Significance
The outer membrane of gram-negative bacteria contains LPS on the cell surface. The presence of LPS creates an effective permeability barrier that protects gram-negative bacteria from small hydrophobic molecules. Because the entire LPS biogenesis pathway, including biosynthesis and transport, is highly conserved, proteins involved are attractive targets for antibiotic discovery. Historically, it has been challenging to target LPS biogenesis since many of the components are membrane proteins with hard-to-assay activities. Utilizing the nonessentiality of this pathway in a gram-negative pathogen, we developed a cell-based screen specific to LPS biogenesis. We identified a small-molecule inhibitor targeting an essential component of the pathway, MsbA, and validated it as an antibacterial target using a combination of genetics, biochemistry, and cellular assays.
Keywords: LPS biogenesis, ABC transporter, high-throughput screening, MsbA inhibitor, Acinetobacter
Abstract
New drugs are needed to treat gram-negative bacterial infections. These bacteria are protected by an outer membrane which prevents many antibiotics from reaching their cellular targets. The outer leaflet of the outer membrane contains LPS, which is responsible for creating this permeability barrier. Interfering with LPS biogenesis affects bacterial viability. We developed a cell-based screen that identifies inhibitors of LPS biosynthesis and transport by exploiting the nonessentiality of this pathway in Acinetobacter. We used this screen to find an inhibitor of MsbA, an ATP-dependent flippase that translocates LPS across the inner membrane. Treatment with the inhibitor caused mislocalization of LPS to the cell interior. The discovery of an MsbA inhibitor, which is universally conserved in all gram-negative bacteria, validates MsbA as an antibacterial target. Because our cell-based screen reports on the function of the entire LPS biogenesis pathway, it could be used to identify compounds that inhibit other targets in the pathway, which can provide insights into vulnerabilities of the gram-negative cell envelope.
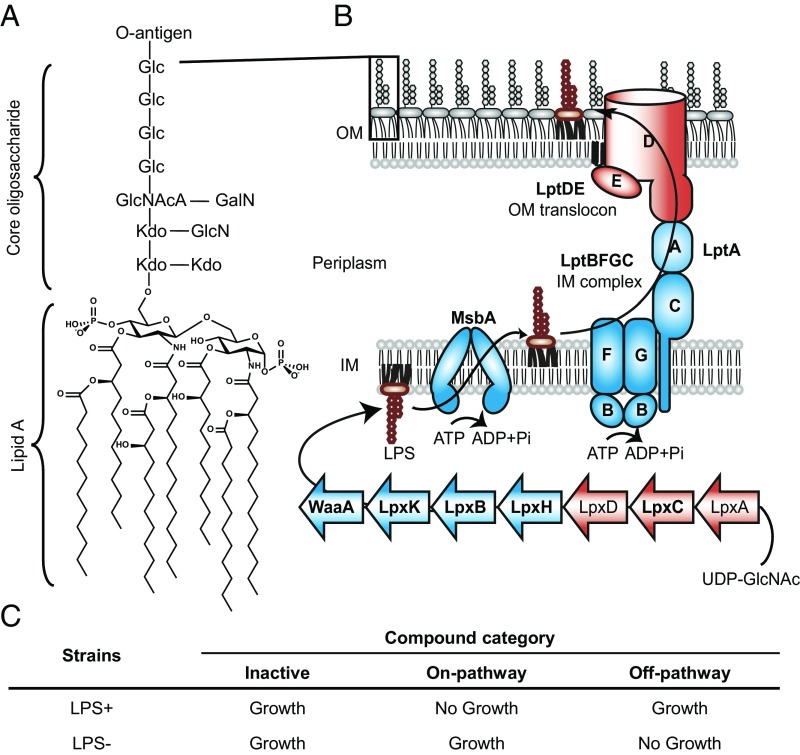
With the increase in antibiotic resistance, treatment of bacterial infections has become a major unmet clinical need today (1, 2). It is more difficult to kill gram-negative bacteria than gram-positive bacteria because they contain a second membrane, the outer membrane, which prevents antibiotics from reaching their targets inside the cell (3). This outer membrane is an asymmetric bilayer containing phospholipids in the inner leaflet and LPS in the outer leaflet (4, 5). LPS is a complex glycolipid containing a highly acylated diglucosamine (lipid A) that is connected to repeating sugars (O-antigen) through core oligosaccharides (Fig. 1A). Adjacent LPS molecules at the cell surface are stabilized by electrostatic interactions with divalent cations and strong lateral interactions between neighboring hydrophobic acyl chains. This tight packing of LPS at the cell surface makes the outer membrane an effective permeability barrier, which makes gram-negative bacteria insensitive to many antibiotics (6).
Fig. 1.
Certain genes in the LPS biogenesis pathway of Acinetobacter are conditionally essential. (A) LPS is a complex glycolipid and consists of hepta-acylated lipid A (in Acinetobacter), core oligosaccharides, and O-antigen. (B) LPS biogenesis starts with biosynthesis in the cytoplasm and transport of LPS from the inner membrane to the outer membrane, followed by assembly at the cell surface. Nonessential genes initiating lipid A biosynthesis (lpxA/C/D, red) and at the final step of the transport process (lptD/E, red), along with conditionally essential intermediate steps (lpxH/B/K, waaA, msbA, and lptA/B/C/F/G, blue) are indicated. Genes (bold) were experimentally verified in A. baylyi (SI Appendix, Fig. S1). The specific acylation pattern and core sugar composition of LPS are based on published reports (45, 46). O-antigen–related proteins are omitted for simplicity. Abbreviations: GalN, galactosamine; Glc, glucose; GlcN, glucosamine; GlcNAcA, 2-acetamido-2-deoxy-glucopyranosyluronic acid; Kdo, 3-deoxy-d-manno-oct-2-ulopyranosonic acid. (C) LPS biogenesis inhibitors are distinguished from off-pathway or inactive compounds based on distinct sensitivities of strains with and without LPS.
The biogenesis of LPS, which includes its biosynthesis and transport, involves hundreds of proteins that are spread across three compartments of gram-negative bacteria (Fig. 1B). The bulk of the LPS molecule is synthesized in the cytoplasm before being translocated across the inner membrane by an ATP-dependent transporter (MsbA) (7, 8). The biosynthesis of LPS is completed at the outer leaflet of the inner membrane following translocation, and subsequently LPS is extracted from the inner membrane and delivered to the cell surface (7, 9, 10). LPS transport to the outer membrane requires a seven-protein LPS transport machine (Lpt) that has been shown to form a transenvelope complex to accomplish periplasmic transit and translocation through the outer membrane (10–13). Strains with mutations that impair LPS biogenesis either are not viable or have a leaky outer membrane (14–16). Therefore, it has been assumed that targeting this pathway may kill gram-negative bacteria or overcome their intrinsic insensitivity to a broad range of antibiotics. Since the proteins involved in LPS biogenesis are highly conserved across gram-negative bacteria, the entire pathway represents an attractive target for novel antibiotic development.
Despite the considerable promise of and interest in developing inhibitors against the LPS pathway (17–19), success has largely been confined to a single biosynthetic target, LpxC (20). LpxC is a zinc-dependent deacetylase that catalyzes the first committed step of lipid A biosynthesis (7). LpxC is a soluble protein with easy-to-assay enzymatic activity, making it a readily tractable target for the design of new antibiotics. In addition to LpxC, there are many other proteins in the LPS biogenesis pathway that may be promising drug targets. However, many of these are membrane proteins operating in complexes that transport LPS without chemically modifying it. This makes it difficult to assay their functions, which is necessary to develop and implement assays robust enough to interrogate large chemical libraries for inhibition in a high-throughput format. Moreover, since the proteins involved in LPS transport are believed to function as a single transenvelope complex, assays using isolated protein components are limited to measuring binding affinity in the absence of function. To look for inhibitors in this pathway, we report here the development of a high-throughput, cell-based screen which allows us to look at all of the targets (biosynthesis and transport) in the LPS pathway in a simple growth-based assay.
Results
Validating Conditional Essentiality of Certain LPS Biogenesis Steps in Acinetobacter.
In many gram-negative organisms, the LPS pathway genes involved in biosynthesis and transport are essential (21). Recently it has been found that several gram-negative pathogens including Acinetobacter can survive without LPS (22–24). Paradoxically, certain steps of the LPS biogenesis pathway appear to remain essential in Acinetobacter despite the fact that LPS is not essential (25–27).
We studied the essentiality of the LPS-pathway genes in Acinetobacter baylyi, an organism that allows for highly efficient genetic manipulation (28) (Fig. 1). Genes encoding LpxC, which performs the first committed step in LPS biogenesis, as well as LptD/E, which carry out the final step of the pathway, could be readily removed without loss of viability. Unlike the lpxC knockout, which grew at the same rate as the WT, the lptD knockout grew more slowly and formed clumps under the microscope (SI Appendix, Fig. S1 A and B). However, these deficiencies disappeared when we knocked out lpxC in the ΔlptD single-deletion background, suggesting that inactivation of LPS biosynthesis resolves problems caused by blockage of LPS assembly at a late stage. In contrast, we could not remove the 10 intermediate genes (lpxH, lpxB, lpxK, waaA, msbA, lptB, lptF, lptG, lptC, and lptA) despite repeated attempts. However, deletions of these 10 genes succeeded in the ΔlpxC strain background. Attempts to reintroduce LpxC on a plasmid into these double knockouts were not successful (SI Appendix, Fig. S1C). Taken together, these data showed intermediate genes of LPS biogenesis to be conditionally essential in A. baylyi, likely because when they are inactivated there is toxic accumulation of LPS intermediates. We leveraged our findings to develop a screen in Acinetobacter baumannii, a species closely related to A. baylyi. A. baumannii is one of the ESKAPE pathogens and has developed multidrug resistance in the clinic (29). We reasoned that small-molecule inhibitors of intermediate steps of the LPS biogenesis pathway would behave similarly to genetic knockouts of the corresponding genes and become nontoxic if flux into the pathway is abolished. Furthermore, because removal of the LPS pathway significantly sensitizes cells to small molecules in general, only inhibitors of a component of the pathway would be expected to inhibit the growth of LPS+ strains in preference to more permeable LPS− strains (Fig. 1C).
Establishing a Pathway-Directed Screen Based on Conditional Essentiality.
To assess the ability of such a screen to distinguish on-pathway compounds from off-pathway compounds, we constructed an LPS-deficient strain by knocking out lpxC. As expected, removal of LPS made the bacterium much more susceptible to a range of antibiotics relative to WT but conferred 32-fold resistance to colistin (30), which acts by binding to LPS (Table 1). We reasoned that such distinct responses could be utilized to establish a selective screen for discovering on-pathway inhibitors, while eliminating off-pathway compounds. Specifically, novel compounds targeting a conditionally essential step of LPS biogenesis should inhibit growth of the WT strain at lower concentrations compared with the LPS-null strain. At the same time, we expect loss of LPS to make off-pathway targets more accessible, allowing library compounds that target other pathways to inhibit the LPS-null strain at lower concentrations than the WT strain.
Table 1.
Acinetobacter strains with or without LPS display distinct susceptibilities to known antibiotics
| Strains | Rifampicin | Bacitracin | Vancomycin | Colistin |
| WT | 0.36 (0.3) | 75 (110) | 86 (128) | 1.6 (2) |
| ΔlpxC | 0.0003 (0.0002) | 0.06 (0.09) | 0.17 (0.25) | 50 (64) |
| Δ5 | 0.07 (0.06) | 9 (13) | 22 (32) | 1.6 (2) |
| Δ5 ΔlpxA | 0.0007 (0.0006) | 0.4 (0.6) | 0.3 (0.5) | 50 (64) |
MICs are measured in micromolar or micrograms per milliliter (shown in parentheses).
The outer membrane and efflux pumps of gram-negative bacteria constitute a significant challenge in whole-cell screening, which is particularly important when screening the broad range of pharmacologically unoptimized library compounds that are typically present in compound collections. To overcome this problem, we engineered a screening strain background by undermining the permeability barrier of WT A. baumannii. Specifically, we removed three major efflux pumps that are known to cause multidrug resistance in A. baumannii (31–33), as well as two LPS-modifying enzymes: a secondary acyltransferase that we found to be important for maintaining the barrier function (SI Appendix, Table S1) and a phosphoethanolamine transferase that confers colistin resistance when activated (34–36). This allowed us to use colistin as a positive control and to avoid cross-resistance to similarly acting hits. Next, we knocked out lpxA to inactivate LPS biosynthesis. We found that ΔlpxA A. baumannii grows significantly more poorly in LB broth than its LPS-containing parent strain, making it difficult to score hits by direct comparison of growth profiles. Therefore, we evolved LPS-null A. baumannii by serial passage in LB broth until its growth became comparable to that of WT (hereinafter Δ5 ΔlpxA). To eliminate the confounding effect of adaptive mutations that arose during passaging, we reintroduced the WT lpxA allele into Δ5 ΔlpxA, producing the otherwise isogenic Δ5 strain. This allowed us to confidently compare the growth of the two strains in the presence of compounds.
To ensure that our engineered background performs as expected, we retested our panel of antibiotics against Δ5 and Δ5 ΔlpxA and compared their sensitivities to those of parental WT and ΔlpxC (Table 1). The Δ5 strain was four- to eightfold more sensitive than the unmodified WT, consistent with the lack of efflux and compromised permeability barrier. Nonetheless, absence of LPS in the isogenic Δ5 ΔlpxA strain more strongly sensitized cells to these compounds, causing 23- to 100-fold increased susceptibility. However, these modifications did not change the colistin sensitivity of our engineered screening strains; Δ5 is as sensitive to colistin as WT and Δ5 ΔlpxA remained highly resistant to colistin. Therefore, we conclude that removing the efflux pumps and LPS-modifying enzymes should increase the sensitivity of our screen to on-pathway compounds, allowing us to screen at low concentrations to minimize potential off-target toxic effects. In the meantime, our engineered screening strains should not erode the ability of the assay to eliminate off-pathway compounds given the significantly impaired barrier function upon loss of LPS, while maintaining selectivity toward on-pathway compounds.
Identifying and Optimizing Positive Hits to Achieve Selectivity and Potency.
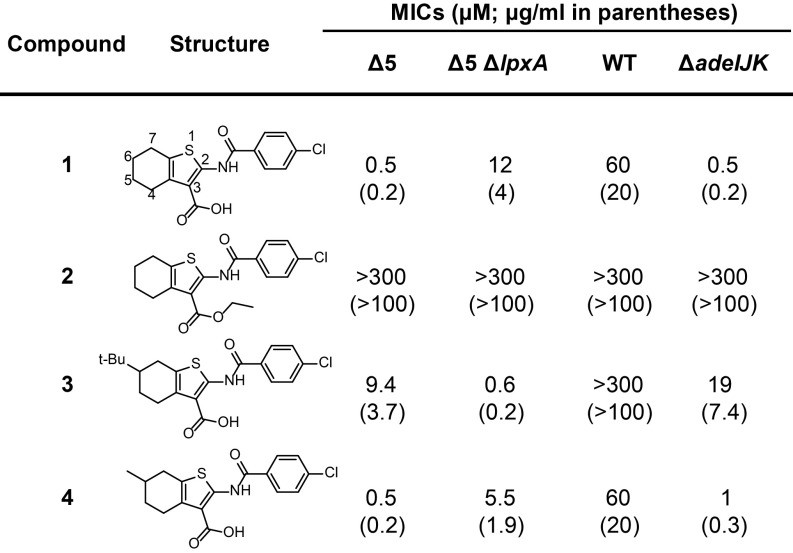
We used our engineered background to screen 150,000 commercially available compounds and bioactives against Δ5 (SI Appendix, Table S2). The top 1,100 compounds that inhibited Δ5 were retested in a counterscreen against Δ5 ΔlpxA. Those compounds (11 in total) that were better inhibitors of Δ5 than of Δ5 ΔlpxA at a single concentration were tested in dose–response against both strains. We thereby identified a tetrahydrobenzothiophene scaffold that demonstrated both selectivity (inhibiting Δ5 24-fold better than Δ5 ΔlpxA) and potency (inhibiting Δ5 at 0.5 µM).
We tested minimal inhibitory concentrations (MICs) of several analogs of the scaffold against a strain panel (Fig. 2). Our best compound 1 displayed 24-fold on-pathway selectivity (0.5 µM and 12 µM for Δ5 and Δ5 ΔlpxA, respectively). The difference in preferential inhibition of the less-permeable Δ5 strain was encouraging given that the actual differential is likely much higher when corrected for intracellular exposure levels. Compound 1 was also active against the parent WT strain. The lack of strong activity against this strain was due to efflux by AdeIJK, since AdeIJK removal is sufficient to achieve the sensitization observed in Δ5. The ethyl ester of 1 (compound 2) had no antibacterial activity, suggesting that the carboxylic acid is important for target engagement. In contrast, a bulky, hydrophobic substituent (t-Bu) at the 6-position of the tetrahydrobenzothiophene ring in compound 3 reversed selectivity. Therefore, 6-tBu erodes on-pathway potency, while increasing off-pathway toxicity. This compound was also completely inactive against WT. When we installed a smaller substituent at the 6-position (Me; compound 4), we found it maintained selectivity and potency against all strains. Hence, relatively moderate changes on the scaffold can cause significant changes in activity, suggesting that this scaffold has a specific cellular target and can be improved.
Fig. 2.
Structure–activity relationship of compounds across four Acinetobacter strains. MIC values in parentheses are in micrograms per milliliter.
We reasoned that if compound 1 targets a conditionally essential step of the LPS pathway, then its antibacterial activity against Δ5 could be modulated in a dose-dependent manner using a small-molecule inhibitor of LpxC. Unsurprisingly, treatment of Δ5 with increasing doses of a known LpxC inhibitor (PF-5081090) (37) protected cells against the antibacterial effect of 1 (SI Appendix, Fig. S2A). A similar pattern was found between colistin and the LpxC inhibitor (SI Appendix, Fig. S2B). These results support the hypothesis that the antibacterial effect of 1 is due to inhibition of a conditionally essential target in the LPS biogenesis pathway.
Identifying Mutations That Inactivate the LPS Pathway to Confer Resistance.
We selected for mutants on plates containing compound 1 at 4 µM, which was significantly above the MIC of the Δ5 strain, but below the MIC of the Δ5 ΔlpxA strain to minimize off-target effects and favor mutations in the LPS biogenesis pathway. Six resistant colonies (frequency of resistance ∼1 × 10−6) were isolated and analyzed by whole-genome sequencing. All contained mutations in the genes encoding LpxA, LpxC, or LpxD, the first three enzymes in lipid A biosynthesis (SI Appendix, Table S3). We found that MICs of the resistant mutants for colistin and rifampicin were in agreement with those of LPS-deficient strains (Table 2). Expressing WT LpxA, LpxC, or LpxD in trans restored their sensitivity profile to that observed in Δ5 (Table 2). Therefore, we conclude that these mutations confer resistance to compound 1 by resulting in loss of function in the pathway.
Table 2.
Two classes of compound 1-resistant mutants display distinct sensitivities to colistin and rifampicin
| Strains | 1 | Colistin | Rifampicin |
| Δ5 | 0.5 (0.2) | 1.6 (2) | 0.07 (0.06) |
| 1R lpxnull | 4.9 (1.6) | 50 (64) | 0.0007 (0.0006) |
| 1R lpxnull + Lpxwt | 0.3 (0.1) | 1.6 (2) | 0.07 (0.06) |
| 1R msbAmissense | 4.9 (1.6) | 1.6 (2) | 0.07 (0.06) |
| Δ5 + MsbAmissense | 8.6 (2.9) | 1.6 (2) | 0.03 (0.02) |
MICs are measured in micromolar or micrograms per milliliter (shown in parentheses).
Although these preliminary experiments validated the principle of the screen, they did not identify the specific target of our compound. Characterizations of additional resistant colonies only yielded more Lpx nulls. This is not unexpected since there are many more possible mutations that remove the pathway than those that preserve the function of the pathway while generating resistance in the target.
Sampling Less Frequently Occurring Mutations on the Specific Target of the LPS Pathway.
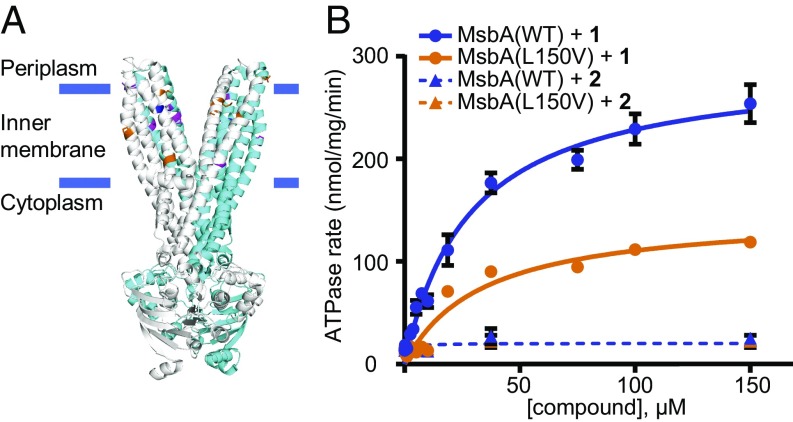
Since we suspected that the frequency of mutations in the putative target of compound 1 is very low, we had to sample a greater number of cells and simultaneously develop an approach that efficiently eliminates the more frequent LPS-deficient mutants (SI Appendix, Fig. S3A). To accomplish this, we plated 20 times more cells (a total of 109–1010) onto the same selection condition (4 µM of 1). Despite the high number of colony-forming units plated, we were able to obtain distinct colonies rather than a bacterial lawn (SI Appendix, Fig. S3B). Colonies were replica plated onto colistin-containing plates to identify LPS-deficient mutants, which should still form colonies since they lack the colistin binding site in the outer membrane. Conversely, we hypothesized that the rare resistant mutants which retained LPS would grow in the presence of 1 but would not grow in the presence of colistin. Seven independent cultures gave rise to 21 resistant colonies of this type (frequency of resistance ∼1 × 10−8). We found that despite being resistant to 1 (MIC 4.9 µM), all isolated mutants exhibited the same susceptibilities to colistin and rifampicin as the parent Δ5 strain, indicating the presence of LPS (Table 2). All these colonies contained missense mutations in msbA, which encodes the ABC transporter that flips LPS across the inner membrane (SI Appendix, Table S4). To better understand how these mutations confer resistance, we mapped them on the crystal structure of Salmonella typhimurium MsbA [Protein Data Bank (PDB) ID code 3B60] (38). Individual mutations are scattered over the transmembrane region of the protein structure (Fig. 3A); the area that they span is sufficiently large to suggest that they do not indicate the inhibitor’s binding site but may impose a conformational change that alters the binding of compound 1.
Fig. 3.
Compound 1 interacts with the LPS flippase MsbA. (A) Mutations (shown in orange and magenta in the two respective monomers) identified in MsbA map to its transmembrane region (structure from S. typhimurium; PDB ID code 3B60). The most prevalent mutation, L150V, is colored blue. (B) Compound 1 stimulates ATPase activity (n = 2; error bars = SE) of WT MsbA in vitro. The L150V mutation attenuates this effect. In contrast, the inactive compound 2 does not affect ATPase activity.
To confirm that the identified msbA point mutations were responsible for resistance to 1, we supplied the most frequently occurring mutant allele, MsbA L150V, in trans and found that it resulted in a 10-fold increase in resistance in the Δ5 strain (MIC 8.6 µM), whereas WT MsbA did not result in substantial resistance (MIC 0.9 µM). This observation indicates that MsbA L150V acts as a dominant resistance allele in the WT background. These results further validate our pathway-directed screening approach for discovering inhibitors of essential steps of LPS biogenesis.
Identifying the Direct Target of Compound 1.
To determine whether MsbA is the direct target of compound 1, we reconstituted the purified WT and the 1-resistant (L150V) MsbA proteins into LPS-containing liposomes and measured ATPase activity under compound treatment (Fig. 3B). We found that 1 stimulated the ATPase activity of WT MsbA in a dose-dependent manner, whereas the effect was significantly weaker on the resistant form (MsbA L150V). Furthermore, we found that the inactive analog 2 produced no such effect. The ability to modulate ATPase activity of MsbA and do so in an allele- and compound-specific manner supports a direct interaction between the protein target and compound 1.
We reasoned that the antibacterial effect of compound 1 could be due to either inhibition or overactivation of LPS flipping. We found that WT MsbA overexpression conferred resistance to 1 (MIC increased to 2.7 µM). This suggests that inhibition, rather than overactivation, of LPS flipping underlies the antibacterial effect of compound 1. We reasoned that if 1 acts by overactivating LPS flipping, overexpression of MsbA would result in sensitization to 1 by enabling even more LPS to be flipped. However, if binding of 1 to MsbA decouples ATP hydrolysis from LPS flipping, overexpression of MsbA would confer resistance by titrating the drug.
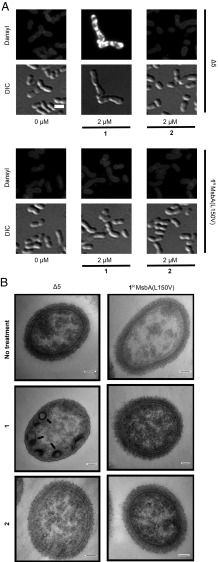
We have recently developed tools to visualize LPS in whole cells using a fluorescent probe, Dansyl-PMBN, a colistin derivative that fluoresces intensely when partitioned into a hydrophobic environment (39). Treating Δ5 cells with 1 resulted in intense fluorescence inside the cell, unlike the moderate staining of LPS at the cell periphery in the untreated control (Fig. 4A). In agreement with our predictions, these effects were not observed when we switched to the resistant mutant (1R MsbA L150V) or treated the two strains with the inactive analog 2. This phenotype is reminiscent of previous observations using a temperature-sensitive MsbA variant in Escherichia coli (39). Interestingly, treating Δ5 cells with 1 led to a moderate but consistent increase in LPS level relative to other conditions, suggesting that this buildup of LPS might be a feature of the antibacterial mechanism of compound 1 (SI Appendix, Fig. S4).
Fig. 4.
Compound treatment causes mislocalization of LPS to the cell interior. (A) Whole-cell staining with Dansyl-PMBN reflected LPS mislocalization in compound 1-treated Δ5 cells. Cells were treated by compound 1 or 2 at 2 µM as indicated for 14 h before being stained by Dansyl-PMBN at 12 µM. DIC, differential interference contrast. (Scale bar: 2 μm.) (B) Transmission electron microscopy showed intracellular accumulation of densely stained extra membrane in compound 1-treated Δ5 cells, a phenotype characteristic of MsbA inhibition. Arrows indicate the buildup of extra membrane materials. Compound 1-treated Δ5 cells also showed a loss of capsular polysaccharides, which could be related to inhibition of LPS targeting to the cell surface (47). The same effects were absent in other conditions. (Scale bar: 100 nm.)
To further probe the mechanism of compound 1, we examined cells by transmission electron microscopy to detect changes caused by compound treatment (Fig. 4B). Consistent with inhibition of LPS flipping, we found that treating Δ5 cells with 1 caused extra membrane buildup in the cytoplasm, a defect reminiscent of disruption of MsbA (40). By contrast, treating resistant mutant (1R MsbA L150V) by 1 or both strains by 2 did not lead to such phenotype. Taken together, these results illustrate that the antibacterial activity of 1 arises from inhibition of MsbA’s flippase activity.
Discussion
In this study, we have established that a subset of the genes responsible for biosynthesis of LPS and its transport to the cell surface are only essential when there is active LPS flux into the pathway. We observed that either genetic inactivation or small-molecule inhibition of essential steps of the pathway can be tolerated when flux into the pathway is abolished. Finally, and importantly, we have exploited this conditional essentiality to develop a cell-based screen that enabled us to discover inhibitors of MsbA, the protein responsible for translocating LPS across the inner membrane during biogenesis. This establishes MsbA as an antibacterial target. We have established that the potency of our compounds can be significantly improved by knocking out a specific efflux pump.
Our compound stimulates ATPase activity of MsbA, while decoupling it from LPS translocation. Similar mechanisms have been reported for inhibitors of other ABC transporters. For example, tariquidar is known to inhibit drug efflux by P-glycoprotein, an efflux pump in mammalian cells, by trapping P-glycoprotein in a closed conformation that results in ATPase activation (41). This effect has also been observed for an inhibitor of a bacterial ABC transporter, LolCDE (42). It is possible that compound 1 locks MsbA in a similar state that stimulates ATP hydrolysis while preventing LPS translocation.
In addition to discovering a small molecule that targets MsbA, our study provides insights into an important question: Why are certain steps of LPS biogenesis essential in Acinetobacter when it is also known that the entire pathway can be removed? (21, 25, 27). Our results show that the organism does not down-regulate LPS biosynthesis upon MsbA inhibition although the most frequenct resistance mechanism we observed is to block LPS biosynthesis. We reason that when essential steps in the pathway are inactivated, the cells are unable to halt LPS biosynthesis. However, aside from lpxACD, inactivation of nonessential steps down-regulates LPS biosynthesis. In particular, it has been reported that inactivation of lptD results in significant decrease of LPS levels (43), suggesting a feedback response that down-regulates LPS biosynthesis. In contrast, our results suggest that MsbA inhibition is unable to induce the same response. We speculate that the elevated levels of LPS at the inner membrane produced by MsbA inhibition alter the properties of the membrane, thereby interfering with one or more of essential processes that rely on its normal state. This may be exacerbated by dissipation of ATP or depletion of common precursors from other essential pathways, such as fatty acid biosynthesis.
The pathway-directed screen developed in this study combines the advantages of traditional cell-based and target-based methods (44). The assay uses bacterial growth as readout; therefore, it is easy and inexpensive to implement. Discovery of LPS biogenesis inhibitors has been difficult because the pathway involves a seven-protein transenvelope complex and complex substrates with limited solubility. By using whole cells, the screen we described here allows for interrogation of all of the possible targets of the LPS biogenesis pathway in a relevant cellular context.
Establishing a cell-based, pathway-directed screen requires a distinct phenotype that is specific to the pathway of interest. A common approach is to screen in cells that have been compromised in the pathway of interest on the assumption that inhibitors of that pathway will further push the weakened strain toward death. However, a potential problem with this approach is that cells weakened in the pathway of interest are also weakened in other ways that increase susceptibility to off-pathway compounds; this can result in a large number of false positives which must be painstakingly triaged using secondary assays. In contrast, in the screen we present here, we demand that compounds be able to kill the relevant WT strain in preference to a more permeable, LPS-deficient strain. The requirement that the removal of the LPS pathway, which significantly impairs general fitness, confers resistance rather than sensitivity is a much more specific criterion to define hits. Therefore, this approach excels at filtering out compounds with off-pathway activities. With such a stringent initial filter, the assay facilitates the discovery of on-pathway small molecules at an early stage. This improvement is particularly important to justify drug discovery efforts at later stages, as taking a large number of primary hits into follow-up studies is both time-consuming and expensive. These unique benefits of the pathway-directed screen thus position it as an ideal approach for drug discovery of LPS biogenesis in gram-negative bacteria.
Materials and Methods
Methods of strain construction and characterization, plasmid construction, high-throughput screening and hit follow-up, isolation of resistant mutants, protein overexpression and purification, ATPase assay, microscopy, and chemical synthesis of analogs are decribed in detail in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank ICCB-Longwood Screening Facility for providing access to their equipment, the Bauer Core Facility at Harvard University for the use of equipment and sequencing services, and Claudio Zambaldo (The Scripps Research Institute) for providing reagents. We also thank Maria Ericsson, Louise Trakimas, and Elizabeth Benecchi at the Harvard Medical School Electron Microscopy Facility for providing transmission electron microscopy services and for helpful discussions. This work was supported by NIH Grants U19 AI109764 and R01 AI081059 (to D.K.) and the Blavatnik Biomedical Accelerator at Harvard University. Fluorescence microscopy was performed at the Nikon Imaging Center at Harvard Medical School. Electron microscopy was performed at the Harvard Medical School Electron Microscopy Facility.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 6530.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804670115/-/DCSupplemental.
References
- 1.Boucher HW, et al. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.Hersh AL, Newland JG, Beekmann SE, Polgreen PM, Gilbert DN. Unmet medical need in infectious diseases. Clin Infect Dis. 2012;54:1677–1678. doi: 10.1093/cid/cis275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bladen HA, Mergenhagen SE. Ultrastructure of Veillonella and morphological correlation of an outer membrane with particles associated with endotoxic activity. J Bacteriol. 1964;88:1482–1492. doi: 10.1128/jb.88.5.1482-1492.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 5.Mühlradt PF, Golecki JR. Asymmetrical distribution and artifactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. Eur J Biochem. 1975;51:343–352. doi: 10.1111/j.1432-1033.1975.tb03934.x. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doerrler WT. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol Microbiol. 2006;60:542–552. doi: 10.1111/j.1365-2958.2006.05130.x. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: The long road of discovery. Nat Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simpson BW, May JM, Sherman DJ, Kahne D, Ruiz N. Lipopolysaccharide transport to the cell surface: Biosynthesis and extraction from the inner membrane. Philos Trans R Soc Lond B Biol Sci. 2015;370:20150029. doi: 10.1098/rstb.2015.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat Rev Microbiol. 2016;14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May JM, Sherman DJ, Simpson BW, Ruiz N, Kahne D. Lipopolysaccharide transport to the cell surface: Periplasmic transport and assembly into the outer membrane. Philos Trans R Soc Lond B Biol Sci. 2015;370:20150027. doi: 10.1098/rstb.2015.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman DJ, et al. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science. 2018;359:798–801. doi: 10.1126/science.aar1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaara M. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1993;37:2255–2260. doi: 10.1128/aac.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onishi HR, et al. Antibacterial agents that inhibit lipid A biosynthesis. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 18.Srinivas N, et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science. 2010;327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 19.Gronenberg LS, Kahne D. Development of an activity assay for discovery of inhibitors of lipopolysaccharide transport. J Am Chem Soc. 2010;132:2518–2519. doi: 10.1021/ja910361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erwin AL. Antibacterial drug discovery targeting the lipopolysaccharide biosynthetic enzyme LpxC. Cold Spring Harb Perspect Med. 2016;6:a025304. doi: 10.1101/cshperspect.a025304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Meredith TC, Kahne D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol. 2013;16:779–785. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steeghs L, et al. Meningitis bacterium is viable without endotoxin. Nature. 1998;392:449–450. doi: 10.1038/33046. [DOI] [PubMed] [Google Scholar]
- 23.Peng D, Hong W, Choudhury BP, Carlson RW, Gu XX. Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect Immun. 2005;73:7569–7577. doi: 10.1128/IAI.73.11.7569-7577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffatt JH, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richie DL, et al. Toxic accumulation of LPS pathway intermediates underlies the requirement of LpxH for growth of Acinetobacter baumannii ATCC 19606. PLoS One. 2016;11:e0160918. doi: 10.1371/journal.pone.0160918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Berardinis V, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei JR, et al. LpxK is essential for growth of Acinetobacter baumannii ATCC 19606: Relationship to toxic accumulation of lipid A pathway intermediates. MSphere. 2017;2:e00199-17. doi: 10.1128/mSphere.00199-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzgar D, et al. Acinetobacter sp. ADP1: An ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 2004;32:5780–5790. doi: 10.1093/nar/gkh881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willyard C. Drug-resistant bacteria ranked. Nature. 2017;543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 30.Velkov T, Thompson PE, Nation RL, Li J. Structure–Activity relationships of polymyxin antibiotics. J Med Chem. 2010;53:1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damier-Piolle L, Magnet S, Brémont S, Lambert T, Courvalin P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother. 2008;52:557–562. doi: 10.1128/AAC.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Périchon B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54:4389–4393. doi: 10.1128/AAC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams MD, et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother. 2009;53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beceiro A, et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arroyo LA, et al. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother. 2011;55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery JI, et al. Pyridone methylsulfone hydroxamate LpxC inhibitors for the treatment of serious Gram-negative infections. J Med Chem. 2012;55:1662–1670. doi: 10.1021/jm2014875. [DOI] [PubMed] [Google Scholar]
- 38.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moison E, et al. A fluorescent probe distinguishes between inhibition of early and late steps of lipopolysaccharide biogenesis in whole cells. ACS Chem Biol. 2017;12:928–932. doi: 10.1021/acschembio.7b00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doerrler WT, Reedy MC, Raetz CR. An Escherichia coli mutant defective in lipid export. J Biol Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- 41.Loo TW, Clarke DM. Tariquidar inhibits P-glycoprotein drug efflux but activates ATPase activity by blocking transition to an open conformation. Biochem Pharmacol. 2014;92:558–566. doi: 10.1016/j.bcp.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Nickerson NN, et al. A novel inhibitor of the LolCDE ABC transporter essential for lipoprotein trafficking in Gram-negative bacteria. Antimicrob Agents Chemother. 2018;62:e02151-17. doi: 10.1128/AAC.02151-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bojkovic J, et al. Characterization of an Acinetobacter baumannii lptD deletion strain: Permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J Bacteriol. 2015;198:731–741. doi: 10.1128/JB.00639-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castoreno AB, Eggert US. Small molecule probes of cellular pathways and networks. ACS Chem Biol. 2011;6:86–94. doi: 10.1021/cb1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boll JM, et al. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. MBio. 2015;6:e00478-15. doi: 10.1128/mBio.00478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vinogradov EV, Duus JO, Brade H, Holst O. The structure of the carbohydrate backbone of the lipopolysaccharide from Acinetobacter baumannii strain ATCC 19606. Eur J Biochem. 2002;269:422–430. doi: 10.1046/j.0014-2956.2001.02647.x. [DOI] [PubMed] [Google Scholar]
- 47.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.