Abstract
Perch are promising species for freshwater aquaculture and, differently from other fish, have not yet been domesticated through artificial selection; therefore, they show a wide genetic variability that is undesirable for aquaculture. In addition to the more traditional methods of aquatic biotechnology, the most recently developed molecular biological techniques can augment the overall efficiency of aquaculture. To help these new molecular techniques find their place in the everyday management of fish farming, we should make an effort to reduce the gap in genomic resources that separates farming species from “model organisms.” We performed single-pass sequencing on 1237 randomly selected clones from a perch liver cDNA expression library, 350 clones of a brain-minus-liver, and 639 clones of a liver-minus-brain subtraction library. The sequences were deposited in the NCBI Expressed Sequence Tags database (www.ncbi.nlm.nih.gov/projects/dbEST). In the three libraries we identified 108, 46, and 104 genes, respectively. EST cataloguing and profiling of perch will provide a basis for functional genomic research in this species, but will also promote studies in comparative and environmental genomics, for identifying polymorphic markers that are useful, for example, to survey the disease resistance of fish and for discovering of new molecular markers of exposure. Using these genomic resources, micro- and macroarrays can be produced that will give immediate and practical benefits in the field of aquaculture, allowing early diagnosis of the fish conditions and helping in the generation of new mechanistic data on the nature of fish responses to different farming conditions.
Key words: cDNA library, Hepcidin, Complement C3, Aquaculture, Marine biotechnology, Transcriptome analysis
INTRODUCTION
Given the projected population growth over the next two decades, it is estimated that at least an additional 40 million tons of aquatic food will be required by 2030 to maintain the current per capita consumption (11). Because fishery support is almost stagnant, freshwater aquaculture is essential and this even gains in importance where food security for the developing world is concerned (10). Among others, both Eurasian (Perca fluviatilis) and yellow (Perca flavescens) perch are promising species for freshwater aquaculture. In contrast to other fish, such as salmonids, perch have not yet been domesticated through artificial selection and, therefore, show a wide genetic variability, which is undesirable for aquaculture. Eurasian perch populations kept under aquaculture conditions, for example, have been shown to present survival and growth performances that greatly depend on their geographic origin (25).
In addition to the more traditional methods of aquatic biotechnology, the most recently developed molecular biological techniques can augment the overall efficiency of aquaculture. Functional genomics, for instance, can provide information on the actual genes responsible for growth, disease resistance, stress, appetite, flesh quality, health conditions, welfare, and other attributes (14,43–45). Moreover, breeding selection incorporating the use of molecular markers for specific traits could help to enhance the desired characteristics (27).
To help these new molecular techniques find their place in the everyday management of fish farming, we need to reduce, as far as genomic resources are concerned, the gap that separates farming species from “model organisms.” EST cataloguing and profiling of perch will promote functional genomic research in this species, comparative (37) and environmental (7) genomics, the identification of polymorphic markers (24), the production of microarrays (35) and macroarrays (31), and the discovery of new molecular markers of exposure (15). With regard to a search for such molecular biomarkers, we selected hepcidin and complement C3, two immunorelated genes that are highly expressed in our EST database, and evaluated their expression after LPS challenge.
MATERIALS AND METHODS
cDNA Library and Mass Excision Protocol
For this study, we constructed a cDNA library from the liver of P. fluviatilis using ZAP-cDNA Gigapack III Gold Cloning Kit (Stratagene, CA, USA) following the manufacturer’s instructions. Briefly, total RNA was extracted with TRIzol (Invitrogen) and mRNA was isolated through oligo(dT)-cellulose columns (Sigma). We then synthesized the cDNAs using oligo(dT) primers containing the XhoI restriction site and Stratascript reverse transcriptase (Stratagene). After the second-strand cDNA was synthesized, the uneven termini of the double-stranded cDNA were filled in with Pfu DNA polymerase, and EcoRI adapters were ligated to the blunt ends. Then the double-stranded cDNA was digested with XhoI restriction enzyme. After ligation of the cDNA with the UNI-ZAP vector, the cDNA library was packaged using Gigapack III Gold Packaging extract (Stratagene).
For the mass excision procedure, 107 pfu of phage particles was combined with 108 XL1Blue MRF′ and 109 pfu of ExAssist helper phage (Stratagene) in falcon tubes and incubated at 37°C for 15 min. Then 20 ml LB broth was added and incubated for 3 h at 37°C. The tube was heated to 70°C for 20 min and subsequently centrifuged at 1000 rcf. In the next step, 1 μl of phage supernatant containing the excise pBluescript was added to 200 μl freshly grown SOLR cells (Stratagene); the mixture was plated onto LB ampicillin agar plates to an appropriate density such that individual colonies could be selected.
Suppression Subtractive Hybridization (SSH)
For SSH, total RNA was isolated from 1 g of brain and liver of P. fluviatilis using TRIzol (Invitrogen), and mRNA was isolated using the Gene elute mRNA miniprep kit (Sigma) according to the manufacturer’s instructions.
The library was prepared by EVROGEN JCS (www.evrogen.com). Differentially expressed genes in liver and brain were analyzed with the Clontech PCR-Select complementary DNA subtraction kit (BD Bio-sciences Clontech) according to the manufacturer’s instructions. For each cDNA synthesis, 2 μg of mRNA was used for the reaction. After the second synthesis, the double-stranded cDNA from both tissues was digested with RsaI. Part of the digested cDNA was ligated with Adapter I and part with Adapter 2R, and the rest was used as a driver in preparation for the hybridization procedure. One library was made by hybridizing adapter-ligated cDNA from liver tissue as the tester in the presence of cDNA from brain tissue as the driver. This library consisted of clones that are specific in the liver tissue. The reverse library was made in the same way, except that adapter-ligated cDNA from the brain tissue served as the tester and was hybridized in the presence of cDNA from liver tissue as the driver. Primary and secondary PCR amplification of these reciprocal subtractions of cDNA from brain and liver produced two SSH libraries enriched in differentially expressed transcripts. Differentially expressed cDNA was cloned using a pGEMT-easy T/A cloning kit (Promega), transformed into XL1blue-competent cells, and plated onto LB with ampicillin, X-Gal, and IPTG.
PCR and DNA Sequencing
Random clones were picked and then resuspended in sterile water. The PCR reactions (in a total volume of 25 μl) were run using a 2-μM solution of primers (T3: 5′-AATTAACCCTCACTAAAGGG-3′, T m = 53.2°C; T7: 5′-TAATACGACTCACTATAGGG-3′, T m = 53.2°C; and SP6: 5′-CATTTAGGTGACACTA TAG-3′, T m = 50.2°C), 10 μl of sample, 0.75 U of Taq DNA polymerase (Biotools), 2.5 μl 10× appropriate buffer (Biotools), and 0.2 mM dNTPs mix at the following conditions: 94°C for 4 min and 30 cycles at 94°C for 30 s, annealing at 53°C for 30 s, and elongation at 72°C for 2 min. Only clones larger than 500 bp were sequenced. These cDNA clones were subjected to a sequencing reaction using T3 or SP6 primers and the MEGABace DYEnamic ET DYE terminator kit (GE Healthcare). The sequencing reactions (in a total volume of 10 μl) were run using 5 pmol of T3 or SP6 primers, 4 μl of sequencing reagent Premix, and 100 ng of DNA at the following condition: 25 cycles at 95°C for 20 s, 50°C for 15 s, and 60°C for 1 min. Randomly selected Perca fluviatilis cDNA clones were sequenced using the MEGA Bace 500 (GE Healthcare).
Sequence Analysis
All sequences were analyzed with Vector NTI 9.0 software. They were first subjected to 5′ trimming to eliminate the vector and, when necessary, to 3′ trimming to eliminate low-quality (rich in undetermined bases) portions. Trimmed sequences were compared with nucleotide and protein sequences by BlastN and BlastX (http://www.ncbi.nlm.nih.gov/BLAST/).
Animal Treatment and Gene Expression Evaluation
Perca fluviatilis were kept at the Tinella experimental station located on the north shore of the Lake of Varese, Italy. Fifteen fish weighing on average 10 g were divided into three groups of 5 each. The animals of the control group (group 1) were given a single IP injection of saline solution and were sacrificed after 48 h. Animals of groups 2 and 3 were given a single IP injection of E. coli lipopolysaccaride (LPS) (15 mg/kg body weight). Animals were sacrificed 24 h (group 2) or 48 h (groups 1 and 3) after the injection. Their livers were removed immediately and kept at −80°C until they were processed for RNA purification.
Semiquantitative and Quantitative (Real-Time) RT-PCR
RNA was extracted from 100 mg of tissue using RNAgents Total RNA Isolation System (Promega) and the first strand of cDNA was synthesized using MMLV reverse transcriptase (Invitrogen) and oligo-dT primers.
The sequences of hepcidin and complement C3 were obtained by assembling ESTs from our database (see Table 2 for the NCBI dbEST accession numbers), and a pair of primers for hepcidin (Hep_left: 5′-GAAGACATTCAGTGTTGCAG-3′, T m = 55.3°C; Hep_right: 5′-CAGATTCTGCAGCACAATC-3′, T m = 54.5°C) and for complement C3 (C3_left: 5′-CAAC CAGCTGCTGTGTCTG-3′, T m = 58.8°C; C3_right: 5′-CTTTATTCAGAGGCACCCAC-3′, T m = 57.3°C) was designed. Normalization was carried out with actin (ACT_left: 5′-GGAGAAGATCTGGCATCACA-3′, T m = 57.3°C; ACT_right: 5′-GCACAGCTTCTC CTTGATGT-3′, T m = 57.3°C).
TABLE 2.
LIST OF THE 10 MOST EXPRESSED GENES IN THE LIVER EXPRESSION LIBRARY WITH THEIR ACCESSION NUMBERS AND FREQUENCIES
For semiquantitative PCRs, the reaction was run in a final volume of 25 μl using 2 μM solution of primers, 1 μl of cDNA, 5 μl of 5× buffer (Promega), 0.2 mM dNTP mix, and 1.25 U of GoTaq Polymerase (Promega) at the following conditions: 94°C for 4 min and 24 cycles at 94°C for 30 s, 54°C for 30 s, and 72°C for 30 s. The PCR products were loaded onto 1% agarose gel and run in TAE 1× buffer at 100 mV for 30 min. Fluorescence of the gel bands was determined for both the target and the reference genes using Gel Doc 2000 (Bio-Rad) equipped with Quantity One software, and the mRNA quantity was expressed as the ratio between the pixels of the target and reference genes. For quantitative PCR assays, the fluorescent reporter SYBR green was used to monitor the reaction. The reaction was run in a final volume of 25 μl using 1 μl of cDNA, 1 jol of forward primer (5 μM), 1 μl reverse primer (5 μM) for target and housekeeping genes, 11.25 of 2.5× Real Master mix/20× SYBR solution (Eppendorf), and 10.75 μl of water. The PCR reactions were run on the ABI Prism 7000 Sequence Detection System thermocycler (Applied Biosystem) at the following conditions: 95°C for 2 min and then 40 cycles at 95°C for 20 s, 56°C for 15 s, and 68°C for 40 s. All the data from quantitative and semiquantitative PCR were analyzed by ANOVA.
Hepcidin Gene
Genomic DNA was amplified by PCR using the specific primers Hep_left and Hep_right and the same conditions already used for RT-PCR (see above). PCR products were analyzed by agarose gel electrophoresis and sequenced (see above).
RESULTS
Sequence Analysis
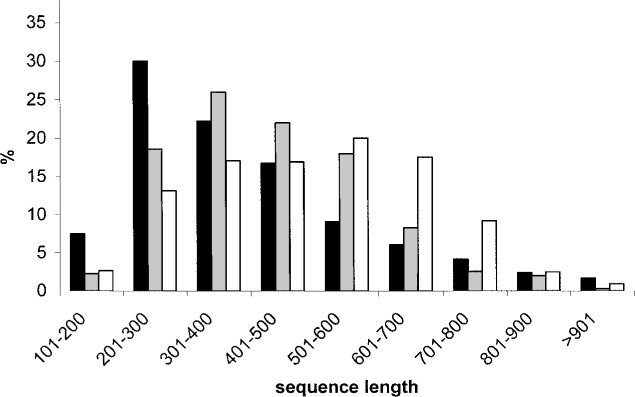
We performed single-pass sequencing on 1237 randomly selected clones from a perch liver cDNA expression library targeting the 5′-terminus of each insert. We also sequenced 350 clones of a brain-minus-liver and 639 clones of a liver-minus-brain subtraction library. For each sequence, we eliminated the vector portion and the 3′-terminus portions if they were rich in undetermined bases. The length distribution of the trimmed sequences for the three libraries is reported in Figure 1. The sequences were deposited in the NCBI Expressed Sequence Tags database (www.ncbi.nlm.nih.gov/projects/dbEST) and are available with the following accession numbers: from DR730572 to DR731129, from DV671023 to DV671392, and from DW985630 to DW985938 for the expression library; from DV752316 to DV752665 for the brain-minus-liver library; and from DT901717 to DT901885 and from DY614961 to DY615430 for the liver-minus-brain library.
Figure 1.
Length distribution after trimming the sequences originating from the liver expression library (black), liver-minus-brain subtractive library (gray), and brain-minus-liver subtractive library (white).
Comparative Analysis
A BlastN and BlastX search on all the sequences of the three libraries and 858 (69.36%), 267 (76.28%), and 293 (45.85%) sequences of the liver expression library, brain-minus-liver library, and liver-minus-brain library, respectively, did not produce any matches [i.e., did not show any significant similarity (their E-value was >e−10)] to the sequences deposited in public databases based on nucleotides or translated peptides. This relatively low percentage of matching is probably due to the chosen threshold, which is quite stringent, and to the possibility that a substantial fraction of the ESTs could not reach the coding sequence and represent untranslated regions (UTRs), which are notoriously less conserved.
In all, 43 (3.5%) of the 1237 sequences of the liver expression library, 13 (3.7%) of the 350 sequences of the brain-minus-liver library, and 40 (6.3%) of the 639 sequences of the liver-minus-brain library were similar to sequences already present in the public databases, but could not be identified. Several of these sequences showed a great similarity with sequences of Tetraodon nigroviridis, a freshwater pufferfish with the smallest known vertebrate genome. A draft sequence of T. nigroviridis genome is available, which has made it possible to carry out a comparative analysis of the human genome, in particular, and the vertebrate genome, in general, and which has provided insights into vertebrate genome evolution (19).
The remaining 336 (27.2%), 70 (20%), and 306 (47.9%) sequences of the liver expression library, brain-minus-liver library, and liver-minus-brain library, respectively, showed sufficient similarity (E-value <e−10) with known genes. Based on our subjective criteria for gene identification, we grouped the 336 sequences of the liver expression library into 43 contigs and 65 singletons, the 70 sequences of the brain-minus-liver library into 15 contigs and 31 singletons, and the 306 sequences of the liver-minus-brain library into 49 contigs and 55 singletons. Therefore, we identified 108, 46, and 104 genes, respectively, expressed in the three libraries of P. fluviatilis and classified them according to the hierarchy of the Human Protein Reference Database available at http://www.hprd.org (30) as shown in Table 1. To a certain degree the classification is arbitrary as several genes may have multiple functions and not all the genes were found in the database.
TABLE 1.
SUMMARY OF ESTs
| Biological Process | Liver Expression Library | Brain-Minus-Liver Library | Liver-Minus-Brain Library | |||
|---|---|---|---|---|---|---|
| Sequences | Genes | Sequences | Genes | Sequences | Genes | |
| Apoptosis | 0 | 0 | 3 | 2 | 2 | 2 |
| Cell growth and/or maintenance | 1 | 1 | 9 | 4 | 2 | 2 |
| Immune response | 75 | 11 | 0 | 0 | 97 | 16 |
| Metabolism, energy pathways | 31 | 18 | 18 | 10 | 61 | 32 |
| Process unknown | 27 | 4 | 1 | 1 | 1 | 1 |
| Protein metabolism | 61 | 41 | 3 | 3 | 56 | 19 |
| Regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolism | 11 | 10 | 4 | 4 | 10 | 3 |
| Signal transduction; cell communication | 4 | 4 | 14 | 10 | 9 | 5 |
| Transport | 99 | 10 | 14 | 9 | 33 | 15 |
| Unclassified | 27 | 9 | 4 | 3 | 35 | 9 |
| Unknown | 901 | 280 | 333 | |||
| Total | 1,237 | 108 | 350 | 46 | 639 | 104 |
In Table 2, we report the 10 most frequently expressed genes of the liver expression library, while in Table 3 we have listed the genes for ribosomal proteins. In Table 4 and Table 5 we list the most frequently expressed genes in the subtraction libraries.
TABLE 3.
LIST OF RIBOSOMAL PROTEINS FROM THE LIVER EXPRESSION LIBRARY WITH THEIR ACCESSION NUMBERS
| Type | Accession Number(s) |
|---|---|
| L12 | DV671294, DV671388 |
| L13a | DR731115, DV671068 |
| L15 | DR730872, DV671178 |
| L18 | DW985718 |
| L18a | DV671215 |
| L21 (40S) | DV671043, DV671081, DV671316 |
| L22 | DV671091, DV671169 |
| L27a | DV671111 |
| L28 | DV671302 |
| L3 | DR730915 |
| L30 | DR731085 |
| L31 | DR730609, DW985842 |
| L36 | DW985666 |
| L36A | DW985913 |
| L37 | DR731075 |
| L37a | DW985800 |
| L4 | DR730707, DV671315 |
| L41p | DR731004 |
| S7 (40S) | DR731064 |
| L5 (60 S) | DR730927 |
| L6 | DR730614 |
| L7 | DR730738 |
| Large P2 | DR730756, DW985921 |
| S12 | DR730797 |
| S13 | DV671370 |
| S14 | DW985691 |
| S15 | DV671236 |
| S16 | DR730632, DR730810 |
| S17 | DV671073 |
| S18 | DR730633, DR731108 |
| S19 | DR730668, DR730735, DR731101 |
| S20 | DW985924 |
| S21 (40S) | DR730729, DR730819, DR730831, DW985827 |
| S25 | DW985892 |
| S27a (40S) | DV671325, DV671180, DR730884 |
| S4 (40S) | DV671135 |
| S5 (40S) | DW985906, DW985835 |
TABLE 4.
LIST OF THE MOST FREQUENTLY EXPRESSED GENES IN THE BRAIN-MINUS-LIVER LIBRARY WITH THEIR ACCESSION NUMBERS
TABLE 5.
LIST OF THE MOST EXPRESSED GENES IN THE LIVER-MINUS-BRAIN LIBRARY WITH THEIR ACCESSION NUMBERS AND FREQUENCIES
Hepcidin Transcript and Gene
We found the sequence for Perca fluviatilis hepcidin in 22 clones of our liver expression library (Table 2). Clone DV671273 contained the entire 261-bp open-reading frame (ORF), a 69-bp 5′-untranslated region (UTR), and an 165-bp 3′-UTR (Fig. 2). The ORF encoded an 87-amino acid prepro-hepcidin. By comparing our prepro-hepcidin sequence with that of other fish species that were found to be highly conserved, we determined a potential cleavage site for the signal peptide between amino acids 25 and 26 in the precursor and a consensus nuclear localization sequence (NLS) at amino acid 62 (RHKR).
Figure 2.
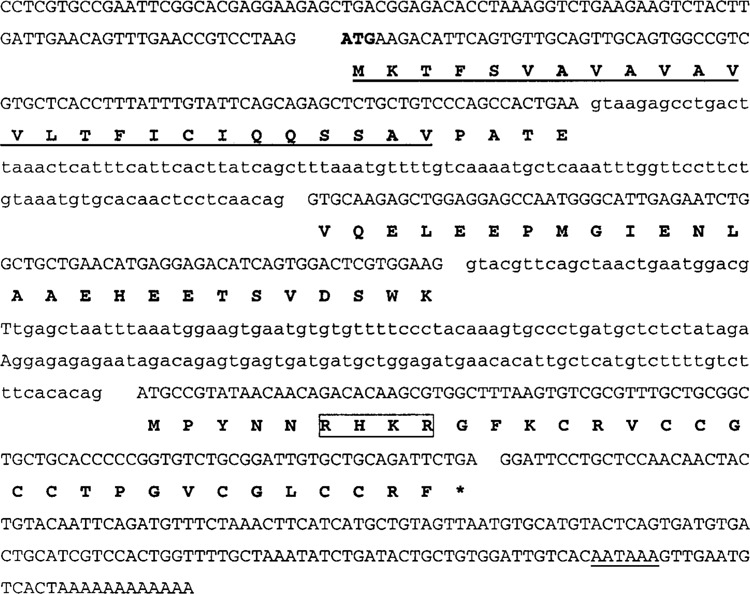
Perca fluviatilis hepcidin gene. The complete nucleotide sequence, including the two introns (small letters), is reported. The putative amino acid sequence of preprohepcidin (bold capital letters) is indicated within the coding sequence, signal peptide is underlined, and the NLS is boxed.
The hepcidin gene was sequenced from Perca fluviatilis genomic DNA. PCR was run using Hep_right and Hep_left primers designed on the start and the stop codons of the perch sequence. The segment of 624 bp was sequenced and deposited in GenBank with the accession number EF602303. Complete sequencing of this fragment and comparing it with the sequence of the transcript revealed the presence of two introns with standard GT/AG splice junctions (Fig. 2).
LPS Challenge
Both semiquantitative and quantitative RT-PCRs for the evaluation of hepcidin expression were run using Hep_right and Hep_left primers and β-actin as a reference gene. The results, reported in the box and whisker plots of Figure 3, indicate that Perca fluviatilis liver hepcidin is upregulated by LPS treatment. After 48 h, a recovery is evident. ANOVA analysis of the data, however, gave statistically significant results only for the semiquantitative experiments because of the large variance in the response to LPS challenge, which was probably due to the large genetic variability present in the wild perch population and to the relatively low size of the experimental groups.
Figure 3.
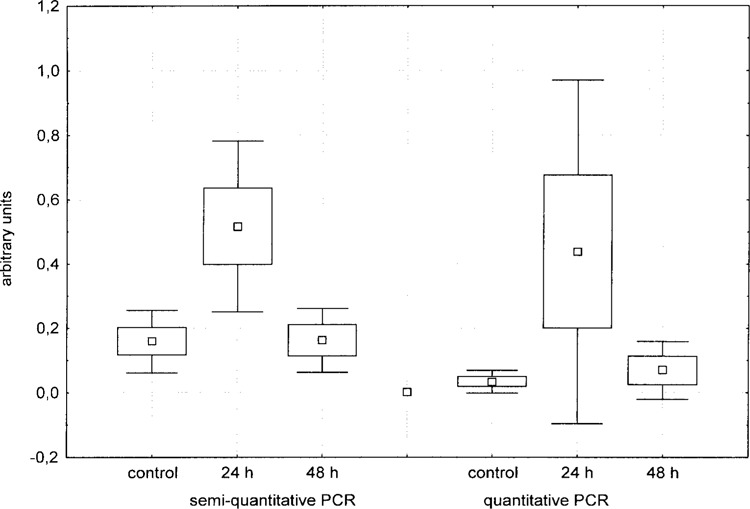
Box and whisker diagrams describing means, SEs, and SDs of hepcidin expression in the liver of control and LPS-treated animals. Hepcidin expression was evaluated by semiquantitative and real-time RT-PCRs and both techniques produced a similar trend.
In contrast, we did not notice any considerable change in complement C3 expression after LPS challenge, except for a slight decrease that did not appear to be statistically significant (data not shown).
DISCUSSION
The beneficial effects that modern biotechnology can bring to the aquaculture industry might be reduced by the scarcity of genomic resources for fish species of commercial interest. Because the availability of nucleotide sequences is a fundamental tool for molecular research, we initiated an EST project on perch by sequencing 2226 randomly selected clones from three different cDNA libraries. To obtain an expression profile of the P. fluviatilis liver, we used an expression (nonnormalized) library, while to obtain brain- and liver-specific genes, we used subtractive libraries. In a subtractive library, in fact, the transcripts present in both samples are reduced, which makes is possible to discover tissue-specific genes (9).
The most redundant clones in the liver expression library (Table 2) were those coding for apolipoprotein A. Apolipoproteins are structural components of plasma lipoproteins (i.e., chylomicrons), very low-density lipoproteins, low-density lipoproteins, and high-density lipoproteins (HDL). HDL, which are particularly abundant in fish, are constituted by apolipoprotein A (I and II). Apolipoprotein A, known to be important in reverse cholesterol transport, has recently also been shown to be involved in innate immunity (6). The second most redundant clones were those coding for transferrin, a glycoprotein that is responsible for the transport and delivery of iron to cells. Transferrin also contributes to the innate immune response of fish (40), and, in particular, it is required for the induction of macrophage antimicrobial response (41). Moreover, transferrin, together with other genes whose expression is abundant in the liver (i.e., apolipoprotein A, fibrinogen beta, fibrinogen gamma, and complement component C3), has been shown to be regulated by estrogens (32); these genes are therefore good candidates for evaluating endocrine disruption in natural fish populations.
A total of 22 sequences correspond to hepcidin, a cationic peptide with a hairpin structure whose two arms are linked by disulfide bridges in a ladder-like configuration. Hepcidin is downregulated by hypoxia and anemia whereas it is induced by iron overload, infection, and inflammation. Hepcidin probably evolved from an antimicrobial peptide to an iron regulatory hormone during vertebrate evolution (38). Interestingly, we only found one sequence (NCBI accession number: DV216833) for hepcidin in the expression library of sea bass liver that we obtained from animals reared in a controlled environment (5).
Hepcidin expression has been shown to be upregulated by infection in several fish species, such as striped bass (22), red sea bream (4), catfish (1,18), Atlantic halibut (29), sea bass (36), zebrafish (23), and rainbow trout (13). We have shown that also the expression of Perca fluviatilis liver hepcidin responds to inflammatory stimuli. Cationic antimicrobial peptides have been shown to have a broad range of immunomodulatory properties and it has been suggested that the selective upregulation of innate immunity should provide a new means of treating infective aggressions that do not induce resistance, as these peptides act more through the innate immune system than directly on bacteria (16). Intensive fish culture is often plagued by microbial infection outbreaks and, because of the acute nature of these infections, fish rely heavily on the innate components of immunity. In this context, hepcidin might offer a new therapeutic approach for the control of disease in aquaculture (2).
The organization of the hepcidin gene with three exons and two introns is well conserved in vertebrates, and Perca fluviatilis is not an exception. These introns, similarly to those of white bass, Atlantic salmon, and Japan sea bass, are shorter than those of other organisms, such as zebrafish, mouse, rat, and human. The deduced amino acid sequence of perch was quite similar to hepcidins of other vertebrates, and particularly similar to turbot (accession number: AJ890336) and Japan sea bass (accession number: AY604195) hepcidins (34).
The complement system of fish is well developed and plays an important role in the innate immune response; like in mammals, complement C3 is the central protein of all three activation pathways. The study of the proteins of the complement in teleosts is important not only to gain a better understanding of the evolution of the innate immune system in vertebrates, but also for disease control in aquaculture (17). After LPS challenge, however, we did not notice any considerable alteration in the expression of complement C3 mRNA, except for a slight decrease that did not appear to be statistically significant. This possible downregulation of complement C3, however, is not surprising as it has already been observed in rainbow trout after inflammatory stimulus (13).
Several sequences correspond to the complement regulatory plasma protein cloned for the first time in teleosts, in the barred sand bass (8). Regulatory proteins of complement activation are of crucial importance to protect the self cells from autologous attack (17). Among the 10 most represented genes, we also found serum amyloid A protein, one of the major proteins produced by the liver in response to inflammation. For this highly conserved protein, several are the suggested physiological roles comprised its cooperation with C-reactive protein to facilitate the rapid endogenous recycling of cell membrane cholesterol and phospholipids during the acute phase response (26). Ten sequences correspond to another immune-relevant gene, chemotaxin, a chemotactic factor released from inflammatory sites to activate neutrophils; this has recently been shown to possess further functional roles. In fish, it has been cloned in carp (12) and in trout (21). The saxitoxin-binding protein (47) is involved in the metabolism of saxitoxin, a very toxic product of dinoflagellates and cyanobacteria, which can accumulate in the tissues of fish products through the food chain and represents a threat for consumer health (33). The tributyltin-binding protein is a novel protein of 191 amino acids found in the serum of the Japanese flounder, Paralichthys olivaceus, which binds tributyltin, a chemical used as an antifoulant in marine environments and accumulates in the serum or plasma of some fishes (39).
Several sequences correspond to genes involved in protein metabolism (Table 1) and in particular in the translation machinery; we have, in fact, sequenced ESTs of several ribosomal proteins (Table 3). As for the expression library of sea bass liver (5), we did not observe the large differences in relative abundance that were reported for catfish brain (20). In fact, as each ribosome contains about 50 distinct proteins that must be available at the same rate, similar expression levels in their mRNAs are expected. In the two subtraction libraries, in contrast, we found virtually no sequences for ribosomal proteins; indeed, a subtraction library is constructed so that the transcripts present in both samples are reduced.
Among other genes, we obtained ependymin and reticulon in the brain-minus-liver subtractive library (Table 4). Ependymin is the predominant protein in the cerebrospinal fluid of many teleosts. Ependymin-related proteins have been identified in mammals, amphibians, and echinoderms and seem to be involved in regenerative processes (42). For neuroregeneration reticulons, a recently isolated new family of proteins that are highly expressed in brain tissues also seem to be important (3). These proteins are primarily localized in the membrane of the tubular, peripheral endoplasmic reticulum and have a role in shaping this organelle (46).
The most frequently found sequences in the liver-minus-brain subtraction library correspond to genes coding for proteins of the complement system, fibrinogen and plasminogen (Table 5).
EST cataloguing and profiling of perch will provide a basis for functional genomic research in this species, but will also promote studies of comparative (19) and environmental (7,7) genomics, for identifying polymorphic markers useful, for example, to survey the disease resistance of fish (28) and for discovering new molecular markers of exposure. Using these genomic resources micro- and macroarrays can be produced that will give immediate and practical benefits in the field of aquaculture, allowing early diagnosis of the fish conditions and helping to generate new mechanistic data on the nature of fish responses to different farming conditions.
ACKNOWLEDGMENT
This research was supported by grant “Fondo d’Ateneo per la Ricerca” to G.B and R.G.
REFERENCES
- 1. Bao B. L.; Peatman E.; Li P.; He C. B.; Liu Z. J. Catfish hepcidin gene is expressed in a wide range of tissues and exhibits tissue-specific upregulation after bacterial infection. Dev. Comp. Immunol. 29:939–950; 2005. [DOI] [PubMed] [Google Scholar]
- 2. Bao B. L.; Peatman E.; Xu P.; Li P.; Zeng H.; He C. B.; Liu Z. J. The catfish liver-expressed antimicrobial peptide 2 (LEAP-2) gene is expressed in a wide range of tissues and developmentally regulated. Mol. Immunol. 43:367–377; 2006. [DOI] [PubMed] [Google Scholar]
- 3. Cai Y. P.; Saiyin H. X.; Lin Q.; Zhang P. Z.; Tang L. S.; Pan X. H.; Yu L. Identification of a new RTN3 transcript, RTN3-A1, and its distribution in adult mouse brain. Mol. Brain Res. 138:236–243; 2005. [DOI] [PubMed] [Google Scholar]
- 4. Chen S. L.; Xu M. Y.; Ji X. S.; Yu G. C.; Liu Y. Cloning, characterization, and expression analysis of hepcidin gene from red sea bream (Chrysophrys major). Antimicrob. Agents Chemother. 49:1608–1612; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chini V.; Rimoldi S.; Terova G.; Saroglia M.; Rossi F.; Bernardini G.; Gornati R. EST-based identification of genes expressed in the liver of adult sea bass (Dicentrarchus labrax, L.). Gene 376:102–106; 2006. [DOI] [PubMed] [Google Scholar]
- 6. Concha M. I.; Smith V. J.; Castro K.; Bastias A.; Romero A.; Amthauer R. J. Apolipoproteins A-I and A-II are potentially important effectors of innate immunity in the teleost fish Cyprinus carpio . Eur. J. Biochem. 271:2984–2990; 2004. [DOI] [PubMed] [Google Scholar]
- 7. Cossins A. R.; Crawford D. L. Opinion—Fish as models for environmental genomics. Nat. Rev. Genet. 6:324–333; 2005. [DOI] [PubMed] [Google Scholar]
- 8. Dahmen A.; Kaidoh T.; Zipfel P. F.; Gigli I. Cloning and characterization of a cDNA representing a putative complement-regulatory plasma protein from barred sand bass (Parablax neblifer). Biochem. J. 301:391–397; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diatchenko L.; Lau Y.; Campbell A.; Chenchik A.; Moqadam F.; Huang B.; Lukyanov S.; Lukyanov K.; Gurskaya N.; Sverdlov E.; Siebert P. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025–6030; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dugan P.; Dey M. M.; Sugunan V. V. Fisheries and water productivity in tropical river basins: Enhancing food security and livelihoods by managing water for fish. Agric. Water Manage. 80:262–275; 2006. [Google Scholar]
- 11. FAO: The state of world fisheries and aquaculture (SOFIA). Rome: Food and Agriculture Organization; 2004. [Google Scholar]
- 12. Fujiki K.; Shin D. H.; Nakao M.; Yano T. Molecular cloning of carp (Cyprinus carpio) leucocyte cell-derived chemotaxin 2, glia maturation factor beta, CD45 and lysozyme C by use of suppression subtractive hybridisation. Fish Shellfish Immunol. 10:643–650; 2000. [DOI] [PubMed] [Google Scholar]
- 13. Gerwick L.; Corley-Smith G.; Bayne C. J. Gene transcript changes in individual rainbow trout livers following an inflammatory stimulus. Fish Shellfish Immunol. 22:157–171; 2007. [DOI] [PubMed] [Google Scholar]
- 14. Gornati R.; Gualdoni S.; Cavaliere R.; Terova G.; Saroglia M.; Bernardini G. Molecular biology and fish welfare: A winning combination. Aquaculture Int. 13:51–55; 2005. [Google Scholar]
- 15. Gornati R.; Papis E.; Rimoldi S.; Chini V.; Terova G.; Prati M.; Saroglia M.; Bernardini G. Molecular markers for animal biotechnology: Sea bass (Dicentrarchus labrax, L.) HMG-CoA reductase mRNA. Gene 344:299–305; 2005. [DOI] [PubMed] [Google Scholar]
- 16. Hancock R. E.; Sahl H. G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557; 2006. [DOI] [PubMed] [Google Scholar]
- 17. Holland M. C. H.; Lambris J. D. The complement system in teleosts. Fish Shellfish Immunol. 12:399–420; 2002. [DOI] [PubMed] [Google Scholar]
- 18. Hu X. Y.; Camus A. C.; Aono S.; Morrison E. E.; Dennis J.; Nusbaum K. E.; Judd R. L.; Shi J. S. Channel catfish hepcidin expression in infection and anemia. Comp. Immunol. Microbiol. Infect. Dis. 30: 55–69; 2007. [DOI] [PubMed] [Google Scholar]
- 19. Jaillon O.; Aury J. M.; Brunet F.; Petit J. L.; Stange-Thomann N.; Mauceli E.; Bouneau L.; Fischer C.; Ozouf-Costaz C.; Bernot A.; Nicaud S.; Jaffe D.; Fisher S.; Lutfalla G.; Dossat C.; Segurens B.; Dasilva C.; Salanoubat M.; Levy M.; Boudet N.; Castellano S.; Anthouard R.; Jubin C.; Castelli V.; Katinka M.; Vacherie B.; Biemont C.; Skalli Z.; Cattolico L.; Poulain J.; de Berardinis V.; Cruaud C.; Duprat S.; Brottier P.; Coutanceau J. P.; Gouzy J.; Parra G.; Lardier G.; Chapple C.; McKernan K. J.; McEwan P.; Bosak S.; Kellis M.; Volff J. N.; Guigo R.; Zody M. C.; Mesirov J.; Lindblad-Toh K.; Birren B.; Nusbaum C.; Kahn D.; Robinson-Rechavi M.; Laudet V.; Schachter V.; Quetier F.; Saurin W.; Scarpelli C.; Wincker P.; Lander E. S.; Weissenbach J.; Crollius H. R. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431:946–957; 2004. [DOI] [PubMed] [Google Scholar]
- 20. Ju Z. L.; Karsi A.; Kocabas A.; Patterson A.; Li P.; Cao D. F.; Dunham R.; Liu Z. J. Transcriptome analysis of channel catfish (Ictalurus punctatus): Genes and expression profile from the brain. Gene 261:373–382; 2000. [DOI] [PubMed] [Google Scholar]
- 21. Kokkinos P. A.; Kazantzi A.; Sfyroera G.; Zarkadis I. K. Molecular cloning of leukocyte cell-derived chemotaxin 2 in rainbow trout. Fish Shellfish Immunol. 18:371–380; 2005. [DOI] [PubMed] [Google Scholar]
- 22. Lauth X.; Babon J. J.; Stannard J. A.; Singh S.; Nizet V.; Carlberg J. M.; Ostland V. E.; Pennington M. W.; Norton R. S.; Westerman M. E. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J. Biol. Chem. 280:9272–9282; 2005. [DOI] [PubMed] [Google Scholar]
- 23. Lin B.; Chen S. W.; Cao Z.; Lin Y.; Mo D.; Zhang H.; Gu J.; Dong M.; Liu Z.; Xu A. Acute phase response in zebrafish upon Aeromonas salmonicida and Staphylococcus aureus infection: Striking similarities and obvious differences with mammals. Mol. Immunol. 44:295–301; 2007. [DOI] [PubMed] [Google Scholar]
- 24. Liu Z. J.; Cordes J. F. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238:1–37; 2004. [Google Scholar]
- 25. Mandiki S. N. M.; Blanchard G.; Melard C.; Koskela J.; Kucharczyk D.; Fontaine P.; Kestemont P. Effects of geographic origin on growth and food intake in Eurasian perch (Perca fluviatilis L.) juveniles under intensive culture conditions. Aquaculture 229: 117–128; 2004. [Google Scholar]
- 26. Manley P. N.; Ancsin J. B.: Kisilevsky R. Rapid recycling of cholesterol: The joint biologic role of C-reactive protein and serum amyloid A. Med. Hypoth. 66:784–792; 2006. [DOI] [PubMed] [Google Scholar]
- 27. Melamed P.; Gong Z. Y.; Fletcher G.; Hew C. L. The potential impact of modern biotechnology on fish aquaculture. Aquaculture 204:255–269; 2002. [Google Scholar]
- 28. Nakao M.; Kondo M.; Yano T. Protein polymorphism of an isoform of the third complement component in the common carp Cyprinus carpio . Fisheries Sci. 70:366–371; 2004. [Google Scholar]
- 29. Park K. C.; Osborne J. A.; Tsoi S. C. M.; Brown L. L.; Johnson S. C. Expressed sequence tags analysis of Atlantic halibut (Hippoglossus hippoglossus) liver, kidney and spleen tissues following vaccination against Vibrio anguillarum and Aeromonas salmonicida . Fish Shellfish Immunol. 18:393–415; 2005. [DOI] [PubMed] [Google Scholar]
- 30. Peri S.; Navarro J. D.; Amanchy R.; Kristiansen T. Z.; Jonnalagadda C. K.; Surendranath V.; Niranjan V.; Muthusamy B.; Gandhi T. K.; Gronborg M.; Ibarrola N.; Deshpande N.; Shanker K.; Shivashankar H. N.; Rashmi B. P.; Ramya M. A.; Zhao Z.; Chandrika K. N.; Padma N.; Harsha H. C.; Yatish A. J.; Kavitha M. P.; Menezes M.; Choudhury D. R.; Suresh S.; Ghosh N.; Saravana R.; Chandran S.; Krishna S.; Joy M.; Anand S. K.; Madavan V.; Joseph A.; Wong G. W.; Schiemann W. P.; Constantinescu S. N.; Huang L.; Khosravi-Far R.; Steen H.; Tewari M.; Ghaffari S.; Blobe G. C.; Dang C. V.; Garcia J. G.; Pevsner J.; Jensen O. N.; Roepstorff P.; Deshpande K. S.; Chinnaiyan A. M.; Hamosh A.; Chakravarti A.; Pandey A. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 13:2363–2371; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peterson J. S. K.; Bain L. J. Differential gene expression in anthracene-exposed mummichogs (Fundulus heteroclitus). Aquatic Toxicol. 66:345–355; 2004. [DOI] [PubMed] [Google Scholar]
- 32. Pinto P. I. S.; Teodosio H. R.; Galay-Burgos M.; Power D. M.; Sweeney G. E.; Canario A. V. M. Identification of estrogen-responsive genes in the testis of sea bream (Sparus auratus) using suppression subtractive hybridization. Mol. Reprod. Dev. 73:318–329; 2006. [DOI] [PubMed] [Google Scholar]
- 33. Poletti R.; Milandri A.; Pompei M. Algal biotoxins of marine origin: New indications from the European Union. Vet. Res. Commun. 27:173–182; 2003. [DOI] [PubMed] [Google Scholar]
- 34. Ren H. L.; Wang K. J.; Zhou H. L.; Yang M. Cloning and organisation analysis of a hepcidin-like gene and cDNA from Japan sea bass, Lateolabrax japonicus . Fish Shellfish Immunol. 21:221–227; 2006. [DOI] [PubMed] [Google Scholar]
- 35. Rise M. L.; von Schalburg K. R.; Brown G. D.; Mawer M. A.; Devlin R. H.; Kuipers N.; Busby M.; Beetz-Sargent M.; Alberto R.; Gibbs A. R.; Hunt P.; Shukin R.; Zeznik J. A.; Nelson C.; Jones S. R. M.; Smailus D. E.; Jones S. J. M.; Schein J. E.; Marra M. A.; Butterfield Y. S. N.; Stott J. M.; Ng S. H. S.; Davidson W. S.; Koop B. F. Development and application of a salmonid EST database and cDNA microarray: Data mining and interspecific hybridization characteristics. Genome Res. 14:478–490; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodrigues P. N. S.; Vazquez-Dorado S.; Neves J. V.; Wilson J. M. Dual function of fish hepcidin: Response to experimental iron overload and bacterial infection in sea bass (Dicentrarchus labrax). Dev. Comp. Immunol. 30:1156–1167; 2006. [DOI] [PubMed] [Google Scholar]
- 37. Roest Crollius H.; Weissenbach J. Fish genomics and biology. Genome Res. 15:1675–1682; 2005. [DOI] [PubMed] [Google Scholar]
- 38. Shi J.; Camus A. C. Hepcidins in amphibians and fishes: Antimicrobial peptides or iron-regulatory hormones? Dev. Comp. Immunol. 30:746–755; 2006. [DOI] [PubMed] [Google Scholar]
- 39. Shimasaki Y.; Oshima Y.; Yokota Y.; Kitano T.; Nakao M.; Kawabata S.; Imada N.; Honjo T. Purification and identification of a tributyltin-binding protein from serum of Japanese flounder, Paralichthys olivaceus . Environ. Toxicol. Chem. 21:1229–1235; 2002. [PubMed] [Google Scholar]
- 40. Stafford J. L.; Belosevic M. Transferrin and the innate immune response of fish: Identification of a novel mechanism of macrophage activation. Dev. Comp. Immunol. 27:539–554; 2003. [DOI] [PubMed] [Google Scholar]
- 41. Stafford J. L.; Wilson E. C.; Belosevic M. Recombinant transferrin induces nitric oxide response in goldfish and murine macrophages. Fish Shellfish Immunol. 17:171–185; 2004. [DOI] [PubMed] [Google Scholar]
- 42. Suarez-Castillo E. C.; Medina-Ortiz W. E.; Roig-Lopez J. L.; Garcia-Arraras J. E. Ependymin, a gene involved in regeneration and neuroplasticity in vertebrates, is overexpressed during regeneration in the echinoderm Holothuria glaberrima. Gene 334:133–143; 2004. [DOI] [PubMed] [Google Scholar]
- 43. Terova G.; Bernardini G.; Binelli G.; Gornati R.; Saroglia M. cDNA encoding sequences for myostatin and FGF6 in sea bass (Dicentrarchus labrax L.) and the effect of fasting and re-feeding on their abundance levels. Domestic Anim. Endocrinol. 30:304–319; 2006. [DOI] [PubMed] [Google Scholar]
- 44. Terova G.; Rimoldi S.; Chini V.; Gornati R.; Bernardini G.; Saroglia M. Cloning and expression analysis of insulin-like growth factor I and II in liver and muscle of sea bass (Dicentrarchus labrax, L.) during long-term fasting and re-feeding. J. Fish Biol. 70(Suppl. B):219–233; 2007. [Google Scholar]
- 45. Terova G.; Rimoldi S.; Bernardini G.; Gornati R.; Saroglia M. Sea bass ghrelin: Molecular cloning and involvement in the control of food intake. Gen. Comp. Endocrinol. in press. [DOI] [PubMed] [Google Scholar]
- 46. Voeltz G. K.; Prinz W. A.; Shibata Y.; Rist J. M.; Rapoport T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573–586; 2006. [DOI] [PubMed] [Google Scholar]
- 47. Yotsu-Yamashita M.; Sugimoto A.; Terakawa T.; Shoji Y.; Miyazawa T.; Yasumoto T. Purification, characterization, and cDNA cloning of a novel soluble saxitoxin and tetrodotoxin binding protein from plasma of the puffer fish, Fugu pardalis . Eur. J. Biochem. 268:5937–5946; 2001. [DOI] [PubMed] [Google Scholar]