Abstract
Motivation
Oxidative stress and protein damage have been associated with over 200 human ailments including cancer, stroke, neuro-degenerative diseases and aging. Protein carbonylation, a chemically diverse oxidative post-translational modification, is widely considered as the biomarker for oxidative stress and protein damage. Despite their importance and extensive studies, no database/resource on carbonylated proteins/sites exists. As such information is very useful to research in biology/medicine, we have manually curated a data-resource (CarbonylDB) of experimentally-confirmed carbonylated proteins/sites.
Results
The CarbonylDB currently contains 1495 carbonylated proteins and 3781 sites from 21 species, with human, rat and yeast as the top three species. We have made further analyses of these carbonylated proteins/sites and presented their occurrence and occupancy patterns. Carbonylation site data on serum albumin, in particular, provides a fine model system to understand the dynamics of oxidative protein modifications/damage.
Availability and implementation
The CarbonylDB is available as a web-resource and for download at http://digbio.missouri.edu/CarbonylDB/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Oxidative stress—an imbalanced state of excessive production of reactive oxygen species (ROS) and diminished ROS scavenging ability in the cell—causes damage to cellular components. While all bio-molecules are prone to oxidative damage, proteins are particularly vulnerable and critical targets because most of the cellular functional-biomass is proteins (de Graff et al., 2016). Mild oxidative stress may cause simple oxidation/hydroxylation like reversible oxidation of methionine to methionine sulfoxide. However, more sustained/severe stress leads to carbonylation—an irreversible oxidative protein ‘damage’—a chemically diverse form of oxidative post-translational modification (PTM) wherein the side-chains of various amino acids are modified to diverse aldehydes, ketones, lactams and conjugates of advanced glycation (or lipid-peroxidation) end-products (AGEs/ALEs; Møller et al., 2011).
Carbonylation triggers protein aggregation due to loss of protein stability, which in turn contributes to aging–it is estimated that half of all proteins are damaged in old individuals. Protein carbonylations are widely recognized as the biomarkers for oxidative stress, protein damage, aging/senescence, age-related diseases and numerous degenerative/other diseases including cancer (de Graff et al., 2016; Fedorova et al., 2014; Ros, 2017).
Given their importance, it is of great interest to identify and document carbonylated proteins in different biological conditions. Further, knowing the exact site of carbonylation is important to understand the nature of oxidative damage, it’s mechanism, and downstream effects. Many studies have been conducted for large-scale identification of carbonylated proteins/sites in different organisms, cells and experimental conditions (Havelund et al., 2017; Maisonneuve et al., 2009). Consequently, it is essential to catalogue the known carbonylated proteins/sites for comparative analyses. A database that provides easy access to information on experimentally-confirmed carbonylated proteins/sites would therefore be a very valuable resource. Surprisingly, no such a database is available in case of protein carbonylation (Rao and Møller, 2011; Weng et al., 2017). We have developed CarbonylDB, a manually-curated resource, to fill this important knowledge gap.
2 Materials and methods
2.1 Curation of protein carbonylation sites
We searched PubMed and Google for publications and their cross-references that might provide information on experimentally-identified protein carbonylation sites. The list of carbonylated protein IDs and location/site information were manually curated from these publications and their (Supplementary Material).
2.2 Implementation
The CarbonylDB provides a user-friendly web-interface for searching, browsing and downloading data. It was implemented using MySQL as the back-end database, PHP as the server-side scripting and JavaScript and jQuery plugins as the front-end interface (Supplementary Material).
3 Results
3.1 CarbonylDB web-interface
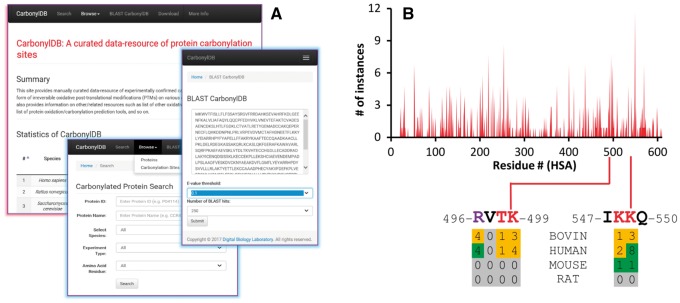
The CarbonylDB web-interface (Fig. 1A) provides access to manually-curated experimentally-confirmed carbonylated proteins/sites. The data-resource can be searched using UniProt ID, protein name or other features or can be browsed for carbonylated protein/site. For each protein, it displays relevant information like species, number of sites, etc.; and for each site it provides residue type/position, 21-mer flanking sequence, experimental information and PubMed reference number. Information on protein name, sequence and list of carbonylated sites with all relevant data, including solvent accessibility, is displayed when any protein ID is clicked. It provides a BLAST search-feature for easy database query using user-specified sequences. The entire resource can be downloaded as a tab delimited flat-file and associated sequences as a Fasta file in the download page. The CarbonylDB also provides additional information on other oxidative modification databases and prediction tools for carbonylated proteins/sites.
Fig. 1.
(A) Composite screenshots of CarbonylDB web-interface. It provides statistics of carbonylated proteins/sites in various species as well as search/browse, BLAST and download options. (B) CarbonylDB provides valuable information on oxidative protein damage, including data on the likelihood of protein carbonylations in serum albumin at specific positions (Supplementary Material)
3.2 Patterns of oxidative protein damage
At present, there are 1495 experimentally-identified carbonylated proteins and 3781 sites from 21 species in the CarbonylDB data-resource. At 40% identity cutoff there are 1170 distinct clusters wherein 14.6% clusters contain two or more sequences with many heat shock/stress-related and/or mitochondrial proteins in top clusters in terms of cluster size. The CarbonylDB provides information on carbonylation sites for serum albumins from four species–human (HSA, 121 sites), bovine (BSA, 176 sites), rat (2 sites) and mouse (4 sites) (Fig. 1B;Supplementary Material).
4 Discussion
As there exists no repository of protein carbonylation sites (Rao and Møller, 2011; Weng et al., 2017), we have done a manual literature search and curated experimentally-confirmed carbonylated proteins/sites. The CarbonylDB is the first/novel data-resource on irreversible oxidative protein damage/carbonylation, and along with other oxidative PTM data-resources like RedoxDB (Sun et al., 2012) and MetOx DB (Jacques et al., 2015), it will help oxidative stress-related research in biology/medicine. The CarbonylDB resource provides a large curated dataset on the carbonylation sites in serum albumin–an important in vitro model protein for the study of oxidative protein damage (Maisonneuve et al., 2009; Ros, 2017). It also provides a much-needed large dataset for better prediction of carbonylation sites, which until now performs rather poorly mainly because of the lack of a good dataset (Weng et al., 2017). In conclusion, we have curated CarbonylDB—a novel and valuable data-resource on protein carbonylations—which will help comparative analyses and hypothesis generation, to select prospective candidates as in vitro models and to develop potential in vivo biomarkers for oxidative protein damage (Supplementary Material).
Funding
This work was partially supported by National Institutes of Health [R01-GM100701 to D.X.] and Danish Council for Independent Research–Technology and Production Sciences [DFF|FTP 4005-00082 to I.M.M.].
Conflict of Interest: none declared.
Supplementary Material
References
- de Graff A.M.R. et al. (2016) Highly charged proteins: the Achilles’ heel of aging proteomes. Structure, 24, 329–336. [DOI] [PubMed] [Google Scholar]
- Fedorova M. et al. (2014) Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom. Rev., 33, 79–97. [DOI] [PubMed] [Google Scholar]
- Havelund J.F. et al. (2017) A biotin enrichment strategy identifies novel carbonylated amino acids in proteins from human plasma. J. Proteomics, 156, 40–51. [DOI] [PubMed] [Google Scholar]
- Jacques S. et al. (2015) Protein methionine sulfoxide dynamics in Arabidopsis thaliana under oxidative stress. Mol. Cell Proteomics, 14, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E. et al. (2009) Rules governing selective protein carbonylation. PLoS ONE, 4, e7269.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller I.M. et al. (2011) Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J. Proteomics, 74, 2228–2242. [DOI] [PubMed] [Google Scholar]
- Rao R.S.P., Møller I.M. (2011) Pattern of occurrence and occupancy of carbonylation sites in proteins. Proteomics, 11, 4166–4173. [DOI] [PubMed] [Google Scholar]
- Ros J. (2017) Protein Carbonylation: Principles, Analysis, and Biological Implications. John Wiley & Sons, Inc., NJ, USA, p. 416. [Google Scholar]
- Sun M.A. et al. (2012) RedoxDB–a curated database for experimentally verified protein oxidative modification. Bioinformatics, 28, 2551–2552. [DOI] [PubMed] [Google Scholar]
- Weng S.L. et al. (2017) Investigation and identification of protein carbonylation sites based on position-specific amino acid composition and physicochemical features. BMC Bioinformatics, 18, 66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.