Significance
The conventional single-agent approach to therapeutics is challenged by the redundancy and feedback in biological networks. The use of multiple drugs in combination might improve desired functional outcomes while reducing toxicity and overcoming drug resistance. However, the complexity and resources required to test many combinations have slowed discovery efforts. Here, we introduce a screening platform that automatically constructs combinations of drugs from nanoliter-scale droplets, greatly simplifying screening logistics and reducing resource consumption. We applied our platform to discover drug pairs that act synergistically against the model pathogen Escherichia coli. Our platform can be further developed to support many types of phenotypic assays in a variety of disease models.
Keywords: high-throughput screening, nanoliter droplet, drug synergy, antibiotics, small molecules
Abstract
Combinatorial drug treatment strategies perturb biological networks synergistically to achieve therapeutic effects and represent major opportunities to develop advanced treatments across a variety of human disease areas. However, the discovery of new combinatorial treatments is challenged by the sheer scale of combinatorial chemical space. Here, we report a high-throughput system for nanoliter-scale phenotypic screening that formulates a chemical library in nanoliter droplet emulsions and automates the construction of chemical combinations en masse using parallel droplet processing. We applied this system to predict synergy between more than 4,000 investigational and approved drugs and a panel of 10 antibiotics against Escherichia coli, a model gram-negative pathogen. We found a range of drugs not previously indicated for infectious disease that synergize with antibiotics. Our validated hits include drugs that synergize with the antibiotics vancomycin, erythromycin, and novobiocin, which are used against gram-positive bacteria but are not effective by themselves to resolve gram-negative infections.
Much of modern drug discovery acts to modulate a specific drug target using a single agent with maximally selective effects, arising from the idea of Paul Ehrlich’s “magic bullet” (1). However, the prevalence of redundancy, feedback, and multifunctionality in biological networks challenges this approach (2–4). Therapeutic strategies comprising multiple drugs in combination have been proposed to exploit network-driven interactions to achieve the desired functional perturbation, to reduce toxicity, and to prevent or overcome drug resistance (2–6). In particular, combination antimicrobial treatments that overcome drug resistance by targeting known resistance elements (e.g., beta-lactamase enzymes) in addition to essential targets make up a substantial fraction of antibiotic treatments in clinical development today (7).
Despite the applicability of novel drug combinations, their identification by high-throughput screening has been slowed by the high complexity, cost, and compound consumption of conventional screening methods (8). For example, testing all pairs of drugs from a modest library of 2,000 drugs (e.g., drugs approved by the US Food and Drug Administration) requires almost 2 million pairwise combinations, and far more if compounds are titrated. Experiments of this scale are currently restricted to specialized laboratories and facilities that can accommodate the large costs and complexity (e.g., total liquid handling steps and logistics of plate layout and workflow design). Additionally, since these screens test each compound across thousands of others, thousands of times the compound quantity is required compared with single-compound screening and can deplete an entire chemical inventory in a single screening experiment. Current methods for combinatorial discovery strive to work around these issues, either through computational predictions of drug synergies to reduce screening scale or by combining multiple tests in pools with subsequent deconvolution (9–11).
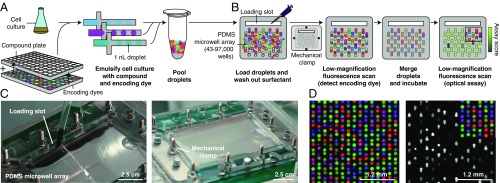
Here, we introduce a strategy for combinatorial drug screening based on droplet microfluidics that unlocks order-of-magnitude improvements in logistical complexity and compound consumption and reduces the need for capital equipment (Fig. 1). Through miniaturization and high-speed processing, advances in droplet microfluidics are making major impacts across the life sciences, such as allowing the measurement of 1,000+ single-cell morphological (12) and transcriptomic phenotypes (13) and high-resolution and low cell-input drug dose-response testing (14–19). Our platform leverages the throughput potential of such microfluidic and microarray systems (12, 20–22), substituting deterministic liquid handling operations needed to construct combinations of compounds with spontaneous merging of random pairs of droplets in parallel inside a microwell array device (Fig. 1) (23). We used this device to screen compound combinations for bacterial growth inhibition in nanoliter droplets, demonstrating its manual operation at high throughput without robotic liquid handling.
Fig. 1.
Droplets platform for combinatorial drug screening. (A) Compounds, cells, and encoding dyes are emulsified into nanoliter droplets and subsequently pooled. (B) A microwell array randomly pairs droplets (SI Appendix, Fig. S1 and Movies S1 and S2). Once loaded, free surfactant is depleted by washing to limit compound exchange. Low-magnification epifluorescence microscopy identifies the compounds carried by each droplet. Pairs of droplets in each microwell are merged and incubated (Movie S3). A second optical scan reads out a phenotypic assay (e.g., cell growth inhibition). (C) Photographs of the microwell array during loading (SI Appendix, Fig. S1 and Movies S1 and S2). Scale bars are approximate due to perspective effect. (D) Three-color fluorescence micrograph of droplets in microwell array paired with a subsequent assay of growth inhibition of E. coli cells, monitored by fluorescence from constitutively expressed GFP. Only 50% of droplet inputs contained cells; therefore, a fraction of microwells do not show GFP fluorescence. PDMS, polydimethylsiloxane.
Results and Discussion
Our platform constructs and assays all pairwise combinations of a set of input compounds (Fig. 1). First, we combine concentrated compounds in well plates with fluorescence barcodes (unique ratios of three fluorescent encoding dyes), cells, and media (Fig. 1A). We then emulsify a sample from each well into 20,000 1-nL aqueous droplets (total volume of 20 μL) in a fluorocarbon oil continuous phase with a fluorosurfactant that stabilizes the emulsion. We use standard micropipettes to pool and load the droplets into a microwell array such that each microwell captures two droplets at random (Fig. 1B, SI Appendix, Fig. S1, and Movies S1 and S2) (23). We then seal the microwell array to the glass substrate to limit evaporation and chemical cross-contamination across microwells and fix the assembly by mechanical clamping. We identify the contents of each droplet by reading the fluorescence of the encoding dyes by low-magnification epifluorescence microscopy (2× or 4× magnification, 6.5 μm per pixel optical resolution) (Fig. 1 B and D) (95 to 99% accuracy; see SI Appendix, Fig. S2) (15, 18). We then merge all pairs of droplets by applying a high-voltage AC electric field (Movie S3) and incubate the microwell array to allow cells to respond to the pair of compounds (24). Last, we image the microwell array to read out the assay result (e.g., cell growth inhibition) and map this measurement to the pair of compounds previously identified in each well (Fig. 1 B and D). Pairwise compound combinations are thus constructed spontaneously by droplet merging, considerably reducing the number of liquid handling steps required (SI Appendix, Fig. S3).
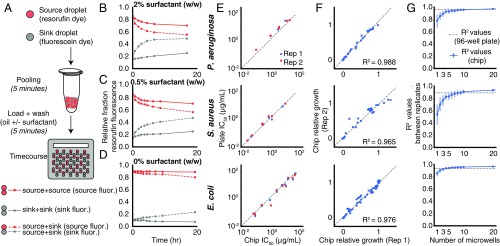
A challenge in droplet microfluidics that has limited cell-based compound screening is the exchange of compounds between droplets on assay-relevant timescales. Exchange is mediated by the dynamic equilibrium of surfactant molecules between the aqueous–oil droplet interface and reverse micelles in the oil phase (see SI Appendix, Supplementary Text) (25–27). In a screening context, this creates the potential both for false negatives resulting from compound loss from droplets and for false positives resulting from the exchange of active compounds among droplets. In our workflow, transport of compounds can occur between droplets during the pooling of droplets, and between microwells after droplets are loaded into the microwell array. Although compound exchange cannot be eliminated in the brief droplet-pooling phase before loading the microwell array (Figs. 1A and 2A), we hypothesized that the microwell array design could limit the extent of false positives caused by cross-contamination during the longer incubation step by (i) depleting free surfactant by an oil wash, and (ii) limiting reverse micelle diffusion between microwells by mechanically sealing the microwell array to a substrate (Fig. 1 B and C and SI Appendix, Fig. S1). To measure compound cross-contamination on our platform, we monitored the transport of the fluorescent dye resorufin (a reference compound commonly used to assess chemical cross-contamination) from “source” droplets to “sink” droplets (encoded by fluorescein, a slow-exchanging dye) (Fig. 2A) (25, 26). We found that compartmentalization alone (without depletion of free surfactant) limited resorufin transport compared with exchange between pairs of droplets in the same microwell (Fig. 2 B and C). Depleting free surfactant by washing the loaded microwell array before sealing further decreased exchange to levels below our detection limit (Fig. 2D). Although some compounds can exchange more quickly than resorufin and may exchange significantly during the droplet-pooling phase before loading (26, 28), these compounds would likely be diluted below active concentrations across many other droplets. The low false-positive rate of the screen (described below) and further analysis of our screening hits show that these fast-exchanging compounds (presumably present in our library of drugs and druglike molecules) do not need to be identified and removed before screening (SI Appendix, Supplementary Text and Figs. S4 and S5).
Fig. 2.
Characterization of droplet platform performance. (A) To model cross-contamination, we tracked the exchange of the fluorescent dye resorufin (carried by source droplets) to empty droplets (sink droplets, encoded by fluorescein). (B–D) Exchange is measured over time by the fraction of fluorescence measured in source droplets (red lines) and sink droplets (gray lines) as a function of surfactant wash concentration (0%, 0.5%, and 2% wt/wt). The effect of compartmentalization is measured by comparing the rates of dye accumulation in sink droplets when cocompartmentalized in microwells with source droplets (dotted lines) or other sink droplets (solid lines). Exchange that occurred before loading was measured by the fraction of fluorescence in sink droplets at the first time point (SI Appendix, Supplementary Text and Fig. S4F). (E) Estimated IC50 for each antibiotic compared between 96-well plate and droplet platform formats (SI Appendix, Figs. S8–S10). Dotted lines show the diagonal. (F) Comparison of two technical replicates on the droplet platform of growth values at particular antibiotic dosages normalized to a media-only control (SI Appendix, Figs. S8–S10). Dotted lines show line of best fit. (G) Relationship between microwell-level replication and technical noise, estimated by bootstrap resampling of the data in F. Error bars represent 10th- to 90th-percentile bootstrapped R2 values. Dotted lines represent R2 values between technical replicates in 96-well plates (SI Appendix, Fig. S11).
As a first application of our platform, we developed fluorescence-based growth-inhibition phenotypic screening assays for three model bacterial pathogens often used in antibiotic discovery: Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli (14, 16). For each organism, we compared growth dynamics, antibiotic drug responses, and reproducibility of the droplet platform with conventional Erlenmeyer flask and 96-well plate broth-culture methods (Fig. 2 E–G and SI Appendix, Figs. S6–S11). Growth dynamics (monitored by constitutive GFP fluorescence) between Erlenmeyer flasks and the droplet platform showed close correspondence, indicating no detectable toxicity or gross physiological impact on the bacteria (SI Appendix, Fig. S6). We chose 6 to 12 antibiotics representing different chemical classes and mechanisms of action and compared IC50 values estimated from five-point dose-response curves measured with the droplet platform and with the same fluorescence assay in a 96-well plate broth-culture format (SI Appendix, Fig. S7). Overall, we found similar potency for each antibiotic and comparable levels of assay noise (R2 values between technical replicates) (Fig. 2 E–G and SI Appendix, Figs. S8–S11).
High-throughput screening is extremely sensitive to assay noise, as hits must be enriched with respect to false positives. In the droplet platform, droplets carrying different compounds are paired randomly in microwells, and noise is suppressed by making multiple measurements of the same compound pair across replicate microwells. The number of replicate microwells is a random variable with an expected value determined by the number of possible unique input droplet-pair combinations and the number of microwells on a given chip (SI Appendix, Fig. S12). To explore this relationship between microwell-level replication and throughput, we down-sampled the number of replicate microwells per antibiotic dose and compared measurements from two technical replicate microwell arrays. We observed diminishing improvements at replication levels past 5 to 10 microwells, which is the approximate level of replication obtained when 64 unique inputs are applied to our standard-size array (Fig. 2G and SI Appendix, Fig. S12).
To evaluate our ability to detect synergy between compound pairs, we tested a canonically synergistic pair—ampicillin (a beta-lactam antibiotic) and sulbactam (a beta-lactamase inhibitor)—against P. aeruginosa (SI Appendix, Fig. S13). Synergy is commonly assessed by crossing a dilution series of each compound in a “checkerboard” assay matrix and quantifying the activity levels via Bliss independence or the fractional inhibitory concentration (FIC) index method (29, 30). Synergy (defined as an FIC index of ≤0.5) was detected in both 96-well plate broth culture (FIC index ≤0.5) and the droplet platform (FIC index ≤0.25) (SI Appendix, Fig. S13).
We next applied our system to identify compounds that can potentiate the activity of antibiotic drugs. In the face of rising antibiotic resistance, efforts to develop new classes of antibiotics have yielded little success (7, 31, 32). Unfortunately, many antibiotics such as vancomycin, erythromycin, and novobiocin cannot be used to treat important gram-negative pathogens such as E. coli, P. aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae due to the impermeability of their outer membranes and numerous efflux systems (7, 32). Previous work suggests that identifying compounds that sensitize drug-resistant pathogens is a promising strategy to broaden the spectrum of existing antibiotics (33–35).
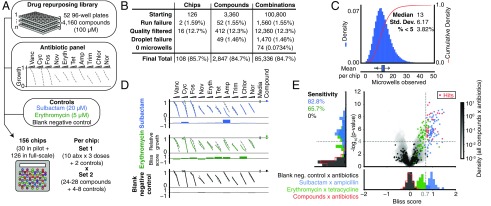
We screened for potentiation of a panel of 10 antibiotics that have diverse mechanisms and biochemical target localizations (each antibiotic titrated across a three-point response curve; see SI Appendix, Table S1) by a “drug repurposing” library of 4,160 compounds against E. coli (Fig. 3A) (34, 36). This curated library is composed of tool and investigational compounds with extensive preclinical and clinical research data, as well as launched drugs (36). We reasoned that hits from screening this library may have potential for expedited translation for use in sensitizing gram-negative pathogens to existing antibiotics (36, 37).
Fig. 3.
Drug repurposing antibiotic potentiation screen. (A) To measure antibiotic potentiation, we generated three-point dose-response curves for 10 different antibiotics in combination (SI Appendix, Table S1) with 4,160 compounds (each at single concentration, 100 μM) from a drug repurposing library, as well as positive controls (sulbactam and erythromycin) and negative controls (blank media). Each chip formulated all pairwise combinations of two input sets: (i) three antibiotics × 10 concentrations + 2 controls (32 total) and (ii) 24 to 28 compounds + 4 to 8 controls (32 total). (B) The final numbers of analyzed combinations in the full-scale screening phase, after accounting for losses and quality filtering (SI Appendix, Fig. S14). (C, Top) The histogram (blue bars) and cumulative distribution (red line) of the number of microwells observed for each compound × antibiotic combination. (C, Bottom) Tukey box plot of mean numbers of microwells of all compound × antibiotic combinations on each chip in full-scale phase of screen. (D) Representative primary screening data are shown for control compounds (Dataset S1). Synergy was identified by comparing antibiotic response curves in the presence of a compound compared with those of the antibiotic alone (gray dotted lines), made quantitative by calculating a Bliss score. At Right, the growth in the presence of compound alone (Compound, indicated by colored points), or the absence of antibiotic and compound (Media, indicated by gray points) are shown. (E, Right) A total of 28 compound × antibiotic combination hits (red points) were determined by thresholding all compound × antibiotic pairs (gray, shaded contours) on effect size (Bliss score >0.7, gray dotted line) and statistical significance (P < 10−4, gray dotted line) (Datasets S2 and S3). (E, Left) Projection of vertical axis. Sensitivity to positive controls (blue indicates sulbactam; green indicates erythromycin) (Dataset S1). (E, Bottom) Projection of horizontal axis. Bliss score distributions of positive and negative (black indicates blank) controls (Dataset S1). Histograms are set to 50% opacity to show overlap. Amp, ampicillin; Chlor, chloramphenicol; Cyc, cycloserine; Eryth, erythromycin; Fos, fosfomycin; Nor, norfloxacin; Nov, novobiocin; Tet, tetracycline; Trim, trimethoprim; Vanc, vancomycin.
This screening effort resulted in the construction of 4+ million total microwell assays across 156 microwell array chips, and was completed in 10.3 d in two phases (pilot phase: 800 compounds, 30 chips, 3.33 d; full-scale phase: 3,360 compounds, 126 chips, 7 d). With a total of 64 unique inputs per microwell array chip (set 1: 10 antibiotics at three dose points + 2 controls; set 2: 24 to 28 compounds + 4 to 8 controls), each chip-run constructed 720 to 840 compound × antibiotic combinations (24 to 28 compounds × 10 antibiotics × three dose points), 276 to 378 compound × compound combinations [0.5 × (24 × 23); 0.5 × (28 × 27)], 120 to 240 control × antibiotic combinations (4 to 8 controls × 10 antibiotics × 3 dose points), and 48 to 56 compound × control combinations (24 to 28 compounds × 2 controls) (Fig. 3A).
Our analysis focused on determining compound × antibiotic synergies by evaluating a shift of a three-point antibiotic dose response with and without compound, quantified by a synergy metric (Bliss score) for each compound × antibiotic-pair scoring deviation from Bliss independence (Fig. 3 A and D). Hits from our screen were then validated in eight-point checkerboard assays and quantified by Bliss independence and the FIC index method.
We evaluated screening performance from our full-scale phase, comprising 126 microwell array chip-runs and 100,800 compound × antibiotic assay points from 3,360 compounds. Dropout can occur due to chip-run failures; failures to produce, load, and classify droplets; or failure to observe any microwells containing a particular compound × antibiotic combination. Of the 126 chip-runs, we had two logistical failures and removed 16 runs due to failed controls to yield a final chip passing rate of 85.7% (108 runs) (Fig. 3B and SI Appendix, Fig. S14). Droplet production, pooling/loading, or fluorescence barcode assignment failed for 49 compounds (Fig. 3B). Overall, of the starting 100,800 compound × antibiotic combinations, 84.7% were successfully measured, with an overall median value of 13 replicate microwells (Fig. 3 B and C).
To assess data quality, each chip-run was performed with a set of positive (sulbactam × ampicillin; erythromycin × tetracycline) and negative (blank media × all antibiotics) controls to determine the expected sensitivity and false-positive rate of the screen (Fig. 3 A and D and Dataset S1). The Bliss score distribution of all blank media × antibiotic pairs was well described by a t distribution, which we used as a null model to calculate P values for each compound × antibiotic pair (SI Appendix, Fig. S15). To measure sensitivity, each run included one or both positive controls: sulbactam × ampicillin (large effect size, expected Bliss score ∼1) and erythromycin × tetracycline (small effect size, expected Bliss score ∼0.5) (Fig. 3D). At an expected false-positive rate of 10−4 (P value threshold), we recovered 82.8% of sulbactam × ampicillin controls (n = 58/70) and 65.7% of erythromycin × tetracycline controls (n = 46/70) (Fig. 3E). To call hits from all of the compound × antibiotic pairs (Dataset S2), we chose a Bliss score effect-size threshold that separated sulbactam × ampicillin controls from erythromycin × tetracycline controls (Bliss score >0.7) (Fig. 3E). Using these thresholds to score all pairs yielded 28 hit compound × antibiotic pairs (0.098% of total 28,470) from 20 distinct compounds (0.70% of total 2,847) (Fig. 3E and Dataset S3). Although we focused analysis on compound × antibiotic pairs, we did identify that one hit compound, pasireotide, also synergized with tedizolid, another compound in the repurposing library run on the same microwell array chip (SI Appendix, Fig. S16).
We selected 17 hit compound × antibiotic pairs from 11 distinct compounds for confirmation in eight-point checkerboard assays measured in 96-well plate broth-culture format (Fig. 4A and SI Appendix, Fig. S17). For comparison, we measured an additional 29 pairs that did not pass Bliss score and P value thresholds in the primary screen, for a total of 46 tested pairs. Of the hit combinations, 14 of 17 (82.4%) scored as synergistic by Bliss independence at the same dosages as in the primary screen (P = 5.8 × 10−4, calculated from a binomial distribution null model where compound × antibiotic pairs randomly score positive with probability P = 19/46, the fraction positive of total tested pairs) (Fig. 4A, SI Appendix, Figs. S17 and S18, and Dataset S4). We investigated whether differences between primary-screening Bliss scores and 96-well plate Bliss scores could be explained by physicochemical properties of compounds, but did not find any systematic effects (SI Appendix, Fig. S18 and Dataset S4).
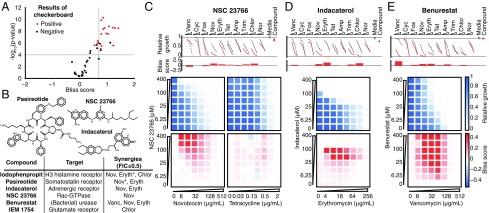
Fig. 4.
Validation of hits from primary screen. (A) In eight-point checkerboard assays with E. coli, we tested a total of 46 compound × antibiotic combinations, of which 17 (11 distinct compounds) scored as hits in the primary screen (SI Appendix, Figs. S17 and S18 and Dataset S4). Combinations that scored positive (red) and negative (black) for Bliss synergy are plotted according to results from the primary screen. Gray dotted lines indicate primary screening thresholds. (B) Target, status, antibiotic synergy set (by FIC index method), and selected structures of validated hits (SI Appendix, Figs. S17, S19, and S20 and Dataset S3). The first four compounds are from the full-scale phase; the last two are from the pilot phase. Antibiotics marked with an asterisk represent additional synergies revealed in validation. Structures were rendered in ChemDraw from SMILES strings. (C, Top) Primary screening data and calculated Bliss scores for NSC 23766. Growth in presence of compounds alone (Compound, indicated by red dots) relative to the absence of antibiotic and compound (Media, indicated by gray dots) is shown. (C, Bottom) The 96-well plate checkerboard assay of NSC 23766 × novobiocin (positive for synergy) (Left) and NSC 23766 × tetracycline (negative for synergy) (Right). For each checkerboard assay in C–E, the relative growth values and Bliss scores are shown (color scaling is indicated at Far Right). (D) Primary screening and checkerboard data for indacaterol × erythromycin (positive for synergy). (E) Primary screening and checkerboard data for benurestat × vancomycin (positive for synergy). Amp, ampicillin; Chlor, chloramphenicol; Cyc, cycloserine; Eryth, erythromycin; Fos, fosfomycin; Nor, norfloxacin; Nov, novobiocin; Tet, tetracycline; Trim, trimethoprim; Vanc, vancomycin.
After applying the more stringent FIC synergy criterion (FIC index ≤0.5) to each checkerboard, we identified six compounds among our hits with synergies with at least one antibiotic (four from full-scale phase, two from pilot phase; see SI Appendix, Figs. S17, S19, and S20). For two hit compounds, we identified additional synergies beyond what was detected in the primary screen, and upon further inspection, we found these additional compound × antibiotic pairs scored close to the thresholds applied in the primary screen (Fig. 4 A and B, SI Appendix, Fig. S17, and Dataset S4). Notably, we found no previous indication of antibacterial activity for five of these six compounds, which constitute a range of chemical structures, characteristics, and known biochemical targets (Fig. 4B). Comparing the primary screening data for each hit across the full panel of antibiotics shows some commonalities and differences that may provide clues as to mechanism (Fig. 4 C–E and Dataset S3) (34). For example, many hits showed common interactions with novobiocin and erythromycin, but divergent effects with vancomycin, ranging from strong synergy to strong antagonism (Fig. 4 C–E).
Conclusion
Here, we demonstrated a nanoliter droplet combinatorial drug screening platform and applied it at scale to discover novel potentiators of antibiotics from a drug repurposing library. Exploring the mechanism of action of these potentiators may guide future efforts to engineer antibiotic adjuvants, an important strategy in the fight against antibiotic resistance in gram-negative pathogens (33–35). By replacing robotic liquid handling with spontaneous random assembly of compound combinations in nanoliter droplets at a large scale, the logistical complexity of our combinatorial screen was reduced such that we could complete this screen using only manual pipetting at the macroscale (SI Appendix, Fig. S3) in just 10 d. This platform is compatible with commercially available laboratory equipment already present in many academic and industrial life science research facilities. Other groups have already demonstrated successful droplet-based culture of a wide range of organisms, including human cell lines (12, 15, 18, 19), and we expect that our platform can be developed to support many types of phenotypic and biochemical assays. The use of optical microscopy for assay readout facilitates extension to a variety of disease-specific models and imaging assays, including gene expression reporters and high-content cell imaging (12, 15). While much work remains, our platform represents an important tool to leverage drug combinations for chemical biology and therapeutics discovery.
Materials and Methods
SI Appendix, Table S2 includes a summary of parameters relevant to methods.
Microwell Array Chip Design and Fabrication.
Each chip (6.2 × 7.2 × 0.64 cm) had 43,000 microwells (two circles with a diameter of 148.6 μm, set at 10% overlap; 100 to 120 μm in height; 50-μm spacing). Silicon wafer molds were created by photolithography (SU8-2050; Microchem). Chips were fabricated by soft lithography from polydimethylsiloxane (Dow Corning Sylgard) and coated with 1.5-μm parylene C (Paratronix).
Cell Culture Preparation.
Plasmid-borne constitutive GFP-expressing strains of S. aureus (LB media), P. aeruginosa PAO1 [cation-adjusted Mueller–Hinton broth (CAMHB) media], and E. coli K-12 MG1655 (CAMHB media) were used. Overnight cultures were diluted 1:1,000 and, upon reaching log phase, normalized in fresh media to OD600 of 0.03 to 0.04 (∼10 cells per 1-nL droplet).
Fluorescence Encoding.
Each input was premixed with a unique ratio of three fluorescent encoding dyes (Alexa Fluor 555, 594, and 647; Thermo-Fisher Scientific; final concentration 1 µM).
Fluorescence Microscopy.
Decoding droplet contents and measuring growth responses were performed on a Nikon Ti-E inverted epifluorescence microscope with excitation from a Lumencor Sola LED illuminator (100% power setting). Images were collected by a Hamamatsu ORCA-Flash 4.0 camera at 6.5 μm per pixel resolution (2× or 4× magnification). Droplet contents were decoded by collecting images in three fluorescence channels corresponding to excitation from the three respective dyes and processed using custom software (SI Appendix, Fig. S2). GFP signal was measured from each microwell at the assay end point. The time required for imaging (∼12 min) was comparable to the scan time, enabling a staggered protocol where each chip was set up while the previous chip was scanning. See SI Appendix, Supplementary Materials and Methods for details on how images were analyzed.
Microwell Array Chip Operation.
A 20 μL aliquot of each input was transferred from a 96-well plate by micropipette to a Bio-Rad QX200 cartridge and emulsified into 20,000 1-nL droplets (continuous phase: fluorocarbon oil 3M Novec 7500, 0.5 to 2% wt/wt RAN Biotech 008-FluoroSurfactant) using the Bio-Rad QX200 instrument or a custom aluminum pressure manifold (Fig. 1A) (20 min to process a 96-well plate).
To set up a chip (total setup time 30 min, with throughput of 18 chips per day by staggering the protocol), the chip was placed inside an acrylic assembly (10 min) and suspended over a hydrophobic glass slide (1.2-mm thickness, Aquapel treated; Brain Research Laboratories) by plastic spacers (250-μm height) (SI Appendix, Fig. S1 and Movies S1 and S2), creating a gap that was then filled with fluorous oil. Using a micropipette, a fraction (e.g., 4 μL) of each set of droplets was pooled into a single well/tube (5 min) (∼200,000 droplets total), and injected into the chip while adding oil to sweep away excess droplets (5 min) (Fig. 1 B and C and Movies S1 and S2). Lastly, the chip was washed with oil (0% wt/wt surfactant), the spacers were removed, and the chip was mechanically clamped to the glass (Fig. 1B).
The chip was imaged by epifluorescence microscopy to identify the fluorescence barcodes (12 min, standard-size chip). The droplets were merged via an AC electric field [4.5 MHz, 10,000- to 45,000-V source underneath the glass supplied by a corona treater (Electro-Technic Products), ∼10-s exposure)] (Movie S3).
The cells were incubated (37 °C, 7 h, without shaking) and their growth was assayed by constitutive GFP fluorescence (Fig. 1D), starting at ∼10 cells per droplet to a saturation of 103 to 104 cells per droplet.
Antibiotic Potentiation Screening Logistics.
For all screening, each chip received droplets containing a total of 64 inputs, with 32 held constant (set 1: 10 antibiotics at three concentrations and controls) and 32 varied across chips (set 2: compounds and controls) (Fig. 3A). See SI Appendix, Supplementary Materials and Methods for a comprehensive breakdown. All drug responses were measured in E. coli K-12 MG1655 (see Cell Culture Preparation above), by evaluating the median GFP intensity across all microwells containing a given droplet pair. In the full-scale phase of screening, chip quality and assay performance were also scored (SI Appendix, Supplementary Materials and Methods).
Bliss Synergy Scoring.
For each compound × antibiotic (single dosage), we computed the deviation of the observed growth inhibition (fAB) from the expectation from Bliss independence (fA + fB − fA fB) (29), where fA and fB represent the growth inhibition of the antibiotic alone and the compound alone, respectively. For each compound, we summed this metric across the three antibiotic dosages to yield a final metric, the Bliss score. To estimate the SE, we computed the SD of Bliss scores obtained by bootstrapping (100 iterations). We then computed a test statistic (Bliss score/SE), modeled with a t distribution fit to blank negative controls (SI Appendix, Fig. S15). Compounds with an fB value >80% were removed from analysis (Dataset S5). See SI Appendix, Supplementary Materials and Methods for details on the estimation of fA, fB, and fAB values and SEs.
Checkerboard Validation.
Compounds were resupplied and tested against E. coli K-12 MG1655 in 64-point matrix checkerboard assays using 96-well V-bottom plates (Costar), in CAMHB media and 2% DMSO (final volume 100 μL), measured at a single end point (7 h, 37 °C, 220 rpm) by GFP signal using a plate reader (SpectraMax; Molecular Devices).
We estimated Bliss synergy using the same method and compound (100 μM) and antibiotic dosages (SI Appendix, Table S1) as in primary screening, scoring positive if the Bliss score summed across antibiotic doses was ≥0.4 (Fig. 4A, SI Appendix, Fig. S18, and Dataset S4).
Fractional Inhibitory Concentration Determination.
We measured the minimum inhibitory concentration (MIC) of antibiotic A (MICA) and compound B (MICB) individually as the first well in the dosage series with <10% growth. For a well in the matrix at dosage point (A: x, B: y) with <10% growth, we calculated FIC index = x/MICA + y/MICB. The interaction was classified as synergistic if the minimum FIC index was ≤0.5 (30).
Compound Exchange Between Microwells.
Source droplets (resorufin, 10 μM in CAMHB) and sink droplets (fluorescein, 5 μM, CAMHB) were made in Bio-Rad QX200 cartridges using 2% wt/wt RAN Biotechnologies 008-FluoroSurfactant in Novec 7500 fluorous oil (3M). Equal volumes of droplets were pooled by micropipette in a microcentrifuge tube (5 min) and loaded into the chip (5 min) (Fig. 2A). We washed the chip with oil containing 2%, 0.5%, or 0% wt/wt surfactant and mechanically clamped the chip to the glass (SI Appendix, Fig. S1). To measure interwell exchange, we compared the mean resorufin signal of source-only wells and sink-only wells (normalized to their sum) with resorufin signal of source droplets and sink droplets in source + sink wells (normalized to their sum) (Fig. 2 B–D).
Supplementary Material
Acknowledgments
We thank Navpreet Ranu and David Feldman for early discussions about microfluidic and encoding strategies; Deepan Thiruppathy and Jameson Kief for assistance; Stew Fisher, Jon Stokes, Jason Yang, and Wes Chen for discussions; members of the Hung laboratory for bacterial samples and discussions; Chris Emig and Tommy Moriarty for assistance with custom pressure manifolds; Josh Bittker, Samuel Figueroa-Lazú, and Anita Vrcic for compound library support; Scott Sassone for video production; and the jupyter, numpy, scipy, scikit-image, scikit-learn, and pandas development teams. This work was supported, in part, by the National Science Foundation Graduate Research Fellowship Program (to A.K. and J.K.), the MIT Institute for Medical Engineering and Science Broshy Fellowship (to A.K.), a Career Award at the Scientific Interface from the Burroughs Wellcome Fund (to P.C.B.), an MIT Deshpande Center Innovation Grant, a Scialog seed grant from the Gordon and Betty Moore Foundation and the Research Corporation for Science Advancement, and a Bridge Project grant from the Dana Farber/Harvard Cancer Center and the Koch Institute for Integrative Cancer Research at MIT.
Footnotes
Conflict of interest statement: Broad Institute and MIT may seek to commercialize aspects of this work; related applications for intellectual property have been filed.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802233115/-/DCSupplemental.
References
- 1.Strebhardt K, Ullrich A. Paul Ehrlich’s magic bullet concept: 100 years of progress. Nat Rev Cancer. 2008;8:473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 3.Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 4.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 5.Roemer T, Boone C. Systems-level antimicrobial drug and drug synergy discovery. Nat Chem Biol. 2013;9:222–231. doi: 10.1038/nchembio.1205. [DOI] [PubMed] [Google Scholar]
- 6.Lehár J, et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol. 2009;27:659–666. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Vilar S, Tatonetti NP. High-throughput methods for combinatorial drug discovery. Sci Transl Med. 2013;5:205rv1. doi: 10.1126/scitranslmed.3006667. [DOI] [PubMed] [Google Scholar]
- 9.Borisy AA, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci USA. 2003;100:7977–7982. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan X, et al. Systematic identification of synergistic drug pairs targeting HIV. Nat Biotechnol. 2012;30:1125–1130. doi: 10.1038/nbt.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Severyn B, et al. Parsimonious discovery of synergistic drug combinations. ACS Chem Biol. 2011;6:1391–1398. doi: 10.1021/cb2003225. [DOI] [PubMed] [Google Scholar]
- 12.Konry T, Golberg A, Yarmush M. Live single cell functional phenotyping in droplet nano-liter reactors. Sci Rep. 2013;3:3179. doi: 10.1038/srep03179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Churski K, et al. Rapid screening of antibiotic toxicity in an automated microdroplet system. Lab Chip. 2012;12:1629–1637. doi: 10.1039/c2lc21284f. [DOI] [PubMed] [Google Scholar]
- 15.Baret JC, Beck Y, Billas-Massobrio I, Moras D, Griffiths AD. Quantitative cell-based reporter gene assays using droplet-based microfluidics. Chem Biol. 2010;17:528–536. doi: 10.1016/j.chembiol.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Baraban L, et al. Millifluidic droplet analyser for microbiology. Lab Chip. 2011;11:4057–4062. doi: 10.1039/c1lc20545e. [DOI] [PubMed] [Google Scholar]
- 17.Miller OJ, et al. High-resolution dose-response screening using droplet-based microfluidics. Proc Natl Acad Sci USA. 2012;109:378–383. doi: 10.1073/pnas.1113324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouzes E, et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci USA. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clausell-Tormos J, et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 20.MacBeath G, Koehler AN, Schreiber SL. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J Am Chem Soc. 1999;121:7967–7968. [Google Scholar]
- 21.Gosalia DN, Diamond SL. Printing chemical libraries on microarrays for fluid phase nanoliter reactions. Proc Natl Acad Sci USA. 2003;100:8721–8726. doi: 10.1073/pnas.1530261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ismagilov RF, Ng JMK, Kenis PJA, Whitesides GM. Microfluidic arrays of fluid-fluid diffusional contacts as detection elements and combinatorial tools. Anal Chem. 2001;73:5207–5213. doi: 10.1021/ac010502a. [DOI] [PubMed] [Google Scholar]
- 23.Fradet E, Bayer C, Hollfelder F, Baroud CN. Measuring fast and slow enzyme kinetics in stationary droplets. Anal Chem. 2015;87:11915–11922. doi: 10.1021/acs.analchem.5b03567. [DOI] [PubMed] [Google Scholar]
- 24.Zagnoni M, Cooper JM. On-chip electrocoalescence of microdroplets as a function of voltage, frequency and droplet size. Lab Chip. 2009;9:2652–2658. doi: 10.1039/b906298j. [DOI] [PubMed] [Google Scholar]
- 25.Skhiri Y, et al. Dynamics of molecular transport by surfactants in emulsions. Soft Matter. 2012;8:10618. [Google Scholar]
- 26.Gruner P, et al. Controlling molecular transport in minimal emulsions. Nat Commun. 2016;7:10392. doi: 10.1038/ncomms10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan M, et al. Fluorinated pickering emulsions impede interfacial transport and form rigid interface for the growth of anchorage-dependent cells. ACS Appl Mater Interfaces. 2014;6:21446–21453. doi: 10.1021/am506443e. [DOI] [PubMed] [Google Scholar]
- 28.Woronoff G, et al. New generation of amino coumarin methyl sulfonate-based fluorogenic substrates for amidase assays in droplet-based microfluidic applications. Anal Chem. 2011;83:2852–2857. doi: 10.1021/ac200373n. [DOI] [PubMed] [Google Scholar]
- 29.Bliss CI. The calculation of microbial assays. Bacteriol Rev. 1956;20:243–258. doi: 10.1128/br.20.4.243-258.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 31.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 32.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 33.Ejim L, et al. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol. 2011;7:348–350. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 34.Farha MA, Brown ED. Chemical probes of Escherichia coli uncovered through chemical-chemical interaction profiling with compounds of known biological activity. Chem Biol. 2010;17:852–862. doi: 10.1016/j.chembiol.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Stokes JM, et al. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat Microbiol. 2017;2:17028. doi: 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corsello SM, et al. The drug repurposing hub: A next-generation drug library and information resource. Nat Med. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburn TT, Thor KB. Drug repositioning: Identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.