Abstract
With the unique property of self-renewal and developmental pluripotency, human embryonic stem cells (hESC) provide an opportunity to study molecular aspects of developmental biology. Understanding gene regulation of hESC pluripotency is a critical step toward directing hESC differentiation for regenerative medicine. However, currently little is known about hESC gene regulation of hESC pluripotency. Applying network analysis to micro-array gene expression profiling data, we compared gene expression profiles from pluripotent hESC to hESC-derived astrocytes and identified potential gene regulation networks. These gene regulation networks suggest that hECS has stringent control of cell cycle and apoptosis. Our data reveal several potential hESC differentiation biomarkers and suggest that IGF2 and A2M could play a role in hESC pluripotency by altering the availability of cytokines at the local environment of hECS. These findings underscore the importance of network analysis among differentially expressed genes, and should facilitate future study for understanding the gene regulation of hESC pluripotency.
Key words: Pluripotent stem cells, Regulation of gene expression, Neuroglia, Cell differentiation, Microarray analysis
INTRODUCTION
Human embryonic stem cells (hESC) derived from inner cell mass of blastocysts can be maintained in vitro without losing their pluripotency and be differentiated into somatic cells (1,41,52). In addition to the promising clinical value for regenerative medicine, hESCs provide a unique in vitro system for studying molecular aspects of human developmental biology, including regulation of stem cell pluripotency. However, despite intensive efforts, the search of hESC pluripotency regulatory genes is far from being complete. One approach is to compare the gene expression profiles from mouse, rat, and human ESC with the assumption that the common orthologous genes are the most relevant for pluripotency (21,60). Recent studies indicate that mouse embryonic stem cells (mESC) are very different from hESCs. Murine leukemia-inhibitory factor (LIF) plays a critical role in maintaining mESC undifferentiated growth in vitro, but has no such effect on hESC (10,14). In addition, bone morphogenetic protein 4 (BMP4) blocks neuroectoderm differentiation of mESC, but promotes the trophectoderm differentiation of hESC (66,68). These studies underscore the difficulty of using mESC as a model to study hESC, and the importance of identifying hESC specific pluripotency regulatory genes.
Microarray gene expression profiling has been rapidly transformed from a novel approach into a common tool in study of gene regulation. However, interpreting the data generated from microarray gene expression profiling remains a challenging task. Differentially expressed genes from microarray studies can be organized into networks based on various relationships such as expression patterns, common promoter sequences, physical location on chromosomes, and the direct binding of gene products. We have utilized some of these gene relationships to analyze microarray gene expression data in a previous study of murine hematopoietic stem cells (72).
In this study we compare the gene expression profile of hESC (H1, WiCell Research Institute, Inc., Madison, WI) to that from astrocytes derived from this hESC line. By comparing cells between the pluripotent stage and the neuronal differentiated stage within the same hESC lines, genetic variation among hESC lines is eliminated from the analysis. A large number of relatively pure cells from each population was used to extract RNA for subsequent microarray gene expression profiling. Literature mining was performed to identify gene networks among differentially expressed genes. Instead of focusing on individual differentially expressed genes, we organized differentially expressed genes into networks based on the direct binding relationships of their encoded proteins. This approach can identify genes that play a critical role in pluripotency regulation, but are not in the differential expression gene list. We found that the expressions of multiple members of the p53 pathway are lower in hESC-derived astrocytes than in hESC. In contrast, the p53 mRNA level shows no significant difference between pluripotent hESC and the astrocytes differentiated from these hESC. p53, a tumor suppressor, has been shown to regulate mESC differentiation (34), but the role of p53 in regulation of pluripotency of human ESC remains unclear. Our data suggest that a posttranscriptional mechanism of p53 may be employed in the regulation of hESC differentiation. We also identified IGF2 and alpha 2-macroglobulin (A2M) as potentially important factors for hESC pluripotency by this approach. Finally, we show that A2M induces cell death in pluripotent hESC without affecting differentiated hESC. These findings underscore the importance of network analysis among differentially expressed genes, and should facilitate future study for understanding the gene regulation of hESC pluripotency.
MATERIALS AND METHODS
hESC Culture and Differentiation
The hESC line (H1) was obtained from the WiCell Research Institute (Madison, WI) and maintained as instructed (52). Undifferentiated hESC were cultured on an irradiated (5500 rads) layer of mouse embryonic fibroblast (MEF) feeder cells in six-well plates or Matrigel-coated plates with MEF conditioned medium. MEF were prepared from the embryos of 13–14-day pregnant CF-1 mice (Charles River Labs) and stocks were cryopreserved until required for culture of hESC. hESC were consistently observed as large clumps that appear on top of the MEF layer, consistent with the published observations of others. Under these conditions, the hESC remained continuously undifferentiated and periodic karyotying has confirmed their human diploid chromosomal character. We have tested the hESC cultured in our laboratory for their potential to form all three embryonic germ layers. These cells produced teratomas after injection into severe combined immunodeficient (SCID) mice; the pathology included tissues of ectodermal, mesodermal, and endodermal origin. Immunostaining of the hESC for alkaline phosphatase and with antibodies specific for SSEA-4, TRA-1-60, TRA-1-81, or Oct-3/4 confirmed the stem cell phenotype of these cells. Prior to induction into astrocytes, hESC were transferred from MEF feeder layers onto six-well plates coated with Matrigel in the same culture medium. The medium was changed every other day. For differentiation, feeder-free hESCs were cultured in differentiation medium comprised of: DMEM/F12, N2 supplement, 0.1 mM nonessential amino acids, 1 mg/ml heparin, 10 ng/ml bFGF, 20 ng/ml EGF. The medium was changed every other day and cells were passaged as required by confluence. After 4 weeks in the differentiation medium, the majority of cells (>90%) differentiated into GFAP+ cells.
Immunofluorescent Staining
The hESC cells grown on cover slips were fixed for 20 min at room temperature in 4% paraformaldehye and then washed three times in PBS. Cells were consequently permeabilized for 20 min in PBS containing 0.1% Triton X at room temperature. Cells were then washed three times with PBS and blocked for 1 h at room temperature in PBS containing 10% goat serum. Subsequently, cells were incubated with a primary antibody (Oct-3/4, 1:100; Santa Cruz) in PBS containing 1% goat serum at 37°C for 1 h. As a negative control, cells were also treated separately with a nonspecific anti-mouse IgG antibody (1:100; Chemicon, USA). After being washed three times in freshly made PBS containing 0.5% Tween 20, cells were then incubated with the secondary antibody (phycoerythrin-conjugated anti-mouse IgG, 1:200; Santa Cruz) in PBS containing 1% goat serum at 37°C for 45 min. The unbound secondary antibodies were removed by washing three times in freshly made PBS containing 0.5% Tween 20. The cells were then washed briefly in PBS before mounting. The hESC-derived astrocytes were fixed with ethanol and labeled with Cy3-conjugated mouse anti-glial fibrillary acidic protein (GFAP) monoclonal antibody (Sigma, USA).
Microarray Gene Expression Profiling
Total RNA was extracted with Trizol (Invitrogen, USA) from pluripotent hESC and astrocytes differentiated from hESC. RNA quality was examined and hybridized to Applied Biosystems (ABI) 1700 chemiluminescent human microarray chips at ABI micro-array facility (Foster City, CA). Triplicate samples were used from each group. A t-test was performed using Matlab (Natick, MA). Differentially expressed genes were selected with the criteria: 1) passed all ABI 1700 data control filters; and 2) at least a threefold difference with p < 0.01 between hESC and astrocytes. The fold changes for individual genes are calculated by dividing the average of normalized expression units from hESC and astrocytes. Literature mining and pathway analysis were performed with Pathwaystudio (Rockville, USA) and GenMAPP (Gladstone Institutes, UCSF, USA). To confirm microarray data, quantitative real-time RT-PCR was performed with Taqman primer sets purchased from ABI (Foster City, CA) on LightCycler (Roche, USA). The fold changes were calculated with the ΔΔCt method.
RESULTS
hESC Culture and Differentiation
To identify genes that are related to neural differentiation, hESCs (H1 line) were differentiated into GFA-positive astrocytes using neural differentiation medium. As shown in Figure 1, before differentiation, hESC grew as colonies on Matrigel and only the undifferentiated pluripotent hESC in the center of the colony expressed Oct-3/4, a marker for pluripotent hESC. The spontaneous differentiated hESC cells at the edge of the colony did not express Oct3/4. After neural cell differentiation treatment, cells no longer grow as colonies. The dispersed neural differentiated cells expressed the astrocytic marker antigen, GFAP, and exhibited a flat morphology, which is typical for cultured astrocytes (Fig. 1B). Total RNAs were extracted from undifferentiated pluripotent hESC and differentiated astrocytes for microarray gene expression profiling. The gene expression profiles between hESC and hESC-derived astrocytes were compared.
Figure 1.
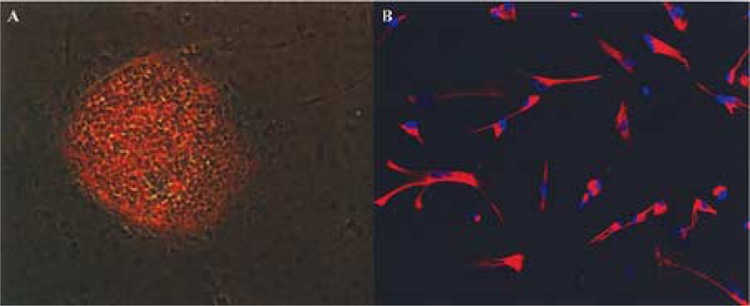
Pluripotent and neuronal differentiated hESC. (A) Merged image of an immunofluorescent-stained (Oct-3/4) and light microscope images from a pluripotent hESC colony. The hESC colony was labeled with mouse α human Oct-3/4 IgG and PE-conjugated rabbit anti-mouse IgG antibodies. Only cells in the center of the hESC colony expressed Oct-3/4. The spontaneous differentiated cells around the colony did not expressed Oct-3/4. (B) hESC-derived astrocytes were labeled with Cy3-conjugated mouse α human GFAP antibody as well as nuclei staining, DAPI (blue). Cells expressed GFAP (an astrocyte marker) and displayed astrocytes morphology.
Expression of Pluripotency-Related Genes
The expression of approximately 28,000 human genes (33,183 transcripts) was interrogated with Applied Biosystem’s expression array system (Fig. 2). After statistical analysis, we identified 684 differentially expressed genes with at least a threefold difference (p < 0.01) between pluripotent hESC and hESC-derived astrocytes. As expected, many known stem cell genes such as Oct3-/4, Nodal, and Rex1(45) were expressed higher in the hESC than in the astrocytes. The expression of Oct-3/4, Nodal, and Rex1 in hESC was 66-fold, 15-fold, and 35-fold of that in astrocytes, respectively. Quantitative RT-PCR confirmed the perturbation of these representative genes. BMP4 has been shown to sustain pluripotency of mESC (68), but induce differentiation of hESC (20,66). BMP4 expression is threefold higher in hESC-derived astrocytes than that in hESC. The expression levels of bFGF (14-fold higher in hESC) and BMP4 agree with a published report that bFGF represses BMP4 in order to maintain hESC pluripotency (67). The high expression of Oct-3/4 and Nodal in hESC agrees with the immunoassay data presented above (Fig. 1), and confirms reports from other investigators (53). Compared to undifferentiated hESC, there wass 43-fold more A2M mRNA expressed in astrocytes than in pluripotent hESC. Quantitative RT-PCR with ΔΔCt estimation confirmed the 43-fold upregulation of A2M in astrocytes (Table 1).
Figure 2.
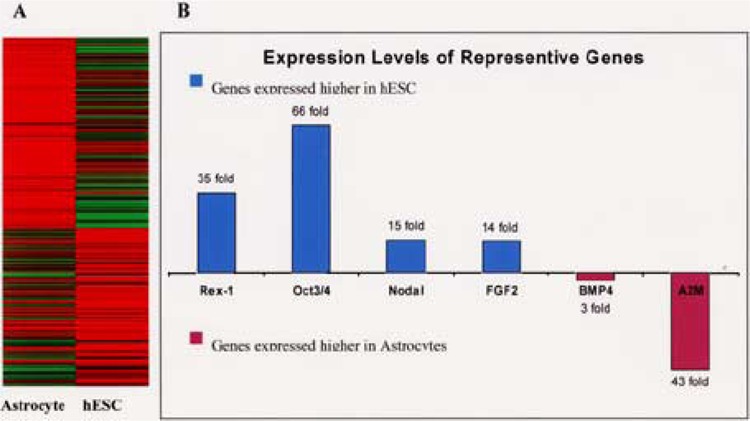
Differential gene expression between pluripotent hESC and hESC-derived astrocytes. (A) There are 684 differentially expressed genes between hESC and astrocytes derived from hESC. The gene expression levels are presented as an expression heatmap. Red: high; yellow: medium; green: low. (B) Expression levels of representative genes are shown with fold difference, which is calculated as described in Materials and Methods.
TABLE 1.
QUANTITATIVE RT’PCR CONFIRMS THE UPREGULATION OF A2M IN hESC-DERIVED ASTROCYTES
| ΔCt (hESC-Astrocyte) | ΔΔCt | Upregulation in Astrocytes | |
|---|---|---|---|
| B2M (reference gene) | −2.8 ± 0.05 | — | — |
| A2M | 2.6 ± 0.03 | 5.4 | 42-fold |
The fold change (estimated with the ΔΔCt method) is very close to the microarray measurement.
Gene Expression Networks Among Differentially Expressed Genes
Although differentially expressed genes may reveal valuable information, it is important to view them from a landscape or network point of view, rather than as individual entities. Cellular events and characteristics including pluripotency are rarely regulated by a single gene, but rather by networks of genes interacting with each other. While some genes are subjected to regulation at the protein or translational level, such regulations would cause mRNA level perturbation at downstream genes within the gene network. Even though gene expression profiling measures the mRNA level and cannot reveal gene regulation information at the translational regulation level (protein synthesis or miRNA regulation), valuable information can be inferred from differential expression genes with network analysis. In contrast to some gene expression profiling studies that only analyze differentially expressed genes at the individual gene level, in this study a data mining process was carried out on peer-reviewed literature with computational software. The network analysis revealed several networks among the differentially expressed genes based on direct-binding relationships of their gene products, the encoded proteins.
The regulatory relationships (e.g., induction, repression/degradation) of genes in published papers are not always consistent. On the other hand, the experimental facts of direct binding (e.g., immunoco-precipitation, yeast two-hybrids assays) are less arguable. Therefore, we based our network analysis on reported direct-binding relationships of proteins. Among the 684 differentially expressed genes, 95 proteins formed 22 networks. Three representative networks with potential significance in astrocyte differentiation (based on high fold changes and significance of p-values) are presented in Figure 3.
Figure 3.
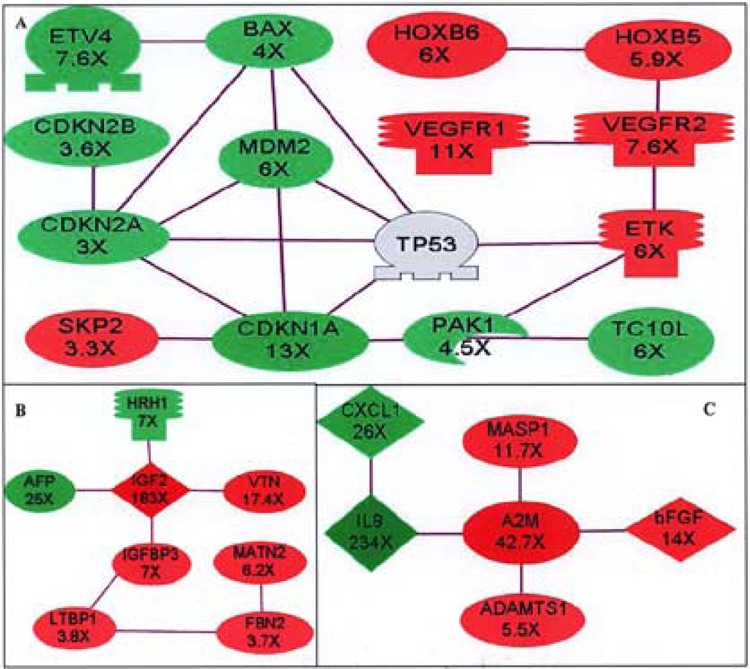
Gene expression networks among differentially expressed genes. Networks are defined by the relationship of protein–protein direct binding among differentially expressed genes. Lines connecting genes represent the protein–protein binding relationships from published peer-reviewed articles. Red: genes are upregulated in hESC-derived astrocytes; green: genes are downregulated in hESC-derived astrocytes. The fold changes between hESC and hESC-derived astrocytes are indicated under each corresponding gene name. (A) p53-related network. The TP53 is not differentially expressed, and is included to show its relationship with other genes in the network.). (B) IGF2-related network. (C) A2M-related network. A2M: alpha-2-macroglobulin; ADAMTS1: a disintegrin-like and metalloprotease with thrombospondin type 1 motif 1; AFP: alpha-fetoprotein; BAX: Bcl2-associated X protein; CDKN1A: cyclin-dependent kinase inhibitor 1A (p21); CDKN2A: cyclin-dependent kinase inhibitor 2A (p16); CDKN2B: cyclin-dependent kinase inhibitor 2B (p15); ETK: ephrin receptor A3; ETV4: ets variant gene 4 (E1AF); FBN2: fibrillin 2; HOXB5: homeo box B5; HOXB6: homeo box B6; HRH1: histamine receptor H1; IGF2: insulin-like growth factor 2; IGFBP3: insulin-like growth factor binding protein 3; IL8: interleukin 8 (CXCL8); LTBP1: latent transforming growth factor beta binding protein 1; MASP1: mannan-binding lectin serine protease 1; MATN2: matrilin 2; MDM2: mouse double minute 2 homolog; PAK1: p21 (CDKN1A)-activated kinase 1; SKP2: S-phase kinase-associated protein 2 (p45); TC10L: TC10-like protein (ras homolog gene family Y, member J); VEGFR1: vascular endothelial growth factor receptor 1(FLT1); VEGFR2: vascular endothelial growth factor receptor 2(KDR); VTN: vitronectin.
While p53 is not among the differentially expressed genes (p53 is 2.2-fold lower at GFAP-positive cells with p = 0.037, and excluded from the differentially expressed gene list by our selection criteria), many members of the p53 pathway are downregulated in astrocytes (Fig. 3A). The downregulated p53-related genes are: CDKN1A (13-fold), CDKN2A (3fold), CDKN2B (3.6-fold), MDM2 (6-fold), and BAX (4-fold). CDKN1A, CDKN2A, and CDKN2B are three cylin-dependent kinase inhibitors that regulate G1/S progression. The promoter of CDKN1A (also called p21 or WAF1) has a p53 binding site and its expression is inducible by p53. Overexpression of p21 in tumor cell lines suppresses colony formation similar to that resulting from p53 overexpression, and this strongly suggests that p21 is a mediator of p53-dependent growth suppression (17). CDKN2A (also called p16) arrests normal diploid cells in late G1 (35). An alternative beta transcript of CDKN2A encodes p14 (ARF), which binds to MDM2 and promotes the rapid degradation of MDM2 (71). MDM2 (downregulated sixfold in astrocytes) is a major regulator of the tumor suppressor p53 by targeting its destruction (25,37). CDNK2B (also called p15) is adjacent to CDKN2A (28), and a potential effector of TGF-β-induced cell cycle arrest in human keratinocytes (24). In addition to lower expression of cylin-dependent kinase inhibitors, the BAX and its regulator ETV4 (also called Pea3) (19) also expressed 4-fold and 7.6-fold lower in hESC-derived astrocytes, respectively. BAX is a proapoptotic protein parent of BCL2, and p53 directly activated BAX in the absence of other proteins to permeabilize mitochondria and engage the apoptotic program (11). While cyclin-dependent kinase inhibitors and apoptotic genes are downregulated, the S-phase kinase-associated protein 2 (SKP 2, also called p45) is upregulated 3.3-fold in hESC-derived astrocytes. Microinjection of specific antibodies to SKP 2 specifically blocked the cell cycle in G1, and suggest that SKP 2 is essential for S-phase entry (69). Together, these data indicate a more constricted regulation of cell cycle and apoptosis in hESC compared to hESC-derived astrocytes. That agrees with our observation of the slow growth and apoptotic susceptibility of hESC in vitro.
As expected, the p21 (CDKN1A)-activated kinase 1 (PAK1) and the gene of its binding protein TC10L (55) are downregulated with CDKN1A in hESC-derived astrocytes. VEGF-related genes are upregulated in astrocytes compared to hESC. These genes include: VEGFR1 (11-fold), VEGFR2 (7.6-fold), ETK (6-fold), HOXB5 (5.9-fold), and HOXB6 (6-fold). ETK directly associated with PAK1 and VEGFR2. Wild-type ETK in noninvasive human breast cancer MCF-7 cells increased proliferation and anchorage-independent growth (3,70), and VEGFR2 (Flk-1) is the earliest marker of cells fated to endothelial and hematopoietic lineages (38). Studies in the murine model showed that HOXB5 is a potent transcriptional regulator of VEGFR2 (65). The relative low expression of HOXB5, HOXB6, VEFGR1, and VEGFR2 in hESC compared to hESC-derived astrocytes suggests they may be involved in the initiation of hESC differentiation.
Another interesting network centers on a growth factor: insulin-like growth factors II (IGF2) (Fig. 3B). IGF2 and its major carrying protein IGFBP3 (18) are upregulated 183-fold and 7-fold, respectively in hESC-derived astrocytes. The latent TGF-β binding protein (LTBP-1), which targets latent complex of TGF-β to the extracellular matrix where the latent complex is activated, is also upregulated 3.8-fold. LTBP-1 also binds IGFBP3 and is a potential connector of IGFBP3 to the TGF family proteins (23). Three extracellular matrix protein genes, Fibrilin 2 (FBN2), matrilin 2 (MATN2), and vitronectin (VTN), are also upregulated in hESC-derived astrocytes. It has been reported that IGF2 binds to VTN with an affinity similar to that for the IGF receptor, and the IGF2/ VTN complex enhances migration of breast cancer cells (40). This network suggests that IGF2/IGFBP3/ LTBP1 complex may target IGF2 to the extracellular FBN2/MATN2/VTN complex in hESC-derived astrocytes. Alpha-fetoprotein (AFP), a major plasma protein produced by the yolk sac and the liver during fetal life, is downregulated in astrocytes. AFP is the fetal counterpart of adult serum albumin, and its lower expression in hESC-derived astrocytes is interesting but not unexpected. Histamine receptor H1 (HRH1), which can mediate the internalization and degradation of IGF2 (22), is downregulated also. The downregulation of HRH1 and the upregulation of IGF2 as well as IGF2 extra cellular binding proteins in hESC-derived astrocytes compared to hESC suggest an important role of IGF2 in the hESC differentiation. We are currently investigating the function of IGF2 protein in hESC pluripotency.
The most interesting network centers on A2M (Fig. 3C). The expression of ADAMTS1 and MASP1 is upregulated 5.5- and 11.7-fold, respectively, with A2M (upregulated 42.7-fold) in hESC-derived astrocytes. ADAMTS1 is a secreted protein that has an N-terminal signal peptide, a zinc metalloprotease domain containing a zinc-binding site, and a cysteine-rich region containing two putative disintegrin loops (54). It is associated with the inflammation response (29). MASP1 is an extracellular serine protease that activates complement components C3 and C2 (51). Because A2M binds to both ADAMTS1 (an active metalloprotease that is associated with the extracellular matrix) (30) and MASP1 (50), it is possible that A2M (a protease inhibitor and cytokine binding protein) is expressed to suppress these two proteases. However, A2M is upregulated to a much higher magnitude than the two proteases. This suggests that it may also play a role in cytokine binding. Interestingly, the IL8 and CXCL1 are downregulated in hESC-derived astrocytes 234-fold and 36-fold, respectively. Structurally, CXCL1 is related to a superfamily of inflammatory proteins that includes IL8 and platelet factor-4 (PF4) (26). Both IL8 and CXCL1 bind to the class II IL-8 receptor (CXCR2) with high affinity during inflammation (33). IL8 and CXCR2 are responsible for the resistance of Fas-induced cell death in astrocytes (46). The upregulation of A2M (which binds IL8) as well as the downregulation of IL8 and CXCL1 suggests that Fas-induced cell death may be a part of normal astrocyte maturation from stem cells.
Effects of A2M on hESC Culture
In the A2M network, A2M is upregulated approximately 43-fold, while IL8 is downregulated 234-fold. A2M has been recognized as a binding protein for various growth factors, particularly those of the TGF-β family (7,12,32). The expression of bFGF, a member of the TGF-β family and key player of hESC pluripotency, is significantly downregulated in hESC-derived astrocytes (Fig. 2), while A2M expression is significantly upregulated in hESC-derived astrocytes. To investigate the role of A2M in hESC pluripotency, we added human A2M (final concentration of 77 μg/ml) into undifferentiated hESC culture. This concentration of A2M is significantly lower than the A2M concentration in human plasma, which is 2–5 μM (1440–3590 μg/ml) (36). Preliminary experiments on 293T cells also showed no affects of adding A2M at this concentration (data not shown). Interestingly, A2M induces hESC colony detachments and cell death on Oct-3/4-positive pluripotent hESC, but has no such effects on Oct-3/4-negative hESC cells (Fig. 4).
Figure 4.
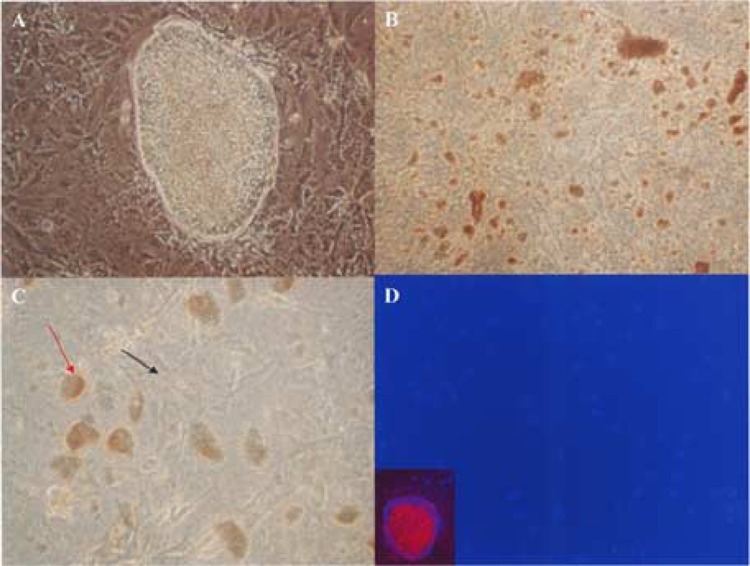
A2M induces detachment and cell death in pluripotent hESC. (A) A2M was added to hESC growing on a Matrigel-coated six-well plate at a final concentration of 77 μg/ml. hESC cell detachment and cell death were observed 72 h after adding A2M. In wells without adding A2M, a hESC colony is surrounded by differentiated hESC. (B) In wells with A2M treatment, hESC colonies disappeared and small clumps of dead cells were observed. (C) Differentiated cells still attached to Matrigel have healthy morphology (black arrow), while detached and fragmented hESC clumps appeared (red arrow). (D) Immunostaining after A2M treatment showed that Oct-3/4-positive cells disappeared, and the remaining cells are Oct-3/4 negative. The areas occupied by hESC colonies have no remaining cells. Immunostaining of wells without A2M treatment showed pluripotency marker Oct-3/4 expressed by cells inside the hESC colony, but not by the differentiated cells around it (inset). Red: Oct-3/4; blue: DAPI.
DISCUSSION
Human embryonic stem cells have the ability to self-renew and differentiate into all tissue types in the human body. Various animal cloning experiments (4,43,47,56) and stem cell fusion experiments 3,48) indicate that somatic cells can be reprogrammed to pluripotent stem cells. Therefore, identifying the genes that control hESC pluripotency and genes that initiate specific linage differentiation is critically important for clinical and research stem cell applications. Once the pluripotency-controlling genes and their interaction networks are known, somatic cells could be reprogrammed to pluripotent stem cells by gene manipulation techniques such as siRNA, micro-RNA, and gene transfer. Reprogramming somatic cells to pluripotent stem cells with gene manipulation rather than with nuclear transfer technique may overcome many difficulties in hESC study such as the ethical issues of using human embryos, and low efficiency of nuclear transfer.
Various attempts have been made to understand stem cell gene regulation. A recent proteomic study conducted by Wang et al. explored the direct binding relationship of proteins in murine stem cells (57). This study demonstrated the network nature of stem cell pluripotency regulation. Comparisons of mouse and human ESC indicate that they are very different (10,14,66,68). It is now known that gene regulation is orchestrated by multiple genes at multiple levels, such as protein modification, micro-RNA interaction, mRNA alternative splicing, and stability. One of the major concerns of microarray gene expression experiment is mRNA expression levels do not always correlate with protein levels. The conventional microarray data analysis approaches such as fold changes, statistical analysis, and gene annotation often overlook valuable information. Rather than focusing on individual differentially expressed genes, in this study we analyzed differentially expressed genes from a network point of view to obtain information beyond conventional approaches. By performing network analysis with differentially expressed genes between hESC and hESC-derived astrocytes, we identified three networks among differentially expressed genes based on the binding relationships of their encoded proteins. These networks support previous published studies of hESC and reveal new information of hESC pluripotency.
In one network, p53-related cylin-dependent kinase inhibitors and proapoptotic genes were expressed at a lower level in hESC-derived astrocytes compared to that in hESC. Interestingly, the expression level of p53 itself did not change significantly in our study and was not on the differentially expressed gene list. It has been reported that p53 expression is higher in mESC than that in hESC (21). Study of mESC showed that p53 induces differentiation of mESC and leads to apoptosis by suppressing Nanog expression (34). But the role of p53 in hESC differentiation is not clear. Our data show that many members of the p53 pathway are expressed significantly lower in hESC-derived astrocytes compared to pluripotent hESC. This supports the idea that p53, a guardian of the genome, prevents cells entering the cell cycle, and could play a role in hESC pluripotency, but with a mechanism different from that in mESC. The down-regulation of both MDM2 and genes of the p53 pathway at first glance is a paradox. However, Schreiber’s group recently demonstrated that p53 accumulated at apoptotic neurons despite the presence of high levels of MDM2 and p53–MDM2 complex (49). They further showed that ubiquitin regulates p53 stability at neurons. Together with the data from Schreiber’s group, the downregulation of MDM2 in our data suggests the co-regulation of p53 and MDM2 at neural lineages. It is generally believed that the principle mechanisms governing p53 activity occur at the protein level, such as posttranslational modifications as well as regulation of the stability and subcellular location of p53 (63). Our data suggest that in hESC, the p53 pathway needs to be downregulated to allow differentiated cells to enter cell cycle; but rather than downregulating p53 mRNA level directly, this is to be accomplished by downregulating expression of other members of the p53 pathway.
In addition to the p53 pathway, we also found that IGF2 may play a role in neural differentiation of hESC toward astrocytes. The IGF2 and its related proteins are expressed significantly higher in astrocytes. Increasing expression of IGF2 at both mRNA and protein levels in tumor tissues has been reported (8,58). The most interesting network is the A2M network. A2M is a 720-kDa homotetrameric serum glycoprotein, and a major plasma and inflammatory fluid proteinase scavenger. Besides binding proteinase, A2M binds to cytokines and growth factors such as CXCL8 (IL8) (31), human tumor necrosis factor-α (TNF-α) (61), platelet-derived growth factor (PDGF) (5,6,44), transforming growth factor (15,59), bFGF (16), and activin (39,42). Because bFGF irreversibly binds to A2M and the resulting complex has diminished ability to recognize either the high- or low-affinity bFGF binding site in cells (2,16,36,64), A2M appears to act as a limiter of bFGF. Activin can bind to both the native and active form of A2M with a higher affinity for the active form (42). Because Activin and bFGF are needed to maintain the pluripotency of hESC (53), A2M could affect hESC pluripotency by removing critical cytokines and growth factors that maintain hESC pluripotency, such as bFGF and activin.
Our gene expression data indicate that A2M is upregulated 43-fold in astrocytes differentiated from hESC and our in vitro study confirms that adding A2M to an hESC culture results in cell death of pluripotent hESC while not having such effects on differentiated hESC. These results agree with our observation that hESC differentiation is difficult to be reversed after differentiation is initiated. A2M may be responsible for removing critical growth factors such as bFGF and activin from the medium once differentiation is initiated. It is worth pointing out that A2M is an extracellular secretory protein; therefore, a high A2M concentration could be built up around the cells producing it. Also, not all cytokines that bind to A2M will lose their function. Some cytokines are protected by A2M from degradation (6,27). Therefore, A2M could create an environment for particular cells to grow while eliminate other cell types such as the pluripotent hESC. This may be the reason why daily medium change is necessary for maintaining pluripotent hESC cultures. As an acute-phase protein in brain, A2M is involved in a variety of neuropathological conditions (9). Examination of postmortem multiple sclerosis (MS) tissues has shown that a large number of mature oligodendrocytes survived after the loss of myelin sheaths throughout the center of the recent MS lesions, but differentiation of oligodendrocyte precursor cells (OPC) is difficult to demonstrate (62). Our in vitro data showed that undifferentiated hESC are susceptible to A2M-induced cell death. If this mechanism occurs in OPC differentiation, A2M could be a potential factor to limit the replenishment of the oligodendrocyte precursor cells from stem cells.
In this study, with an approach of network analysis on differentially expressed genes, we identify three potentially significant gene regulation networks related to hESC pluripotency and neural differentiation. These gene regulation networks should provide useful information for better understanding of the initiation of hESC differentiation. However, it is important to know the limitation of such network analysis. Current knowledge of protein binding relationships is far from complete. Therefore, the networks we identified are not complete and need to be studied further. It is expected that these gene regulation networks will expand with increasing proteomic knowledge of genes. Although protein–protein binding is currently the most reliable evidence for gene regulation networks, the binding of two proteins does not equal to regulatory relationship. Network analysis is a powerful tool to identify potential gene regulation networks, but these networks must be confirmed with functional experiments such as gene manipulation. We are in the process to further investigate these identified networks with functional study.
ACKNOWLEDGMENTS
This study was supported by the Nancy Davis Foundation (Race to Erase MS) and a NIH grant (NIH 1RO1 HG002644-01A1).
REFERENCES
- 1. Amit M.; Carpenter M. K.; Inokuma M. S.; Chiu C. P.; Harris C. P.; Waknitz M. A.; Itskovitz-Eldor J.; Thomson J. A. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev. Biol. 227:271–278; 2000. [DOI] [PubMed] [Google Scholar]
- 2. Asplin I. R.; Wu S. M.; Mathew S.; Bhattacharjee G.; Pizzo S. V. Differential regulation of the fibroblast growth factor (FGF) family by alpha(2)-macroglobulin: Evidence for selective modulation of FGF-2-induced angiogenesis. Blood 97:3450–3457; 2001. [DOI] [PubMed] [Google Scholar]
- 3. Bagheri-Yarmand R.; Mandal M.; Taludker A. H.; Wang R. A.; Vadlamudi R. K.; Kung H. J.; Kumar R. Etk/Bmx tyrosine kinase activates Pak1 and regulates tumorigenicity of breast cancer cells. J. Biol. Chem. 276:29403–29409; 2001. [DOI] [PubMed] [Google Scholar]
- 4. Baguisi A.; Behboodi E.; Melican D. T.; Pollock J. S.; Destrempes M. M.; Cammuso C.; Williams J. L.; Nims S. D.; Porter C. A.; Midura P.; Palacios M. J.; Ayres S. L.; Denniston R. S.; Hayes M. L.; Ziomek C. A.; Meade H. M.; Godke R. A.; Gavin W. G.; Overstrom E. W.; Echelard Y. Production of goats by somatic cell nuclear transfer. Nat. Biotechnol. 17:456–461; 1999. [DOI] [PubMed] [Google Scholar]
- 5. Bonner J. C.; Badgett A.; Hoffman M.; Lindroos P. M. Inhibition of platelet-derived growth factor-BB-induced fibroblast proliferation by plasmin-activated alpha 2-macroglobulin is mediated via an alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein-dependent mechanism. J. Biol. Chem. 270:6389–6395; 1995. [DOI] [PubMed] [Google Scholar]
- 6. Bonner J. C.; Badgett A.; Osornio-Vargas A. R.; Hoffman M.; Brody A. R. PDGF-stimulated fibroblast proliferation is enhanced synergistically by receptor-recognized alpha 2-macroglobulin. J. Cell Physiol. 145:1–8; 1990. [DOI] [PubMed] [Google Scholar]
- 7. Borth W. Alpha 2-macroglobulin, a multifunctional binding protein with targeting characteristics. FASEB J. 6:3345–3353; 1992. [DOI] [PubMed] [Google Scholar]
- 8. Cariani E.; Lasserre C.; Seurin D.; Hamelin B.; Kemeny F.; Franco D.; Czech M. P.; Ullrich A.; Brechot C. Differential expression of insulin-like growth factor II mRNA in human primary liver cancers, benign liver tumors, and liver cirrhosis. Cancer Res. 48:6844–6849; 1988. [PubMed] [Google Scholar]
- 9. Cavus I.; Koo P. H.; Teyler T. J. Inhibition of long-term potentiation development in rat hippocampal slice by alpha 2-macroglobulin, an acute-phase protein in the brain. J. Neurosci. Res. 43:282–288; 1996. [DOI] [PubMed] [Google Scholar]
- 10. Chambers I.; Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene 23:7150–7160; 2004. [DOI] [PubMed] [Google Scholar]
- 11. Chipuk J. E.; Kuwana T.; Bouchier-Hayes L.; Droin N. M.; Newmeyer D. D.; Schuler M.; Green D. R. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303:1010–1014; 2004. [DOI] [PubMed] [Google Scholar]
- 12. Chu C. T.; Pizzo S. V. alpha 2-Macroglobulin, complement, and biologic defense: Antigens, growth factors, microbial proteases, and receptor ligation. Lab. Invest. 71:792–812; 1994. [PubMed] [Google Scholar]
- 13. Cowan C. A.; Atienza J.; Melton D. A.; Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309:1369–1373; 2005. [DOI] [PubMed] [Google Scholar]
- 14. Daheron L.; Opitz S. L.; Zaehres H.; Lensch W. M.; Andrews P. W.; Itskovitz-Eldor J.; Daley G. Q. LIF/ STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells 22:770–778; 2004. [DOI] [PubMed] [Google Scholar]
- 15. Danielpour D.; Sporn M. B. Differential inhibition of transforming growth factor beta 1 and beta 2 activity by alpha 2-macroglobulin. J. Biol. Chem. 265:6973–6977; 1990. [PubMed] [Google Scholar]
- 16. Dennis P. A.; Saksela O.; Harpel P.; Rifkin D. B. Alpha 2-macroglobulin is a binding protein for basic fibroblast growth factor. J. Biol. Chem. 264:7210–7216; 1989. [PubMed] [Google Scholar]
- 17. el-Deiry W. S.; Tokino T.; Velculescu V. E.; Levy D. B.; Parsons R.; Trent J. M.; Lin D.; Mercer W. E.; Kinzler K. W.; Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825; 1993. [DOI] [PubMed] [Google Scholar]
- 18. Ferry R. J. Jr.; Cerri R. W.; Cohen P. Insulin-like growth factor binding proteins: new proteins, new functions. Horm. Res. 51:53–67; 1999. [DOI] [PubMed] [Google Scholar]
- 19. Firlej V.; Bocquet B.; Desbiens X.; de Launoit Y.; Chotteau-Lelievre A. Pea3 transcription factor cooperates with USF-1 in regulation of the murine bax transcription without binding to an Ets-binding site. J. Biol. Chem. 280:887–898; 2005. [DOI] [PubMed] [Google Scholar]
- 20. Gerami-Naini B.; Dovzhenko O. V.; Durning M.; Wegner F. H.; Thomson J. A.; Golos T. G. Trophoblast differentiation in embryoid bodies derived from human embryonic stem cells. Endocrinology 145:1517–1524; 2004. [DOI] [PubMed] [Google Scholar]
- 21. Ginis I.; Luo Y.; Miura T.; Thies S.; Brandenberger R.; Gerecht-Nir S.; Amit M.; Hoke A.; Carpenter M. K.; Itskovitz-Eldor J.; Rao M. S. Differences between human and mouse embryonic stem cells. Dev. Biol. 269:360–380; 2004. [DOI] [PubMed] [Google Scholar]
- 22. Grimme S.; Honing S.; von Figura K.; Schmidt B. Endocytosis of insulin-like growth factor II by a mini-receptor based on repeat 11 of the mannose 6-phosphate/insulin-like growth factor II receptor. J. Biol. Chem. 275:33697–33703; 2000. [DOI] [PubMed] [Google Scholar]
- 23. Gui Y.; Murphy L. J. Interaction of insulin-like growth factor binding protein-3 with latent transforming growth factor-beta binding protein-1. Mol. Cell. Biochem. 250:189–195; 2003. [DOI] [PubMed] [Google Scholar]
- 24. Hannon G. J.; Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371:257–261; 1994. [DOI] [PubMed] [Google Scholar]
- 25. Haupt Y.; Maya R.; Kazaz A.; Oren M. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299; 1997. [DOI] [PubMed] [Google Scholar]
- 26. Horuk R.; Yansura D. G.; Reilly D.; Spencer S.; Bourell J.; Henzel W.; Rice G.; Unemori E. Purification, receptor binding analysis, and biological characterization of human melanoma growth stimulating activity (MGSA). Evidence for a novel MGSA receptor. J. Biol. Chem. 268:541–546; 1993. [PubMed] [Google Scholar]
- 27. Huang S. S.; O’Grady P.; Huang J. S. Human transforming growth factor beta.alpha 2-macroglobulin complex is a latent form of transforming growth factor beta. J. Biol. Chem. 263:1535–1541; 1988. [PubMed] [Google Scholar]
- 28. Kamb A.; Gruis N. A.; Weaver-Feldhaus J.; Liu Q.; Harshman K.; Tavtigian S. V.; Stockert E.; Day R. S. 3rd; Johnson B. E.; Skolnick M. H. A cell cycle regulator potentially involved in genesis of many tumor types. Science 264:436–440; 1994. [DOI] [PubMed] [Google Scholar]
- 29. Kuno K.; Kanada N.; Nakashima E.; Fujiki F.; Ichimura F.; Matsushima K. Molecular cloning of a gene encoding a new type of metalloproteinase-disintegrin family protein with thrombospondin motifs as an inflammation associated gene. J. Biol. Chem. 272:556–562; 1997. [DOI] [PubMed] [Google Scholar]
- 30. Kuno K.; Terashima Y.; Matsushima K. ADAMTS-1 is an active metalloproteinase associated with the extracellular matrix. J. Biol. Chem. 274:18821–18826; 1999. [DOI] [PubMed] [Google Scholar]
- 31. Kurdowska A.; Carr F. K.; Stevens M. D.; Baughman R. P.; Martin T. R. Studies on the interaction of IL-8 with human plasma alpha 2-macroglobulin: Evidence for the presence of IL-8 complexed to alpha 2-macroglobulin in lung fluids of patients with adult respiratory distress syndrome. J. Immunol. 158:1930–1940; 1997. [PubMed] [Google Scholar]
- 32. LaMarre J.; Wollenberg G. K.; Gonias S. L.; Hayes M. A. Reaction of alpha 2-macroglobulin with plasmin increases binding of transforming growth factors-beta 1 and beta 2. Biochim. Biophys. Acta 2:197–204; 1991. [DOI] [PubMed] [Google Scholar]
- 33. Lee J.; Horuk R.; Rice G. C.; Bennett G. L.; Camerato T.; Wood W. I. Characterization of two high affinity human interleukin-8 receptors. J. Biol. Chem. 267:16283–16287; 1992. [PubMed] [Google Scholar]
- 34. Lin T.; Chao C.; Saito S.; Mazur S. J.; Murphy M. E.; Appella E.; Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 7:165–171; 2005. [DOI] [PubMed] [Google Scholar]
- 35. Lukas J.; Parry D.; Aagaard L.; Mann D. J.; Bartkova J.; Strauss M.; Peters G.; Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 375:503–506; 1995. [DOI] [PubMed] [Google Scholar]
- 36. Mathew S.; Arandjelovic S.; Beyer W. F.; Gonias S. L.; Pizzo S. V. Characterization of the interaction between alpha2-macroglobulin and fibroblast growth factor-2: Te role of hydrophobic interactions. Biochem. J. 374:123–129; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Michael D.; Oren M. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 13:49–58; 2003. [DOI] [PubMed] [Google Scholar]
- 38. Ng E. S.; Davis R. P.; Azzola L.; Stanley E. G.; Elefanty A. G. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood 106:1601–1603; 2005. [DOI] [PubMed] [Google Scholar]
- 39. Niemuller C. A.; Randall K. J.; Webb D. J.; Gonias S. L.; LaMarre J. Alpha 2-macroglobulin conformation determines binding affinity for activin A and plasma clearance of activin A/alpha 2-macroglobulin complex. Endocrinology 136:5343–5349; 1995. [DOI] [PubMed] [Google Scholar]
- 40. Noble A.; Towne C.; Chopin L.; Leavesley D.; Upton Z. Insulin-like growth factor-II bound to vitronectin enhances MCF-7 breast cancer cell migration. Endocrinology 144:2417–2424; 2003. [DOI] [PubMed] [Google Scholar]
- 41. Odorico J. S.; Kaufman D. S.; Thomson J. A. Multi-lineage differentiation from human embryonic stem cell lines. Stem Cells 19:193–204; 2001. [DOI] [PubMed] [Google Scholar]
- 42. Phillips D. J.; McFarlane J. R.; Hearn M. T.; de Kretser D. M. Inhibin, activin and follistatin bind preferentially to the transformed species of alpha 2-macroglobulin. J. Endocrinol. 155:65–71; 1997. [DOI] [PubMed] [Google Scholar]
- 43. Polejaeva I. A.; Chen S. H.; Vaught T. D.; Page R. L.; Mullins J.; Ball S.; Dai Y.; Boone J.; Walker S.; Ayares D. L.; Colman A.; Campbell K. H. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature 407:86–90; 2000. [DOI] [PubMed] [Google Scholar]
- 44. Raines E. W.; Bowen-Pope D. F.; Ross R. Plasma binding proteins for platelet-derived growth factor that inhibit its binding to cell-surface receptors. Proc. Natl. Acad. Sci. USA 81:3424–3428; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rogers M. B.; Hosler B. A.; Gudas L. J. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development 113:815–824; 1991. [DOI] [PubMed] [Google Scholar]
- 46. Saas P.; Walker P. R.; Quiquerez A. L.; Chalmers D. E.; Arrighi J. F.; Lienard A.; Boucraut J.; Dietrich P. Y. A self-defence mechanism of astrocytes against Fas-mediated death involving interleukin-8 and CXCR2. Neuroreport 13:1921–1924; 2002. [DOI] [PubMed] [Google Scholar]
- 47. Shin T.; Kraemer D.; Pryor J.; Liu L.; Rugila J.; Howe L.; Buck S.; Murphy K.; Lyons L.; Westhusin M. A cat cloned by nuclear transplantation. Nature 415:21; 2002. [DOI] [PubMed] [Google Scholar]
- 48. Tada M.; Takahama Y.; Abe K.; Nakatsuji N.; Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11:1553–1558; 2001. [DOI] [PubMed] [Google Scholar]
- 49. Tan Z.; Qu W.; Tu W.; Liu W.; Baudry M.; Schreiber S. S. p53 accumulation due to down-regulation of ubiquitin: Relevance for neuronal apoptosis. Cell Death Differ. 7:675–681; 2000. [DOI] [PubMed] [Google Scholar]
- 50. Terai I.; Kobayashi K.; Matsushita M.; Fujita T.; Matsuno K. alpha 2-Macroglobulin binds to and inhibits mannose-binding protein-associated serine protease. Int. Immunol. 7:1579–1584; 1999. [DOI] [PubMed] [Google Scholar]
- 51. Thiel S.; Vorup-Jensen T.; Stover C. M.; Schwaeble W.; Laursen S. B.; Poulsen K.; Willis A. C.; Eggleton P.; Hansen S.; Holmskov U.; Reid K. B.; Jensenius J. C. A second serine protease associated with mannan-binding lectin that activates complement. Nature 386:506–510; 1997. [DOI] [PubMed] [Google Scholar]
- 52. Thomson J. A.; Itskovitz-Eldor J.; Shapiro S. S.; Waknitz M. A.; Swiergiel J. J.; Marshall V. S.; Jones J. M. Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147; 1998. [Erratum appears in Science 282:1827; 1998] [DOI] [PubMed] [Google Scholar]
- 53. Vallier L.; Alexander M.; Pedersen R. A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118:4495–4509; 2005. [DOI] [PubMed] [Google Scholar]
- 54. Vazquez F.; Hastings G.; Ortega M. A.; Lane T. F.; Oikemus S.; Lombardo M.; Iruela-Arispe M. L. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J. Biol. Chem. 274:23349–23357; 1999. [DOI] [PubMed] [Google Scholar]
- 55. Vignal E.; De Toledo M.; Comunale F.; Ladopoulou A.; Gauthier-Rouviere C.; Blangy A.; Fort P. Characterization of TCL, a new GTPase of the rho family related to TC10 and Ccdc42. J. Biol. Chem. 275:36457–36464; 2000. [DOI] [PubMed] [Google Scholar]
- 56. Wakayama T.; Perry A. C.; Zuccotti M.; Johnson K. R.; Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394:369–374; 1998. [DOI] [PubMed] [Google Scholar]
- 57. Wang J.; Rao S.; Chu J.; Shen X.; Levasseur D. N.; Theunissen T. W.; Orkin S. H. A protein interaction network for pluripotency of embryonic stem cells. Nature 444:364–368; 2006. [DOI] [PubMed] [Google Scholar]
- 58. Wang Z.; Ruan Y. B.; Guan Y.; Liu S. H. Expression of IGF-II in early experimental hepatocellular carcinomas and its significance in early diagnosis. World J. Gastroenterol. 9:267–270; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Webb D. J.; Weaver A. M.; Atkins-Brady T. L.; Gonias S. L. Proteinases are isoform-specific regulators of the binding of transforming growth factor beta to alpha 2-macroglobulin. Biochem. J. 320:551–555; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wei C. L.; Miura T.; Robson P.; Lim S. K.; Xu X. Q.; Lee M. Y.; Gupta S.; Stanton L.; Luo Y.; Schmitt J.; Thies S.; Wang W.; Khrebtukova I.; Zhou D.; Liu E. T.; Ruan Y. J.; Rao M.; Lim B. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells 23:166–185; 2005. [DOI] [PubMed] [Google Scholar]
- 61. Wollenberg G. K.; LaMarre J.; Rosendal S.; Gonias S. L.; Hayes M. A. Binding of tumor necrosis factor alpha to activated forms of human plasma alpha 2 macroglobulin. Am. J. Pathol. 138:265–272; 1991. [PMC free article] [PubMed] [Google Scholar]
- 62. Wolswijk G. Oligodendrocyte survival, loss and birth in lesions of chronic-stage multiple sclerosis. Brain 123:105–115; 2000. [DOI] [PubMed] [Google Scholar]
- 63. Woods D. B.; Vousden K. H. Regulation of p53 function. Exp. Cell Res. 264:56–66; 2001. [DOI] [PubMed] [Google Scholar]
- 64. Wu S. M.; Patel D. D.; Pizzo S. V. Oxidized alpha2-macroglobulin (alpha2M) differentially regulates receptor binding by cytokines/growth factors: Implications for tissue injury and repair mechanisms in inflammation. J. Immunol. 161:4356–4365; 1998. [PubMed] [Google Scholar]
- 65. Wu Y.; Moser M.; Bautch V. L.; Patterson C. HoxB5 is an upstream transcriptional switch for differentiation of the vascular endothelium from precursor cells. Mol. Cell. Biol. 23:5680–5691; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu R. H.; Chen X.; Li D. S.; Li R.; Addicks G. C.; Glennon C.; Zwaka T. P.; Thomson J. A. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20:1261–1264; 2002. [DOI] [PubMed] [Google Scholar]
- 67. Xu R. H.; Peck R. M.; Li D. S.; Feng X.; Ludwig T.; Thomson J. A. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat. Methods 2:185–190; 2005. [DOI] [PubMed] [Google Scholar]
- 68. Ying Q. L.; Nichols J.; Chambers I.; Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115:281–292; 2003. [DOI] [PubMed] [Google Scholar]
- 69. Zhang H.; Kobayashi R.; Galaktionov K.; Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell 82:915–925; 1995. [DOI] [PubMed] [Google Scholar]
- 70. Zhang R.; Xu Y.; Ekman N.; Wu Z.; Wu J.; Alitalo K.; Min W. Etk/Bmx transactivates vascular endothelial growth factor 2 and recruits phosphatidylinositol 3-kinase to mediate the tumor necrosis factor-induced angiogenic pathway. J. Biol. Chem. 278:51267–51276; 2003. [DOI] [PubMed] [Google Scholar]
- 71. Zhang Y.; Xiong Y.; Yarbrough W. G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725–734; 1998. [DOI] [PubMed] [Google Scholar]
- 72. Zhong J. F.; Zhao Y.; Sutton S.; Su A.; Zhan Y.; Zhu L.; Yan C.; Gallaher T.; Johnston P. B.; Anderson W. F.; Cooke M. P. Gene expression profile of murine long-term reconstituting vs. short-term reconstituting hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 102:2448–2453; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


