Abstract
Retinoic acid (RA) upregulates expression of PDGF ligands and receptors in neonatal rat lung fibroblasts, a process likely to promote maturation of the lung alveolus and possibly microstructures of other organs. A mutational analysis of the gene encoding the PDGF-A ligand has identified a complex retinoic acid response element (RARE) located far upstream of the transcription start site, in a 5′-distal enhanceosome region previously shown to harbor basal and vitamin D-inducible enhancer activity. Maximal RA responsiveness (fourfold) was conferred by nucleotide sequence located between −7064 and −6787, with a variety of deletion and point mutations revealing the importance of at least three nuclear receptor half-sites within the enhancer region (−6851 to −6824), as well as nucleotides located further upstream. Recombinant human retinoic acid receptor/retinoid-X receptor heterodimers bound with high affinity and sequence specificity to multiple regions within the RARE, as demonstrated by electrophoretic mobility shift and DNase I footprinting assays. The addition of RARE activity to previously described functions of the 5′-distal enhanceosome underscores the importance of this region as a key integration point for regulatory control of PDGF-A expression.
Key words: Platelet-derived growth factor, Transcription, Enhancer, Retinoic acid response element, Nuclear receptor
INTRODUCTION
Platelet-derived growth factors (PDGFs) are glycoprotein mitogens consisting of various dimeric combinations of four distinct subunits, denoted PDGF-A, PDGF-B, PDGF-C, and PDGF-D. Genetic manipulation of cultured cells and, more recently, transgenic mice have revealed critical roles of PDGF ligands and receptors (PDGFRs) in embryonic development (4). Disruption of the PDGF-A gene causes impaired septation of lung alveoli and an emphysema-like condition in neonatal mice (6). This defect was attributed to a requirement for epithelial production of PDGF-A in the proliferation and directional migration of fibroblasts during the pseudoglandular stage of alveolarization (6,21). PDGF appears to mediate, at least in part, the alveolarization-promoting activity of all-trans retinoic acid (RA) in neonatal rats (22,26) by stimulating fibroblast proliferation via an autocrine mechanism (20). Induction of PDGF growth loops by RA in the developing lung involves upregulation of transcription for genes encoding PDGF-A and PDGFRs α and β (20), as well as in F9 embryonal carcinoma cells (39) and the murine osteoblast-like cell line, MC3T3-E1 (38).
While considerable progress has been made in defining key regulatory elements within the PDGF-A promoter and flanking regions of the gene (17), the mechanisms underlying upregulation of PDGF-A gene transcription by RA have yet to be elucidated. RA exerts its biological actions by binding and activating heterodimers of the retinoic acid receptor (RAR) and retinoid-X receptor (RXR), which associate with specific DNA sequences in RA target genes and promote chromatin decondensation and recruitment of transcriptional activators (3,23). Retinoic acid response elements (RAREs) are typically composed of two directly repeated motifs (PuGG/TTCA) that are separated by 1 bp (DR1), 2 bp (DR2), or 5 bp (DR5). Noncanonical RAREs have also been described, such as an inverted repeat motif (IR0) in the murine TR2-11 gene (19), a 30-bp element (TGRRE1) in the transglutaminase promoter that contains overlapping DR7 and DR5 half-sites (28), and an element in the retinol binding protein gene consisting of two degenerate half-sites separated by 30 bp (29). A complex enhanceosome located far upstream of the PDGF-A promoter (−6852 to −6787) contains at least four directly repeated half-sites for nuclear receptor binding, suggesting potential as a RARE that could mediate the stimulatory effects of RA (27). Two of the half-sites are present as a DR3 motif, which has been shown to serve as a binding platform for the vitamin D receptor (VDR)-RXR heterodimer and to be required for both vitamin D-dependent (31) and vitamin D-independent (32) enhancer activity. In the current study, we demonstrate that RA inducibility involves three of the half-sites within the enhanceosome region, as well as interactions with one or more additional motifs located at least 142 bp further upstream.
MATERIALS AND METHODS
Plasmid Construction
pAC261luc contains a PDGF-A promoter fragment extending from −261 to +8 in fusion with the luciferase reporter gene; J+/pAC261luc contains a 507-bp fragment from the 5′-distal region of the PDGF-A gene (−7294 to −6787) inserted upstream of the PDGF-A promoter; pAC261-ACE66luc harbors an insertion of the 66-bp ACE enhancer (−6852 to −6787); pAC-CAT7.3 and pAC-CAT6.8 contain large 5′-flanking fragments of the PDGF-A gene (−7294 to +8 and −6787 to +8, respectively) in fusion with the chloramphenicol acetyltransferase (CAT) reporter gene. Construction of the plasmids above has been described previously (27). pBL-TK-βRE-luc is a luciferase reporter plasmid containing a DR5 retinoic acid response element from the RARβ gene in fusion with a minimal promoter fragment from the thymidine kinase gene of herpes simplex virus (generously provided by D. Noonan, University of Kentucky).
The following plasmids were constructed for these studies. Two subfragments of the −7294 to −6787 region were obtained by digestion with restriction endonucleases and inserted into pAC261luc: −7199 to −6787 (SstI/EcoRI) and −7000 to −6787 (BspMI/EcoRI). All other subfragments and point mutations of nuclear receptor-binding half-sites were obtained by the overlap extension modification of polymerase chain reaction (PCR) (14), followed by insertion into the TA-cloning vector, pCR1000 (Invitrogen, Carlsbad, CA). Guanine residues were substituted into the fourth and fifth positions (NNNccN or NNNtcN to NNNggN) of half-sites A-C and the putative upstream half-site a (−6952 to −6947), as described previously (32). PCR-constructed inserts in pCR1000 were sequenced by automated methods (ACGT, Wheeling, IL), then excised with NdeI and inserted upstream of the PDGF-A promoter in pAC261luc.
Cell Culture, Transfections, and Luciferase Assays
JEG-3 and CV-1 cells were obtained from the American Type Culture Collection and maintained as described previously (27) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 10 nM nonessential amino acids, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Primary rat lung fibroblasts were isolated according to the method of Al-Jumaily and Bruce (2) and cultured in a 1:1 mixture of DMEM and Ham’s F12 media, supplemented with 10% FBS. For treatment with RA, lung fibroblasts were made quiescent by preincubation for 24 h in a serum-free medium formulation (Keratinocyte SFM). All culture media and media additions were obtained from GibcoBRL (Grand Island, NY).
Plasmids used in transient transfection experiments were purified by a batch ion-exchange chromatographic procedure (Qiagen, Valencia, CA). JEG-3 and CV-1 cells were transfected by a modification of calcium phosphate/DNA coprecipitation (13) as described previously (27). Briefly, 2 × 105 cells were plated per 60-mm plastic dish, 24 h prior to transfection. Transfections were performed in duplicate dishes and in at least three independent experiments. Each dish received precipitate containing 1.5 μg of an experimental reporter plasmid, 1.5 μg of the control plasmid pCMV β-gal, and 0.3μg of plasmids directing expression of VDR (pCMV-VDR), RARα (pCMV-RARα), or parent vector alone (pL2) (31). Sixteen hours later, precipitates were removed and cells were treated for 24 h with fresh DMEM supplemented with 5% charcoal-stripped FBS and 10−7 M 1,25-dihydroxyvitamin D3 [1,25(OH2D3; generously provided by L. Binderup, Leo Pharmaceuticals, Ballerup, Denmark], 10−6 M all-trans retinoic acid (RA), or solvent alone (ethanol or isopropanol). Cells were harvested for measurement of firefly and Renilla luciferase, CAT, or β-galactosidase activity, as described previously (27).
Real-Time Quantitative PCR
Total cellular RNA was isolated from neonatal rat fibroblasts cells using an RNA Easy kit (Invitrogen) and stored at −80°C. RNA (1 μg) was reverse transcribed (SuperScript first strand synthesis kit; Invitrogen), and the cDNA product was used to generate a standard curve (1 pg to 100 ng). A master mix (40 μl/sample) was prepared containing a primer/ probe combination for either rat GAPDH or PDGF-A (Assay-on-Demand, Applied Biosystems, Foster City, CA) at final concentrations of 0.9 μM primer and 0.25 μM probe. Standard PCR cycle conditions were employed for amplification using an ABI Prism Sequence Detection System 7700 Analyzer (Applied Biosystems). Initial RNA concentrations were extrapolated from Ct values and the relative standard curve, with PDGF-A values expressed relative to those obtained for GAPDH.
Expression and Preparation of Recombinant Human RARα and RXRα
Recombinant human RARαΔAB (aa 84–462) and mouse RXRαΔAB (aa 133–467) were expressed as histidine-tagged fusion proteins under lacZ induction in the Escherichia coli strain BL21 (DE3) pLysS using the vector pET3b (gift of H. Gronemeyer, Strasbourg, France). Both receptors lack the N-terminal A/B region but retain DNA binding, ligand binding, and dimerization activities (18). Freshly transformed bacteria were used to inoculate 5 ml of liquid LB medium containing ampicillin and chloramphenicol and grown for 16 h at 37°C. The culture (2 μl) was used to inoculate 200 ml of LB medium containing ampicillin. When the A600 reached 0.6, protein expression was induced with isopropyl-1-thio-β-d-galactopyranoside (IPTG; 1 mM). After 3 h of IPTG induction, cells were harvested by centifugation and resuspended in 19 ml of binding buffer (20 mM Tris, 500 mM sodium chloride, 5 mM imidazole, 0.5 mM dithiothreitol and phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin and antipain, pH 8.0) containing 100 μg/ml lysozyme. Cells were lysed by freeze-thawing with sequential incubations in liquid nitrogen and ice water. Further cell disruption was accomplished by sonication for 1 min using alternating 1-s pulses and rests (Branson Model 1026, 30% amplitude), followed by dilution of the lysate with an equal volume of binding buffer and an additional 1-min cycle of sonication. Lysates were cleared by centrifugation (70,000 × g, 30 min), followed by purification of recombination proteins using nickel-affinity chromatography (HiTrap Chelating HP column, Amersham Biosciences). Bound proteins were eluted using an imidazole gradient (10–500 mM) in 20 mM sodium phosphate, 500 mM sodium chloride, pH 7.4. Peak fractions containing the receptors were concentrated and transferred into storage buffer (20 mM HEPES, 100 mM potassium chloride, 0.2 mm EDTA, 0.5 mM dithiothreitol, 20% glycerol, pH 7.9) by centrifugal ultrafiltration (Centricon-10, Millipore, Billerica, MA). Protein concentrations were quantitated by the method of Bradford (7) and the proteins were stored at −80°C until use.
Electrophoretic Mobility Shift Assays and DNase I Protection Footprinting
Electrophoretic mobility shift assays (EMSAs) were conducted as described previously (27). The −7064 to −6787 fragment employed as a DNA binding substrate was gel purified after excision from pAC261luc with NdeI, and 5′-end-labeled with Klenow fragment of DNA polymerase I and [32P]dCTP (3000 Ci/mmol, Amersham Biosciences, Piscataway, NJ). RXRαΔAB and RARαΔAB (300 ng) were pre-incubated on ice with poly(dI-dC) (0.5 mg) for 30 min, followed by incubation with radiolabeled DNA for 15 min at 20–22°C. For antibody supershift/inhibition studies, rabbit anti-RARα, anti-RXRα, or irrelevant IgGs (Santa Cruz, Santa Cruz, CA) were added after the initial binding reaction and incubated for 60 min at 2–4°C. DNA-protein complexes were resolved by electrophoresis on 6% nondenaturing polyacrylamide gels (0.25× TBE) and visualized by phosphorimaging (Storm 840, Molecular Dynamics, Sunnyvale, CA). Competitor DNAs containing internal mutations within individual nuclear receptor-binding half-site motifs of the −7064 to −6787 region of the PDGF-A gene were synthesized by overlap extension PCR, as described previously (14).
DNase I protection footprinting was carried out using the Core Footprinting System (Promega) according to the manufacturer’s instructions. The −7064 to −6787 fragment was selectively 32P-labeled on either the coding or noncoding strand as described previously (31). The DNA probe was incubated with heterodimeric RARαΔAB/RXRαΔAB as described above for EMSA, followed by addition of 0.75 U of RNase-free DNase I for 1 min at 22–24°C. The cleavage reaction was terminated by addition of stop solution, followed by DNA extraction with phenol:chloroform: isoamyl alcohol (25:1:24) and ethanol precipitation. DNA fragments were resolved on 12% sequencing gels and visualized by phosphorimaging.
RESULTS
A Potent Retinoic Acid Response Element Is Located in the Vicinity of a 5′-Distal Enhanceosome of the PDGF-A Gene
We showed previously that mitogenic activity of RA in neonatal rat fibroblasts (NRFs) was mediated by activation of an autocrine PDGF growth loop (20). To measure the extent to which RA induces expression of the endogenous PDGF-A gene, PDGF-A mRNA levels were determined by quantitative PCR (Q-PCR) following 24 h of treatment over a range of RA concentrations (10−9 to 10−7 M). Maximal induction was obtained with 10−7 M (25-fold) (Fig. 1), with significant stimulation (fivefold) seen at the lowest concentration of RA administered (10−9 M). This response to RA was more robust than those reported for many other RA target genes (10,16,33).
Figure 1.
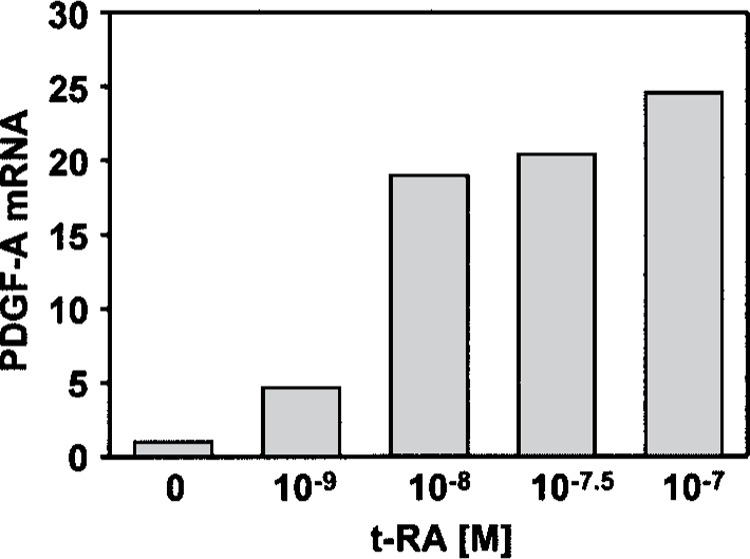
Retinoic acid induces expression of the endogenous PDGF-A gene. Neonatal rat lung fibroblasts were treated with all-trans retinoic acid for 24 h at the concentrations indicated. Values represent the fold-change in the ratio of PDGF-A to GAPDH mRNA concentrations following RA treatment, as determined by real-time PCR and described in Materials and Methods. The results shown are representative of duplicate experiments that demonstrate retinoic acid inducibility of PDGF-A mRNA expression.
To identify RA response elements (RAREs) mediating RA inducibility of the PDGF-A gene, a large −7294 to +8 fragment of the PDGF-A gene was linked to a luciferase reporter gene and analyzed by transient transfection in CV-1 cells. The fragment mediated a significant RA-induced increase in transcriptional activity when either RARα (eightfold) or RARβ (approx. sixfold) was coexpressed (Fig. 2), indicating the presence of RARE activity. Basal transcription activity of the fragment was inhibited significantly upon overexpression of RARa (∼70%) and, to a lesser extent, RARβ. A 5′-deletion of the region between −7294 and −6787, which contains the ACE66 basal enhancer (27,32) and its four nuclear receptor-binding half-site motifs (sites A-D) (Fig. 3) resulted in loss of RA responsiveness as well as reduced basal transcription activity. Taken together, these results indicate the presence of RARE activity over a span of 7294 bp of 5′-flanking sequence and localize the response to a 5′-distally situated region between −7294 and −6787. The inhibitory effects of RARα and RARβ on ACE66 enhancer activity could suggest competition with basal enhancer transcription factors for DNA motifs in this region.
Figure 2.
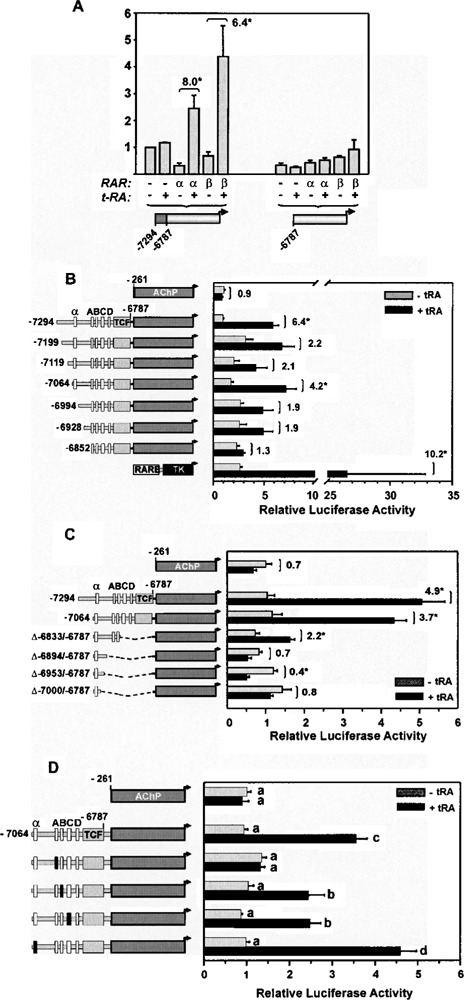
Localization of a retinoic acid response element (RARE) between nucleotides −7064 and −6787 in the 5′-distal enhanceosome region of the PDGF-A gene. All results were obtained by transient transfection of CV-1 cells and represent relative luciferase activity obtained after correction for interwell variation in transfection efficiency with a cotransfected plasmid directing consitutive expression of Renilla luciferase. Results are expressed as the mean (±SEM) of at least six replicate transfected wells obtained over three independent transfection experiments. (A) CV-1 cells were transfected with luciferase reporter plasmids containing either a −7294 to +8 fragment of the PDGF-A gene, or a 5′-truncated fragment (−6787 to +8), as shown below the figure. Other treatments indicated are the presence or absence (+ or −) of expression vectors for RARβ and RARP, and RA administration (10−6 M) for 24 h. Fold-induction in luciferase activity is presented for treatment pairs in which a significant induction (p < 0.05, denoted with asterisk) from RA treatment was observed. (B) 5′-Endpoint deletional analysis of the −7064 to −6787 region. Represented at left is the series of 5′-endpoint deletions employed, within which four nuclear receptor-binding half-sites (A–D) and the ternary complex factor (TCF) binding motif within the ACE66 enhancer are represented with open and gray-filled boxes, respectively. An additional nuclear receptor-binding half-site (denoted α) located further upstream is also represented. Also shown is the construct pBL-TK-PRE, which contains an RARE from the RARP gene in fusion with the minimal HSV thymidine kinase (TK) promoter. (C) 3′-Endpoint deletional analysis of the −7064 to −6787 region, with deleted areas represented by dotted lines. (D) Mutational analysis of nuclear receptor-binding half-sites. Half-site motifs that contain inactivating mutations are identified with black boxes. Transfection efficiency was assessed by cotransfection with a β-galactosidase expression vector. Means not sharing an identical superscript are significantly different, as determined by ANOVA and the Fisher’s PLSD post hoc test for mean separation (p < 0.05).
Figure 3.
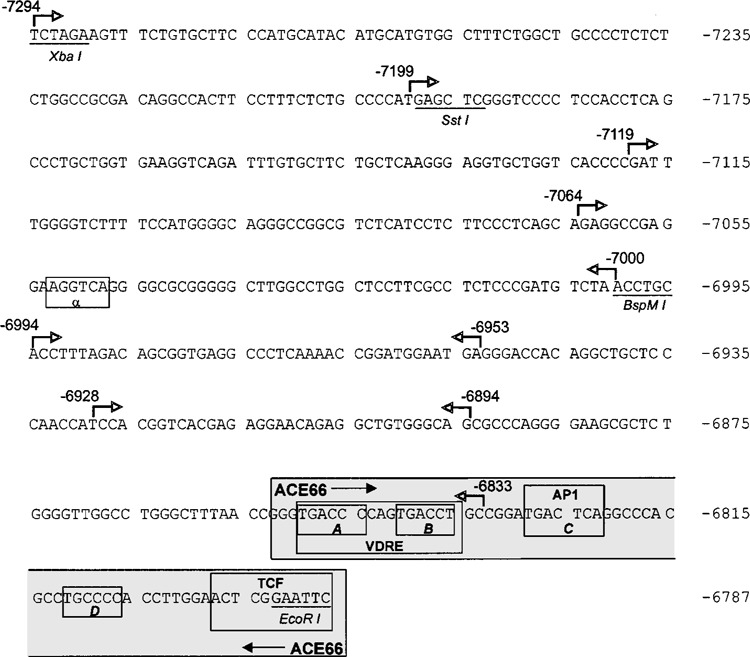
Nucleotide sequence of the 5′-distal enhanceosome and neighboring regions from the PDGF-A gene. Shown is the sequence spanning the region between −7294 and −6787, which contains the basal enhancer element ACE66 (−6852 to −6787, outlined in gray box) at the 3′-terminus. Nucleotides are numbered relative to the transcription start site as determined previously (5,38). Relevant restriction endonuclease sites (XbaI, SstI, BspMI, and EcoRI) are underlined, while half-site motifs for nuclear receptor binding (a, A, B, C, and D), and binding sites for VDR (VDRE), AP1 and ternary complex factors (TCF) are boxed. Arrows denote locations of 5′-endpoints (left arrows) and 3′-endpoints (right arrows) employed in construction of deletion mutants for localization of the RARE.
To localize the RARE in finer detail, an array of deletions and point mutations were introduced into the −7294 and −6787 region (Fig. 3) and relocated upstream of a minimal PDGF-A promoter fragment (−261 to +8). The intact −7294 to −6787 subfragment mediated a sixfold induction following RA administration and forced RARα expression (Fig. 2B), equivalent to that obtained with the full-length PDGF-A genomic fragment (−7294 to +8). A 5′-endpoint deletion to −7199 reduced RARE activity to twofold, a loss attributable to a threefold increase in basal activity rather than a decrease in RA-stimulated activity per se. Interestingly, RARE activity was recovered partially (fourfold) by further 5′-deletion to −7064, while progressive 5′-deletions to −6994 and −6928 reduced RARE activity to twofold. The ACE66 enhancer alone (−6852 to −6928) was unresponsive to RA treatment, an unexpected finding in light of the four nuclear receptor half-sites present in the element. Nevertheless, the enhancer remained resistant to RA treatment even when presented as three tandem copies (data not shown), an arrangement that provides marked synergism of both basal and vitamin-D-inducible enhancer activity (27,31). Overall, this series of 5′-endpoint deletions revealed that RA inducibility was mediated by at least three sequences distributed over the −7294 and −6787 region, located between −7294 and −7119, −7064 and −6994, and −6928 and −6852.
Next, a series of 3′-endpoint deletions were introduced into the −7064 to −6787 subfragment to determine the 3′ border of the RARE. An approximately fourfold induction obtained with the −7064 to −6787 fragment was blunted significantly by a 3′-deletion to −6833 that removed a TCF binding site, and nuclear receptor-binding half-sites C and D (2.2-fold) (Fig. 2C). An additional 3′-deletion to −6894, which excised half-sites A and B, resulted in loss of the remaining RARE activity. To determine which of the nuclear receptor-binding half-sites contributed to the response, each was disrupted individually with dinucleotide substitutions shown previously to disrupt basal enhancer and VDRE activity (32). Mutation of half-site A resulted in a complete loss of RA inducibility, while disruption of sites B and C caused partial reductions in the response (∼50%) (Fig. 2D). Disruption of an additional perfectly conserved upstream half-site (−7054 to −7047; half-site α) had no effect. Taken together, these mutagenic analyses demonstrate that site A is obligatory for RARE activity, while sites B and C are also required for optimal responsiveness. Moreover, these sites were shown to be necessary but not sufficient for RARE activity, also requiring additional DNA sequences located further upstream of the ACE66 enhancer.
Transcription-Inducing Effect of RA on the 5′-Distal Region of the PDGF-A Gene Dominates Over That of 1,25-(OH)2D3: Lack of Transcriptional Synergy Between Overlapping RARE and VDRE Elements
The overlap of RARE activity with a previously described VDRE motif (DR3, sites A and B) in the ACE66 enhancer suggested the potential for cooperativity in response to the respective ligand activators, RA and 1,25-(OH)2D3. To investigate this possibility, transcriptional activity of the −7294 to −6787 fragment was measured by transient transfection analysis in CV-1 cells following administration of RA and 1,25-(OH)2D3, either alone or in combination (Fig. 4). The minimal PDGF-A promoter fragment was essentially unresponsive to treatment with either agent, regardless of RAR or VDR status. In this series of experiments, the −7294 to −6787 fragment mediated significant enhancement in response to either RA (3.5-fold) and 1,25-(OH)2D3 (4.7-fold) in the absence of heterologous overexpression of the cognate receptors RARα and VDR. This response would appear to be the result of sporadic expression of endogenous RARs and VDR in the CV-1 cell line, and reinforces the potential for physiological relevance of these regulatory events. Heterologous overexpression of RARα and VDR, however, boosted significantly the response to RA (4.7-fold) and 1,25-(OH)2D3 (9.1-fold). When both RARα and VDR were overexpressed, the induction from RA treatment was nearly the same as that observed when RARα was expressed alone. In contrast, coexpression of RARα and VDR blunted the transcriptional response to 1,25-(OH)2D3 (2.5-fold), probably the result of competition between unliganded, overexpressed RARα and liganded VDR for binding to half-sites A and B. Concurrent administration of RA and 1,25-(OH)2D3 in the presence of heterologous expression of both RARα and VDR induced activity no greater than that obtained with RA administration alone. This lack of transcriptional synergy was not due to use of maximally stimulatory concentrations of RA and 1,25-(OH)2D3, as downward titration of both ligands also failed to reveal cooperativity (data not shown). The lack of transcriptional synergy between RA and 1,25-(OH)2D3 and their cognate response elements is consistent with the notion that binding of RAR to this upstream region dominates over that of VDR, both in the presence or absence of RA stimulation.
Figure 4.
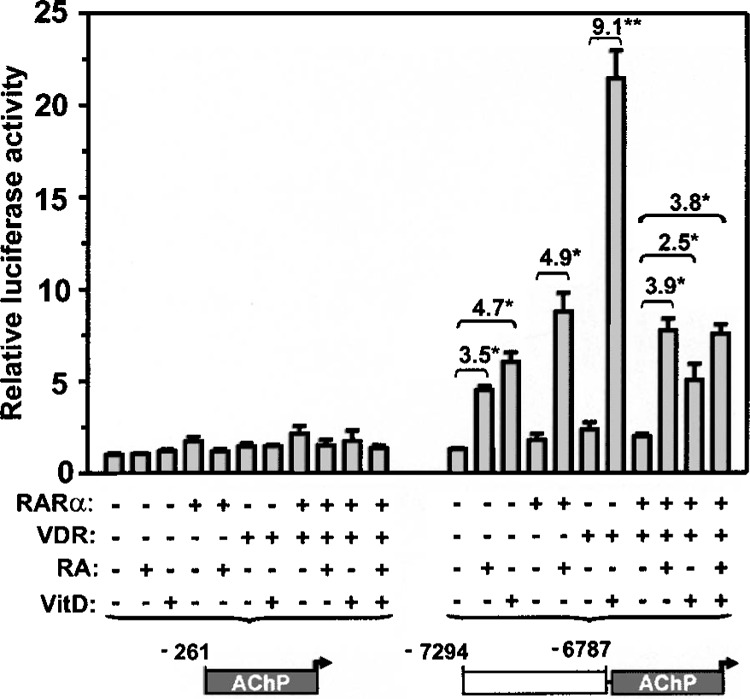
Induction of 5′-distal enhanceosome activity by retinoic acid is not cooperative with vitamin D inducibility. Results represent fold-change in relative luciferase activity obtained with the −7294 to −6787 region in response to the treatments summarized below the graph. Treatments consisted of plasmid-directed overexpression of RARα and/or VDR, and administration of 1,25-(OH)2D3 (VitD, 10−7 M) and/or RA (10−6 M).
RARα/RXRα Heterodimers Bind With High Affinity and Nucleotide Sequence Specificity to the 5′-Distal Enhanceosome via Interactions With Multiple Sequence Motifs
The above-described reporter gene analyses indicated that RARE activity is distributed over the entire −7294 to −6787 region of the PDGF-A gene. To verify this region is indeed a bona fide binding site for the human RARα/RXRα heterodimer, the proteins were expressed in E. coli as histidine-tagged fusion proteins and prepared in near homogeneous form by nickel-affinity chromatography (Fig. 5A). When RARα and RXRα were incubated with a double-stranded, 32P-endlabeled form of the −7064 to −6787 fragment, EMSA revealed formation of a DNA-protein complex (Fig. 5B). Titration of increasing amounts of RARα (2–75 ng) into a fixed quantity of RXRα (12.5 ng) led to increasing amounts of the complex, which was saturated at 19 ng. No complex was formed in the absence of RXRα, strongly suggesting that the complex represented the RARα/RXRα heterodimer. This conclusion was further supported by the efficient formation of DNA-protein complexes when binding reactions were conducted with small, equivalent amounts of RARα and RXRα (Fig. 5C). As concentrations of the two proteins were increased, larger protein complexes of reduced mobility were obtained, probably representing higher order multimers. Either RARα or RXRα alone was much less efficient in eliciting complex formation than were the proteins in combination.
Figure 5.
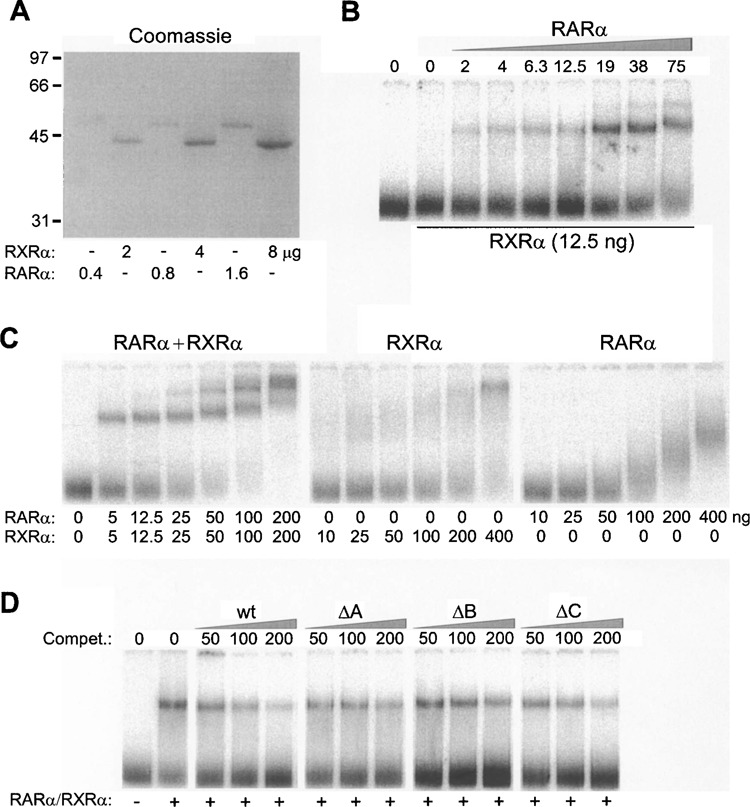
Retinoic acid receptor α/retinoid-X receptor α (RARα/RXRα) heterodimers bind with high affinity and nucleotide sequence specificity to the 5′-distal RARE: electrophoretic mobility shift assay (EMSA). (A) SDS-PAGE analysis of affinity-purified recombinant RARα and RXRα. (C) Binding of RARα/RXRα heterodimers to the −7064 to −6787 fragment of the PDGF-A gene: titration of increasing RARα protein (0−75 ng) into a fixed concentration of RXRα (12.5 ng). EMSA was conducted with a fixed amount (15 pmol, 20,000 cpm) of 32P-labeled −7064 to −6787 probe (Pr). Binding reactions were conducted in the presence of bovine serum albumin (BSA) to minimize inert binding of recombinant proteins in very low concentrations to plastic surfaces. (C) RARα/RXRα heterodimers bind to the −7064 to −6787 fragment with much higher affinity than either protein alone. (D) Mutations in nuclear receptor-binding motifs A, B, and C result in diminished binding affinity for the RARα/RXRα heterodimer in vitro. Assays were conducted with 12.5 ng of both RARα and RXRα, 32P-labeled −7064 to −6787 probe, and the indicated amounts (50–200-fold molar excess relative to probe) of unlabeled competitor DNAs (Compet.) corresponding to the −7064 to −6787 region. Competitors were constructed in wild-type (wt) form or containing substitution mutations within consensus nuclear receptor-binding motifs A–C as indicated.
To compare the relative contributions of nuclear receptor half-sites A–C to binding of the RARα/RXRα heterodimer, synthetic DNAs corresponding to the −7064 to −6787 fragment and harboring mutations within motifs A, B, or C were created by polymerase chain reaction and analyzed for their ability to compete for binding to the radiolabeled, unmodified sequence. Unlabeled, homologous DNA achieved half-maximal competition for DNA-protein complex formation at an approximately 50-fold excess relative to the probe (IC50 ∼12.5 nM) (Fig. 5D), reflecting a high affinity of the receptor for this sequence. A twofold lower, but nevertheless, high affinity (i.e., IC50 ∼25 nM) was observed for each of the three mutant DNAs, suggesting that the presence of two intact half-sites is sufficient to maintain occupancy of the −7064 to −6787 fragment by the RARα/RXRα heterodimer.
To localize specific points of contact between the RARα/RXRα heterodimer and the −7064 to −6787 region at single nucleotide resolution, DNA footprinting was conducted using the DNase I protection approach. Incubation of recombinant RARα and RXRα with a −7064 to −6787 probe that was selectively radiolabeled on the coding strand revealed the presence of multiple regions of binding over a region spanning between −7060 and −6850 (Fig. 6). Of particular note were pronounced footprints between −7060 and −7047, which was comprised of the half-site α and eight additional 5′-flanking nucleotides. The location of these footprints corresponded well with nucleotide sequence (−7064 to −6994) shown in transient transfection analyses to be critical for RARE activity. At least five other smaller footprints were also observed 3′ to this site, most of which were accompanied by regions of DNase I hypersensitivity. DNase I hyper-sensitivity is often a hallmark of protein–DNA binding, resulting from projection of torsional stress and disruption of double helical structure at adjacent DNA. Similarly, footprinting analyses conducted with the noncoding strand of the −7064 to −6787 DNA fragment demonstrated multiple sites of binding by the RARα/RXRα heterodimer (Fig. 6). Foot-printing was localized clearly to each of the four nuclear receptor binding half-site motifs (A–D) of the ACE66 enhancer, with marked DNase I hypersensitivity observed 3′ to sites A, C, and D. Taken together, the EMSA and footprinting studies demonstrate that the −7064 to −6787 region represents a high-affinity binding site for the RARα/RXRα heterodimer. Moreover, the distribution of multiple binding sites over this region corroborates the finding that several sequence motifs found in the vicinity of the 5′-distal enhanceosome contribute to RARE activity.
Figure 6.
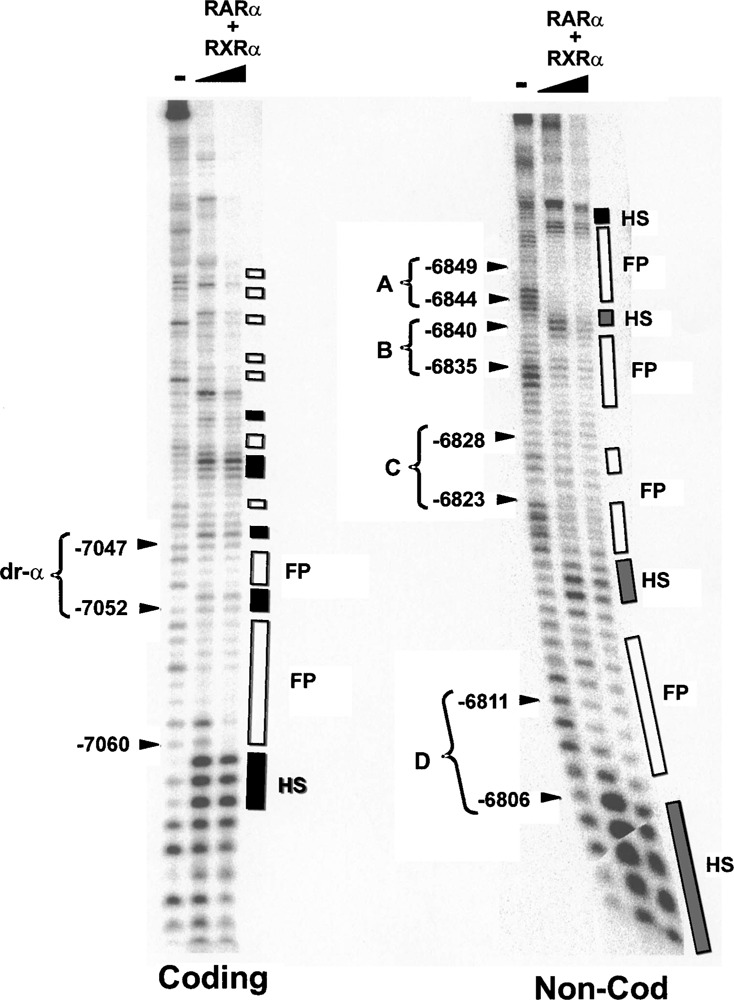
RARα/RXRα binds to multiple sites within the −7064 to −6787 fragment: DNase I footprinting analysis. Analyses were conducted in the absence (−) or presence (0.4 or 1.0 μg) of RARα/RXRα. Double-stranded, 32P-labeled −7064 to −6787 fragment was used as the binding substrate for analyses performed with labeled coding (left panel) or noncoding (right panel) strands. Identified with arrows to the left of each panel are mobilities of bands corresponding to nuclear receptor binding sites A–D and α, while regions of footprinting (FP, open boxes) and DNase I hypersensitivity (HS, shaded boxes) are indicated to the right of each panel.
DISCUSSION
The primary goal of these studies was to identify and localize DNA elements mediating induction of the PDGF-A gene expression by RA, a phenomenon we observed initially in cultures of neonatal lung fibroblasts (NRFs) (20). Our analysis of a large 7294-bp region of 5′-flanking sequence in the PDGF-A gene revealed the presence of a single RA-responsive domain, 507 bp in length and located approximately 7 kb from the transcription start site. The domain functioned as a potent RARE both from its natural 5′-distal location and when relocated next to a minimal PDGF-A promoter fragment. RARE activity of the 507-bp region was stimulated modestly in the absence of exogenous RAR (i.e., by endogenous receptors) by twofold in the mouse lung epithelial cell line, MLE-12 (data not shown). Localization of RARE activity in close proximity to previously identified basal (ACE) and vitamin D-inducible enhancers further underscores the importance of the 5′-distal enhanceosome region as a key integration site for signals regulating PDGF-A gene transcription. The multiplicity of DNA motifs mediating RARE activity, as well as their proximity to binding sites within the ACE enhancer for such MAPK-inducible factors as AP1, suggests the potential for cooperativity in the PDGF-A response to RA.
The RARE identified in this study appears to be one of the most complex described to date, although noncanonical RAREs also have been described in other genes. For example, the mouse transglutaminase gene possesses an RARE (mTGRRE1) consisting of three RAR binding half-sites separated by two internal spacer sequences of 5 and 7 bp each (28). While the two 5′-most half-sites are sufficient for binding of RAR in vitro, RARE activity requires the full tripartite element. Moreover, additional complexity is evident in the obligate requirement for interactions between the mTGRRE1 element and an additional, non-RAR binding element located further downstream. Other noncanonical RAREs include another tripartite element in the rat growth hormone gene (36), and a composite of two adjacent DR2 and DR1 motifs that cooperate to provide full RA responsiveness of the mouse cellular retinoic acid binding protein II promoter (12). Multiplicity of RARE elements may in fact prove to be more often the rule, rather than rare exceptions to prototypical single direct repeat motifs (24), serving to enhance recruitment of transcription factors and coactivators via increased protein interaction surfaces. A recent X-ray crystallographic analysis of the interferon-β enhanceosome also indicates that transcriptional synergy is dependent on transactions of bound factors with the DNA double helix, contributions that may even outweigh those provided by protein-protein interactions (30). In this regard, it is noteworthy that binding of RARα/RXRα heterodimers to the PDGF-A RARE projects significant torsional stress to DNA sequences adjacent to the primary points of contact, evident in footprinting analyses of this study as regions of strong DNase I hypersensitivity (Fig. 6). This raises the interesting possibility that occupancy of the PDGF-A RARE by RAR at multiple sites elicits transcriptional induction not only by increasing direct recruitment of coactivator mass, but by influencing binding of factors at other nearby DNA sequences as well. Also relevant is the finding that mutational disruption of any single half-site A, B, or C has only minimal impact on binding of the heterodimer to the RARE in vitro, but results in very significant dimunition of RARE-mediated transcriptional induction.
RA and 1,25-(OH)2D3 failed to provide synergistic or even additive effects on enhancer activity, despite the joint presence of RARE and VDRE elements in the 5′-distal enhanceosome region. When these ligands were administered concurrently, either at maximal or submaximal concentrations, ligand inducibility was limited to that obtained with RA alone. Given the well-known ability of unliganded RAR to bind to DNA and recruit such corepressor molecules as SMRT and NCoR (9,15), it will be of interest to determine whether such repression is operative and, if so, how this repression is relieved in cells that exhibit elevated ACE activity. The ACE element does confer modest ACE repression (twofold) in MCF-7 breast carcinoma and U87 glioblastoma cells (unpublished observations), although it is inactive in most cell lines. Our previous studies (32) have shown that ACE activity is dependent to a large extent upon MAPK signaling to proteins that bind to some of the same half-sites involved in RARE and VDRE activity, suggesting these signals may play an important role in optimal activation of both RAR and VDR. The current study provides further evidence of the 5′-distal region of the PDGF-A gene as a site of considerable overlapping as well as discrete sites for basal and ligand-induced enhancement of transcription.
Retinoids such as RA play vital roles in development of the vertebrate embryo and maintenance of the differentiated state in adult tissues (34). A particularly interesting property of these compounds is their ability to stimulate regeneration of complex structures that are either damaged or missing (22). Examples include the RA-induced “supergeneration” of extra limb structures following amputation in Urodele amphibians, and promotion of development of fully developed alveoli (alveolarization) in the rodent lung (22,26). Such pleiotropic effects of RA are likely to be mediated via modulation of such master regulators of developmental patterning as the homeobox (Hox) genes (25). Many Hox and other genes possess RAREs and are likely to be part of an immediate response to retinoids as so-called “first-order dependence” genes (34). Our identification of a potent RARE in the PDGF-A gene strongly suggests PDGF-A is also a direct transcriptional target of RA. This conclusion is supported by the rapidity of RA-induced increases in PDGF-A mRNA (<24 h), as well as the requirement for binding of RARα and RARβ for full RARE activity. The RA/PDGF-A regulatory axis appears to be particularly relevant in the process of alveolarization in the lung, in which PDGF-A has been shown to provide a critical stimulus for the proliferation and migration of alveolar myofibroblasts during septal formation in mice (6). A direct demonstration of the RA/PDGF-A axis has been obtained recently in RARβ-null mice, which exhibit an alveolar deficit and decreased expression of PDGF-A, as predicted by our current study (35). The extensive roles for retinoids and PDGFs in the development of multiple organ structures suggest the RA/ PDGF-A axis may have other important settings as well. While many other first-order dependence genes for RA have been identified to date, few possess the profound relationship with developmental processes as do the PDGFs and PDGF receptors (4). Growth factors such as EGF, TGF-β, and FGF have also been implicated as participants in the morphogenic and teratogenic activities of RA (1,8,11), but the transcriptional mechanisms mediating these effects are not fully elucidated. Thus, the RARE in the PDGF-A gene provides new insights into mechanisms underlying the morphogenic activity of RA, particularly in the lung. As such, it may also represent a novel target for treatment of lung diseases characterized by alveolar insufficiency, such as bronchopulmonary dysplasia in infants and emphysema in adults.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (HL 62877, CA 83237) and the Kentucky Lung Cancer Research Board to D.M.K. We would also like to thank H. Gronemeyer, D. Noonan, and J. Whitsett for providing plasmids and cell lines essential to these studies.
REFERENCES
- 1. Abbott B. D.; Best D. S.; Narotsky M. G. Teratogenic effects of retinoic acid are modulated in mice lacking expression of epidermal growth factor and transforming growth factor-alpha. Birth Defects Res. A Clin. Mol. Teratol. 73:204–217; 2005. [DOI] [PubMed] [Google Scholar]
- 2. Al Jumaily W.; Bruce M. C. The postnatal age of rat lung fibroblasts influences G1/S phase transition in vitro. In Vitro Cell Dev. Biol. Anim. 35:410–416; 1999. [DOI] [PubMed] [Google Scholar]
- 3. Bastien J.; Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328:1–16; 2004. [DOI] [PubMed] [Google Scholar]
- 4. Betsholtz C. Insight into the physiological functions of PDGF through genetic studies in mice. Cytokine Growth Factor Rev. 15:215–228; 2004. [DOI] [PubMed] [Google Scholar]
- 5. Bonthron D. T.; Morton C. C.; Orkin S. H.; Collins T. Platelet-derived growth factor A-chain: Gene structure, chromosomal location, and basis for alternative mRNA splicing. Proc. Natl. Acad. Sci. USA 85:1492–1496; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bostrom H.; Willetts K.; Pekny M.; Leveen P.; Lindahl P.; Hedstrand H.; Pekna M.; Hellstrom M.; Gebre-Medhin S.; Schalling M.; Nilsson M.; Kurland S.; Tornell J.; Heath J. K.; Betsholtz C. PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85:863–873; 1996. [DOI] [PubMed] [Google Scholar]
- 7. Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254; 1976. [DOI] [PubMed] [Google Scholar]
- 8. Butts S. C.; Liu W.; Li G.; Frenz D. A. Transforming growth factor-beta(1) signaling participates in the physiological and pathological regulation of mouse inner ear development by all-trans retinoic acid. Birth Defects Res. A Clin. Mol. Teratol. 73:218–228; 2005. [DOI] [PubMed] [Google Scholar]
- 9. Chen J. D.; Evans R. M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454–457; 1995. [DOI] [PubMed] [Google Scholar]
- 10. Costet P.; Lalanne F.; Gerbod-Giannone M. C.; Molina J. R.; Fu X.; Lund E. G.; Gudas L. J.; Tall A. R. Retinoic acid receptor-mediated induction of ABCA1 in macrophages. Mol. Cell. Biol. 23:7756–7766; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diez del Corral R.; Storey K. G. Opposing FGF and retinoid pathways: A signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays 26:857–869; 2004. [DOI] [PubMed] [Google Scholar]
- 12. Durand B.; Saunders M.; Leroy P.; Leid M.; Chambon P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell 71:73–85; 1992. [DOI] [PubMed] [Google Scholar]
- 13. Graham F. L.; Van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467; 1973. [DOI] [PubMed] [Google Scholar]
- 14. Ho S. N.; Hunt H. D.; Horton R. M.; Pullen J. K.; Pease L. R. Site-directed mutagenesis by overlap extension using the PCR. Gene 77:51–59; 1989. [DOI] [PubMed] [Google Scholar]
- 15. Horlein A. J.; Naar A. M.; Heinzel T.; Torchia J.; Gloss B.; Kurokawa R.; Ryan A.; Kamei Y.; Soderstrom M.; Glass C. K. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397–404; 1995. [DOI] [PubMed] [Google Scholar]
- 16. Jacobson A.; Johansson S.; Branting M.; Melhus H. Vitamin A differentially regulates RANKL and OPG expression in human osteoblasts. Biochem. Biophys. Res. Commun. 322:162–167; 2004. [DOI] [PubMed] [Google Scholar]
- 17. Kaetzel D. M. Transcription of the platelet-derived growth factor A-chain gene. Cytokine Growth Factor Rev. 14:427–446; 2003. [DOI] [PubMed] [Google Scholar]
- 18. Kato S.; Sasaki H.; Suzawa M.; Masushige S.; Tora L.; Chambon P.; Gronemeyer H. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol. Cell. Biol. 15:5858–5867; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee C. H.; Wei L. N. Characterization of an inverted repeat with a zero spacer (IR0)-type retinoic acid response element from the mouse nuclear orphan receptor TR2-11 gene. Biochemistry 38:8820–8825; 1999. [DOI] [PubMed] [Google Scholar]
- 20. Liebeskind A.; Srinivasan S.; Kaetzel D. M.; Bruce M. Retinoic acid stimulates immature lung fibroblast growth via a PDGF-mediated autocrine mechanism. Am. J. Physiol. Lung Cell. Mol. Physiol. 279:L81–L90; 2000. [DOI] [PubMed] [Google Scholar]
- 21. Lindahl P.; Karlsson L.; Hellstrom M.; Gebre-Medhin S.; Willetts K.; Heath J. K.; Betsholtz C. Alveogenesis failure in PDGF-A-deficient mice is coupled to a lack of distal spreading of alveolar smooth muscle cell progenitors during lung development. Development 124:3943–3953; 1997. [DOI] [PubMed] [Google Scholar]
- 22. Maden M.; Hind M. Retinoic acid in alveolar development, maintenance and regeneration. Philos. Trans. R Soc. Lond. B Biol. Sci. 359:799–808; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mangelsdorf D. J.; Evans R. M. The RXR heterodimers and orphan receptors. Cell 83:841–850; 1995. [DOI] [PubMed] [Google Scholar]
- 24. Mangelsdorf D. J.; Umesono K.; Evans R. M. In: Sporn M. B.; Roberts A. B.; Goodman D. S., eds. The retinoids. New York: Raven Press; 1994:319–350. [Google Scholar]
- 25. Marshall H.; Morrison A.; Studer M.; Popperl H.; Krumlauf R. Retinoids and Hox genes. FASEB J. 10:969–978; 1996. [PubMed] [Google Scholar]
- 26. Massaro G. D.; Massaro D. Postnatal treatment with retinoic acid increases the number of pulmonary alveoli in rats. Am. J. Physiol. 270:L305–L310; 1996. [DOI] [PubMed] [Google Scholar]
- 27. Maul R. S.; Zhang H. X.; Reid J. D.; Pedigo N. G.; Kaetzel D. M. Identification of a cell type-specific enhancer in the distal 5′-region of the platelet-derived growth factor A-chain gene. J. Biol. Chem. 273:33239–33246; 1998. [DOI] [PubMed] [Google Scholar]
- 28. Nagy L.; Saydak M.; Shipley N.; Lu S.; Basilion J. P.; Yan Z. H.; Syka P.; Chandraratna R. A.; Stein J. P.; Heyman R. A.; Davies P. J. Identification and characterization of a versatile retinoid response element (retinoic acid receptor response element-retinoid X receptor response element) in the mouse tissue trans-glutaminase gene promoter. J. Biol. Chem. 271:4355–4365; 1996. [DOI] [PubMed] [Google Scholar]
- 29. Panariello L.; Quadro L.; Trematerra S.; Colantuoni V. Identification of a novel retinoic acid response element in the promoter region of the retinol-binding protein gene. J. Biol. Chem. 271:25524–25532; 1996. [DOI] [PubMed] [Google Scholar]
- 30. Panne D.; Maniatis T.; Harrison S. C. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J. 23:4384–4393; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pedigo N.; Zhang H.; Koszewski N. J.; Kaetzel D. M. A 5′-distal element mediates vitamin D-inducibility of PDGF-A gene transcription. Growth Factors 21:151–160; 2003. [DOI] [PubMed] [Google Scholar]
- 32. Pedigo N. G.; Zhang H.; Bruno E. C. M.; Kaetzel C. S.; Dugan A.; Shanehsaz P.; Hennigan R. F.; Xing Z.; Koszewski N. J.; Kaetzel D. M. A 5′-distal enhanceosome in the PDGF-A gene is activated in choriocarcinoma cells via ligand-independent binding of vitamin D receptor and constitutive jun kinase signaling. Oncogene 24:2654–2666; 2005. [DOI] [PubMed] [Google Scholar]
- 33. Qin P.; Haberbusch J. M.; Soprano K. J.; Soprano D. R. Retinoic acid regulates the expression of PBX1, PBX2, and PBX3 in P19 cells both transcriptionally and post-translationally. J. Cell Biochem. 92:147–163; 2004. [DOI] [PubMed] [Google Scholar]
- 34. Ross S. A.; McCaffery P. J.; Drager U. C.; De Luca L. M. Retinoids in embryonal development. Physiol. Rev. 80:1021–1054; 2000. [DOI] [PubMed] [Google Scholar]
- 35. Snyder J. M.; Jenkins-Moore M.; Jackson S. K.; Goss K. L.; Dai H. H.; Bangsund P. J.; Giguere V.; McGowan S. E. Alveolarization in retinoic acid receptor-beta-deficient mice. Pediatr. Res. 57:384–391; 2005. [DOI] [PubMed] [Google Scholar]
- 36. Sugawara A.; Yen P. M.; Chin W. W. 9-cis retinoic acid regulation of rat growth hormone gene expression: Potential roles of multiple nuclear hormone receptors. Endocrinology 135:1956–1962; 1994. [DOI] [PubMed] [Google Scholar]
- 37. Takimoto Y.; Wang Z.; Kobler K.; Deuel T. F. Promoter region of the human platelet-derived growth factor A-chain gene. Proc. Natl. Acad. Sci. USA 88:1686–1690; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsukamoto T.; Matsui T.; Takaishi T.; Ito M.; Fukase M.; Chihara K. Retinoic acid differentially affects platelet-derived growth factor and epidermal growth factor-regulated cell growth of mouse osteoblast-like cells. Cell Growth Differ. 5:207–212; 1994. [PubMed] [Google Scholar]
- 39. Wang C.; Kelly J.; Bowen-Pope D. F.; Stiles C. D. Retinoic acid promotes transcription of the platelet-derived growth factor alpha-receptor gene. Mol. Cell. Biol. 10:6781–6784; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


