs were delivered into target cells by spinoculation for 2 hr at 1,600 × g at 20°C (30 min at 1,000 × g and 24°C for human iPSCs), followed by overnight incubation at 37°C. J-Lat-A1 cells were induced by TPA (Sigma-Aldrich) and analyzed for genome editing 3 days after the last transduction or, for EGFP reduction, at least 7 days after the treatment. For VEGFA and CXCR4 loci, triple transduction experiments were performed. Cells were trypsinized 24 hr after each transduction; 2/3 of cells were collected for genomic analysis and 1/3 was subcultured for following treatments, one every 48 hr. Cells were collected for final analysis 3 days after the last treatment.
SpCas9-sgRNA Electroporation
Recombinant SpCas9 protein was produced in bacteria according to Gagnon et al.51 Briefly, the pET-28b-Cas9-His plasmid (a gift from Alex Schier, 47327) was used to express SpCas9 in E. coli Rosetta cells (Novagen), which were grown for 12 hr at 37°C, followed by 24 hr induction at 18°C. Purification was performed on his-tag resin (G-Biosciences), and the column was washed with 20 mM Tris (pH 8), 30 mM imidazole, and 500 mM NaCl and eluted with 20 mM Tris (pH 8), 500 mM imidazole, and 500 mM NaCl. After dialysis into 20 mM Tris, 200 mM KCl, and 10 mM MgCl2, single-use aliquots were stored at −80°C. To produce in vitro-transcribed sgRNAs, we PCR-amplified pVAX-T7-sgEGFPBi and pVAX-T7-sgCtr with primers T7 promoter forward (fw) and gRNA end reverse (rev) (Table S2). This PCR product, containing the T7 promoter and the complete sequence of the sgRNA, was used for in vitro transcription using the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs) following the manufacturer’s instructions. TRIzol (Invitrogen)-purified sgRNAs, precipitated with isopropanol and washed with 75% ethanol, were analyzed by acrylamide gel electrophoresis and quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Purified sgRNAs were mixed with recombinant SpCas9 immediately before electroporation by incubating 12.4 μg of SpCas9 with 3.1 μg of sgRNA (or as indicated with a 4:1 mass ratio between protein and RNA) in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.5), 150 mM KCl, 1 mM MgCl2, 10% (v/v) glycerol, and 1 mM DTT at 37°C for 10 min. 2.5 × 105 HEK293-EGFP cells were nucleofected using the Q001 protocol in 120 mM K2HPO4/KH2PO4 (pH 7.2), 15 mM MgCl2, 10 mM glucose, and 5 mM KCl using Lonza Nucleofector II. Cells were analyzed for EGFP loss 7 days after electroporation.
Viability Assay
48 hr following VEsiCas spinoculation or RNP electroporation, HEK293-EGFP cells were collected, and the viability was measured with the Vybrant MTT Cell Proliferation Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Detection of SpCas9-Induced Mutations
EGFP expression was analyzed with the Invitrogen Tali image-based cytometer. In the comparative analysis with electroporated RNPs, analysis of EGFP expressing cells was performed by FACS (FACSCanto, BD Biosciences). To detect indels in the CXCR4 and VEGFA loci, genomic DNA was isolated using the DNeasy Blood & Tissue Kit (QIAGEN). PCRs on purified genomic DNA were performed using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific). Samples were amplified using the oligonucleotides listed in Table S2. Purified PCR products were analyzed by sequencing and applying the TIDE tool.38 Detection of the deletion in the EGFP locus after Multi-VEsiCas treatment was revealed by PCR amplification using oligonucleotides EGFP fw and EGFP rev.
GUIDE-Seq
2 × 105 HEK293T cells were transfected with 750 ng of SpCas9-expressing plasmid together with 250 ng of VEGFA site3 sgRNA-coding plasmid or an empty pUC19 plasmid, 10 pmol of the bait double-stranded oligodeoxynucleotide (dsODN) containing phosphorothioate bonds at both ends (designed according to the original GUIDE-seq protocol39), and 50 ng of a pEGFP-IRES-Puro plasmid expressing both EGFP and the puromycin resistance gene. VEsiCas targeting VEGFA site3 were delivered by spinoculation 6 hr following transfection of dsODN and pEGFP-IRES-Puro coding plasmids. The following day, cells were trypsinized and replated. The procedure was repeated every 48 hr. After the last treatment, cells were detached and selected with 2 μg/mL of puromycin for 48 hr. Cells were then collected, and genomic DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN) following the manufacturer’s instructions and sheared to an average length of 500 bp with the Bioruptor Pico sonication device (Diagenode). Library preparations were performed with the original adapters and primers according to previous work.39 Libraries were quantified with the Qubit dsDNA High Sensitivity Assay Kit (Invitrogen) and sequenced with the MiSeq sequencing system (Illumina) using an Illumina Miseq Reagent kit V2, 300 cycles (2 × 150 bp paired end). Raw sequencing data (FASTQ files) were analyzed using the GUIDE-seq computational pipeline.39 After demultiplexing, putative PCR duplicates were consolidated into single reads. Consolidated reads were mapped to the human reference genome GrCh37 using BWA-MEM37; reads with a mapping quality lower than 50 were filtered out. Upon identification of the genomic regions integrating dsODNs in aligned data, off-target sites were retained when at most eight mismatches against the target were present and absent in the background controls. Visualization of aligned off-target sites is available as a color-coded sequence grid. Off-target sites detected are listed in Table S3.
Western Blots
Collected cells or supernatants containing VSV-G vesicles were lysed in NEHN buffer (20 mM HEPES [pH 7.5], 300 mM NaCl, 0.5% NP40, 1 mM EDTA, and 20% glycerol) supplemented with 1% of protease inhibitor cocktail (Pierce). Protein extracts were separated by SDS-PAGE using the PageRuler Plus Protein Standards as the standard molecular mass markers (Thermo Fisher Scientific). After electrophoresis, samples were transferred onto 0.22 μm PVDF membranes (GE Healthcare). The membranes were incubated with mouse anti-FLAG (Sigma) for detecting Gag-Cas9, MinGag-Cas9, and SpCas9 and mouse anti-α-tubulin (Sigma) and with the appropriate horseradish peroxidase (HRP)-conjugated goat anti-mouse (Kirkegaard & Perry Laboratories [KPL]) secondary antibody for enhanced chemiluminescence (ECL) detection. The Guide-it Cas9 monoclonal antibody (clone TG8C1) (Clontech) was used to quantify SpCas9 by western blotting using recombinant SpCas9 as a reference. Images were acquired and bands were quantified using the UVItec Alliance detection system.
In Vivo Delivery of VEsiCas
Animal care and treatments were conducted in conformity with institutional guidelines in compliance with national and international laws and policies (European Economic Community [EEC] Council Directive 86/609, OJL 358, December 12, 1987 and D.lgs 116/92) upon approval by the International Centre for Genetic Engineering and Biotechnology (ICGEB) Ethical Committee for Animal Experimentation and by the Italian Ministry of Health. Transgenic mice (males) expressing EGFP (C57BL/6-Tg(CAG-EGFP)1Osb/J from The Jackson Laboratory) on post-natal day 5 were anesthetized by hypothermia on ice for ∼3–5 min, placing a gauze below the pup to avoid direct contact with ice. A transverse skin incision across the lower half of the chest was performed using small scissors, followed by gentle separation of the skin from underlying muscle by using blunt dissection. A lateral tracheotomy was created by making a small incision at the fourth intercostal space to visualize the heart. VEsiCas (5 μL, corresponding to 4 μg of SpCas9) carrying sgCtr or a guide targeting EGFP (sgEGFPBi) were injected into the left ventricular anterior wall using a 31G needle (n = 5 per group). The ribs and the skin were sutured together using an 8-0 nonabsorbable prolene suture to seal the chest wall incision. The neonates were warmed rapidly under a heat lamp for several minutes until recovery and re-introduced to the mother. After 10 days, the hearts were collected, fixed in 4% paraformaldehyde, and snap-frozen in isopentane and liquid nitrogen for fluorescence microscopy analysis.
Immunofluorescence and Image Analysis
Frozen sections were washed 3 times in PBS, permeabilized in 0.5% Triton X-100 for 30 min, and blocked in 10% goat serum for 1 hr. Sections were stained overnight at 4°C with anti-sarcomeric α-actinin antibodies (Abcam) 1:100 in 5% goat serum. After 2 washing steps of 5 min in 0.5% Triton X-100 at room temperature, sections were incubated 1 hr in 1:200 anti-mouse secondary antibody conjugated to Alexa Fluor 594 (Life Technologies) in 10% goat serum for 45 min at room temperature. Nuclei were stained with DAPI solution (Sigma). Quantification of EGFP-positive cardiomyocytes was performed on 10× images of coronal heart sections, corresponding to a 2 mm2 area of the left ventricular anterior wall, surrounding the injection site. At least 3 different sections and 10 images per animal were used for the quantification (50 images per group). Data are expressed as the percentage of EGFP-positive cardiomyocytes over the total number of α-actinin-labeled cardiomyocytes.
Data Availability
All relevant data are available from the authors upon request.
Author Contributions
C.M., G.P., A. Casini, G.M., G.M.F., and I.Z. performed the experiments in cell lines. C.M., G.P., and A. Casini designed the experiments and collected and analyzed the data. C.M., G.P., A. Casini, and A.C. conceived and designed the study. L.C. contributed to human iPSC experiments. F.A. and O.R.B. contributed in setting up the producing system and provided reagents. S.Z., L.Z., and M.G. contributed to the experiments in the mouse model. A.C. was responsible for the coordination of the study. All authors read, corrected, and approved the final manuscript.
Conflicts of Interest
The technology reported in this manuscript is under consideration for patent filing.
Acknowledgments
We thank K. Conzelmann (Ludwig-Maximilians-Universität, Munich, Germany) for the BSR-T7/5 cell line and the LaBSSAH – CIBIO Next Generation Sequencing Facility of the University Trento for sequencing samples. The plasmid encoding a minimal HIV-1 Gag fused to RSV p2b (D-Zwt-p2b) was a kind gift from Heinrich Gottlinger. G.P.’s fellowship was supported by the CARITRO Foundation. This work was supported by intramural funding from the University of Trento.
Footnotes
Supplemental Information includes six figures, three tables, and sequences and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.05.010.
Contributor Information
Gianluca Petris, Email: gianluca.petris@unitn.it.
Anna Cereseto, Email: anna.cereseto@unitn.it.
Supplemental Information
References
- 1.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao H.-K., Gu Y., Diaz A., Marlett J., Takahashi Y., Li M., Suzuki K., Xu R., Hishida T., Chang C.J. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 6.Varshney G.K., Pei W., LaFave M.C., Idol J., Xu L., Gallardo V., Carrington B., Bishop K., Jones M., Li M. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015;25:1030–1042. doi: 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amabile A., Migliara A., Capasso P., Biffi M., Cittaro D., Naldini L., Lombardo A. Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell. 2016;167:219–232.e14. doi: 10.1016/j.cell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diao Y., Li B., Meng Z., Jung I., Lee A.Y., Dixon J., Maliskova L., Guan K.L., Shen Y., Ren B. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 2016;26:397–405. doi: 10.1101/gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klann T.S., Black J.B., Chellappan M., Safi A., Song L., Hilton I.B., Crawford G.E., Reddy T.E., Gersbach C.A. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 2017;35:561–568. doi: 10.1038/nbt.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romanel A., Garritano S., Stringa B., Blattner M., Dalfovo D., Chakravarty D., Soong D., Cotter K.A., Petris G., Dhingra P. Inherited determinants of early recurrent somatic mutations in prostate cancer. Nat. Commun. 2017;8:48. doi: 10.1038/s41467-017-00046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casini A., Olivieri M., Petris G., Montagna C., Reginato G., Maule G., Lorenzin F., Prandi D., Romanel A., Demichelis F. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018;36:265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai S.Q., Joung J.K. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petris G., Casini A., Montagna C., Lorenzin F., Prandi D., Romanel A., Zasso J., Conti L., Demichelis F., Cereseto A. Hit and go CAS9 delivered through a lentiviral based self-limiting circuit. Nat. Commun. 2017;8:15334. doi: 10.1038/ncomms15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S., Kim D., Cho S.W., Kim J., Kim J.-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramakrishna S., Kwaku Dad A.B., Beloor J., Gopalappa R., Lee S.K., Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H., Maeder M.L., Joung J.K., Chen Z.Y., Liu D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mout R., Ray M., Yesilbag Tonga G., Lee Y.W., Tay T., Sasaki K., Rotello V.M. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mout R., Ray M., Lee Y.-W., Scaletti F., Rotello V.M. In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges. Bioconjug. Chem. 2017;28:880–884. doi: 10.1021/acs.bioconjchem.7b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chick H.E., Nowrouzi A., Fronza R., McDonald R.A., Kane N.M., Alba R., Delles C., Sessa W.C., Schmidt M., Thrasher A.J., Baker A.H. Integrase-deficient lentiviral vectors mediate efficient gene transfer to human vascular smooth muscle cells with minimal genotoxic risk. Hum. Gene Ther. 2012;23:1247–1257. doi: 10.1089/hum.2012.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakai H., Yant S.R., Storm T.A., Fuess S., Meuse L., Kay M.A. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A., Ando D., Urnov F.D., Galli C., Gregory P.D. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 24.Zacchigna S., Zentilin L., Giacca M. Adeno-associated virus vectors as therapeutic and investigational tools in the cardiovascular system. Circ. Res. 2014;114:1827–1846. doi: 10.1161/CIRCRESAHA.114.302331. [DOI] [PubMed] [Google Scholar]
- 25.Choi J.G., Dang Y., Abraham S., Ma H., Zhang J., Guo H., Cai Y., Mikkelsen J.G., Wu H., Shankar P., Manjunath N. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 2016;23:627–633. doi: 10.1038/gt.2016.27. [DOI] [PubMed] [Google Scholar]
- 26.Voelkel C., Galla M., Maetzig T., Warlich E., Kuehle J., Zychlinski D., Bode J., Cantz T., Schambach A., Baum C. Protein transduction from retroviral Gag precursors. Proc. Natl. Acad. Sci. USA. 2010;107:7805–7810. doi: 10.1073/pnas.0914517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramqvist T., Andreasson K., Dalianis T. Vaccination, immune and gene therapy based on virus-like particles against viral infections and cancer. Expert Opin. Biol. Ther. 2007;7:997–1007. doi: 10.1517/14712598.7.7.997. [DOI] [PubMed] [Google Scholar]
- 28.Kaczmarczyk S.J., Sitaraman K., Young H.A., Hughes S.H., Chatterjee D.K. Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16998–17003. doi: 10.1073/pnas.1101874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangeot P.-E., Dollet S., Girard M., Ciancia C., Joly S., Peschanski M., Lotteau V. Protein transfer into human cells by VSV-G-induced nanovesicles. Mol. Ther. 2011;19:1656–1666. doi: 10.1038/mt.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim K., Park S.W., Kim J.H., Lee S.H., Kim D., Koo T., Kim K.E., Kim J.H., Kim J.S. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 2017;27:419–426. doi: 10.1101/gr.219089.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchholz U.J., Finke S., Conzelmann K.K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Habjan M., Penski N., Spiegel M., Weber F. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J. Gen. Virol. 2008;89:2157–2166. doi: 10.1099/vir.0.2008/002097-0. [DOI] [PubMed] [Google Scholar]
- 33.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D.-H., Longo M., Han Y., Lundberg P., Cantin E., Rossi J.J. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 35.Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Webe F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 36.Accola M.A., Strack B., Göttlinger H.G. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chew W.L., Tabebordbar M., Cheng J.K., Mali P., Wu E.Y., Ng A.H., Zhu K., Wagers A.J., Church G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson C.E., Gersbach C.A. Engineering Delivery Vehicles for Genome Editing. Annu. Rev. Chem. Biomol. Eng. 2016;7:637–662. doi: 10.1146/annurev-chembioeng-080615-034711. [DOI] [PubMed] [Google Scholar]
- 43.Deyle D.R., Russell D.W. Adeno-associated virus vector integration. Curr. Opin. Mol. Ther. 2009;11:442–447. [PMC free article] [PubMed] [Google Scholar]
- 44.Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., Marco E., Gotta G.M., Maeder M.L., Kennedy E.M. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truong D.-J.J., Kühner K., Kühn R., Werfel S., Engelhardt S., Wurst W., Ortiz O. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43:6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gabriel R., Lombardo A., Arens A., Miller J.C., Genovese P., Kaeppel C., Nowrouzi A., Bartholomae C.C., Wang J., Friedman G. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Wang Y., Wu X., Wang J., Wang Y., Qiu Z., Chang T., Huang H., Lin R.J., Yee J.K. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 48.Komor A.C., Badran A.H., Liu D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jordan A., Bisgrove D., Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casini A., Olivieri M., Vecchi L., Burrone O.R., Cereseto A. Reduction of HIV-1 infectivity through endoplasmic reticulum-associated degradation-mediated Env depletion. J. Virol. 2015;89:2966–2971. doi: 10.1128/JVI.02634-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gagnon J.A., Valen E., Thyme S.B., Huang P., Akhmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the authors upon request.