Abstract
Background
Cardiac hypertrophy is a well-known risk factor for heart failure and sudden cardiac death (SCD). On the other hand, physiological cardiac hypertrophy is often observed in young healthy men, and it is difficult to predict SCD in cardiac hypertrophy subjects who do not show symptoms of heart failure. MicroRNAs (miRNAs) widely regulate biological activity and play pivotal roles in heart failure progression. In this study, we investigated whether miRNA expression is altered in SCD with cardiac hypertrophy (SCH).
Methods
Cardiac tissues were sampled at autopsy from SCH patients, compensated cardiac hypertrophy (CCH) subjects who died of causes other than heart failure, and control cases without cardiac hypertrophy or heart failure. After histopathological examination, we performed deep sequencing and quantitative PCR of cardiac miRNAs.
Results and discussion
Although SCH and CCH showed indistinguishable histological features, their miRNA expression signatures were distinct. Among the 240 miRNAs stably detected in the heart, 8 were differentially expressed between SCH and CCH. Specifically, miR-221 increased in SCH compared to CCH and control cases. The significant elevation of cardiac miR-221 in SCH patients is correlated with lethal outcomes. Thus, our results indicate that an elevated miR-221 level is potentially associated with an increased risk of SCD in subjects with cardiac hypertrophy.
Keywords: Cardiology, Pathology, Medicine, Clinical genetics
1. Introduction
Cardiac hypertrophy occurs as an adaptive response to increased body size because the mammalian myocardium has limited ability to proliferate after birth [1]. Physiological cardiac hypertrophy maintains normal cardiac function and sometimes exhibits enhanced pumping capacity, such as in athletes performing systematic training [2, 3]. However, prolonged and abnormal loading conditions promote pathological cardiac hypertrophy, accompanied by decreased cardiac function [4, 5, 6]. Maladaptive cardiac remodeling provokes conductive disturbance and arrhythmia, leading to sudden cardiac death (SCD) [7]. However, it is clinically difficult to predict SCD with cardiac hypertrophy (SCH) in the absence of preceding symptoms of heart failure.
Hypertrophic cardiomyopathy (HCM) is a well-studied type of pathological cardiac hypertrophy that is caused by mutations in genes encoding sacromeric proteins. In most cases, the patient inherited the condition by autosomal dominant transmission [8, 9]. HCM patients are at a high risk for SCD, mostly because of ventricular arrhythmias that are associated with a characteristic pathology such as myofiber disarray and interstitial fibrosis [10, 11]. However, although SCD is the major cause of sudden unexpected death in people, HCM is rarely identified among SCH victims by forensic autopsy because HCM is often diagnosed premortem. SCD in HCM patients is typically considered a natural death and, therefore, rarely the subject of a forensic autopsy. Alternatively, it may be examined by pathological autopsy. Most SCH cases at forensic autopsy lack a family history of SCD and histological characteristics of HCM; typically, they show symmetric ventricular hypertrophy accompanied by prolonged hypertension or severe sclerosis of the coronary artery or aortic valve. Although this common type of SCH has not yet been systematically studied, it can be categorized into subtypes such as hypertensive heart failure and secondary cardiomyopathy. In this study, we focused on the epigenetic changes in common SCD cases with cardiac hypertrophy.
MicroRNAs (miRNAs) are short noncoding RNAs that widely regulate gene expression through the suppression of target mRNAs [12]. The miRNAs affect cell differentiation and metabolic activities and are involved in many pathophysiological processes [13, 14, 15]. To date, several miRNAs have been reported to play pivotal roles in heart failure progression [16, 17, 18]. Because of their small size and structural features, miRNAs are relatively tough and stable in blood or tissue. We have previously shown that quantification of cardiac miRNAs is applicable in postmortem examination of SCD patients [19, 20]. In this study, we performed miRNA profiling of tissue samples from SCH and compensated cardiac hypertrophy (CCH) subjects, with the aim of identifying changes in miRNA expression related to SCH and the discovery of novel biomarkers to be used in the diagnosis of patients at risk of SCH.
2. Methods
2.1. Tissue sampling
All the subjects were kept refrigerated at 4 °C until autopsy after death, which ranged from half a day to four days. In this study, we defined cardiac hypertrophy as heart weight exceeding 400 g and an adjusted heart index (heart weight/body height, g/cm) greater than 2.5 [21,22]. The SCH subjects were defined as suffering from cardiac hypertrophy in association with hypertension, aortic stenosis, coronary atherosclerosis or post-myocardial infarction. They could perform normal daily activities at least 24 hours prior to death. CCH subjects were defined as showing physiological cardiac hypertrophy without heart failure symptoms. Cases of HCM were not considered for this study because the condition is rarely identified following a forensic autopsy. Control cases did not exhibit cardiac hypertrophy or heart failure. The clinical characteristics of the subjects are summarized in Table 1.
Table 1.
Clinical characteristics of the study cases.
| Group | Cause of death (n) | Age (years) | Sex (m/f) | BMI (kg/m2) | BNP (pg/mL) |
|---|---|---|---|---|---|
| SCH | SCD: AMI (5), OMI (1), HHF (3), AS (1) |
63 ± 5.2 | 8/2 | 22.5 ± 1.1 | 105 ± 43.3* |
| CCH | Accident (7), Pneumonia (1) |
67 ± 3.6 | 7/1 | 23.7 ± 1.1 | 9.2 ± 2.8 |
| Con | Accident (5), Hepatic failure (2), Pneumonia (1) |
59 ± 3.4 | 7/1 | 20.4 ± 1.2 | 5.4 ± 1.7 |
SCH: sudden cardiac death with cardiac hypertrophy (n = 10), CCH: compensated cardiac hypertrophy (n = 8), Con: control (n = 8), BMI: body mass index [body weight/(body height)2], BNP: B-type natriuretic peptide, SCD: sudden cardiac death, AMI: acute myocardial infarction, OMI: old myocardial infarction, HHF: hypertensive heart failure, AS: aortic stenosis *: P < 0.05 versus control by Steel test.
Cardiac tissue and blood samples were obtained following a forensic autopsy. Small tissue samples from the left ventricular free wall were immediately immersed in liquid nitrogen and stored at –80 °C until RNA isolation. Other parts of the heart were fixed with 10% formalin for histopathological examination. This study was approved by the Ethics Committee of Tokai University (#14I06): informed consent was obtained from the bereaved relatives of all patients, and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki.
2.2. Histopathological analysis
Formalin-fixed paraffin-embedded tissues were stained with hematoxylin and eosin dyes. We examined the whole area of each heart to evaluate pathological changes. Microscopic measurements were performed in the middle layer of the left ventricular free wall, where the myocardium showed a uniform alignment of longitudinal sections. Square-shaped myocardia, in which the nuclei were at the centers of the cells, were selected, and the myocardial diameter was measured at the nuclear point. Ten fields per case were observed at 200-fold magnification, and the values were averaged.
2.3. RNA extraction
RNase-free water and equipment were used throughout this study. RNA was isolated from approximately 100 mg frozen tissue using a mirVana miRNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer's protocol. The total RNA concentration and purity were measured with a spectrophotometer, and the integrity of the RNA was assessed using microcapillary electrophoresis on a 2100 Bioanalyzer with an RNA kit (Agilent Technologies, Santa Clara, CA, USA). All RNA samples were stored at −80 °C until further analysis.
2.4. miRNA deep sequencing and data processing
Small RNA libraries were prepared from 4 SCH patients (1 AMI, 2 HHF, and 1 AS) and 4 CCH subjects (all accidents). Total RNA (1 μg) from each cardiac sample was used for TruSeq Small RNA Library Prep Kit (Illumina, San Diego, CA, USA) in accordance with the manufacturer's instructions. Briefly, 3′ and 5′ adapters were hybridized to RNAs, followed by reverse transcription. After amplification, cDNAs were purified and size-selected using gel electrophoresis. Final yields and size distributions of the amplified cDNAs were assessed using a 2100 Bioanalyzer with a High Sensitivity DNA kit (Agilent Technologies). The barcoded small RNA libraries were pooled to equimolar amounts for the final library for sequencing. In total, 10 pM of the library was sequenced with 50-bp single-end reads using a MiSeq instrument (Illumina).
The 3′ adaptor sequences were trimmed from raw reads using Cutadapt (version 1.11); reads with trimmed lengths of less than 14 bp were discarded. Filtered reads were mapped to the reference genome (hg38) using bwa (version 0.7.12) with the following parameters: bwa, aln; -n, 1; -o, 0; -e, 0; -l, 10; -k, 1; -t, 8. Calculation of the raw read counts was performed using htseq-count (version 0.6.0) with the human miRNA database miRBase v21.
Coverage depth data were analyzed on the CLC Genomics Workbench v6.0.1 (Qiagen, Valencia, CA, USA). The read counts of each known miRNA were normalized to the library size and presented as reads per million mapped reads. Only miRNAs with normalized read counts of more than 10 in at least one sample were subjected to further analysis.
2.5. Quantitative polymerase chain reaction
For 10 SCH, 8 CCH, and 8 control cases, the cDNA templates were synthesized using a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems). Specific primers for hsa-miR-193a-5p (TaqMan 002281), hsa-miR-221-3p (TaqMan 000524), hsa-miR-222-3p (TaqMan 002276), hsa-miR-424-5p (TaqMan 000604), hsa-miR-1180-3p (TaqMan 002847), and U6 snRNA (TaqMan 001973) were used, and PCR was performed following the manufacturer's protocol. Serially diluted cDNAs were used as the template for each primer set, and standard curves were generated. Triplicate Ct values were averaged and used for quantification of target miRNAs by applying the ΔΔCq method, which included U6 snRNA values as endogenous controls.
2.6. Statistics
All data are presented as means ± standard errors. Differential expression analysis of the sequencing data was performed with Baggerley's test, followed by a false discovery rate correlation. Comparisons in the histological analysis, quantitative PCR, and immunoblotting were performed using Steel tests. Differences with P values of less than 0.05 were considered statistically significant.
3. Results
3.1. Histopathology
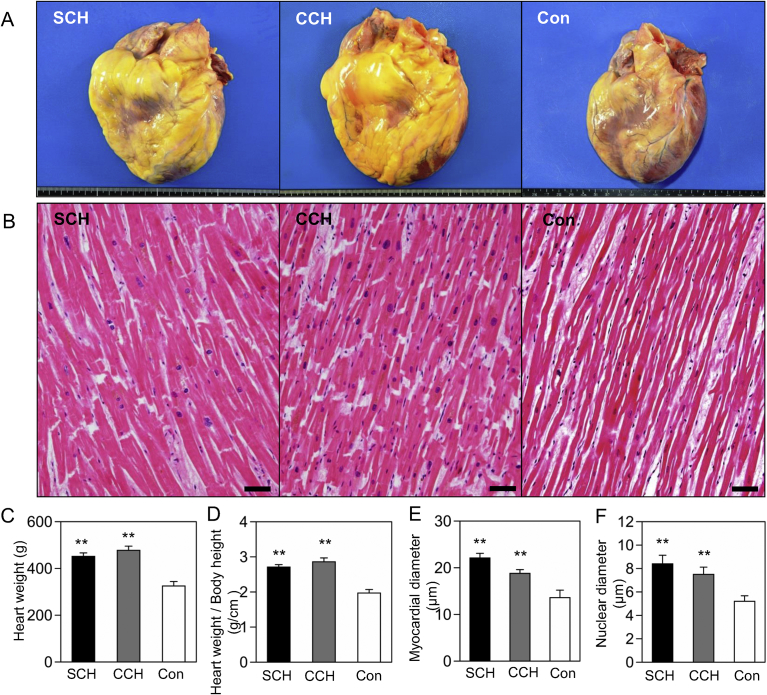
The hearts of patients with SCH and CCH were concentrically enlarged and showed similar gross morphology (Fig. 1A). The heart weight, myocardial diameter, and nuclear diameter were significantly increased in the SCH and CCH groups compared with the control group, whereas no differences were observed between the two hypertrophic groups (Fig. 1B–F). In the SCH, CCH, and control groups, heart weights were 450 ± 15, 478 ± 18, and 325 ± 19 g, respectively; the adjusted heart indexes were 2.7 ± 0.1, 2.9 ± 0.1, and 2.0 ± 0.1 g/cm, respectively; myocardial diameters were 22.1 ± 1.0, 18.8 ± 0.8, and 13.6 ± 1.6 μm, respectively; and nuclear diameters were 8.4 ± 0.7, 7.5 ± 0.6, and 5.2 ± 0.5 μm, respectively. In the SCH group, the OMI subject showed focal fibrosis and the AS subject showed a middle level of diffuse fibrosis in the left ventricles. The other SCH subjects did not present significant myocardial fibrosis, and none of the CCH and control subjects had extensive myocardial fibrosis or coronary atherosclerosis. All subjects lacked a significant accumulation of inflammatory cells.
Fig. 1.
Histopathological changes in the hearts of patients with SCH and CCH. (A) Gross features of the hearts. Horizontal scale = 20 cm. (B) Microscopical changes in the myocardia. Bar = 50 μm. (C) Heart weight. (D) Heart weight/body height. (E) Myocardial diameter. (F) Nuclear diameter. **P < 0.001 versus the control by Steel test. SCH: sudden cardiac death with cardiac hypertrophy (n = 10), CCH: compensated cardiac hypertrophy (n = 8), Con: control (n = 8).
3.2. MicroRNA deep sequencing
In total, 62,062,810 reads were sequenced, 52,140,437 reads (83.2%) were aligned to the human genome sequence, and 21,152,383 reads (31.4%) were mapped to known miRNAs. On average, 2,644,048 ± 662,624 human miRNA reads were sequenced in each library.
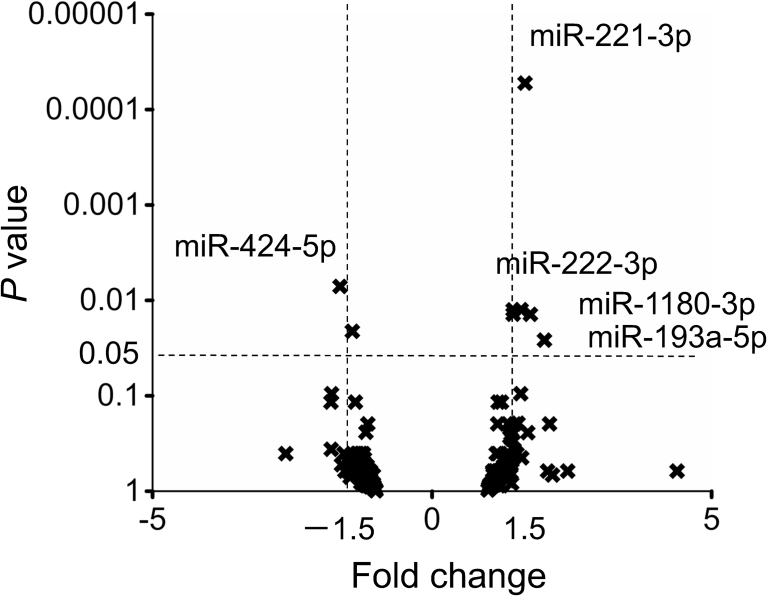
Among the 240 miRNAs that showed more than 10 normalized read counts in at least one library, 8 miRNAs were significantly differentially expressed between CCH and SCH (Supplemental Table 1, Fig. 2, Table 2). Subsequently, we performed real-time PCR to validate the expression differences for the 5 miRNAs that showed a fold change >1.5 in deep sequencing.
Fig. 2.
Alterations in cardiac miRNAs in patients with SCH and CCH. In total, 240 miRNAs were mapped according to fold change (SCH/CCH) and P value with Baggerley's test, followed by a false discovery rate correlation. SCH: sudden cardiac death with cardiac hypertrophy (n = 4), CCH: compensated cardiac hypertrophy (n = 4).
Table 2.
Expression differences in cardiac miRNAs in patients with SCH and CCH.
| miRNA | SCH read counts | CCH read counts | Fold change (SCH/CCH) | P value |
|---|---|---|---|---|
| hsa-miR-221-3p | 3,061 | 1,835 | 1.67 | 0.00005 |
| hsa-miR-424-5p | 107 | 176 | −1.64 | 0.007 |
| hsa-miR-222-3p | 835 | 523 | 1.60 | 0.013 |
| hsa-let-7d-5p | 1,252 | 860 | 1.46 | 0.013 |
| hsa-miR-1307-3p | 181 | 125 | 1.45 | 0.014 |
| hsa-miR-1180-3p | 27 | 15 | 1.76 | 0.014 |
| hsa-miR-218-5p | 61 | 86 | −1.42 | 0.021 |
| hsa-miR-193a-5p | 155 | 77 | 2.02 | 0.026 |
SCH: sudden cardiac death with cardiac hypertrophy (n = 4), CCH: compensated cardiac hypertrophy (n = 4). The read counts were normalized and presented as reads per million mapped reads. P values were determined using Baggerley's test followed by a false discovery rate correlation.
3.3. Quantitative PCR
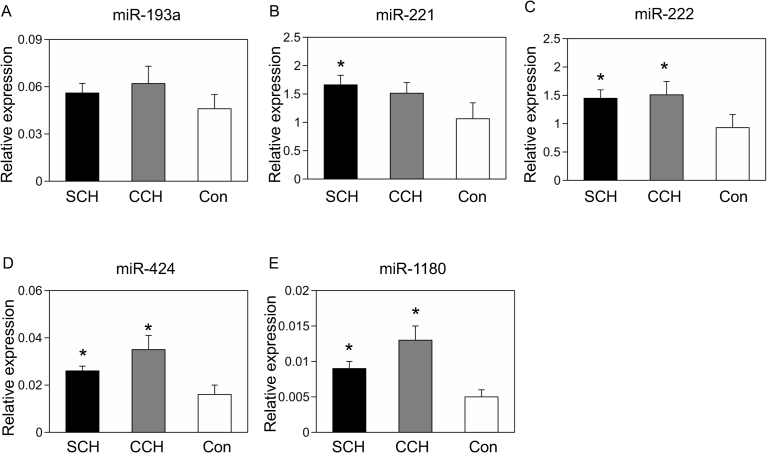
Real-time PCR revealed significant overexpression of miR-221 in SCH (P = 0.030) and a moderate increase in CCH (P = 0.14, Fig. 3B). The gradual elevation of cardiac miR-221 may be negatively correlated with patient outcomes. In contrast, the expression level of miR-222 was equally increased in SCH (P = 0.037) and CCH (P = 0.042, Fig. 3C), which may reflect the similar levels of hypertrophic growth. MiR-424 and miR-1180 were also increased in SCH and CCH compared with controls (Fig. 3D–E); however, their expression levels were relatively higher in CCH (P = 0.019) than in SCH (P = 0.046).
Fig. 3.
Validation of miRNA deep sequencing. Quantitative PCR was performed for miR-193a-5p (A), miR-221-3p (B), miR-222-3p (C), miR-424-5p (D), and miR-1180-3p (E) with U6 snRNA as an endogenous control in human hearts. *P < 0.05 versus the control by Steel test. SCH: sudden cardiac death with cardiac hypertrophy (n = 10), CCH: compensated cardiac hypertrophy (n = 8), Con: control (n = 8).
4. Discussion
To characterize changes in miRNA expression associated with the transition from cardiac hypertrophy to SCD, we compared human heart tissues from patients with SCH and CCH. The histopathological features of SCH and CCH were indistinguishable, such as enlargement of the myocardium and nucleus. Although the miRNA expression profiles of SCH and CCH were largely overlapping, we identified several important differences, allowing us to distinguish between SCH and CCH.
Human miR-221 and miR-222 are located on the X chromosome and share the same gene clusters [23]. Previous studies using transgenic mice have shown that miR-221/222 inhibits autophagy and promotes heart failure [24, 25]. Autophagy is an indispensable clearance system that removes damaged organelles and protein aggregates to maintain cellular homeostasis [26]. In the heart, mitochondrial autophagy plays a protective role against pressure overload [27], and inhibition of autophagy induces age-related cardiac hypertrophy and heart failure [28]. Interestingly, circulating miR-221/222 levels are negatively correlated with cardiac function in patients with heart failure [29, 30]. In this study, we found that cardiac miR-221 expression was significantly elevated in SCH, but that this increase was low to moderate in CCH. The stepwise elevation of miR-221 may reflect not only the hypertrophic growth of the myocardium but also the lethality of heart failure. Moreover, miR-221/222 is known to be involved in the regulation of anti-angiogenic genes, which inhibit tube formation, migration, and wound healing in endothelial cells [31, 32, 33]. Although coronary artery disease is not necessarily complicated with SCH in this study, endothelial dysfunction in small vessels in the myocardial layer can exacerbate heart failure in cardiac hypertrophy, where the oxygen demand is increased, and the heart becomes vulnerable to ischemic stress. In contrast, the expression level of miR-222 was equally increased in CCH and SCH in our study, which mainly reflected the enlargement of the heart and was weakly correlated with the lethal outcomes. Although the precise functional difference between miR-221 and miR-222 in hypertrophic progression is unknown [24, 25, 32, 33], our study on subjects with cardiac hypertrophy showed that miR-221 may have a stronger correlation with SCD than miR-222.
In this study, the expression level of cardiac miR-424 increased significantly in CCH and decreased slightly in SCH. MiR-424 is enriched in cardiac progenitor cells and drives cardiomyocyte specification [34]. Moreover, miR-424 is increased during the early phase of cardiac hypertrophy [35, 36]. Our study revealed, for the first time, that miR-424 was downregulated at the lethal phase of cardiac hypertrophy. The decrease in miR-424 in SCH hearts was consistent with another report showing that miR-424 was downregulated in the coronary sinus blood of patients with congestive heart failure [37]. The expression of miR-424 is elevated during compensated cardiac growth, and such overexpression is thought to be limited by the progression of heart failure at the chronic phase. Therefore, miR-424 may be useful to monitor heart failure progression but seems unsuitable for prediction of SCD.
The molecular changes observed in the process of pathological cardiac hypertrophy are known to resemble those observed during fetal cardiac development [38, 39]. For example, B-type natriuretic peptide (BNP) is abundantly expressed in fetal hearts and is clinically used as a biomarker for adult heart failure [40, 41]. However, the level of BNP is not always elevated in patients, particularly in cases of SCD. In this study, circulating BNP levels stayed in the normal range in about half of SCH patients. Thus, it is practically difficult to predict SCH using classical peptide biomarkers that mainly reflect chronic heart failure. As well, some fetal miRNAs, such as miR-21 and miR-208b, are also increased in failing human hearts [38, 42, 43]. Although overexpression of the fetal miRNA miR-424 was observed in SCH and CCH in our study, the evaluation did not allow us to clearly distinguish SCH from CCH. Therefore, it may also be difficult to predict SCD only by quantifying levels of fetal miRNAs.
Our study has some limitations. Because our current goal was to identify miRNA changes related to SCD, we analyzed autoptic cases and collected clinical information from the families following patients' death. Therefore, some information, including premortem symptoms, may be lacking, which is a major limitation of postmortem examinations. Specifically, because the serum BNP levels were elevated in some subjects of the SCH group, we cannot exclude the possibility that some perseverant patients, who had suffered premortem from an unrecognized heart failure symptom, were diagnosed postmortem with SCH. Moreover, as we excluded ambiguous cases, such as individuals living alone for whom precise clinical information was lacking, the sample size was limited. Although the increase in cardiac miR-221 was significant in our SCH subjects, many published data indicate that its up-regulation also occurs in various human malignancies, including breast cancer, liver cancer, gastric cancer, and glioma [44, 45]. Therefore, if miR-221 is used as a biomarker in SCD, it should be examined whether its expression change is affected by other malignant diseases as well.
In this study, elevated levels of cardiac miR-221 were detected in all SCH subjects using miRNA deep sequencing and qPCR. In cardiac hypertrophy, many compensatory pathways are constitutively activated, and the breakdown of such pathways can trigger SCD. Generally, a single miRNA can target several mRNAs, and individual mRNAs are mediated by several miRNAs [46, 47]; consistent with this, miR-221 has multiple roles in failing hearts as described above. This pleiotropic capacity makes miR-221 a potentially useful novel biomarker for prediction and diagnosis of SCH.
5. Conclusion
Cardiac hypertrophy is a risk factor for SCD, and epigenetic changes may exacerbate the progression from CCH to SCH. In this study, we performed miRNA profiling using human CCH and SCH tissues. Among the 240 miRNAs that were stably expressed in the hearts, miR-221 exhibited the most significant increase in SCH patients. This is consistent with the previously established role of miR-221, induction of heart failure by inhibition of autophagy and endothelial dysfunction. Our results support a potential association between miR-221 levels and SCD in patients with cardiac hypertrophy. Further studies with larger cohorts may assess the utility of miR-221 as a predictor of SCD.
Declarations
Author contribution statement
Yu Kakimoto: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Masayuki Tanaka: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Hideki Hayashi, Keiko Yokoyama: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Motoki Osawa: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by a JSPS KAKENHI grant (no. 16K19300).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Assistance with histopathological staining, miRNA sequencing, data processing, and qPCR was provided by the Support Center for Medical Research and Education at Tokai University.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Cardiac miRNA profiles in cardiac hypertrophy patients.
References
- 1.Soonpaa M.H., Kim K.K., Pajak L. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 1996;271(5):2183–2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 2.Maron B.J., Pelliccia A. The heart of trained athletes: cardiac remodeling and the risks of sports, including sudden death. Circulation. 2006;114(15):1633–1644. doi: 10.1161/CIRCULATIONAHA.106.613562. [DOI] [PubMed] [Google Scholar]
- 3.Tao L., Bei Y., Zhang H. Exercise for the heart: signaling pathways. Oncotarget. 2015;6(25):20773–20784. doi: 10.18632/oncotarget.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu I., Minamino T. Physiological and pathological cardiac hypertrophy. J. Mol. Cell. Cardiol. 2016;97:245–262. doi: 10.1016/j.yjmcc.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Abel E.D., Doenst T. Mitochondrial adaptations to physiological vs. pathological cardiac hypertrophy. Cardiovasc. Res. 2011;90(2):234–242. doi: 10.1093/cvr/cvr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMullen J.R., Jennings G.L. Differences between pathological and physiological cardiac hypertrophy: novel therapeutic strategies to treat heart failure. Clin. Exp. Pharmacol. Physiol. 2007;34(4):255–262. doi: 10.1111/j.1440-1681.2007.04585.x. [DOI] [PubMed] [Google Scholar]
- 7.Shenasa M., Shenasa H. Hypertension, left ventricular hypertrophy, and sudden cardiac death. Int. J. Cardiol. 2017;237:60–63. doi: 10.1016/j.ijcard.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Ho C.Y., Seidman C.E. A contemporary approach to hypertrophic cardiomyopathy. Circulation. 2006;113(24):e858–e862. doi: 10.1161/CIRCULATIONAHA.105.591982. [DOI] [PubMed] [Google Scholar]
- 9.Mathew J., Zahavich L., Lafreniere-Roula M. Utility of genetics for risk stratification in pediatric hypertrophic cardiomyopathy. Clin. Genet. 2018;93(2):310–319. doi: 10.1111/cge.13157. [DOI] [PubMed] [Google Scholar]
- 10.Atteya G., Lampert R. Sudden cardiac death in genetic cardiomyopathies. Card. Electrophysiol. Clin. 2017;9(4):581–603. doi: 10.1016/j.ccep.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Peterson D.F., Siebert D.M., Kucera K.L. Etiology of sudden cardiac arrest and death in US competitive athletes: a 2-year prospective surveillance study. Clin. J. Sport Med. 2018 doi: 10.1097/JSM.0000000000000598. [DOI] [PubMed] [Google Scholar]
- 12.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A., Slack F.J. Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Rottiers V., Naar A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Leva G., Croce C.M. miRNA profiling of cancer. Curr. Opin. Genet. Dev. 2013;23(1):3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bostjancic E., Zidar N., Stajer D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology. 2010;115(3):163–169. doi: 10.1159/000268088. [DOI] [PubMed] [Google Scholar]
- 17.Rao P.K., Toyama Y., Chiang H.R. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ. Res. 2009;105(6):585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akat K.M., Moore-McGriff D., Morozov P. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc. Natl. Acad. Sci. U. S. A. 2014;111(30):11151–11156. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakimoto Y., Kamiguchi H., Ochiai E. MicroRNA stability in postmortem FFPE tissues: quantitative analysis using autoptic samples from acute myocardial infarction patients. PLoS One. 2015;10(6):e0129338. doi: 10.1371/journal.pone.0129338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakimoto Y., Tanaka M., Kamiguchi H. MicroRNA deep sequencing reveals chamber-specific miR-208 family expression patterns in the human heart. Int. J. Cardiol. 2016;211:43–48. doi: 10.1016/j.ijcard.2016.02.145. [DOI] [PubMed] [Google Scholar]
- 21.Sawabe M., Saito M., Naka M. Standard organ weights among elderly Japanese who died in hospital, including 50 centenarians. Pathol. Int. 2006;56(6):315–323. doi: 10.1111/j.1440-1827.2006.01966.x. [DOI] [PubMed] [Google Scholar]
- 22.Michiue T., Sogawa N., Ishikawa T. Cardiac dilatation index as an indicator of terminal central congestion evaluated using postmortem CT and forensic autopsy data. Forensic Sci. Int. 2016;263:152–157. doi: 10.1016/j.forsciint.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Lim L.P., Glasner M.E., Yekta S. Vertebrate microRNA genes. Science. 2003;299(5612):1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 24.Su M., Wang J., Wang C. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ. 2015;22(6):986–999. doi: 10.1038/cdd.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su M., Chen Z., Wang C. Cardiac-specific overexpression of miR-222 induces heart failure and inhibits autophagy in mice. Cell. Physiol. Biochem. 2016;39(4):1503–1511. doi: 10.1159/000447853. [DOI] [PubMed] [Google Scholar]
- 26.Levine B., Klionsky D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6(4):463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 27.Shirakabe A., Zhai P., Ikeda Y. Drp1-dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure. Circulation. 2016;133(13):1249–1263. doi: 10.1161/CIRCULATIONAHA.115.020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taneike M., Yamaguchi O., Nakai A. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010;6(5):600–606. doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- 29.Lok S.I., de Jonge N., van Kuik J. MicroRNA expression in myocardial tissue and plasma of patients with end-stage heart failure during LVAD support: comparison of continuous and pulsatile devices. PLoS One. 2015;10(10):e0136404. doi: 10.1371/journal.pone.0136404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coskunpinar E., Cakmak H.A., Kalkan A.K. Circulating miR-221-3p as a novel marker for early prediction of acute myocardial infarction. Gene. 2016;591(1):90–96. doi: 10.1016/j.gene.2016.06.059. [DOI] [PubMed] [Google Scholar]
- 31.Poliseno L., Tuccoli A., Mariani L. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Song Y.H., Li F. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem. Biophys. Res. Commun. 2009;381(1):81–83. doi: 10.1016/j.bbrc.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dentelli P., Rosso A., Orso F. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler. Thromb. Vasc. Biol. 2010;30(8):1562–1568. doi: 10.1161/ATVBAHA.110.206201. [DOI] [PubMed] [Google Scholar]
- 34.Shen X., Soibam B., Benham A. miR-322/-503 cluster is expressed in the earliest cardiac progenitor cells and drives cardiomyocyte specification. Proc. Natl. Acad. Sci. U. S. A. 2016;113(34):9551–9556. doi: 10.1073/pnas.1608256113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y., Matkovich S.J., Hecker P.A. Epitranscriptional orchestration of genetic reprogramming is an emergent property of stress-regulated cardiac microRNAs. Proc. Natl. Acad. Sci. U. S. A. 2012;109(48):19864–19869. doi: 10.1073/pnas.1214996109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirt M.N., Werner T., Indenbirken D. Deciphering the microRNA signature of pathological cardiac hypertrophy by engineered heart tissue- and sequencing-technology. J. Mol. Cell. Cardiol. 2015;81:1–9. doi: 10.1016/j.yjmcc.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Marques F.Z., Vizi D., Khammy O. The transcardiac gradient of cardio-microRNAs in the failing heart. Eur. J. Heart Fail. 2016;18(8):1000–1008. doi: 10.1002/ejhf.517. [DOI] [PubMed] [Google Scholar]
- 38.Dirkx E., da Costa Martins P.A., De Windt L.J. Regulation of fetal gene expression in heart failure. Biochim. Biophys. Acta. 2013;1832(12):2414–2424. doi: 10.1016/j.bbadis.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Gaggin H.K., Januzzi J.L., Jr. Biomarkers and diagnostics in heart failure. Biochim. Biophys. Acta. 2013;1832(12):2442–2450. doi: 10.1016/j.bbadis.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Berger R., Huelsman M., Strecker K. B-type natriuretic peptide predicts sudden death in patients with chronic heart failure. Circulation. 2002;105(20):2392–2397. doi: 10.1161/01.cir.0000016642.15031.34. [DOI] [PubMed] [Google Scholar]
- 41.Fonarow G.C., Horwich T.B. Combining natriuretic peptides and necrosis markers in determining prognosis in heart failure. Rev. Cardiovasc. Med. 2003;4(Suppl. 4):S20–S28. PMID: 14564225. [PubMed] [Google Scholar]
- 42.Nandi S.S., Mishra P.K. Harnessing fetal and adult genetic reprograming for therapy of heart disease. J. Nat. Sci. 2015;1(4) PMID: 25879081. [PMC free article] [PubMed] [Google Scholar]
- 43.Thum T., Galuppo P., Wolf C. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116(3):258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 44.Di Martino M.T., Rossi M., Caracciolo D. Mir-221/222 are promising targets for innovative anticancer therapy. Expert Opin. Ther. Targets. 2016;20(9):1099–1108. doi: 10.1517/14728222.2016.1164693. [DOI] [PubMed] [Google Scholar]
- 45.Song J., Ouyang Y., Che J. Potential value of miR-221/222 as diagnostic, prognostic, and therapeutic biomarkers for diseases. Front. Immunol. 2017;8:56. doi: 10.3389/fimmu.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cardiac miRNA profiles in cardiac hypertrophy patients.