Significance
The first phosphodiester linkage made during DNA replication is in the context of a short RNA primer synthesized by the dedicated RNA polymerase, primase. Primase, after synthesizing a defined short oligoribonucleotide, then transfers this primer to a DNA polymerase for extension. The mechanisms by which primer length is constrained remain elusive. Here, we dissect the basis of “counting” by an archaeal primase and propose a caliper-like mechanism to account for length determination.
Keywords: Archaea, primase, DNA replication, DNA polymerase, Sulfolobus
Abstract
The cellular replicative DNA polymerases cannot initiate DNA synthesis without a priming 3′ OH. During DNA replication, this is supplied in the context of a short RNA primer molecule synthesized by DNA primase. The primase of archaea and eukaryotes, despite having varying subunit compositions, share sequence and structural homology. Intriguingly, archaeal primase has been demonstrated to possess the ability to synthesize DNA de novo, a property shared with the eukaryotic PrimPol enzymes. The dual RNA and DNA synthetic capabilities of the archaeal DNA primase have led to the proposal that there may be a sequential hand-off between these synthetic modes of primase. In the current work, we dissect the functional interplay between DNA and RNA synthetic modes of primase. In addition, we determine the key determinants that govern primer length definition by the archaeal primase. Our results indicate a primer measuring system that functions akin to a caliper.
DNA replication is critically dependent on the synthesis of short RNA primers by DNA primase (1). An RNA primer is required to initiate leading strand synthesis, and primers are also required for every Okazaki fragment synthesized on the lagging strand. Typically, primase initiates synthesis, extends the primer to a defined length of between 9 and 16 nt, and then disengages to allow association of a DNA polymerase with the primer’s 3′-OH end. In the hyperthermophilic archaeon, Sulfolobus solfataricus, DNA primase is a heterotrimeric assembly, PriSLX (2, 3). The PriS and PriL subunits bear clear sequence homology to the eukaryotic PriS and PriL orthologs (4, 5). PriX is essential for viability, but has a restricted phyletic distribution and possesses no clear sequence homology to the eukaryotic subunits (2). However, despite the lack of sequence conservation, PriX possesses functionally relevant structural homology to the N-terminal portion of the C-terminal domain of eukaryotic PriL (PriL-CTD) (2, 3).
PriS contains the active site of the primase, which represents the site of primer elongation (6, 7). The PriL-CTD plays a critical role in primer synthesis by enabling primase to bind simultaneously a second nucleotide and to synthesize the first di-nucleotide linkage (8, 9). We have recently demonstrated that PriX also possesses nucleotide triphosphate binding activity (3). Mutations that reduce nucleotide binding by PriX (D70A, R72A, or R74A), when incorporated into reconstituted PriSLX heterotrimers, impair the ability of primase to initiate primer synthesis de novo. Notably, however, the nucleotide-binding and initiation-defective PriX-mutant primase retains the ability to elongate preformed oligo ribonucleotide primers. Thus, the nucleotide-binding site in PriX serves as an initiation site for primer synthesis, delivering the initiating nucleotide to the active site within PriS for formation of the initial di-nucleotide. Di-nucleotide formation is the rate-limiting step in primer formation. Importantly, mutation of an analogous residue (R306 in human PriL) in the structurally related N-terminal portion of the C-terminal domain of eukaryotic PriL causes an analogous initiation defect, suggesting conservation of the initiation mechanism in archaeal and eukaryotic domains of life (7, 10).
Structural studies by Tahirov and colleagues have revealed that the PriX-related portion of eukaryotic PriL can interact with the triphosphate at the 5′-end of a RNA oligonucleotide in the context of a RNA/DNA heteroduplex (10). Our work with archaeal PriSLX has demonstrated that PriX solely contacts the nucleotide triphosphate via the phosphate moieties. In addition, PriX’s position relative to the PriS catalytic site is only loosely determined, as judged by cross-linking studies. We also observed that although PriSLX assemblies containing PriX D70A, R72A, or R74A are elongation competent, they generate a distinct distribution of product sizes compared with those synthesized by the wild-type enzyme (10). Taken together, these observations lead us to speculate that PriX, in addition to being crucial for initiation of primer synthesis, may also play an important role in determining primer length.
In eukaryotes, PriS and PriL are contained within a heterotetrameric complex with DNA polymerase α and its B subunit (11). Thus, once a primer has reached its required length, there is a tightly coupled internal hand-off to DNA polymerase alpha for primer extension with DNA (12). Archaea do not possess orthologs of Polα or the B subunit; however, it has been demonstrated that archaeal primases are able to synthesize DNA as well as RNA (4, 13). This has led to the proposal that there may be an internal switch-off between RNA and DNA synthetic modes in the archaeal enzyme (2).
In the current work, we reveal that, at physiologically relevant ratios of ribo- and deoxyribo-nucleotides, primase functions as a RNA synthetic enzyme. Further, we observe that the interaction of PriX with the 5′ triphosphate of the initiating nucleotide, in addition to being essential for initiation, plays key roles in governing the length of primers synthesized by primase and in stabilizing the primer on the DNA template before hand-off to the DNA polymerase.
Results
Nucleotide Triphosphate Binding by Primase Subunits.
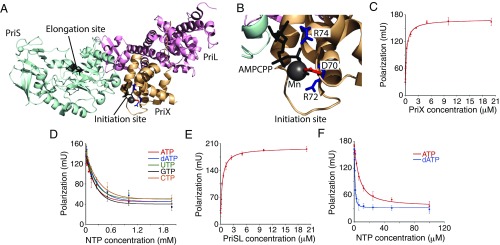
The S. solfataricus primase contains two nucleotide binding sites: an initiation site in PriX and an elongation site in PriS (Fig. 1A). The crystal structure of PriSLX bound to the ATP analog AMPCPP (3) reveals that the principal contacts between PriX and the nucleotide are mediated by the triphosphate moiety and the side chains of PriX amino acids D70, R72, and R74 (Fig. 1B), with no direct contacts between PriX and either sugar or base of the initiating nucleotide. Furthermore, previous studies have demonstrated that the archaeal primases can act as both DNA and RNA polymerases (2, 4, 13). Accordingly, we tested whether there was any intrinsic preference for deoxyribonucleotide triphosphates (dNTP) or ribonucleotide triphosphate (rNTP) binding in the initiation site of the primase. We performed fluorescence anisotropy binding assays, using purified recombinant PriX and fluorescein-12-ATP, and confirmed our previous observation of interaction between these two molecules (Fig. 1C). Next, we performed competition assays in which increasing concentrations of unlabeled rNTPs or dATP were titrated into the assay at fixed PriX and fluorescein-12-ATP concentrations (Fig. 1D). No significant difference was observed between the abilities of any of the four rNTPs or dATP to compete for binding of the fluorescein-12-ATP.
Fig. 1.
Nucleotide triphosphate binding by primase subunits. (A) Crystal structure of PriSLX. The two nucleotide-binding sites in Sulfolobus primase are indicated. The figure was generated using PDB file 5OF3.pdb, using Pymol. (B) Close view of the PriX nucleotide-binding residues. (C) ATP binding by PriX, measured by fluorescence polarization. PriX concentration ranges from 0 to 20 µM. (D) Nucleotide competition binding by PriX. Fluorescein-12-ATP and PriX are at fixed concentrations of 20 nM and 2.5 µM, respectively. Concentrations of unlabeled rNTPs/dATP range from 0 to 2 mM. (E) ATP binding by PriSL, measured by fluorescence polarization. PriSL concentration ranges from 0 to 20 µM. (F) Nucleotide competition binding by PriSL. Fluorescein-12-ATP and PriSL are at fixed concentrations of 20 nM and 2 µM, respectively. Concentrations of unlabeled ATP/dATP range from 0 to 100 µM. The experiments in C–F were performed in triplicate; the error bars are the standard deviation.
Next, we tested whether the elongation site of the primase, located in PriS, had nucleotide selectivity. To do this, we performed the ATP binding assay with reconstituted primase lacking the PriX subunit (3). We could readily detect binding of fluorescein-12-ATP to PriS with a Kd of ∼440 nM (Fig. 1E). Next, we performed competition assays with ATP and dATP. Strikingly, we observed that dATP outcompetes ATP for fluorescein-12-ATP binding to PriS (Fig. 1F). We also performed analogous assays using PriSLX containing an R72A substitution in PriX. This substitution abrogates nucleotide binding by the initiation site in PriX, leaving the only functional binding site in the catalytic center of PriS. Similar results were obtained with this mutant (SI Appendix, Fig. S1).
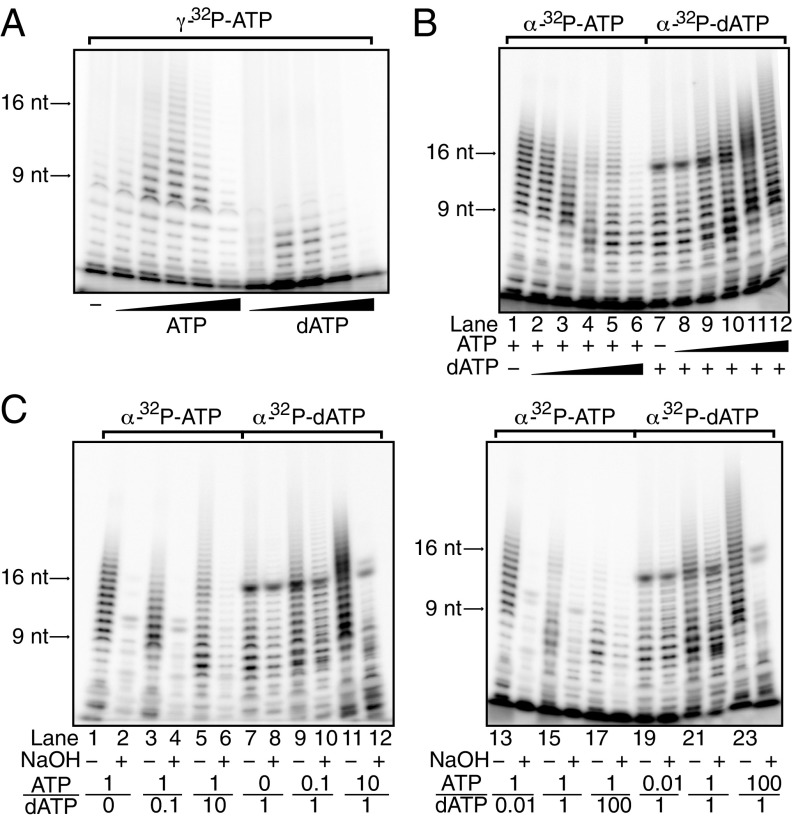
Our nucleotide binding studies were performed using isolated proteins or with the initiation-defective PriX R72A-containing primase. We were concerned that the observed nucleotide preferences might not accurately represent the architecture, and thus discriminatory power, of the juxtaposed PriX initiation and PriS elongation sites during primer synthesis. To address whether there is a preference for initiation with dATP or rATP during de novo primer synthesis on a poly(dT) template, we performed primase assays using gamma-32P-labeled ATP. As the beta and gamma phosphates of the incoming (d)NTPs are removed during nucleic acid polymerization, retention of gamma-[32P]ATP serves as an indicator for the nature of the 5′ end of the primer. We therefore performed assays with either nonradiolabeled ATP or dATP to compete for the gamma-[32P]ATP in initiation. As can be seen in Fig. 2A, inclusion of either nucleotide competes for overall yield of radiolabeled primer with similar efficiencies, indicating that there is little discrimination in the initiation site for ATP over dATP. Notably, however, the inclusion of dATP resulted in a general decrease of primer length, suggesting that dATP, although binding with higher affinity than ATP to the elongation site (Fig. 1F), does so in a manner that is suboptimal for phosphodiester bond formation.
Fig. 2.
Poly(dT) template primer synthesis by primase using ATP or dATP. (A) ATP/dATP initiation competition assay. Each reaction contained 165 nM [γ-32P] ATP and 10-fold dilutions of cold ATP or dATP ranging from 100 nM to 1 mM. Selected product sizes are indicated on the left. (B) ATP/dATP elongation competition assay. Reactions from lanes 1–6 contained 33 nM [α-32P] ATP, 10 µM cold ATP, and 10-fold dilutions of cold dATP ranging from 100 nM to 1 mM; reactions from lane 7–12 contained 33 nM [α-32P] dATP, 10 µM cold dATP, and 10-fold dilutions of cold ATP ranging from 100 nM to 1 mM. (C) ATP/dATP elongation competition assay followed by alkaline treatment. ATP:dATP concentration ratios are indicated at the bottom of the gel, where “1” represents a concentration of 10 µM.
To further investigate nucleotide preference during primer elongation, we performed primase assays using a poly(dT) template with varying relative concentrations of dATP or ATP. In addition, we monitored incorporation of either radio-labeled dAMP or AMP into product from α-32P-labeled dATP or ATP, respectively. As can be seen in Fig. 2B, reactions with ATP produce a majority of products in the size range of 9–20 nt. In contrast, reactions with solely dATP as substrate primarily generate a lower yield of predominantly shorter products (Fig. 2B, lane 7). We have recently determined that the intracellular concentration of ATP in Sulfolobus is at least 100-fold higher than that of dATP (14). In reactions with a 1:100 ratio of dATP:ATP, there is little effect on the incorporation of radiolabeled AMP into primers (Fig. 2B, lane 2). However, titration of dATP into reactions with radiolabeled ATP result in a dATP concentration-dependent reduction in primer size and yield. At nonphysiological 1:1 or greater ratios of dATP:ATP, the migration of the products is altered compared with the radiolabeled ATP-only lane, indicating the incorporation of dAMP into the radiolabeled AMP-containing products (Fig. 2B, lane 4–7).
In reactions with radiolabeled dATP, the inclusion of ATP results in an increase in the yield and size of the radiolabeled product species. We note that the appearance of hybrid dAMP•AMP-containing product species is detectable at a 10-fold molar excess of dATP over ATP (Fig. 2B, lane 9). Taken together, our data indicate that primase is capable of generating hybrid DNA•RNA primers. We next sought to test whether these molecules resulted from the sequential synthesis by primase of RNA followed by DNA. Accordingly, we monitored synthesis of DNA-containing primers with radiolabeled dATP in the presence of varying concentrations of ATP (Fig. 2C, lanes 7–12 and 19–24). We then divided reactions into two aliquots, one of which was treated with sodium hydroxide. The presence of base will catalyze hydrolysis via the 2′-OH group of incorporated AMPs. If the hybrid primers possessed a contiguous tract of RNA followed by one of DNA, we would anticipate base treatment to yield a clear ladder of DNA products shorter than the non-base-treated sample. In contrast, if AMP and dAMP moieties are randomly incorporated in the products of the primase reaction, we would expect a complex range of short products containing either purely DNA or DNA appended at the 3′ end with a ribonucleotide possessing a 2′ or 3′ phosphate. The complex pattern of products that we observe in Fig. 2C therefore supports the latter model of stochastic incorporation of dAMP and AMP during primer synthesis.
Next, we performed an analogous experiment monitoring RNA synthesis with radiolabeled ATP (Fig. 2C, lane 1–6 and 13–18). At physiologically relevant ATP:dATP concentrations, we observe near complete base-mediated hydrolysis of the RNA species (Fig. 2C, lane 13–14). Under nonphysiological conditions of 100-fold excess of dATP over ATP, we observe the existence of base-resistant species (Fig. 2C, lane 17–18). These can be explained by incorporation of radiolabeled AMP at the end of a tract of DNA. As described here, base-mediated cleavage will result in the tagging of the DNA molecule with a single radiolabeled ribonucleotide 2′ or 3′ phosphate. Taken together, we do not find any compelling evidence for an ordered sequential synthesis of first RNA and then DNA by the primase. Rather, at physiologically relevant relative concentrations of nucleotide triphosphates, we observe almost exclusively RNA products.
Primer Length Determination.
Primers synthesized by primase are restricted to a narrow size distribution. We wished to determine how the primase counts primer length. Two models could be envisaged. In the first, primer length determination is simply a manifestation of the inherent processivity of the enzyme; after initiation, the primase will synthesize for a short length and then stochastically dissociate from the primer•template. In the second model, the primase could contain an internal ruler that defines primer length.
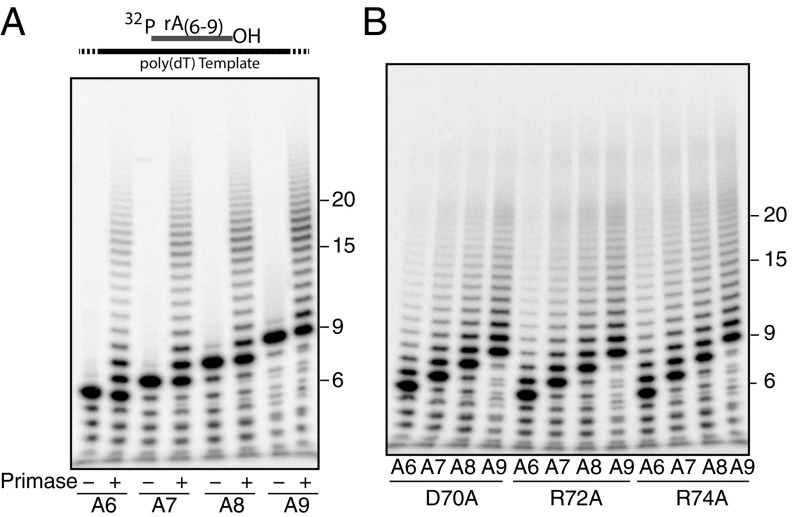
To test these models, we first supplied the primase with 5′-32P-labeled oligoribonucleotides of 6, 7, 8, or 9 nt in length annealed to a poly(dT) template. If the intrinsic processivity model dictates primer length, we would anticipate the primase extending for the same distance from each of these different-size primers, yielding increasing product sizes for longer oligoribonucleotides. In contrast, if the ruler model holds, then elongation from all four substrates should produce the same spectrum of products. As seen in Fig. 3A and SI Appendix, Fig. S2, the most abundant product of the various reactions is a +1 step; thereafter, product yield diminishes as the product grows longer until primers reach 13 nt in length, when we observe an increase in yield of products between 14 and 18 nt, followed by a reduction in the levels of larger products. This peak of products between 14 and 18 nt shows very similar distribution, regardless of the length of oligoribonucleotide priming the reaction, and thus is supportive of a ruler-like model in governing primer synthesis.
Fig. 3.
Poly(dT) template elongation synthesis with different primer length. (A) Different-sized primer elongation by wild-type primase. Substrate information is illustrated at the top. Primers were end-labeled with [γ-32P] ATP. Selected product sizes in nucleotides are indicated on the right. (B) Primer elongation by primase PriSLX containing the indicated PriX initiation site mutants (D70A, R72A, and R74A).
Given that PriX plays a role in delivering the initiating nucleotide to the primase active site, we speculated that PriX could retain a grip on the 5′ end of the growing RNA chain. Accordingly, we performed assays analogous to those described earlier with primase containing derivatives of PriX (D70A, R72A or R74A) that are impaired for nucleotide binding (Fig. 3B). Although these mutant forms of primase are impaired for initiation, they are fully capable of elongating preformed primers. Notably, reactions containing these primase mutants no longer generate the peak of products between 14 and 18 nt in length, suggesting Prix may play a role in determining primase product size distribution.
The Role of the 5′ Triphosphate in Primer Length Determination.
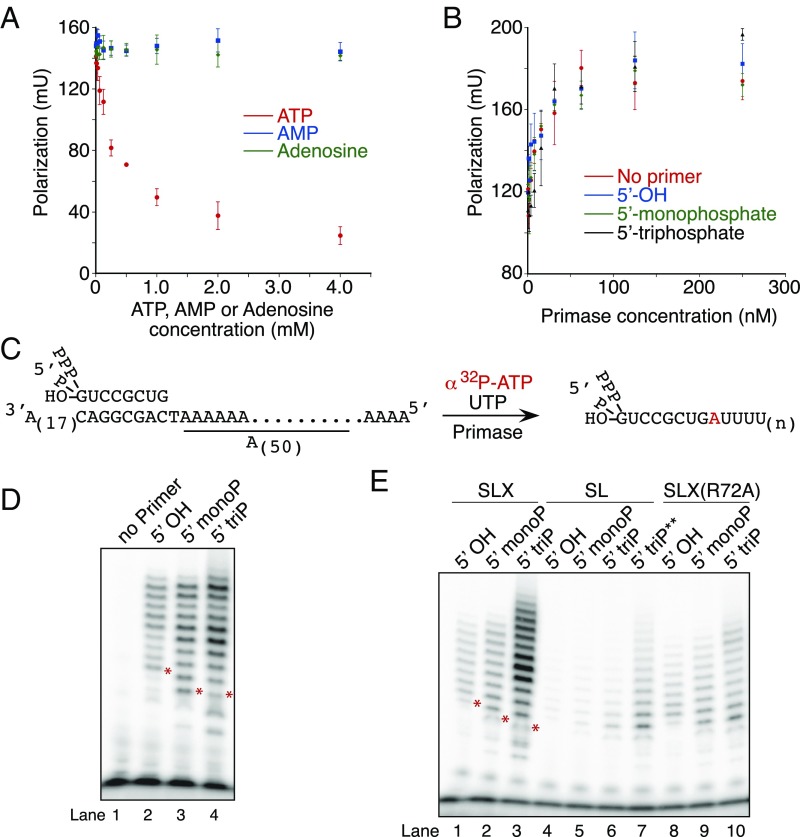
The change in primer product length observed with primase containing PriX mutants deficient in NTP binding could conceivably arise via a reduction in the affinity for the single-stranded DNA (ssDNA) substrate. Using fluorescence anisotropy assays, we were unable to detect any reduction in ssDNA binding between wild-type and mutant primase; indeed, the primase containing PriX D70A showed an increase in affinity for ssDNA (SI Appendix, Fig. S3). The experiments described in Fig. 3 were performed with 5′-32P monophosphate-labeled oligoribonucleotides to ensure we were solely visualizing the products of elongation rather than a mixture of initiation and elongation events. The crystal structure of PriX bound to the ATP analog AMPCPP reveals that PriX contacts with the nucleotide are principally with the 5′ phosphates (Fig. 1B; ref. 3). We tested the importance of these interactions for nucleotide binding by PriX, using fluorescence anisotropy measurements with fluorescein-12-ATP and performing competition experiments with ATP, AMP, or adenosine. As can be seen in Fig. 4A, although ATP competes for binding of fluorescein-12-ATP to PriX, neither AMP nor adenosine show any competition for binding over the concentration range tested.
Fig. 4.
The role of the 5′ triphosphate in primer length determination. (A) Nucleotide competition binding by PriX, measured by fluorescence polarization. Fluorescein-12-ATP and PriX are at fixed concentrations of 20 nM and 2.5 µM, respectively. Concentrations of unlabeled ATP, AMP, or adenosine range from 0 to 2 mM. (B) Primase substrate binding measured by fluorescence polarization. 5′ FAM-labeled template (with or without primer) is at fixed concentration of 10 nM. Primase concentrations range from 0 to 250 nM. The experiments in A and B were performed in triplicate; the error bars are the standard deviation. (C) Experimental design of the primase assay used to test the 5′ end effect on primer length determination. (D and E) Primer extension by wild-type primase, PriSL or PriSLXR72A. Note that the products run differently for 5′OH primer and 5′ monophosphate or triphosphate primers because of the charge difference. *The 9-nt product position for each primer is indicated. **PriSL concentration is 500 nM in this reaction.
In addition, we tested whether the chemical nature of the 5′ end of an eight-nucleotide oligoribonucleotide primer affected the affinity of the primase for interaction with a preformed RNA-DNA heteroduplex, using fluorescence anisotropy assays. We tested primase binding to heteroduplexes with RNA molecules with 5′-OH, 5′-monophosphate, or 5′-triphosphate, and also examined binding to single-stranded template DNA. As can be seen in Fig. 4B, there was no significant effect of the presence of RNA, nor of its 5′-end composition on the affinity of primase for the substrate.
To test the effect of the nature of the 5′ end of the primer on product yield and size, we devised a substrate in which the template possessed the sequence (3′-CAGGCGAC-5′) to which we could anneal a chemically synthesized ribo-octanucleotide with 5′ OH, 5′-monophosphate, or 5′-triphosphate. The first templating nucleotide available for extension is a thymidine, followed by a homopolymeric run of deoxyadenosines (Fig. 4C). This unique thymidine in the template allowed incorporation of alpha-32P-labeled AMP on +1 extension of the primer by primase, with subsequent elongation requiring uridine triphosphate only. Thus, to ensure we were uniquely labeling elongation products, we performed primase reactions with this substrate in the presence of ATP and UTP only. Radiolabeled product could, in principle, be generated by initiation with ATP from the unique thymidine in the template. However, in control experiments, we could not detect significant initiation at this site (Fig. 4D, lane 1); presumably, the presence of the RNA primer immediately adjacent to this residue sterically interfered with initiation at this site. In addition, the concentration of the radiolabeled ATP in these assays was 33 nM, whereas UTP was at 100 µM. As we have shown that PriX does not discriminate between NTPs (Fig. 1D), these conditions favor UTP’s interaction with the initiation site in PriX.
As can be seen in Fig. 4D, extension of primers possessing a 5′-OH results in a distribution of products ranging from 9 to 18 nt in length (Fig. 4D, lane 2). Extension of a primer with a 5′ monophosphate shows a modestly increased yield of product with a discernable enhancement of products of 13, 14, 17, and 18 nt in length (Fig. 4D, lane 3). The primer with a 5′-triphosphate gave rise to a further increased yield of products and a distinctive set of size distributions (Fig. 4D, lane 4). More specifically, the most abundant products were of 13 and 14 nt, with an additional secondary peak of products of 17 and 18 nt in length. To test the importance of PriX in determining the product size and yield, we performed assays with reconstituted primase lacking the PriX subunit (Fig. 4E). Similar to the PriX NTP binding site mutant-containing primase, the PriSL assembly is highly impaired relative to PriSLX in de novo initiation assays (SI Appendix, Fig. S4, see also ref. 2). In elongation assays, we observed that the activity of PriSL is significantly lower than that of the intact PriSLX (Fig. 4E, lanes 1–3 and 4–7), and in addition, the spectrum of product sizes shows little dependence on the nature of the 5′-end of the primer. We also tested the effect of the PriX R72A mutant in reconstituted PriSLX (Fig. 4E, lanes 8–10). Similar to the effect seen in absence of PriX, this mutant primase, compared with wild-type, no longer shows the peak of products between 12 and 14 nt with the 5′ triphosphate primer. We note that there is still a modest enhancement of products of 16 and 17 nt in reactions with the 5′ triphosphate.
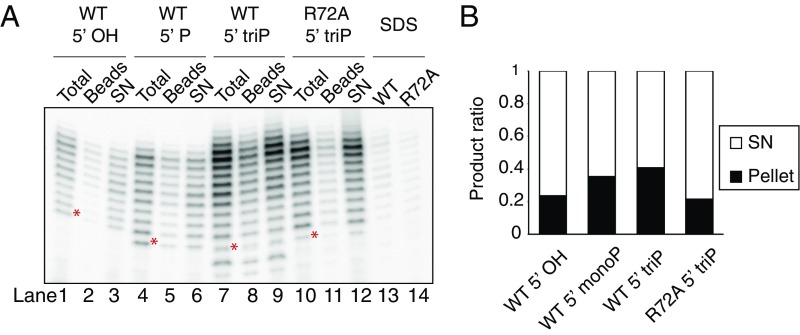
In our in vitro conditions, we observe a primer length distribution of between 9 and 20 nt. This in agreement with the range of primer sizes observed in vivo in archaea (15). Our in vitro assays are performed at 75 °C, which corresponds to the growth temperature of Sulfolobus. The predicted melting temperature of a 20-nt product in our elongation assay is 36 °C (16). Given the short length of primers, we wished to determine whether the primers we detect remain base-paired to the template strand. Accordingly, we immobilized the template DNA via a 5′ biotin moiety to streptavidin-coated magnetic beads. After performing primase assays, we removed an aliquot as “Total” (Fig. 5A), and boiled that in loading dye. We then recovered the template-associated material from the sample by magnetic separation at 75 °C and electrophoresed aliquots of supernatant and bead-associated material on a denaturing polyacrylamide gel (Fig. 5A and quantified in Fig. 5B). The proportion of bead-associated material increases as the 5′ moiety of the primer substrates changes from 5′-OH to monophosphate and to triphosphate. Furthermore, use of the primase with the PriX R72A substitution reduces the bead-retained material to a level equivalent to that in reactions with a 5′-OH RNA primer. Importantly, treatment of reactions with 1% SDS before pull down dramatically reduced the yield of bead-associated primers. These data indicate that interaction of the 5′ triphosphate with PriX helps to stabilize the RNA–DNA heteroduplex.
Fig. 5.
Primer-template duplex stabilized by primase. (A) Immobilized elongation assay with wild-type primase or PriSLXR72A. Beads, the product associated with the immobilized template; SN, the product in the supernatant. Lanes 13 and 14 show the bead-associated product after SDS treatment. (B) Quantified product distribution. For each reaction, total product intensity was normalized to 1. *The 9-nt product position for each primer is indicated.
Discussion
Our data demonstrate that Sulfolobus primase can synthesize DNA, RNA, and mixed DNA–RNA primers in vitro. It has previously been proposed that this promiscuity could reflect an ability of the archaeal primase to function akin to the eukaryotic DNA Pol α complex, which first synthesizes a short RNA primer and then extends it as DNA (2). By varying the relative concentrations of dATP and ATP in our assays, we reveal that, at physiologically relevant ratios of substrate, the primase is almost exclusively a RNA synthetic enzyme. Indeed, dATP exhibits an inhibitory effect on RNA synthesis, in agreement with dATP’s higher affinity for the elongation site. We propose that dATP adopts a configuration in the elongation site that blocks primer elongation. It is conceivable that this serves as a mechanism to prevent inappropriate DNA synthesis by primase. Without such a mechanism, unchecked DNA synthesis by primase, given its lack of proofreading function, could contribute a significant mutagenic load during DNA replication.
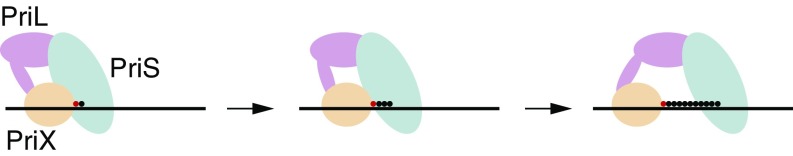
We have previously demonstrated that nucleotide binding by PriX plays a pivotal role in initiation of primer synthesis (3). More specifically, PriX interacts with the 5′ triphosphate of the initiating nucleotide, a chemical group that is retained during primer elongation. Here we reveal that ongoing interactions between PriX and the 5′ triphosphate play roles in several events during primer elongation. Interestingly, the nature of the 5′ end of the primer does not make discernable contributions to the overall affinity of primase to a primer•template complex. However, the presence of a 5′ triphosphate on the primer both elevates overall primer yield and modulates the size distribution of products. Our data thus indicate that PriX retains a hold of the 5′ end of the primer during elongation. As the 3′ end of the primer occupies the catalytic site in PriS during primer elongation, primase grips both ends of the growing primer. This suggests a simple model to govern primer length distribution, in which primase acts as a caliper to “measure” the length of the growing primer (Fig. 6). Primer length would therefore be dictated by the maximum length of RNA that can be accommodated between the PriX and PriS nucleotide binding sites. PriX shows structural and functional conservation with the N-terminal portion of the C-terminal domain of eukaryotic PriLs. Furthermore, structural data support interaction of this portion of eukaryal PriL with the 5′-end of RNA in a short heteroduplex (10). It is likely, therefore, that the caliper model will apply to both archaea and eukaryotes. At present, it is not known what conformation the primer•template heteroduplex adopts during synthesis. Formation of an A-form double helix favored by RNA•DNA hybrids would require considerable conformational changes in primase to accommodate ∼1.5 helical turns while retaining a grip on both ends of the RNA chain during synthesis of a 16-nt primer. We note that in our crystal structure of PriSLX bound to AMPPCP (3), the two nucleotide-binding sites in PriS and PriX are separated by ∼40 Å, a distance that could accommodate 15 bp of A-form RNA•DNA hybrid double helix.
Fig. 6.
Model of primer length determination by Sulfolobus primase. The initiating nucleotide, which is bound by PriX and retains its 5′ triphosphate, is shown as a red circle; subsequent nucleotides, added via the PriS polymerization active site, are shown as black circles. We propose that PriX remains engaged with the 5′ end of the growing RNA chain during primer synthesis. The primer length is thus defined by the maximum spatial distance that can be accommodated between the PriX and PriS nucleotide-binding sites.
Finally, we observe that, by holding on to the 5′ end of the primer, PriX contributes to the overall stability of the RNA•DNA hybrid. This will be particularly significant in Sulfolobus species, in which the growth temperature of the organisms far exceeds the melting temperature of the primer•template heteroduplex. Presumably, the primase, by interacting with its product heteroduplex, helps stabilize it in double-stranded form before facilitating hand-off to the replicative DNA polymerase holoenzyme.
Methods
Expression and Purification of Primase.
PriSLX complex was expressed and purified essentially as previously described (3). Cells expressing PriSL were lysed in 10 mM Hepes at pH 7.5, 150 mM NaCl, 14 mM β-mercaptoethanol, and EDTA-free protease inhibitor mini tablets (Thermo Fisher Scientific), using French press. The lysate was centrifuged at 34, 957 × g for 30 min, and the supernatant was heat-treated at 75 °C for 20 min before further centrifugation. The supernatant was purified over a HiTrip Heparin column (GE Healthcare), and the protein was eluted with a NaCl gradient from 150 to 1,000 mM in 10 mM Hepes at pH 7.5 buffer. The protein was further purified over a HiLoad 26/600 Superdex 200 column (GE Healthcare) in 10 mM Hepes at pH 7.5, 150 mM NaCl.
Cells expressing PriX were lysed in 10 mM Hepes at pH 7.5, 100 mM NaCl, and 1 mM DTT, using French press. The lysate was centrifuged at 34, 957 × g for 30 min, and the supernatant was heat-treated at 75 °C for 20 min before further centrifugation. The supernatant was purified over a HiTrip Heparin column, and the protein was eluted with a NaCl gradient from 150 mM to 1,000 mM in 10 mM Hepes at pH 7.5 buffer. The protein was further purified over a HiLoad 26/600 Superdex 200 column in 10 mM Hepes at pH 7.5, 150 mM NaCl.
Fluorescence Anisotropy.
Nucleotide binding and competition studies were performed in 25 mM Hepes 7.2, 150 mM NaCl, 2 mM MnCl2, 1 mM TCEP, and 0.1 mg/mL BSA with 20 nM fluorescein-12-ATP (PerkinElmer), and a serial dilution of PriX ranging from 0 to 20 µM concentration was assembled into 96-well plates. After incubation at 50 °C for 5 min, polarization was measured in a Synergy 2 plate reader.
Nucleic acid binding studies were performed in 50 mM Mes at pH 6.0, 10 mM MnCl2, 10 µM ZnCl2, 0.1 mg/mL BSA, 10 nM substrate (5′-FAM-AAAAAAAAAAATCAGCGGACAAAAAAAAAAAA-3′) with or without primer (5′-rGrUrCrCrGrCrUrG-3′, 5′-[phos]rGrUrCrCrGrCrUrG-3′ or 5′-[ppp]rGrUrCrCrGrCrUrG-3′), and a serial dilution of PriSLX ranging from 0 to 250 nM. Data were plotted using Kaleidagraph version 4.5. Further details are given SI Appendix, SI Materials and Methods.
Primase Assays.
Primase assays were performed as described (3) previously in 50 mM Mes-NaOH at pH 6.0, 10 mM MnCl2, 10 µM ZnCl2, and 0.1 mg/mL BSA. Further details are given SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
Work in S.D.B.’s lab is supported by The College of Arts and Sciences, Indiana University and NIH Grant 1R01GM125579-01. L.P. is supported by a Wellcome Trust Investigator Award (104641/Z/14/Z), and S.H. was a recipient of a PhD fellowship from the Boehringer-Ingelheim Fonds and awards from the Janggen-Pöhn-Stiftung and the Swiss National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806351115/-/DCSupplemental.
References
- 1.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Liu B, et al. A primase subunit essential for efficient primer synthesis by an archaeal eukaryotic-type primase. Nat Commun. 2015;6:7300. doi: 10.1038/ncomms8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holzer S, Yan J, Kilkenny ML, Bell SD, Pellegrini L. Primer synthesis by a eukaryotic-like archaeal primase is independent of its Fe-S cluster. Nat Commun. 2017;8:1718. doi: 10.1038/s41467-017-01707-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lao-Sirieix SH, Bell SD. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J Mol Biol. 2004;344:1251–1263. doi: 10.1016/j.jmb.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Lao-Sirieix SH, Nookala RK, Roversi P, Bell SD, Pellegrini L. Structure of the heterodimeric core primase. Nat Struct Mol Biol. 2005;12:1137–1144. doi: 10.1038/nsmb1013. [DOI] [PubMed] [Google Scholar]
- 6.Baranovskiy AG, et al. Crystal structure of the human primase. J Biol Chem. 2015;290:5635–5646. doi: 10.1074/jbc.M114.624742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kilkenny ML, Longo MA, Perera RL, Pellegrini L. Structures of human primase reveal design of nucleotide elongation site and mode of Pol α tethering. Proc Natl Acad Sci USA. 2013;110:15961–15966. doi: 10.1073/pnas.1311185110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol. 2007;14:875–877. doi: 10.1038/nsmb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Baranovskiy AG, Tahirov TH, Pavlov YI. The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase α-primase complex. J Biol Chem. 2014;289:22021–22034. doi: 10.1074/jbc.M114.570333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baranovskiy AG, et al. Insight into the human DNA primase interaction with template-primer. J Biol Chem. 2016;291:4793–4802. doi: 10.1074/jbc.M115.704064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellegrini L. The Pol α-primase complex. Subcell Biochem. 2012;62:157–169. doi: 10.1007/978-94-007-4572-8_9. [DOI] [PubMed] [Google Scholar]
- 12.Sheaff RJ, Kuchta RD, Ilsley D. Calf thymus DNA polymerase alpha-primase: “communication” and primer-template movement between the two active sites. Biochemistry. 1994;33:2247–2254. doi: 10.1021/bi00174a035. [DOI] [PubMed] [Google Scholar]
- 13.Bocquier AA, et al. Archaeal primase: Bridging the gap between RNA and DNA polymerases. Curr Biol. 2001;11:452–456. doi: 10.1016/s0960-9822(01)00119-1. [DOI] [PubMed] [Google Scholar]
- 14.Liew LP, et al. Hydroxyurea-mediated cytotoxicity without inhibition of Ribonucleotide reductase. Cell Rep. 2016;17:1657–1670. doi: 10.1016/j.celrep.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsunaga F, Norais C, Forterre P, Myllykallio H. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 2003;4:154–158. doi: 10.1038/sj.embor.embor732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumousseau M, Rodriguez N, Juty N, Le Novère N. MELTING, a flexible platform to predict the melting temperatures of nucleic acids. BMC Bioinformatics. 2012;13:101. doi: 10.1186/1471-2105-13-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.