Significance
CD8+ T cell recognition of tumor cells is typically based on the detection of specific MHC–peptide complexes, while natural killer (NK) cell recognition relies on the detection of NK ligands by an array of NK receptors. In this study we uncovered a distinct small population of CD8+ T cells expressing NKp30, a potent activating NK receptor, on peripheral blood from healthy donors. Those innate-like CD8+ T cells, coexpressing FcεRIγ and PZLF, could be generated and differentiated from a population of peripheral blood CD8+ T cells as result of IL-15–driven acquisition of broad innate features. This unique effector population could potently control the growth of tumors in an NK-like manner, making it promising for cancer immunotherapy by its dual target-recognition potential.
Keywords: CD8+ T cells, NKp30, innate, IL-15, anti-tumor
Abstract
CD8+ T cells are considered prototypical cells of adaptive immunity. Here, we uncovered a distinct CD8+ T cell population expressing the activating natural killer (NK) receptor NKp30 in the peripheral blood of healthy individuals. We revealed that IL-15 could de novo induce NKp30 expression in a population of CD8+ T cells and drive their differentiation toward a broad innate transcriptional landscape. The adaptor FcεRIγ was concomitantly induced and was shown to be crucial to enable NKp30 cell-surface expression and function in CD8+ T cells. FcεRIγ de novo expression required promoter demethylation and was accompanied by acquisition of the signaling molecule Syk and the “innate” transcription factor PLZF. IL-15–induced NKp30+CD8+ T cells exhibited high NK-like antitumor activity in vitro and were able to synergize with T cell receptor signaling. Importantly, this population potently controlled tumor growth in a preclinical xenograft mouse model. Our study, while blurring the borders between innate and adaptive immunity, reveals a unique NKp30+FcεRIγ+CD8+ T cell population with high antitumor therapeutic potential.
Immune cells are classically distinguished between adaptive and innate. While adaptive immune cells rely on high specificity, clonal expansion, and memory formation, innate cells are in the first line of defense, displaying rapid immune responses. CD8+ T cells express specific T cell receptors (TCRs), and their effector functions classically are strictly based on the recognition of MHC class I presenting specific antigens. In contrast, natural killer (NK) cell function is regulated by a balance of activating and inhibitory signals delivered by their receptors (1). Inhibitory NK receptors, including killer cell immunoglobulin-like receptors (KIRs) and NKG2A, generally recognize MHC class I or MHC-related ligands, while activating receptors, such as NKG2D and natural cytotoxicity receptors (NCRs), have cognate ligands that are mainly absent on healthy cells and are induced on tumor or virus-infected cells (1). NCRs, including NKp46, NKp44, and NKp30, were originally identified as activating NK receptors that can immediately trigger killing of transformed cells (2–5). NCRs need to couple via charged residues in their transmembrane domain to immunoreceptor tyrosine-based activation motif (ITAM)-bearing adaptors for signal transduction. In NK cells, CD3ζ and FcεRIγ can form homodimers or heterodimers and couple to NKp46 and NKp30, while DAP12 associates with NKp44 (1, 6). NCR ligands expressed on tumor cells are still poorly characterized. However, B7-H6, a B7-family member, was recently described as a cognate tumor-associated ligand of NKp30 in humans (7). B7-H6 expression was shown to be up-regulated on the cell surface of several tumor cell lines and in tumor tissues (8), underscoring an important role of the NKp30/B7-H6 axis in antitumor immune responses.
IL-15 is a pleiotropic cytokine, crucial for NK and invariant NKT cell (iNKT cell) development and maintenance, and for the survival and proliferation of γδ T cells, tissue-resident T cells, and memory CD8+ T cells (9–11). IL-15 signals through a heterotrimeric receptor including the common gamma chain (γc), the IL-2/15 β chain, and the private high-affinity IL-15Rα subunit (10, 11). IL-15–expressing cells can transpresent IL-15 bound to IL-15Rα at the cell surface. IL-15/IL-15Rα complexes can be internalized and recycled back to the cell surface, allowing a constitutive local expression (10–12). This particular feature allows immune cells to be constantly exposed to IL-15 both during homeostasis and in pathological conditions (11–14). Moreover, it is well established that IL-15 has the capacity to enhance antitumoral immunity both by administration of soluble IL-15 and as a complex with IL-15Rα (15–20). Tumor cells frequently express low levels of MHC class I and escape from the classical TCR-mediated recognition by CD8+ T cells. However, tumors often express ligands for activating NK receptors, rendering them susceptible to killing by cells carrying tumor-reactive activating NK receptors. NKG2D is expressed by all human CD8+ T cells and can function as a costimulatory receptor (21). Moreover, it has been known for a long time that minor fractions of human αβCD8+ T lymphocytes can express KIRs and NKG2A (22, 23). However, so far, the expression of NCRs, potent activating NK receptors, is still widely accepted as being restricted to innate cell populations, and their expression and function on peripheral blood CD8+ T cells, along with their antitumor potential, are mainly unknown. Here, we uncover a population of innate-like CD8+ T cells circulating in the peripheral blood of healthy donors, expressing NKp30, and displaying a diverse TCR repertoire. This CD8+ T cell population could be induced by IL-15, requiring acquisition of the FcεRIγ adaptor after promoter demethylation. IL-15–induced NKp30+CD8+ T cells coexpress other activating NK receptors and a functional TCR, exhibiting potent NK-like antitumor activity both in vitro and in vivo in a preclinical xenograft melanoma mouse model. Our data reveal that IL-15 can initiate and drive a broad reprogramming process of CD8+ T cells toward the generation of a NKp30+CD8+ T cell population with potent NK-like function that is highly relevant for the design of novel antitumor immunotherapeutic strategies.
Results
Identification and Characterization of a Distinct NKp30+CD8+ T Cell Population in Human Peripheral Blood.
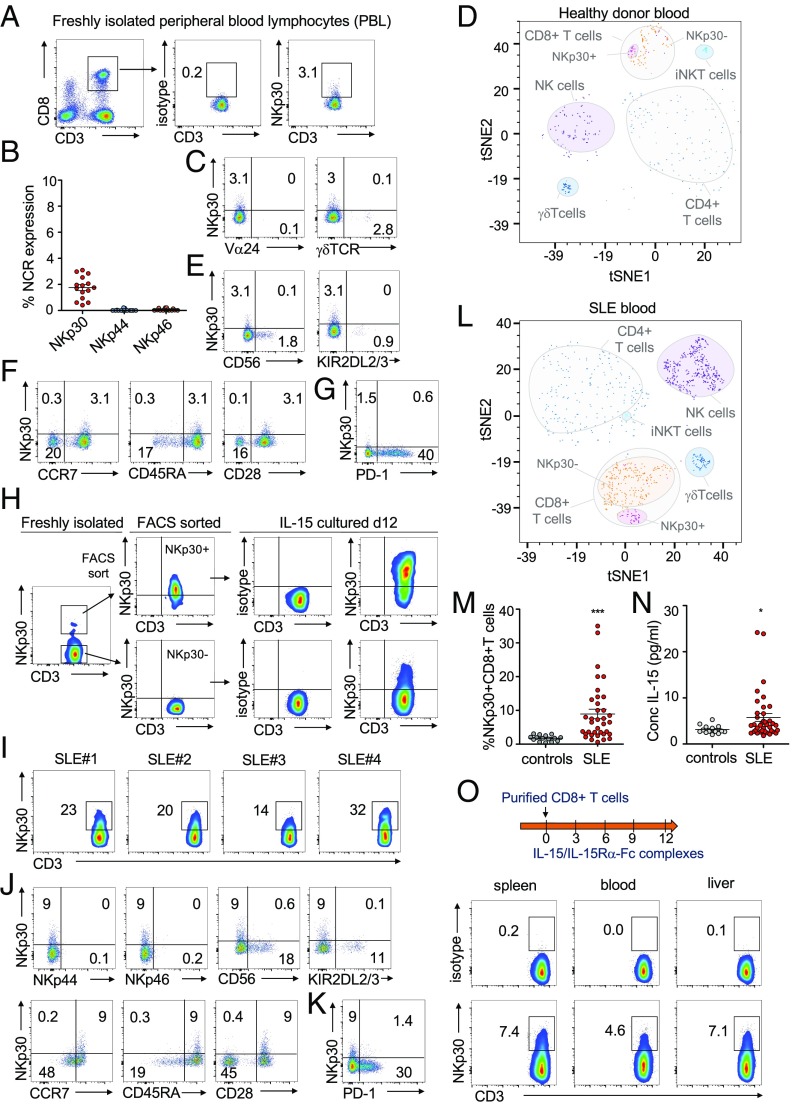
NCR expression is thought to be mainly specific for innate lymphocytes. In this study, by analyzing peripheral blood CD8+ T cells, we could observe low NKp30 expression on a significant percentage of CD8+ T cells of healthy donors (Fig. 1 A and B). Those NKp30+CD8+ T cells, although present at a small percentage, could be distinguished as a clearly defined CD8+ T cell subpopulation distinct from iNKT or γδ T cells (Fig. 1 C and D). Unlike NKp30, NKp46 and NKp44 expression was virtually absent on circulating CD8+ T cells (Fig. 1B). NKp30+CD8+ T cells coexpressed neither the prototypical NK receptor CD56 nor the most commonly expressed KIRs, KIR2DL2/3 (Fig. 1E), reinforcing them as a population distinct from other NK-like CD8+ T cells. Moreover, previously described NK receptor-expressing CD8+ T cells are often characterized by an effector memory phenotype, being enriched within virus-specific T cells (24). In contrast, the NKp30+CD8+ T cell population in peripheral blood displayed a CCR7+CD45RA+CD28+ expression indicative of a naive phenotype (Fig. 1F). Accordingly, in all six healthy donors, the NKp30+CD8+ T cell population stained negative for tetramers detecting virus [cytomegalovirus (CMV), Epstein–Barr virus, or influenza virus]-specific or tumor (Mart-1)-specific CD8+ T cells (SI Appendix, Fig. S1 A and B). Moreover, NKp30+CD8+ T cells were mainly negative for the checkpoint inhibitor molecule PD-1 (Fig. 1G), further supporting their naive phenotype. By analyzing the TCR usage by freshly isolated naive NKp30+CD8+ T cells, we observed that those cells displayed a broad, diverse TCR repertoire similar to the naive NKp30−CD8+ T cells, without an obvious skewing in clonal size distributions (SI Appendix, Fig. S1C). A diverse TCR repertoire further reinforces them as bona-fide naive CD8+ T cells, distinct from γδ T cells or iNKT cells. Next, we sorted naive CD8+ T cells from freshly isolated peripheral blood mononuclear cells and stimulated them via NKp30 with plate-bound mAbs. In these experiments we did not observe any significant levels of degranulation upon NKp30 triggering (SI Appendix, Fig. S1D), in agreement with previous studies showing that, in resting NK cells, NCR responses also depended on prior activation with cytokines (25).
Fig. 1.
Identification and characterization of a distinct CD8+ T cell population expressing NKp30. (A–H) Peripheral blood lymphocytes were isolated from healthy donors. (A) Representative FACS plots showing NKp30 on gated CD8+CD3+ cells. (B) Percentage of NCR+CD8+ T cells; n = 15, mean ± SEM. (C) Dot plots of gated CD8+CD3+ cells showing the absence of NKp30 costaining with Vα24 or γδTCR. (D) t-SNE visualization showing NKp30+CD8+ T cells as a distinctive population based on a multiparametric staining (CD3, CD8, CD4, CD56, γδTCR, Vα24, NKp44, NKp46, and PD-1). (E–G) Representative FACS plots showing NKp30 staining and CD56 and KIR2DL2/3 (E), CCR7, CD45RA, and CD28 (F), and PD-1 (G) on gated CD8+CD3+ cells. (H) Peripheral blood CD8+ T cells were FACS-sorted into NKp30+ and NKp30− populations and were cultured with IL-15 for 12 d. FACS plots show NKp30 expression before and after IL-15 culture. (I–N) Peripheral blood lymphocytes from SLE patients. (I) Flow cytometry plots showing NKp30 expression on CD8+ T cells from four representative SLE patients (SLE#1–4), pregated as 7AAD−CD45+CD3+CD8+ cells. (J and K) Representative FACS plots showing NKp30 and NKp44, NKp46, CD56, KIR2DL2/3, CCR7, CD45RA, and CD28 (J) and PD-1 (K) staining. (L) t-SNE visualization showing a distinctive NKp30+CD8+ T cell population in SLE patients based on multiparametric staining (CD3, CD8, CD4, γδTCR, Vα24, NKp44, NKp46, and PD-1). (M) Graph showing the percentage of NKp30+CD8+ T cells from the blood of control donors (n = 15) and SLE patients (n = 38). Data are shown as mean ± SEM; ***P ≤ 0.001; Mann–Whitney test. (N) Graph showing levels of IL-15 in plasma from control donors (n = 15) and SLE patients (n = 38). ***P ≤ 0.001, *P ≤ 0.05; Mann–Whitney test. (O) Highly purified human CD8+ T cells (1 × 106) were injected into NSG mice. IL-15/IL-15Rα-Fc complexes were supplied every 3 d. On day 12, CD8+ T cells were reisolated from mice and analyzed by flow cytometry. Plots show human NKp30+CD8+ T cells in spleen, blood, and liver, pregated as CD45+CD3+CD8+ cells. Representative data of one of two independent experiments from different donors (each with three mice per group) are shown.
IL-15 has been shown to be able to up-regulate the expression of NK receptors on CD8+ T cells (26, 27). To investigate if IL-15 could have a role in inducing NKp30 expression on peripheral blood CD8+ T cells, freshly isolated peripheral blood lymphocytes were stringently FACS-sorted into NKp30+CD8+ and NKp30−CD8+ T cells and were cultured in vitro with recombinant IL-15 (Fig. 1H). We observed that sorted NKp30+CD8+ T cells maintained and even increased NKp30 expression after 12 d, indicating it as a stable population (Fig. 1H). Notably, IL-15 could drive the FACS-sorted NKp30− population to de novo express NKp30 (Fig. 1H). Next, we investigated whether we could observe an increased frequency of NKp30+CD8+ T cells in peripheral blood in a pathological situation reportedly accompanied by increased systemic IL-15 levels, such as systemic lupus erythematosus (SLE) (28–31). Remarkably, we observed a higher percentage of NKp30+CD8+ T cells in SLE patients than in age-matched healthy blood donors (Fig. 1 I and M and SI Appendix, Fig. S1E). Moreover, while IL-15 was mainly undetectable in plasma from healthy donors, we confirmed its presence in plasma from SLE patients (Fig. 1N) (28–30). The NKp30+CD8+ T cell population in SLE was distinct from iNKT cells or γδ T cells (SI Appendix, Fig. S1E), as also illustrated by t-distributed stochastic neighbor embedding (t-SNE) visualization (Fig. 1L), and showed mainly a CD45RA+CCR7+CD28+CD56−NCR−PD1− naive phenotype similar to NKp30+CD8+ T cells found in healthy peripheral blood (Fig. 1 J and K). To further address if IL-15 could have a role in inducing NKp30 expression on CD8+ T cells in vivo, we adoptively transferred freshly purified human CD8+ T cells into NSG mice and injected IL-15/IL-15Rα-Fc complexes (Fig. 1O). Twelve days upon adoptive transfer, a fraction of CD8+ T cells from blood, spleen, and liver acquired NKp30 expression (Fig. 1O). Together, our findings revealed the presence of an NKp30-expressing population within naive circulating CD8+ T cells and uncovered IL-15 as a signal capable of driving NKp30 induction on CD8+ T cells both in vitro and in vivo.
NKp30 Induction on a Population of Human Peripheral Blood CD8+ T Cells.
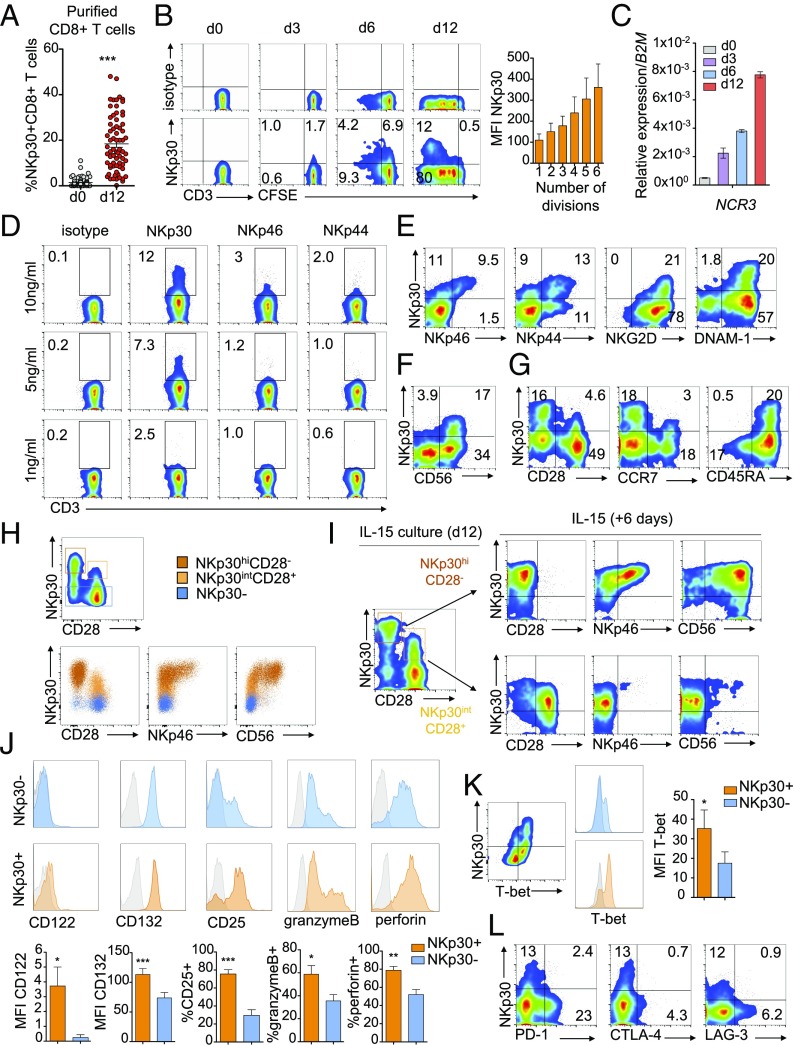
To further investigate the role of IL-15 in NKp30 induction on peripheral blood CD8+ T cells, we cultured highly purified CD8+ T cells with IL-15 for 12 d (SI Appendix, Fig. S2A). As expected, a percentage of the initial CD8+ T cell population showed a low expression of NKp30 on the cell surface (Fig. 2A, day 0). However, after 12 d of culture with IL-15, a significant increase in the percentage of NKp30+CD8+ T cells was observed (Fig. 2A, day 12). NKp30 expression on CD8+ T cells increased over time during IL-15 culture at both the protein and transcriptional levels (Fig. 2 B and C), with higher NKp30 expression associating with increased proliferation (Fig. 2B). It is noteworthy that IL-15 also was able to drive de novo induction of NKp46 and NKp44 (SI Appendix, Fig. S2 B–E) in a dose-dependent manner (Fig. 2D) and to up-regulate NKG2D and DNAM-1 levels (SI Appendix, Fig. S2F). While NKp30 induction consistently occurred at lower IL-15 doses (Fig. 2D) and at earlier time points of IL-15 culture (SI Appendix, Fig. S2E), prolonged in vitro culture with IL-15 could also drive induction of NKp46 and NKp44. Accordingly, IL-15–induced NKp30+CD8+ T cells expressing high levels of NKp30 (NKp30hi) also coexpressed NKp46 and NKp44 (Fig. 2E) and higher levels of the activating receptors NKG2D and DNAM-1 (Fig. 2E). NKp30hiCD8+ T cells also coexpressed CD56 (Fig. 2F) and mainly exhibited a CD45RA+CCR7−CD28− phenotype reminiscent of effector memory CD45RA+ T cells (TEMRA), while a subset with intermediate NKp30 expression (NKp30int) displaying a CD45RA+CCR7+CD28+ naive phenotype (TN) could also be observed (Fig. 2G). NKp30hiCD28− T cells proliferated more than NKp30intCD28+ cells, suggestimg a further differentiation stage (SI Appendix, Fig. S2G). We hypothesized that IL-15–driven induction of NKp30 would follow a differentiation pattern from NKp30−CD8+ T cells → NKp30low/intCD28+NCR−CD8+ T cells → NKp30hiCD28int/−NCR+ T cells. Indeed, the NKp30hiCD28−CD8+ T cell population coexpressed NKp46 and CD56, while the NKp30intCD28+ CD8+ T cell subset appeared in a less differentiated stage (Fig. 2H). Thus, we FACS-sorted IL-15–induced NKp30+CD8+ T cells into NKp30hiCD28− and NKp30intCD28+ T cells and subsequently cultured them with IL-15 for six more days. NKp30intCD28+CD8+ T cells could further differentiate into NKp30hiCD28−CD8+ T cells, while NKp30hiCD28−CD8+ T cells remained with a similar phenotype (Fig. 2I). Moreover, when we FACS-sorted the NKp30+CD8+ T cell population from resting CD8+ T cells (NKp30lowCD28+CD8+ T cells) and cultured them with IL-15 for 12 d, we could observe differentiation toward an NKp30hiCD28−NKp46+ phenotype (SI Appendix, Fig. S2H). Thus, we were capable not only of generating NKp30+CD8+ T cells with IL-15 in vitro but also of further driving their differentiation toward an effector memory phenotype with concomitant acquisition of other NCRs. In addition, compared with the NKp30−CD8+ T cells, NKp30+CD8+ T cells showed a higher expression of granzyme B/perforin and increased levels of IL-15 receptor subunits (Fig. 2J) and T-bet (Fig. 2K). Interestingly, inhibitory receptors known to be expressed in memory cell populations and to impair CD8+ T cell function, such as PD-1, CTLA-4, and LAG-3 (32), showed low expression in NKp30+CD8+ T cells (Fig. 2L). Similar to NK cells (33), IL-15–induced CD8+ T cells expressed different isoforms of NKp30, with NKp30b and NKp30a being the isoforms predominantly expressed, while NKp30c isoform expression was low (SI Appendix, Fig. S2I). Moreover, blockage of IL-4 during the culture period with IL-15 did not affect the induction of NKp30 on the CD8+ T population (SI Appendix, Fig. S2J), indicating that our population was distinct from the recently described IL-4–induced innate CD8+ T cells (34). Notably, results obtained using specific inhibitors suggested that the IL-15–induced PI3K-signaling pathway, rather than STAT5, was involved in the acquisition of NKp30 by CD8+ T cells (SI Appendix, Fig. S3). Taken together, our data show the existence of a NKp30+CD8+ T cell population in human peripheral blood, which can be induced by IL-15 and further differentiated into an effector memory NKp30hiCD8+ T cell population displaying an NK-like phenotype with increased effector potential and low expression of exhaustion markers.
Fig. 2.
IL-15 can induce the generation of a NKp30+CD8+ T cell population coexpressing other NCRs and low levels of exhaustion markers. Highly purified human peripheral blood CD8+ T cells were cultured for 12 d with IL-15. (A) Percentage of NKp30 receptor-expressing CD8+ T cells after isolation (day 0) and on day 12, analyzed by flow cytometry. n = 64–67; data are shown as mean ± SEM; ***P ≤ 0.001; paired Student’s t test. (B, Left) Representative plots showing NKp30 receptor expression on day 0 and kinetics with carboxyfluorescein succinimidyl ester (CFSE) dilution on days 3, 6, and 12. (Right) Graph showing the increase in NKp30 MFI expression with the division number (according with CFSE dilution) on gated NKp30+CD8+ T cells. n = 3; data are shown as mean ± SEM. (C) Representative qPCR analysis of NCR3 expression (relative to B2M) on days 0, 3, 6, and 12. Data are shown as mean ± SEM. (D) FACS plots showing NKp30, NKp46, and NKp44 expression after culture with the indicated amounts of IL-15. (E and F) Flow cytometry plots showing the coexpression of other NK receptors with NKp30 on day 12. (G) Plots showing NKp30, CD28, CCR7, and CD45RA expression on day 12. (H) FACS plots showing NKp30, CD28, NKp46, and CD56 expression on day 12 FACS-sorted NKp30−, NKp30intCD28+, or NKp30hiCD28− CD8+ T cell populations. (I) Day 12 FACS-sorted NKp30hiCD28− or NKp30intCD28+ CD8+ T cell populations were cultured with IL-15 for six additional days, and NKp30, CD28, NKp46, and CD56 expression was analyzed by FACS. (J, Upper) Representative histograms showing expression of IL-2/15 receptor molecules, granzyme B, and perforin on day 12, gated on NKp30+ or NKp30− CD8+ T cells. (Lower) Graphs show mean ± SEM of n = 3–7 independent donors. (K) Representative FACS plot (Left) and corresponding histograms (Center) gated on NKp30+ or NKp30− CD8+ T cells, showing T-bet and NKp30 expression on day 12. (Right) Graph showing mean ± SEM; n = 3 independent donors. (J and K) ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05; paired Student’s t test. (L) Expression of NKp30 and PD-1, CTLA-4, and LAG-3 on day 12. Data representative of three or more independent experiments from different donors are shown.
IL-15–Induced NKp30+CD8+ T Cells Coexpress Functional NCRs and TCRs That Interplay in Degranulation and Cytokine Expression.
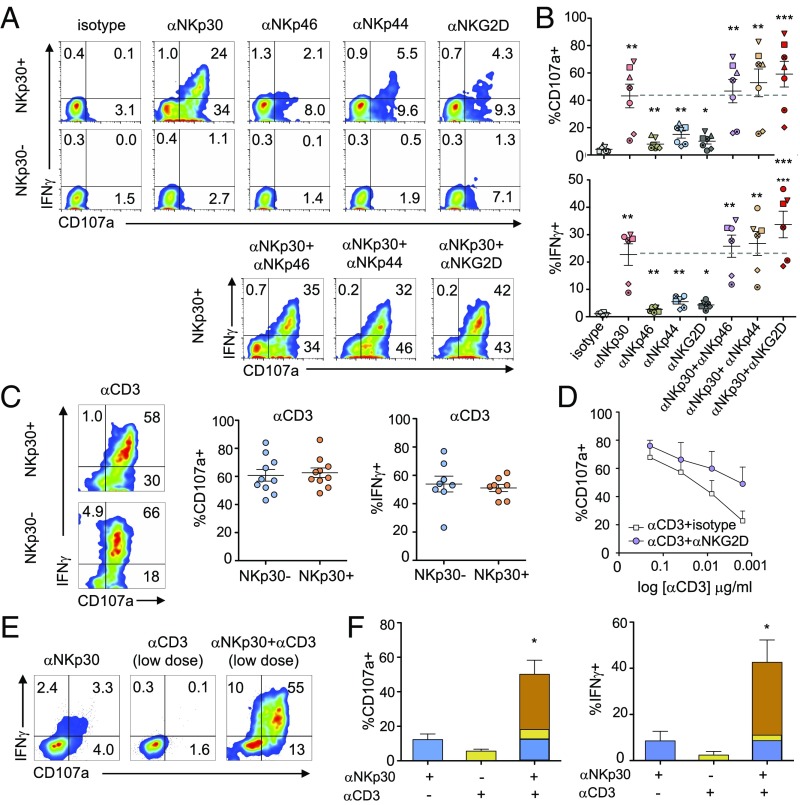
To determine whether the activating NK receptors expressed on the generated NKp30+CD8+ T cell population were functional, we FACS-sorted the IL-15–cultured CD8+ T cells into NKp30+ and NKp30− populations (SI Appendix, Fig. S4). NKp30 stimulation by plate-bound antibodies led, by itself, to a pronounced increase in degranulation and IFN-γ expression in the sorted NKp30+CD8+ T cells, contrary to sorted NKp30−CD8+ T cells (Fig. 3 A and B). Triggering of NKp46, NKp44, or NKG2D induced a lower, but significant, increase in degranulation and IFN-γ expression. Triggering of NKp30 in combination with other NCRs resulted in an additive effect, whereas NKp30 engagement together with NKG2D led to a synergistic increase in the responses (Fig. 3 A and B). CD3 stimulation induced high amounts of degranulation and IFN-γ expression in both NKp30+ and NKp30− CD8+ T cells, indicating that TCR-related signaling was equally functional (Fig. 3C). NKG2D triggering acted synergistically with CD3 stimulation, as previously described (Fig. 3D) (27, 35, 36). Most importantly, when CD3 was engaged with suboptimal concentrations of mAb, NKp30 costimulation could synergistically increase degranulation and IFN-γ expression in NKp30+CD8+ T cells (Fig. 3 E and F). Together, our results reveal that stimulation of NKp30 per se can induce effector function of CD8+ T cells. Moreover, NKp30 could synergistically enhance CD8+ T cell responses when cotriggered with low levels of CD3 stimulation, suggesting that NKp30 costimulation can lower the TCR activation threshold of CD8+ T cells.
Fig. 3.
IL-15–induced NKp30+CD8+ T cells express functional NCRs that can synergize with TCR activation in degranulation and IFNγ production. Purified CD8+ T cells were cultured with IL-15 and were FACS-sorted on day 12 into NKp30+ and NKp30− CD8+ T cells. Sorted cells were then stimulated with the indicated plate-bound mAbs for 4 h. (A) Representative flow cytometry plots showing degranulation (CD107a) and intracellular IFNγ expression by sorted NKp30+ or NKp30− CD8+ T cells after triggering with plate-bound mAb (5 μg/mL). (B) Graphs showing the percentage of CD107a+ (Upper) and IFNγ+ (Lower) cells from six or seven independent donors. Data are shown as mean ± SEM; ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05; paired Student’s t test. Each independent donor is represented by a different symbol. (C, Left) FACS plots representing CD107a and intracellular IFNγ expression by sorted populations after anti-CD3 (0.4 μg/mL) plate-bound stimulation. (Center and Right) Corresponding graphs showing the percentage of CD107a+ and IFNγ+ cells from 9 or 10 independent donors. Data are shown as mean ± SEM. (D) Graph showing the percentage of CD107a+ cells after triggering of sorted NKp30+CD8+ T cells with sequential dilutions of plate-bound anti-CD3 mAb (starting with 2 μg/mL) alone or with anti-NKG2D mAb (5 μg/mL). Data are shown as the mean ± SEM for six independent donors. (E) FACS plots showing sorted NKp30+CD8+ T cell responses (percent CD107a+ and percent IFNγ+) to triggering with low dose of anti-CD3 mAb (0.016 μg/mL) alone or with anti-NKp30 mAb (5 μg/mL). (F) Corresponding graphs showing the percentages of CD107a+ (Left) and IFNγ+ (Right) cells from three independent donors. Data are shown as the mean ± SEM; *P ≤ 0.05; paired Student’s t test.
NKp30+CD8+ T Cells Exhibit a Distinct and Unique Innate-Like Gene-Expression Profile.
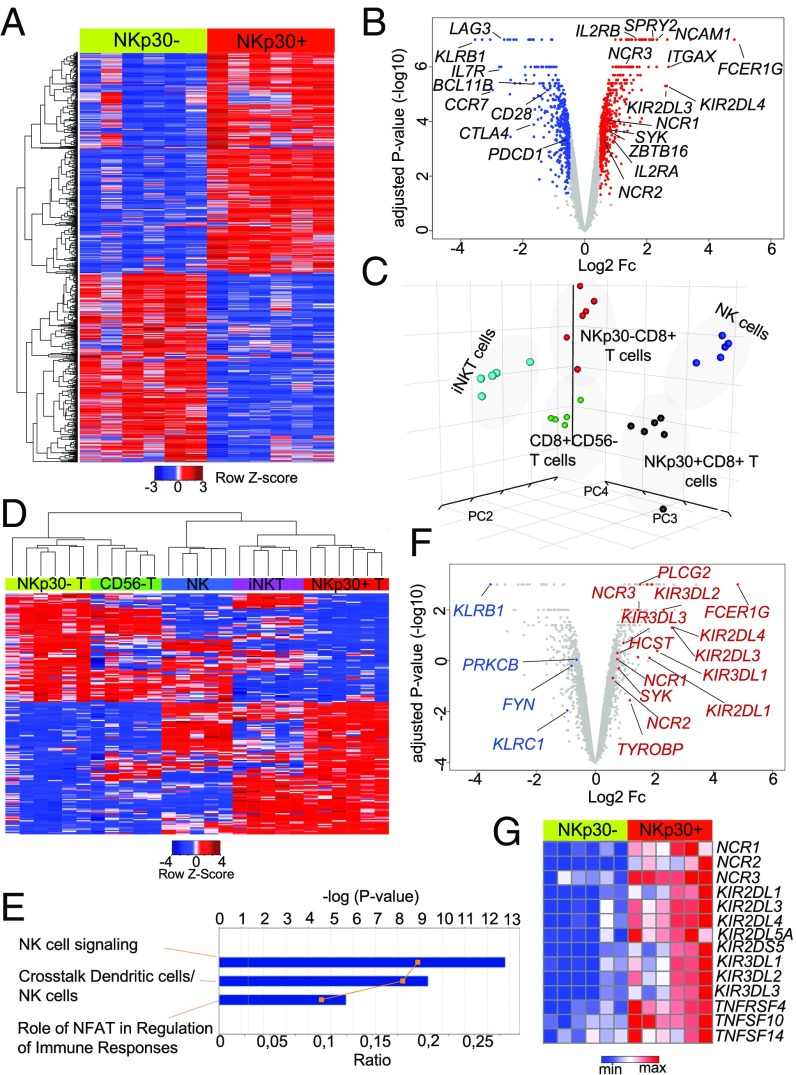
Next, we analyzed the gene-expression profile of IL-15–induced NKp30+(hi)CD8+ T cells, compared with NKp30−CD8+ T cells, by genome-wide microarray. As shown in the heatmap and volcano plot (Fig. 4 A and B), we could observe a distinct transcription profile of the NKp30+ compared with the NKp30− CD8+ T cell population. Importantly, beyond the expression of NK receptors, a wide range of genes was differentially expressed in these populations (Fig. 4 A and B), highlighting the existence of broader transcriptional changes in the NKp30+CD8+ T cells. Those results prompted us to perform a wider comparative analysis of the NKp30+CD8+ T cell population with other populations of lymphocytes. For that, a cross-platform principal component analysis (PCA) (37) was performed comparing our sorted IL-15–induced NKp30+ and NKp30− CD8+ T cells with CD8+CD56− T cells, iNKT cells, and NK cells from a published database (38). Interestingly, NKp30+CD8+ T cells clustered as an independent population, distant from both NKp30−CD8+ T cells and resting CD8+ T cells, which grouped closely together (Fig. 4C). Moreover, NKp30+CD8+ T cells clustered distantly from conventional iNKT cells, reinforcing their identity as a distinct population. Remarkably, the NKp30+CD8+ T cell population clustered close to the NK cell population, emphasizing similarities in their transcription profiles (Fig. 4C). Subsequently, hierarchical clustering from a variance analysis within the five different populations indicated that the gene clusters up-regulated in the NKp30+CD8+ T cell population, compared with NKp30−CD8+ T cells, were also shared with the innate immune cell populations, NK and iNKT cells (Fig. 4D), further underscoring a skew toward an enriched innate-like profile. Additionally, pathway analysis further revealed NK cell signaling as the top up-regulated canonical pathway in the NKp30+CD8+ T cell population, compared with NKp30−CD8+ T cells (Fig. 4 E and F). In addition to the NK receptor genes mentioned (Fig. 4 B, F, and G), several KIRs and NK-related genes indicative of increased effector function emerged as up-regulated, such as TNFRSF4, TNFSF14, and TNFSF10 encoding for OX40, LIGHT, and TRAIL, respectively (Fig. 4G). HCST and TYROBP, encoding for DAP10 and DAP12 adaptor molecules, respectively, were also up-regulated in NKp30+CD8+ T cells (Fig. 4F and SI Appendix, Fig. S5). Remarkably, Sprouty RTK signaling antagonist 2 (SPRY2), described recently as expressed exclusively by innate immune cells (39), was up-regulated (Fig. 4B), while the transcription factor Bcl11b, reported to protect T cell identity (40), was down-regulated in NKp30+CD8+ T cells (Fig. 4B). Together, these data indicate that IL-15 can induce reprogramming in a population of peripheral blood CD8+ T cells, leading to the acquisition of a unique transcriptional expression profile, distinct from other immune cell populations, and toward the acquisition of innate NK-like properties.
Fig. 4.
IL-15–induced NKp30+CD8+ T cells exhibit a distinctive innate-like transcription profile. Highly purified CD8+ T cells from six donors, cultured for 12 d with IL-15, were FACS-sorted into NKp30+ and NKp30− CD8+ T cell populations, and whole-genome expression analysis was performed using Illumina arrays. (A) Heatmap of genes differentially expressed in sorted NKp30+ and NKp30− CD8+ T cells (cutoff: absolute log2 fold change ≥0.5; adjusted P value < 0.05; Euclidian distance metric and average-linkage clustering was used for row reordering). (B) Volcano plot showing the differential gene expression in sorted NKp30+ and NKp30− CD8+ T cells. Genes significantly up-regulated are shown in red; genes significantly down-regulated are shown in blue (cutoff: absolute log2 fold change ≥0.5, adjusted P value < 0.05). (C) PCA of the gene-expression data from the indicated cell populations. (D) Heatmap showing hierarchical clustering of the genes differentially expressed in sorted NKp30+ and NKp30− CD8+ T cells across the indicated cell populations (cutoff: absolute log2 fold change ≥1; Kendall’s tau rank was used together with average-linkage clustering). (E) Top canonical pathways affected by differential gene expression in sorted NKp30+ and NKp30− CD8+ T cells, analyzed by Ingenuity pathway analysis. (F) Volcano plot showing NK-signaling pathway genes from E labeled in red (up-regulated) and blue (down-regulated), according to a cutoff absolute log2 fold change ≥0.5; adjusted P value < 0.05. (G) Heatmap showing NK-related genes up-regulated on NKp30+CD8+ T cells compared with NKp30−CD8+ T cells, generated by GENE-E (Broad Institute) (cutoff: absolute log2 fold change ≥0.5; adjusted P value < 0.05; Euclidian distance metric and average-linkage clustering).
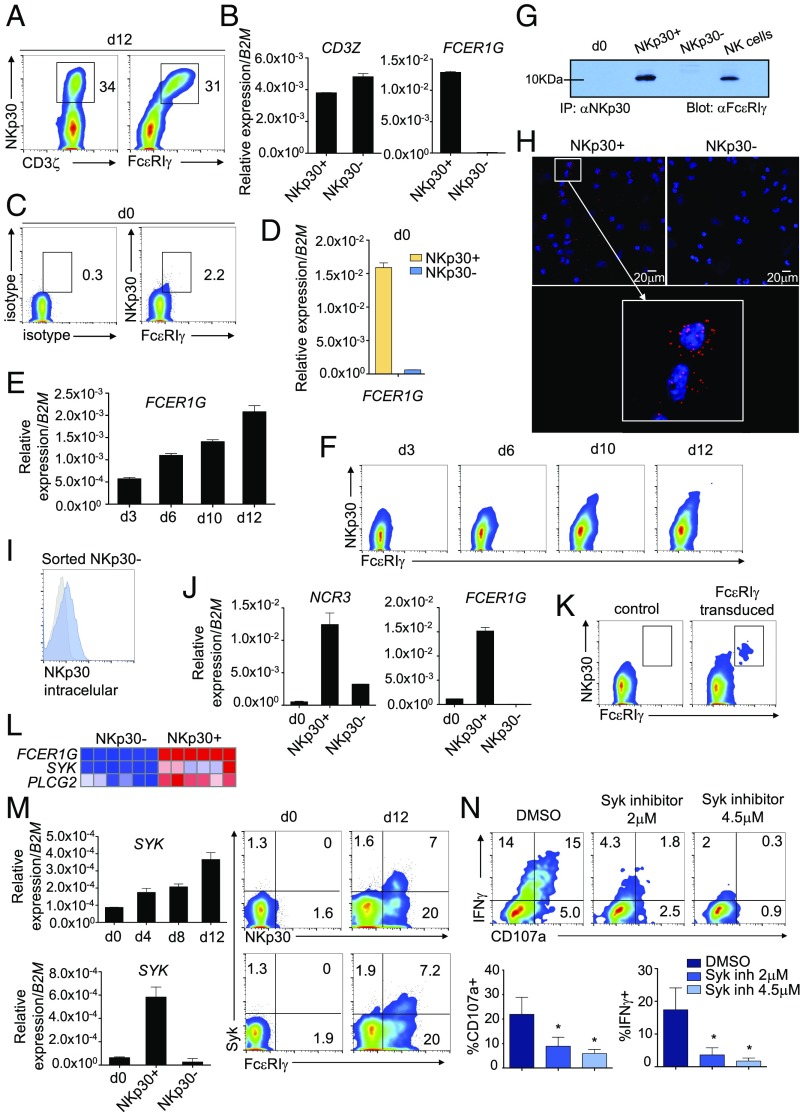
FcεRIγ Is Exclusively Induced in NKp30+CD8+ T Cells and Interacts Directly with NKp30, Enabling Its Surface Expression and Function.
Our microarray data analysis revealed the ITAM domain-containing molecule FcεRIγ as the clear topmost up-regulated gene in the NKp30+CD8+ T cell population (Fig. 4B). FcεRIγ, as well as CD3ζ, is described as an adaptor molecule able to couple to NKp30 in NK cells. Accordingly, we found that FcεRIγ was exclusively expressed in NKp30+CD8+ T cells after IL-15 culture, both at the transcriptional and protein levels, while CD3ζ expression did not differ between the NKp30+ and NKp30− CD8+ T cell population (Fig. 5 A and B). Noteworthy, on freshly isolated CD8+ T cells (day 0), although FcεRIγ expression was mainly unnoticeable, we could observe FcεRIγ and NKp30 coexpression in the small existing NKp30+CD8+ T cell population (Fig. 5C). Indeed, we could detect expression of the FCER1G transcript exclusively in NKp30+CD8+ T cells sorted from circulating resting CD8+ T cells (Fig. 5D). Upon IL-15 culture, FcεRIγ expression on CD8+ T cells increased in a time-dependent manner at both mRNA and protein levels (Fig. 5 E and F). Importantly, the kinetics of FcεRIγ up-regulation correlated with NKp30 surface expression (Fig. 5F), with NKp30hiCD8+ T cells displaying the highest levels of FcεRIγ (Fig. 5 A and F). Indeed, we observed that FcεRIγ expression is higher in the NKp30hiCD28−CD8+ T cell population than in the NKp30intCD28+CD8+ T cell population (SI Appendix, Fig. S6A). It is noteworthy that FcεRIγ expression could also be observed on NKp30+CD8+ T cells generated in vivo in NSG mice (SI Appendix, Fig. S6B) and increased in peripheral blood NKp30+CD8+ T cells from SLE patients (SI Appendix, Fig. S6C). Furthermore, by both coimmunoprecipitation experiments (Fig. 5G) and microscopy proximity ligation assay (Fig. 5H), we demonstrated that FcεRIγ interacts directly with NKp30 at the protein level but does not interact with CD56 (SI Appendix, Fig. S6D). Intriguingly, we observed that a fraction of IL-15–treated NKp30−CD8+ T cells expressed NKp30 protein intracellularly, along with mRNA, while being negative for FcεRIγ expression (Fig. 5 I and J). Thus, to ascertain if FcεRIγ could have a role in enabling NKp30 cell-surface expression, sorted NKp30−CD8+ T cells were transduced with FcεRIγ-expressing retroviral plasmids. Remarkably, the NKp30−CD8+ T cell fraction ectopically expressing FcεRIγ became highly NKp30 cell surface-positive (Fig. 5K), suggesting a nonredundant role for FcεRIγ in supporting high NKp30 cell-surface expression despite the expression of CD3ζ. In addition, our microarray analysis revealed that NKp30+CD8+ T cells exhibited up-regulated expression of genes encoding for signaling molecules downstream of FcεRIγ, such as spleen tyrosine kinase (SYK) and 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 (PLCG2) (Fig. 5L). We confirmed that Syk was up-regulated in CD8+ T cells after culture with IL-15, being expressed mainly in the NKp30+ fraction (Fig. 5M). Moreover, pretreatment of sorted NKp30+CD8+ T cells with a Syk inhibitor resulted in a remarkable dose-dependent decrease in degranulation and IFN-γ expression upon NKp30 triggering (Fig. 5N). Similarly, treatment with PLCγ or PI3K inhibitors, known to be downstream of Syk signaling (41), led to a substantial impairment in degranulation and IFN-γ expression (SI Appendix, Fig. S7). Moreover, we observed that only the NKp30hiCD28−CD8+ T cell population, expressing high levels of FcεRIγ, displayed high effector potential when cross-linked with anti-NKp30 (SI Appendix, Fig. S6E). Together, our results demonstrate that FcεRIγ can be induced in the NKp30+CD8+ T cell population by IL-15, being crucial for supporting the expression and the function of the NKp30 cell-surface receptor on CD8+ T cells.
Fig. 5.
FcεRIγ is exclusively induced in NKp30+CD8+ T cells and interacts directly with NKp30, allowing its surface expression and function. (A and B) Purified CD8+ T cells were cultured for 12 d with IL-15. (A) Representative FACS plots showing CD3ζ, FcεRIγ, and NKp30 expression on day 12. (B) Relative expression of CD3Z and FCER1G mRNA in day 12 FACS-sorted NKp30+ or NKp30− CD8+ T cells. Data are shown as mean ± SEM. (C) FACS plots showing NKp30 and FcεRIγ staining on freshly purified CD8+ T cells (day 0). (D) Relative expression of FCER1G mRNA from FACS-sorted NKp30+ or NKp30− CD8+ T cells on day 0. (E–N) Purified CD8+ T cells were cultured for 12 d with IL-15. (E) Relative expression of FCER1G mRNA on days 3, 6, 10, and 12. Data are shown as mean ± SEM. (F) FACS plots showing expression of FcεRIγ and NKp30 at the indicated time points. (G) Immunoblot showing NKp30 and FcεRIγ coimmunoprecipitation for the indicated cells. (H) PLA showing NKp30 protein interaction with FcεRIγ (red dots) in NKp30+CD8+ T cells (blue, DAPI). (I) Expression of intracellular NKp30 in day 12 sorted NKp30−CD8+ T cells. (J) Relative mRNA expression of NCR3 and FCER1G in day 0 CD8+ T cells and day 12 sorted NKp30+ and NKp30− CD8+ T cells. Data are shown as mean ± SEM. (K) Plots showing FcεRIγ and NKp30 expression on day 12 sorted NKp30−CD8+ T cells transduced with a FcεRIγ-expressing vector or control. (L) Heatmap generated in GENE-E showing differential expression of FCER1G, SYK, and PLCG2 in NKp30− and NKp30+ CD8+ T cells. (M, Left) Relative mRNA expression of SYK in CD8+ T cells at the indicated time points (Upper) and in day 12 sorted NKp30+ and NKp30− CD8+ T cells (Lower). Data are shown as mean ± SEM. (Right) FACS plots showing expression of Syk, FcεRIγ, and NKp30 on day 0 and day 12. Data in A–M are representative of three or more independent experiments. (N) NKp30+CD8+ T cells pretreated with Syk inhibitor (BAY 61-3606) or DMSO were stimulated with plate-bound anti-NKp30 mAb for 4 h. (Upper) Representative FACS plots showing CD107a and IFNγ expression. (Lower) Data from four independent donors are shown in the graphs. Data are shown as mean ± SEM; *P ≤ 0.05; paired Student’s t test.
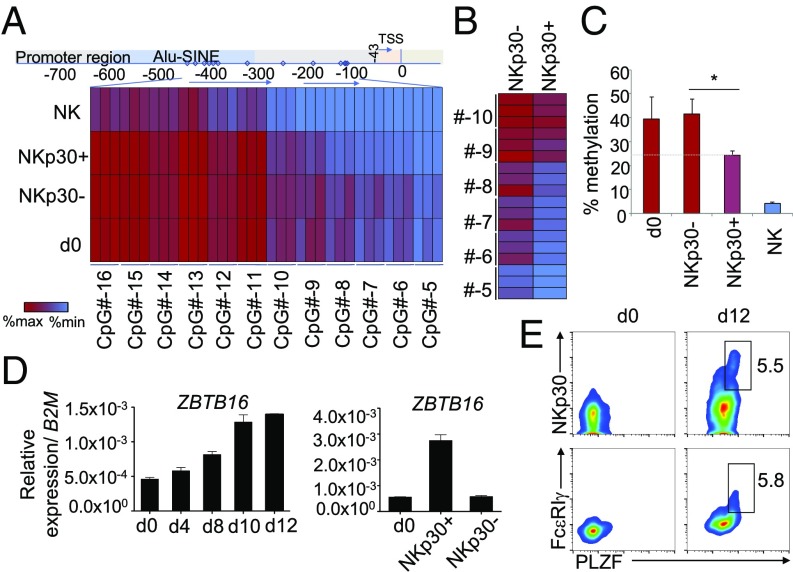
IL-15 Coordinately Induces FCER1G Promoter Demethylation and PLZF Expression in the NKp30+CD8+ T Cell Population.
It has been previously reported that FcεRIγ expression can be regulated by promoter methylation in other cell types (42, 43). To ascertain whether methylation could affect FcεRIγ expression in CD8+ T cells, CpG methylation analysis of the FCER1G promoter was performed by pyrosequencing. As shown in Fig. 6 A–C, the FCER1G promoter of both freshly isolated CD8+ T cells and IL-15–cultured NKp30−CD8+ T cells showed a higher level of methylation than the FCER1G promoter of NKp30+CD8+ T cells. As expected, the FCER1G promoter of NK cells was highly demethylated (Fig. 6A). The results are particularly clear when focusing on the CpG sites outside the short interspersed nuclear elements (SINE) region of the promoter, where about a 40% decrease in the percentage of methylation was observed in NKp30+CD8+ T cells compared with NKp30−CD8+ T cells or freshly isolated CD8+ T cells (day 0) (Fig. 6 B and C). Accordingly, our data indicate that FcεRIγ expression in CD8+ T cells can be epigenetically controlled by methylation of CpG sites in its promoter. IL-15–induced FCER1G promoter demethylation was found exclusively in the NKp30+CD8+ T cell population, indicating demethylation of the FCER1G promoter as a required event preceding FcεRIγ induction in NKp30+CD8+ T cells. The zinc finger and BTB domain-containing protein 16 (ZBTB16) gene encodes the promyelocytic leukemia zinc finger (PLZF). PLZF is expressed by several innate lymphocyte populations but normally is not expressed by resting αβCD8+ T cells (44–46). Recently, it was reported that PLZF interacts with the FCER1G promoter (43). Here, we show that IL-15 can induce a population of CD8+ T cells to express ZBTB16 transcripts (Fig. 6D). Importantly, we observed that PLZF was up-regulated in NKp30+CD8+ T cells while being mainly absent in resting CD8+ T cells (day 0) or in sorted NKp30−CD8+ T cells (Fig. 6 D and E). Those results suggest a potential role of the PLZF transcription factor in the induction of FcεRIγ in CD8+ T cells that concomitantly express cell-surface NKp30. In accordance, we could detect the exclusive expression of ZBTB16 transcript in freshly isolated circulating NKp30+CD8+ T cells (day 0) sorted from the peripheral blood of healthy individuals (SI Appendix, Fig. S8A). In addition, we could observe a significant positive correlation between ZBTB16 and NCR3 expression in the IL-15–induced NKp30+CD8+ T cell population (SI Appendix, Fig. S8B). Moreover, NKp30hiCD28− CD8+ T cells expressed higher ZBTB16 levels than the NKp30intCD28+ CD8+ T cell population (SI Appendix, Fig. S8C), further supporting the suggested differentiation program model: NKp30−/intCD28+FcεRIγ−/lowPLZF−/low → NKp30hiCD28−FcεRIγ+PLZF+. Taken together, our results indicate that IL-15 can drive reprogramming on a fraction of CD8+ T cells, inducing FCER1G promoter demethylation, leading to its enhanced transcription and protein expression, and concurrent expression of the transcription factor PLZF, resulting in the generation of NKp30+CD8+ T cells.
Fig. 6.
Demethylated FCER1G promoter in IL-15–induced NKp30+CD8+ T cells. Highly purified CD8+ T cells were cultured for 12 d with IL-15. (A) Heatmap showing relative methylation levels of the depicted CpG sites (−16 to −5) of the FCER1G promoter for day 0 CD8+ T cells, day 12 sorted NKp30+ and NKp30− CD8+ T cells, and NK cells. Three different donors are shown. TSS, transcriptional start site. (B) Heatmap showing the relative CpG methylation levels outside of the SINE region of the promoter (−10 to −5) for day 12 sorted NKp30+ and NKp30− CD8+ T cells. (C) Graph showing the mean percentage methylation of the CpG sites (−10 to −5) for day 0 CD8+ T cells, day 12 sorted NKp30+ and NKp30− CD8+ T cells, and NK cells from three different donors. Data are shown as mean ± SD; *P ≤ 0.05; paired Student’s t test. (D) Relative mRNA expression of ZBTB16 at the indicated time points (Left) and in day 0 CD8+ T cells and in day 12 sorted NKp30+ and NKp30− CD8+ T cells (Right). (E) FACS plots showing protein expression of PLZF, FcεRIγ, and NKp30 by day 0 CD8+ T cells and day 12 CD8+ T cells cultured with IL-15. Data are representative of three or more independent experiments from different donors.
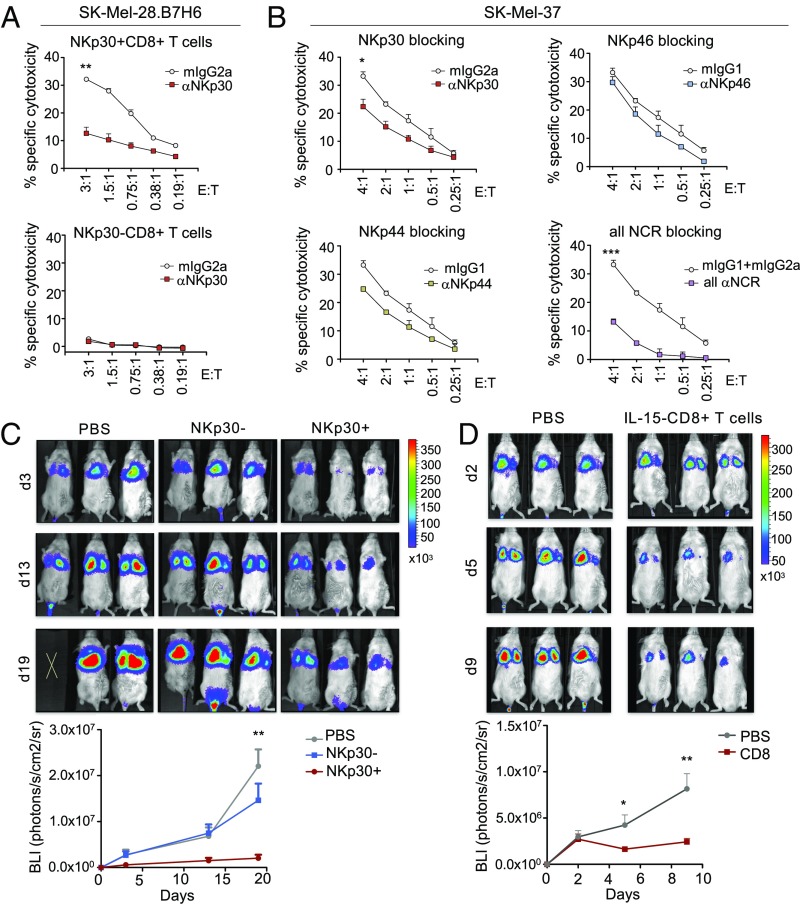
IL-15–Induced NKp30+CD8+ T Cells Display High NK-Like in Vitro Cytotoxicity and Potent Antitumor Activity After Adoptive Transfer in a Xenograft Tumor Model.
To investigate if NKp30+(hi)CD8+ T cells could directly kill tumor cells, 51Cr-release cytotoxicity assays were performed against the melanoma cell line SK-MEL-28.B7-H6, overexpressing B7-H6, and the endogenously B7-H6–expressing cell line SK-MEL-37 (see NK-ligand expression levels in SI Appendix, Fig. S9A). NKp30+CD8+ T cells were able to effectively kill both cell lines in an NKp30-dependent manner, even at low effector cell-to-target cell (E:T) ratios (Fig. 7 A and B). In contrast, NKp30−CD8+ T cells did not show any cytotoxicity toward those cell lines at those ratios (Fig. 7A). In addition, when all NCRs were concomitantly blocked, the observed cytotoxicity was largely abrogated (Fig. 7B). In sum, these results show that the NCRs expressed on IL-15–induced NKp30+CD8+ T cells are functional by themselves and can interplay, resulting in effective killing in an NK-like manner. Next, we examined if IL-15–induced NKp30+CD8+ T cells were capable of exhibiting in vivo killing capacity in a xenograft mouse model. NSG mice were injected i.v. with SK-MEL-28.B7-H6 cells together with sorted IL-15–cultured NKp30+CD8+ or NKp30−CD8+ T cells or PBS as a control. We observed a remarkable decrease in tumor load in the group of mice injected with sorted NKp30+CD8+ T cells, while injection with NKp30−CD8+ T cells gave results similar to those with the PBS control (Fig. 7C and SI Appendix, Fig. S9B), indicating that only the IL-15–induced NKp30+CD8+ T cell population could inhibit tumor growth. Next, we analyzed if our effector cells were able to control a pre-established tumor. To use a more clinically applicable approach, we adoptively transferred bulk IL-15–cultured CD8+ T cells as effector cells without previously sorting them into NKp30+ or NKp30− populations. Interestingly, injection of IL-15–cultured bulk CD8+ T cells greatly diminished the tumor load in comparison with the PBS control (Fig. 7D). Together, our results show that IL-15 can drive the generation of potent NKp30+CD8+ T cells capable of efficiently inhibiting tumor growth in a xenograft mouse model.
Fig. 7.
IL-15–induced NKp30+CD8+ T cells display potent in vitro and in vivo activity against tumor cells. (A and B) Highly purified CD8+ T cells were cultured with IL-15 and on day 12 were FACS-sorted into NKp30+ and NKp30− CD8+ T cells. (A) Percentage of specific cytotoxicity of NKp30+ or NKp30− CD8+ T cells against SK-Mel-28.B7-H6 cells in the presence of neutralizing anti-NKp30 mAbs or the corresponding isotype control (mIgG2a), for the indicated E:T ratios. (B) Percentage of specific cytotoxicity of NKp30+CD8+ T cells against SK-Mel-37 cells in the presence of neutralizing anti-NCR antibodies, or the corresponding isotype controls. (A and B) Data are shown as mean ± SEM; ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05; unpaired Student’s t test, for the highest ratio. (C, Upper) Sorted NKp30+ or NKp30− CD8+ T cells (1 × 106) or PBS were i.v. injected into NSG mice together with 0.25 × 106 luciferase-expressing SK-Mel-28.B7-H6.luci cells. (Lower) Bioluminescence intensity (BLI) values are shown in the graph for each day of measurement, showing the mean ± SEM of a representative experiment. Another independent experiment from a different donor is shown in SI Appendix, Fig. S9B. (D, Upper) SK-Mel-28.B7-H6.luci cells (0.25 × 106) were injected into NSG mice. BLI was measured 2 d afterward, and mice were injected with 30 × 106 bulk IL-15–cultured CD8+ T cells or PBS. (Lower) Graph showing data from two independent experiments from different donors. n = 6; data are shown as mean ± SEM. (C and D) **P ≤ 0.01, *P ≤ 0.05; unpaired Student’s t test, for the specific time points.
Discussion
CD8+ T cells are prototypical adaptive immune cells. Here, we uncovered a distinct population of CD8+ T cells expressing NKp30 in the peripheral blood of healthy individuals. This population exhibited a naive phenotype, did not coexpress CD56, KIR2DL2/3, or other NCRs, and displayed a broad, diverse TCR repertoire, being distinct from other previously described circulating NK receptor-expressing CD8+ T cell populations such as KIR+CD8+ T cells (24). We revealed IL-15 as a signal capable of de novo inducing NKp30 on CD8+ T cells and driving the acquisition of a potent antitumor activity. In previous studies, Correia et al. (26, 27) showed that IL-15 could induce KIRs, NKG2A, and NKp46 on peripheral blood CD8+ T cells. A parallel study reported similar effects on NCR induction exclusively on umbilical cord blood (47), and low NCR levels were reported in a small fraction of cytokine-induced killer cells (48). NKp46 expression was shown on intraepithelial lymphocytes in patients with celiac disease (49) and on NKT cells in patients with T cell large granular lymphocytic leukemia (50). However, the mechanisms of NCR acquisition and surface expression and their functional relevance on CD8+ T cells remained rather unexplored to date. Here we not only identified a population of human peripheral blood NKp30+CD8+ T cells but also showed that this population could be induced by IL-15 from NKp30−CD8+ T cells and further differentiated into a population of NKp30+(hi)CD8+ T cells coexpressing other activating NK receptors. Importantly, we showed that this population not only commenced to express an array of prototypical NK receptors but also underwent a broad molecular reprogramming involving the induction of several genes characteristic of innate immune cells. IL-15–induced NKp30+CD8+ T cells displayed a transcriptional landscape close to an NK cell transcription profile. It would be interesting also to compare IL-15–cultured NK cells in this analysis, since IL-15 might induce common responses/pathways in both cell types. Among the genes differentially regulated in IL-15–induced NKp30+ and NKp30− CD8+ T cell populations, FCER1G appeared unequivocally as the top up-regulated gene. FcεRIγ and CD3ζ are ITAM-bearing adaptor molecules described to couple with NKp30 (1, 4). We showed that IL-15 could drive FcεRIγ induction on CD8+ T cells and revealed it as crucial to enable NKp30 cell-surface expression and functionality. It is noteworthy that, although the CD3ζ adaptor is expressed constitutively in CD8+ T cells, it did not seem capable of supporting surface expression of NKp30, at least in the absence of FcεRIγ. Moreover, we showed that NKp30+CD8+ T cells up-regulated signaling molecules downstream of FcεRIγ, such as Syk and PLCγ2, that were important for NKp30-mediated signaling. Furthermore, our data suggest the involvement of PI3K-signaling pathways not only in the function but also in the generation of IL-15–induced NKp30+CD8+ T cells. Similar to NK cells (25), NKp30 receptor stimulation did not trigger response on resting naive NKp30+CD8+ T cells in the absence of cytokines. According to our data, IL-15 stimulation of CD8+ T cells appears to be required to induce effector potential by up-regulating NKp30 expression, inducing FcεRIγ, and enhancing the expression of additional downstream signaling molecules. In fact, the IL-15–induced reprogramming resulted in an effective NK-like function in this population. NKp30+CD8+ T cells were able to mount a rapid response to tumor targets bearing activating NK receptor ligands without the need of recognition of a specific antigen. Indeed, the NCRs expressed by IL-15–induced NKp30+CD8+ T cells were functional by themselves. Additionally, NKp30 showed the ability to synergize in inducing T cell function, suggesting a role for NCRs in decreasing the TCR activation threshold, which is particularly relevant for detecting and killing tumor cells. Tumor escape mechanisms, such as MHC class I down-regulation, frequently compromise the efficiency of classic T cell adoptive transfer therapies. Our data uncover a high therapeutic potential of the adoptive transfer of IL-15–induced NKp30+(hi)CD8+ T cells for immunotherapy of cancer, not only by the demonstrated capacity to eliminate tumor cells in a NK-like manner, but also by the promising prospect of combining it with TCR specificity, e.g., by ectopically expressing tumor-reactive TCRs. In the future, it will be important to further confirm the antitumor activity of the NKp30+CD8+ T cell population expanded from patients against autologous tumor targets. Additionally, by demonstrating the possibility of generating NKp30+CD8+ T cells in vivo by IL-15/IL-15Rα complex injections, this work opens a perspective on the already promising potential of IL-15 injection for antitumor therapy (11, 20), namely in clinical trials (51, 52). In these studies, NKp30 expression on CD8+ T cells was not monitored. Our results suggest that, upon IL-15 therapy, if high enough doses are reached in patients, NKp30+CD8+ T cells might be generated and expanded, contributing for the control of tumors expressing NK ligands. Although only a small percentage of NKp30+CD8+ T cells can be found in peripheral blood of healthy individuals, we believe that this population might play a relevant physiological role in the presence of increased levels of IL-15 during an immune response. Indeed, a scenario in which unstimulated naive cells require the presence of signals provided by cytokines to be able to respond would make sense. Moreover, according to our results, not only high levels of IL-15 but also the constitutive presence of the cytokine might be needed for the expansion and acquisition of function of this population. In fact, IL-15 expression has been associated with local tissue microenvironments and, although its expression is tightly regulated, IL-15 up-regulation has been reported in several autoimmune disorders (10, 11, 13, 14). Here, we have revealed increased frequencies of NKp30+CD8+ T cells in the blood of SLE patients, indicating that this population can be also generated under pathophysiological conditions associated with increased levels of IL-15. It is noteworthy that FcεRIγ-expressing T cells were reported in SLE, reinforcing our finding (53). Our data suggest an IL-15–induced CD8+ T cell differentiation program from NKp30−CD28+CD56−NCR−FcεRIg− → NKp30intCD28+CD56−NCR−/lowFcεRIγlow → NKp30hiCD28−CD56+NCR+FcεRIγ+, with concomitant acquisition of innate-like features and function. It would be of interest to evaluate other pathologies associated with dysregulated IL-15 production by addressing the presence and role of NKp30-expressing CD8+ T cells, not only systemically, but particularly in the affected organs and tissues with high local IL-15 levels.
The presented concept that a classic adaptive immune cell can acquire innate characteristics blurs the distinction between the strict dichotomic classifications of adaptive and innate. Indeed, in the last years, the notion that NK cells can also acquire adaptive and “memory-like” features has been rising (54–57). Schlums et al. (43) recently described that “NK-like memory cells” found in human CMV-seropositive donors displayed reduced levels of FcεRIγ, correlating with FCER1G promoter hypermethylation. Also, Syk and the transcription factor PLZF were shown to be down-regulated in those adaptive NK cells (43, 57, 58). Here we uncover IL-15 as a signal capable of inducing FCER1G promoter demethylation in the NKp30+CD8+ T cell population. The expression of FcεRIγ was concomitantly accompanied by Syk and PLZF expression in our IL-15–induced innate-like CD8+ T cells. Notably, we could also detect FCER1G and ZBTB16 transcripts in the circulating resting NKp30+CD8+ T cell population. This remarkable parallel between adaptive NK cells downregulating FcεRIγ and innate CD8+ T cells acquiring FcεRIγ makes it appealing to place FcεRIγ as a key molecule bridging innate and adaptive immunity. By showing that IL-15 can induce a wide reprogramming on CD8+ T cells toward the acquisition of innate characteristics, we smudge the distinction between what is idiosyncratically innate or adaptive and reinforce the concept that CD8+ T cells are intrinsically plastic, capable of changing even beyond the borders of innate and adaptive immunity. Also, by uncovering this level of plasticity, we were able to generate a unique population of innate-like NKp30hiCD8+ T cells, a promising prospect for adoptive cell transfer immunotherapy.
Materials and Methods
CD8+ T cells were isolated using the Human CD8+ T Cell Isolation Kit, followed by CD8 MicroBeads (both from Miltenyi Biotec). Recombinant human IL-15 was purchased from R&D Systems. Cell lines, transductions, functional assays, qPCR, microarray analysis, PLA microscopy, pyrosequencing analysis, and statistics are described in detail in SI Appendix, SI Materials and Methods. NSG mice were used in animal experiments, approved by the “Regierungspräsidium Karlsruhe, Germany,” and all procedures are detailed in SI Appendix, SI Materials and Methods. The list of the differentially expressed genes can be found on Dataset S1.
Supplementary Material
Acknowledgments
We thank Annette Arnold for technical assistance. We are thankful to the FACS core facility from DKFZ for valuable support. We thank the microarray unit of the German Cancer Research Center (DKFZ) Genomics and Proteomics Core Facility for providing the Illumina Whole-Genome Expression Beadchips and related services. Support by the DKFZ Light Microscopy Facility is gratefully acknowledged. We thank the DKFZ animal facility staff for great assistance. We thank Prof. Dr. Schäkel (University of Heidelberg) for organizing healthy control blood collection. We are thankful to Dr. Maike Hoffman and Prof Dr. Robert Thimme (Universitätsklinikum Freiburg) and Prof. Dr. David A. Price (Cardiff University) for generously providing the HLA-A2-peptide loaded tetramers. B.B. and D.J. received funding from the German Federal Ministry of Research and Education (BMBF) Grant 01ZX1307B. This study was supported by the Deutsche Krebshilfe Grants 70112233 and 110442.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720564115/-/DCSupplemental.
References
- 1.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sivori S, et al. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J Exp Med. 1997;186:1129–1136. doi: 10.1084/jem.186.7.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vitale M, et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pende D, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moretta L, Bottino C, Cantoni C, Mingari MC, Moretta A. Human natural killer cell function and receptors. Curr Opin Pharmacol. 2001;1:387–391. doi: 10.1016/s1471-4892(01)00067-4. [DOI] [PubMed] [Google Scholar]
- 6.Kruse PH, Matta J, Ugolini S, Vivier E. Natural cytotoxicity receptors and their ligands. Immunol Cell Biol. 2014;92:221–229. doi: 10.1038/icb.2013.98. [DOI] [PubMed] [Google Scholar]
- 7.Brandt CS, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206:1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiegler N, et al. Downregulation of the activating NKp30 ligand B7-H6 by HDAC inhibitors impairs tumor cell recognition by NK cells. Blood. 2013;122:684–693. doi: 10.1182/blood-2013-02-482513. [DOI] [PubMed] [Google Scholar]
- 9.Fehniger TA, Caligiuri MA. Interleukin 15: Biology and relevance to human disease. Blood. 2001;97:14–32. doi: 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- 10.Budagian V, Bulanova E, Paus R, Bulfone-Paus S. IL-15/IL-15 receptor biology: A guided tour through an expanding universe. Cytokine Growth Factor Rev. 2006;17:259–280. doi: 10.1016/j.cytogfr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Waldmann TA. The biology of interleukin-2 and interleukin-15: Implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 12.Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 in trans to neighboring cells. Immunity. 2002;17:537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- 13.Jabri B, Abadie V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat Rev Immunol. 2015;15:771–783. doi: 10.1038/nri3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waldmann TA. Targeting the interleukin-15/interleukin-15 receptor system in inflammatory autoimmune diseases. Arthritis Res Ther. 2004;6:174–177. doi: 10.1186/ar1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klebanoff CA, et al. Determinants of successful CD8+ T-cell adoptive immunotherapy for large established tumors in mice. Clin Cancer Res. 2011;17:5343–5352. doi: 10.1158/1078-0432.CCR-11-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu P, Steel JC, Zhang M, Morris JC, Waldmann TA. Simultaneous blockade of multiple immune system inhibitory checkpoints enhances antitumor activity mediated by interleukin-15 in a murine metastatic colon carcinoma model. Clin Cancer Res. 2010;16:6019–6028. doi: 10.1158/1078-0432.CCR-10-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, et al. Interleukin-15 combined with an anti-CD40 antibody provides enhanced therapeutic efficacy for murine models of colon cancer. Proc Natl Acad Sci USA. 2009;106:7513–7518. doi: 10.1073/pnas.0902637106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoklasek TA, Schluns KS, Lefrançois L. Combined IL-15/IL-15Ralpha immunotherapy maximizes IL-15 activity in vivo. J Immunol. 2006;177:6072–6080. doi: 10.4049/jimmunol.177.9.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epardaud M, et al. Interleukin-15/interleukin-15R alpha complexes promote destruction of established tumors by reviving tumor-resident CD8+ T cells. Cancer Res. 2008;68:2972–2983. doi: 10.1158/0008-5472.CAN-08-0045. [DOI] [PubMed] [Google Scholar]
- 20.Waldmann TA. The shared and contrasting roles of IL2 and IL15 in the life and death of normal and neoplastic lymphocytes: Implications for cancer therapy. Cancer Immunol Res. 2015;3:219–227. doi: 10.1158/2326-6066.CIR-15-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer S, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 22.Mingari MC, Moretta A, Moretta L. Regulation of KIR expression in human T cells: A safety mechanism that may impair protective T-cell responses. Immunol Today. 1998;19:153–157. doi: 10.1016/s0167-5699(97)01236-x. [DOI] [PubMed] [Google Scholar]
- 23.Mingari MC, et al. Cytolytic T lymphocytes displaying natural killer (NK)-like activity: Expression of NK-related functional receptors for HLA class I molecules (p58 and CD94) and inhibitory effect on the TCR-mediated target cell lysis or lymphokine production. Int Immunol. 1995;7:697–703. doi: 10.1093/intimm/7.4.697. [DOI] [PubMed] [Google Scholar]
- 24.Pita-López ML, Pera A, Solana R. Adaptive memory of human NK-like CD8+ T-cells to aging, and viral and tumor antigens. Front Immunol. 2016;7:616. doi: 10.3389/fimmu.2016.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryceson YT, March ME, Ljunggren HG, Long EO. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol Rev. 2006;214:73–91. doi: 10.1111/j.1600-065X.2006.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Correia MP, et al. Hepatocytes and IL-15: A favorable microenvironment for T cell survival and CD8+ T cell differentiation. J Immunol. 2009;182:6149–6159. doi: 10.4049/jimmunol.0802470. [DOI] [PubMed] [Google Scholar]
- 27.Correia MP, Costa AV, Uhrberg M, Cardoso EM, Arosa FA. IL-15 induces CD8+ T cells to acquire functional NK receptors capable of modulating cytotoxicity and cytokine secretion. Immunobiology. 2011;216:604–612. doi: 10.1016/j.imbio.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Park YB, Kim DS, Lee WK, Suh CH, Lee SK. Elevated serum interleukin-15 levels in systemic lupus erythematosus. Yonsei Med J. 1999;40:343–348. doi: 10.3349/ymj.1999.40.4.343. [DOI] [PubMed] [Google Scholar]
- 29.Aringer M, et al. Serum interleukin-15 is elevated in systemic lupus erythematosus. Rheumatology (Oxford) 2001;40:876–881. doi: 10.1093/rheumatology/40.8.876. [DOI] [PubMed] [Google Scholar]
- 30.Robak E, et al. Proinflammatory interferon-gamma–Inducing monokines (interleukin-12, interleukin-18, interleukin-15)–Serum profile in patients with systemic lupus erythematosus. Eur Cytokine Netw. 2002;13:364–368. [PubMed] [Google Scholar]
- 31.Steel JC, Waldmann TA, Morris JC. Interleukin-15 biology and its therapeutic implications in cancer. Trends Pharmacol Sci. 2012;33:35–41. doi: 10.1016/j.tips.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delahaye NF, et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med. 2011;17:700–707. doi: 10.1038/nm.2366. [DOI] [PubMed] [Google Scholar]
- 34.Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011;32:50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meresse B, et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 36.Roberts AI, et al. NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J Immunol. 2001;167:5527–5530. doi: 10.4049/jimmunol.167.10.5527. [DOI] [PubMed] [Google Scholar]
- 37.Warnat P, Eils R, Brors B. Cross-platform analysis of cancer microarray data improves gene expression based classification of phenotypes. BMC Bioinformatics. 2005;6:265. doi: 10.1186/1471-2105-6-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan WK, et al. Multiplex and genome-wide analyses reveal distinctive properties of KIR+ and CD56+ T cells in human blood. J Immunol. 2013;191:1625–1636. doi: 10.4049/jimmunol.1300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bezman NA, et al. Immunological Genome Project Consortium Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 2012;13:1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang K, et al. Syk regulation of phosphoinositide 3-kinase-dependent NK cell function. J Immunol. 2002;168:3155–3164. doi: 10.4049/jimmunol.168.7.3155. [DOI] [PubMed] [Google Scholar]
- 42.Liang Y, et al. Demethylation of the FCER1G promoter leads to FcεRI overexpression on monocytes of patients with atopic dermatitis. Allergy. 2012;67:424–430. doi: 10.1111/j.1398-9995.2011.02760.x. [DOI] [PubMed] [Google Scholar]
- 43.Schlums H, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 2015;42:443–456. doi: 10.1016/j.immuni.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alonzo ES, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Q, et al. Umbilical cord blood T cells express multiple natural cytotoxicity receptors after IL-15 stimulation, but only NKp30 is functional. J Immunol. 2008;181:4507–4515. doi: 10.4049/jimmunol.181.7.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pievani A, et al. Dual-functional capability of CD3+CD56+ CIK cells, a T-cell subset that acquires NK function and retains TCR-mediated specific cytotoxicity. Blood. 2011;118:3301–3310. doi: 10.1182/blood-2011-02-336321. [DOI] [PubMed] [Google Scholar]
- 49.Meresse B, et al. Reprogramming of CTLs into natural killer-like cells in celiac disease. J Exp Med. 2006;203:1343–1355. doi: 10.1084/jem.20060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu J, et al. NKp46 identifies an NKT cell subset susceptible to leukemic transformation in mouse and human. J Clin Invest. 2011;121:1456–1470. doi: 10.1172/JCI43242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Floros T, Tarhini AA. Anticancer cytokines: Biology and clinical effects of interferon-α2, interleukin (IL)-2, IL-15, IL-21, and IL-12. Semin Oncol. 2015;42:539–548. doi: 10.1053/j.seminoncol.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conlon KC, et al. Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. J Clin Oncol. 2015;33:74–82. doi: 10.1200/JCO.2014.57.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Enyedy EJ, et al. Fc epsilon receptor type I gamma chain replaces the deficient T cell receptor zeta chain in T cells of patients with systemic lupus erythematosus. Arthritis Rheum. 2001;44:1114–1121. doi: 10.1002/1529-0131(200105)44:5<1114::AID-ANR192>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 54.O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 55.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Sullivan TE, Sun JC, Lanier LL. Natural killer cell memory. Immunity. 2015;43:634–645. doi: 10.1016/j.immuni.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol. 2016;16:112–123. doi: 10.1038/nri.2015.9. [DOI] [PubMed] [Google Scholar]
- 58.Lee J, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. 2015;42:431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.