Significance
The acquisition of agricultural techniques during the so-called Neolithic revolution has been one of the major steps forward in human history. Using next-generation sequencing and ancient-DNA techniques, we directly test whether Neolithization in North Africa occurred through the transmission of ideas or by demic diffusion. We show that Early Neolithic Moroccans are composed of an endemic Maghrebi element still retained in present-day North African populations, resembling the genetic component observed in Later Stone Age communities from Morocco. However, Late Neolithic individuals from North Africa are admixed, with a North African and a European component. Our results support the idea that the Neolithization of North Africa involved both the development of Epipaleolithic communities and the migration of people from Europe.
Keywords: ancient DNA, North Africa, Neolithic transition, paleogenomics
Abstract
The extent to which prehistoric migrations of farmers influenced the genetic pool of western North Africans remains unclear. Archaeological evidence suggests that the Neolithization process may have happened through the adoption of innovations by local Epipaleolithic communities or by demic diffusion from the Eastern Mediterranean shores or Iberia. Here, we present an analysis of individuals’ genome sequences from Early and Late Neolithic sites in Morocco and from Early Neolithic individuals from southern Iberia. We show that Early Neolithic Moroccans (∼5,000 BCE) are similar to Later Stone Age individuals from the same region and possess an endemic element retained in present-day Maghrebi populations, confirming a long-term genetic continuity in the region. This scenario is consistent with Early Neolithic traditions in North Africa deriving from Epipaleolithic communities that adopted certain agricultural techniques from neighboring populations. Among Eurasian ancient populations, Early Neolithic Moroccans are distantly related to Levantine Natufian hunter-gatherers (∼9,000 BCE) and Pre-Pottery Neolithic farmers (∼6,500 BCE). Late Neolithic (∼3,000 BCE) Moroccans, in contrast, share an Iberian component, supporting theories of trans-Gibraltar gene flow and indicating that Neolithization of North Africa involved both the movement of ideas and people. Lastly, the southern Iberian Early Neolithic samples share the same genetic composition as the Cardial Mediterranean Neolithic culture that reached Iberia ∼5,500 BCE. The cultural and genetic similarities between Iberian and North African Neolithic traditions further reinforce the model of an Iberian migration into the Maghreb.
One of the greatest transitions in human history was the transition from the hunter-gatherer lifestyle to farming. How farming traditions expanded from their birthplace in the Fertile Crescent has been a matter of contention. Two models have been proposed: one involving the movement of people; the other based on the transmission of ideas. Over the last decade, paleogenomics has been instrumental in settling long-disputed archaeological questions (1), including those surrounding the Neolithic revolution (2). Compared with the extensive genetic work done on Europe and the Near East, the Neolithic transition in North Africa, including the Maghreb, remains largely uncharacterized. Archaeological evidence suggests that some of the major innovations associated with the Neolithic, such as farming and pottery production, could have been introduced into northern Morocco through sea voyaging by people from Iberia or the central Mediterranean as early as around 5,400 BCE (3, 4). In fact, some of the Neolithic pottery recorded in North Africa strongly resembles that of European cultures, like the Cardial Early Neolithic (the Mediterranean early farmer culture located in Iberia; ref. 5). However, other innovations, such as some pottery traditions and bone and lithic technical customs, could be the result of in situ development from Epipaleolithic communities, indicating a strong continuity in the local population since the Late Pleistocene (6–10).
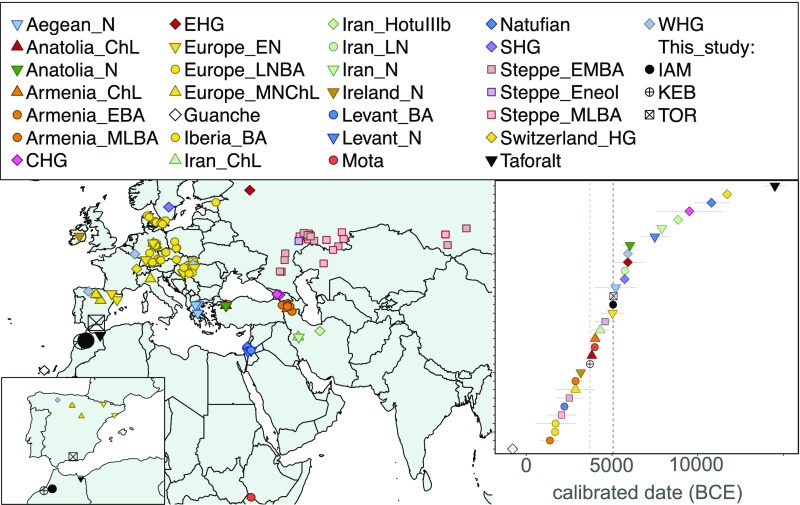
Genetic data from present-day populations (11–13) suggests that North African ancestry has contributions from four main sources: (i) an autochthonous Maghrebi component related to a back migration to Africa ∼12,000 y ago from Eurasia; (ii) a Middle Eastern component probably associated with the Arab conquest; (iii) a sub-Saharan component derived from trans-Saharan migrations; and (iv) a European component that has been linked to recent historic movements. Paleogenomic studies have begun to provide insights into North African prehistory (14–16), including the analysis of Later Stone Age samples from Morocco (17); however, no research to date has tested whether the Neolithic transition in the Maghreb was driven by local populations who adopted cultural and technological innovations or by the migration of people. Here, we perform genome-wide analysis of remains from the Early Neolithic site of Ifri n’Amr or Moussa (IAM) (∼5,000 BCE; n = 7) and the Late Neolithic site of Kelif el Boroud (KEB) (∼3,000 BCE; n = 8) (SI Appendix, Supplementary Note 1). To test possible migrations through the Strait of Gibraltar, we also analyzed human remains from the southern Iberian Early Neolithic site of El Toro (TOR) (∼5,000 BCE; n = 12) (Fig. 1). This Iberian Early Neolithic culture resembles certain early Maghrebi traditions (e.g., similar pottery decoration and similar bone and lithic tool productions), suggesting a North African influence (18) (SI Appendix, Supplementary Note 1). Including these southern Iberian samples in our analysis enabled a direct test of this hypothesis.
Fig. 1.
Geographical location (Left) and calibrated radiocarbon date (Right) of the samples included in this study, as well as other ancient DNA samples from the literature.
Results and Discussion
We sequenced 38 Illumina pair-end libraries from 27 individuals and selected the best-conserved libraries for subsequent analyses. Endogenous DNA content was generally low (2.88% on average) (SI Appendix, Supplementary Note 2). Depth of coverage was consistently improved when samples were enriched using baits targeting specific sites for the Multiethnic Genotyping Array (MEGA) (∼100×), compared with whole-genome capture (∼15×) (SI Appendix, Supplementary Note 2). Following enrichment, we generated 13 low-coverage genomes (five from IAM, four from KEB, and four from TOR), with MEGA coverage ranging from 0.04× to 1.72× depth and genome-wide coverage ranging from 0.01× to 0.74× depth (SI Appendix, Table S1). All samples considered in this study met the standard ancient DNA (aDNA) authentication criteria, including observation of DNA fragmentation (∼46 bp average read length) and damage patterns due to cytosine deamination toward the 5′ ends of molecules (SI Appendix, Supplementary Note 3).
Mitochondrial DNA (mtDNA) and Y chromosome haplogroups obtained for IAM (Moroccan Early Neolithic) and KEB (Moroccan Late Neolithic) suggest either a population replacement or an important genetic influx into Morocco between 5,000 and 3,000 BCE. IAM samples belong to the mtDNA haplogroups U6a and M1—both of which are associated with the back migration to Africa from Eurasia in Upper Paleolithic times (19, 20) and observed in Moroccan Later Stone Age individuals (17)—whereas KEB samples belong to haplogroups K1, T2, and X2, which are prominently found in Anatolian and European Neolithic samples (2, 21) (SI Appendix, Supplementary Note 4). Regarding the paternal lineages, IAM individuals carry Y chromosomes distantly related to the typically North African E-M81 haplogroup, while the Y chromosome from KEB belongs to the T-M184 haplogroup; although scarce and broadly distributed today, this haplogroup has also been observed in European Neolithic individuals (16) (SI Appendix, Supplementary Note 5). Both mtDNA and Y chromosome lineages (K1, J2, and T2 haplogroups and G-M201 haplogroup, respectively) for samples from TOR (Iberian Early Neolithic) are similar to those observed in Europe during Neolithic times (21).
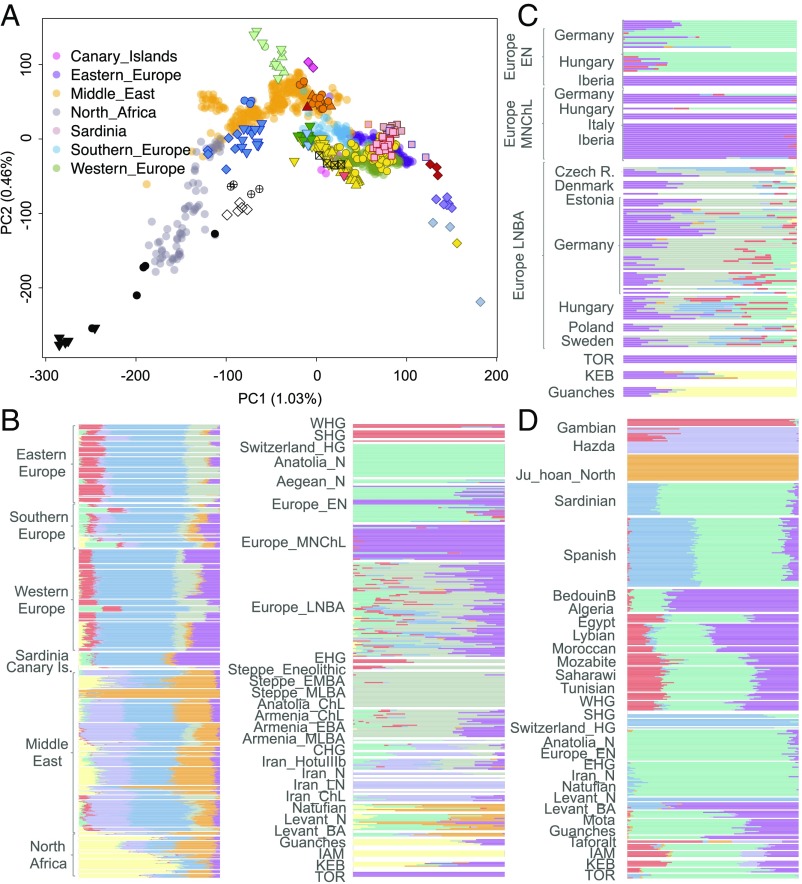
Ancient samples were projected on a principal components analysis (PCA) space, which was built using sub-Saharan African, North African, European, and Middle Eastern populations from the Human Genome Diversity Project (HGDP) dataset genotyped with MEGA. In the PCA, IAM samples are placed close to Mozabites (Berber group inhabiting the Mʾzab oases of southern Algeria), while Iberian Neolithic samples fall close to southern European populations (SI Appendix, Supplementary Note 6). As suspected from the mtDNA and Y chromosome data, KEB samples do not cluster with IAM and are placed in an intermediate position between IAM and TOR. We further explored the genetic structure of these samples using the program ADMIXTURE (22) (SI Appendix, Supplementary Note 7). At K = 5, TOR is composed of the component associated with the European Early Neolithic and IAM is composed of the North African component observed in Mozabites. KEB is placed in an intermediate position, with ∼50% each of European Early Neolithic and North African ancestries. It is worth mentioning that, compared with current North African samples, IAM and KEB do not show any sub-Saharan African ancestry in the MEGA-HGDP ADMIXTURE analysis, suggesting that trans-Saharan migrations occurred after Neolithic times. This could be in agreement with the analysis of present-day genome-wide data from Morocco, which estimated a migration of western African origin into Morocco only ∼1,200 y ago (11).
West Eurasian populations can be modeled as the admixture of four different ancestral components (2): Eastern and Western European hunter-gatherer and Iranian and Levantine Neolithic. We explored the placement of Moroccan and Southern Iberian Neolithic samples in this context and compared their genetic affinities to ancient and present-day West Eurasian and Levant populations in the Human Origins panel, as well as to other available aDNA population data. Interestingly, PCA revealed that IAM individuals are similar to North African Later Stone Age samples from the Taforalt site in Morocco, dated ∼15,000 y ago (Fig. 2 and SI Appendix, Supplementary Note 6). When projected, IAM samples are halfway between Taforalt and modern North Africans, in the Levantine corner of the PCA space (Fig. 2). Southern Iberian Neolithic individuals from TOR cluster with Sardinians and with other Anatolian and European Neolithic samples. Moreover, KEB samples are placed halfway between the IAM and Anatolian/European farmer clusters, in close proximity to Levant aDNA samples and also to Guanche samples (16) (from the indigenous population of the Canary Islands known to have a Berber origin; ref. 23). When compared using ADMIXTURE (SI Appendix, Supplementary Note 7), IAM samples possess ∼100% of a component partially shared by aDNA samples from the Levant (Fig. 2). This IAM-like component is observed mainly in modern North African individuals, following a west-to-east cline, and in the Guanches. Interestingly, the Early Neolithic individuals from Iberia form a different cluster from the Anatolian, Aegean, and European Early Neolithic samples, sharing their main component with Middle Neolithic/Chalcolithic samples. Lastly, KEB can be explained as having both IAM-like and Iberian Early Neolithic components (Fig. 2). The same admixture profile is observed in the Guanche samples, but the amount of IAM ancestry is consistently higher in all of the samples. Given that the Guanches could have originated in a different area of the Maghreb, this result might suggest that the European Neolithic impact in North Africa was heterogeneous.
Fig. 2.
Ancestry inference in ancient samples from North Africa and the Iberian Peninsula (ancient population codes are as in Fig. 1). (A) PCA analysis using the Human Origins panel (European, Middle Eastern, and North African populations) and LASER projection of aDNA samples. (B) ADMIXTURE analysis using the Human Origins dataset (European, Middle Eastern, and North African populations) for modern and ancient samples (K = 8). (C) Detailed ADMIXTURE plot for European Neolithic samples from previous analysis (K = 8). (D) Detail of ADMIXTURE analysis using the Human Origins dataset (European, Middle Eastern, North African, and sub-Saharan African populations) for modern and ancient samples, including Taforalt.
Recent aDNA analysis of Moroccan Later Stone Age samples from the Taforalt site indicates that at least one-third of Taforalt ancestry derives from sub-Saharan African populations. When Taforalt and sub-Saharan African samples are included in the unsupervised clustering analysis (SI Appendix, Supplementary Note 7), we observe that IAM and Taforalt cluster together at K = 7, as observed for the MEGA-HGDP ADMIXTURE analysis. However, as reported by van de Loosdrecht et al. (17) for Taforalt, we can detect both West African (maximized in Gambians and Yoruba) and East African (maximized in Hazda) components in IAM at lower K values. In contrast, TOR does not show any sub-Saharan African ancestry and KEB is again in an intermediate position, with lower sub-Saharan African ancestry than IAM.
To compare our samples directly to the genomes of ancient and modern populations, we calculated pair-wise fixation index (FST) distances, which, unlike PCA and global ancestry analyses, are insensitive to the inclusion of large numbers of individuals from modern populations. FST values (noted in parentheses) indicate that the IAM samples are as differentiated from all other populations as Yoruba are from non-Africans (SI Appendix, Supplementary Note 9), with the exceptions of Taforalt (0.049) and, to a lesser extent, KEB (0.090; similar to the distance between Yoruba and Mbuti), the Guanches (0.125; similar to the distance between Somali and Mbuti), and modern North African populations (0.115 to 0.138). The same pattern is observed for Taforalt samples, which shows its lower FST distance with IAM, followed again by KEB (0.129), the Guanches (0.150), and modern North Africans (0.130 to 0.149). Given the relatively low heterozygosity and high identity-by-descent proportions observed in IAM (SI Appendix, Supplementary Note 8), this differentiation could be driven by isolation and genetic drift. IAM is divergent from the other populations, with the exception of populations that either could have shared genetic drift with them or received genetic influx from them. This raises the possibility that Taforalt/IAM people were isolated in North Africa since Paleolithic times, when a back migration from Eurasia brought mtDNA haplogroups M1 and U6 to the Maghreb (19). Although IAM is clearly more similar to KEB than most populations, the converse is not true. KEB has lower FST distances with ancient Anatolian (0.032), Armenian (0.025 to 0.053), European (0.036 to 0.041, excluding European hunter-gatherers), Levantine (0.020 to 0.079), and Iranian (0.081 to 0.081) populations, as compared with IAM. In the modern DNA reference panel, KEB is similar to North African, European, and Middle Eastern populations. Among the ancient populations, TOR is more similar to Middle Neolithic/Chalcolithic Europeans (0.029 to 0.031), and, among modern populations, TOR is more similar to those from Spain, North Italy, and Sardinia.
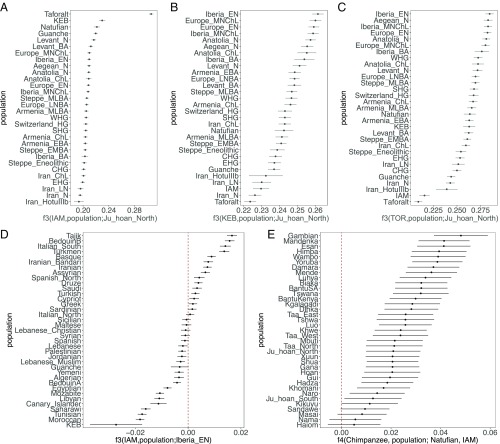
To further investigate the genetic affinities of IAM, KEB, and TOR samples, we conducted outgroup f3-statistic analysis (24). When using an outgroup population that has not experienced any postdivergence gene flow with either one of the compared populations, the f3-statistic calculated in the form f3(PopA, PopB; outgroup) allows us to determine the amount of shared drift between two populations, PopA and PopB. We selected the Ju\ʼhoan population as the outgroup, and compared our three ancient populations (IAM, KEB, and TOR) against all of the ancient populations in the Human Origins panel, as well as the Taforalt and Guanche samples. The highest f3 values for IAM are observed for Taforalt (0.3060) and KEB (0.2296). Consistent with previous results, when Taforalt, KEB, and Guanches (f3 = 0.2196) are excluded, IAM shares more drift with ancient Levantine populations, such as Epipaleolithic communities (Natufians; f3 = 0.2296) and Pre-Pottery Neolithic individuals (f3 = 0.2167) (Fig. 3 and SI Appendix, Supplementary Note 10), than with any other ancient population. This confirms previous results for Taforalt (17) indicating a Levantine intrusion in North Africa in Paleolithic times. To explore further the connection between IAM and Levantine populations, we performed an f4-statistic analysis to test whether IAM shares more alleles with any other population in the Human Origins panel (2, 25) than with ancient populations from the Levant (SI Appendix, Supplementary Note 10). When calculated in the form f4(IAM, chimpanzee; Levantine population, African population), a positive f4-statistic value would indicate that the test population shares more alleles with Levantine than with African populations. Consistently, all comparisons produced significant positive values and indicated a higher similarity of IAM with Natufians and Levantine farmers. This suggests that most of IAM ancestry originates from an out-of-Africa source, as IAM shares more alleles with Levantines than with any sub-Saharan Africans, including the 4,500-y-old genome from Ethiopia (14). To further test the hypothesis that IAM is more closely related to out-of-Africa populations, we determined whether we could detect Neanderthal ancestry in IAM, which is typical of non-African populations. A signal of Neanderthal ancestry has been detected in modern North African populations (26). A lack of Neanderthal ancestry in IAM would imply that the signal observed today is a product of more recent migration into North Africa from the Middle East and Europe in historical times. Compared with the Neanderthal high coverage genome sequence from Altai (27) and the low-coverage sequence from Vindija Cave (28), and using the S statistic (24), we detected a Neanderthal introgression signal into IAM, suggesting derivation from the same event shared by non-African populations (SI Appendix, Supplementary Note 10).
Fig. 3.
Outgroup f3-statistic for IAM (A), KEB (B), and TOR (C); admixture f4-statistic for IAM (D) and admixture f3-statistic for KEB (E).
FST and outgroup-f3 distances indicate a high similarity between IAM and Taforalt. As observed for IAM, most Taforalt sample ancestry derives from Epipaleolithic populations from the Levant. However, van de Loosdrecht et al. (17) also reported that one third of Taforalt ancestry was of sub-Saharan African origin. To confirm whether IAM individuals show a sub-Saharan African component, we calculated f4(chimpanzee, African population; Natufian, IAM) in such a way that a positive result for f4 would indicate that IAM is composed both of Levantine and African ancestries. Consistent with the results observed for Taforalt, f4 values are significantly positive for West African populations, with the highest value observed for Gambian and Mandenka (Fig. 3 and SI Appendix, Supplementary Note 10). Together, these results indicate the presence of the same ancestral components in ∼15,000-y-old and ∼7,000-y-old populations from Morocco, strongly suggesting a temporal continuity between Later Stone Age and Early Neolithic populations in the Maghreb. However, it is important to take into account that the number of ancient genomes available for comparison is still low and future sampling can provide further refinement in the evolutionary history of North Africa.
Both FST and outgroup-f3 statistical analyses indicate that KEB shares ancestry with IAM, as well as more genetic drift with Neolithic and Chalcolithic populations from Anatolia and Europe, with the highest shared genetic drift appearing in Iberian Early Neolithic samples (Fig. 3 and SI Appendix, Supplementary Note 10). This pattern and the result from ADMIXTURE could be explained if the KEB population was a mixture between IAM-related and European Neolithic groups. To formally test this hypothesis, we used an admixture-f3 test (24). When calculated in the form f3(PopA, PopB; target), a negative value of the f3-statistic is indicative of the target population being a mixture of both PopA and PopB populations. For our analysis, we used KEB as the test population, IAM as a reference population, and one of the Anatolian and European Neolithic and Chalcolithic populations as the second reference population. All comparisons produced negative values of the f3-statistic, which suggests that the KEB population can be modeled as a mixture of IAM and Anatolian/European Neolithic.
TOR has more shared ancestry with Iberian Early Neolithic samples and other Neolithic and Chalcolithic populations from Europe. Archaeological studies have suggested that there was an Andalusian Early Neolithic culture with North African influences before the Cardial expansion into the Western Mediterranean basin (29). However, we observe that TOR samples have a similar genetic composition to that of Cardial individuals from Iberia, evidencing a common origin and ruling out an Andalusian Early Neolithic distinct from Cardial culture.
Lastly, although limited by low coverage, phenotypic predictions based on genetic variants of known effects agree with our estimates of global ancestry. IAM people did not possess any of the European SNPs associated with light pigmentation, and most likely had dark skin and eyes. IAM samples contain ancestral alleles for pigmentation-associated variants present in SLC24A5 (rs1426654), SLC45A2 (rs16891982), and OCA2 (rs1800401 and 12913832) genes. On the other hand, KEB individuals exhibit some European-derived alleles that predispose individuals to lighter skin and eye color, including those on genes SLC24A5 (rs1426654) and OCA2 (rs1800401) (SI Appendix, Supplementary Note 11).
Conclusion
Genetic analyses have revealed that the population history of modern North Africans is quite complex (11). Based on our aDNA analysis, we identify an Early Neolithic Moroccan component that is (i) restricted to North Africa in present-day populations (11); (ii) the sole ancestry in IAM samples; and (iii) similar to the one observed in Later Stone Age samples from Morocco (17). We conclude that this component, distantly related to that of Epipaleolithic communities from the Levant, represents the autochthonous Maghrebi ancestry associated with Berber populations. Our data suggests that human populations were isolated in the Maghreb since Upper Paleolithic times. Our hypothesis is in agreement with archaeological research pointing to the first stage of the Neolithic expansion in Morocco as the result of a local population that adopted some technological innovations, such as pottery production or farming, from neighboring areas.
By 3,000 BCE, a continuity in the Neolithic spread brought Mediterranean-like ancestry to the Maghreb, most likely from Iberia. Other archaeological remains, such as African elephant ivory and ostrich eggs found in Iberian sites, confirm the existence of contacts and exchange networks through both sides of the Gibraltar strait at this time. Our analyses strongly support that at least some of the European ancestry observed today in North Africa is related to prehistoric migrations, and local Berber populations were already admixed with Europeans before the Roman conquest. Furthermore, additional European/Iberian ancestry could have reached the Maghreb after KEB people; this scenario is supported by the presence of Iberian-like Bell-Beaker pottery in more recent stratigraphic layers of IAM and KEB caves. Future paleogenomic efforts in North Africa will further disentangle the complex history of migrations that forged the ancestry of the admixed populations we observe today.
Materials and Methods
Measures to avoid and monitor contamination from modern DNA were applied, at all times, during sample manipulation. aDNA was extracted from teeth or bone, built into double-stranded indexed libraries, and sequenced on an Illumina NextSeq 500 (SI Appendix, Supplementary Note 2). Due to the environmental conditions of the burial sites, we expected to recover low proportions of endogenous DNA from these ancient remains. To overcome limitations due to DNA degradation, we applied two different capture methods to enrich for human reads (SI Appendix, Supplementary Note 2), with one targeting the whole genome (30) and one targeting the variants of the MEGA array (Illumina).
Reads were trimmed and adapters removed using AdapterRemoval (31) and then mapped to the human reference genome (hg19) using Burrows-Wheeler Aligner (32). Low-quality (mapping quality value <30) and duplicate reads were removed using SAMtools (33). MapDamage (34) was used to visualize misincorporation and fragmentation patterns and to rescale the quality of bases likely affected by postmortem damage. Confidence intervals of sex determination were calculated following Skoglund et al. (35). mtDNA haplogroups were determined using HaploGrep (36). Y chromosome haplogroup inference was carried out as in Schroeder et al. (37). As the reference panel, we used both the Human Origins panel (2) and the HGDP dataset genotyped with MEGA-ex (Illumina). For PCA, we projected the aDNA samples on the PCA space built with the modern dataset, using LASER (38) and the lsqproject option from smartpca (39). For PCA projection using LASER, analysis was performed without SNP calling. Briefly, sequence data for each reference population was simulated to match the coverage of the aDNA bam file, and the PCA space was built based on the simulated reference dataset along with the aDNA sample. Next, the low-coverage clustering was projected onto the full-coverage reference PCA space using Procrustes analysis. For PCA projection using smartpca and subsequent analyses, SNP calling was performed with SAMtools pileup to obtain the list of variants present on either HGDP or the Human Origins panel, filtering bases with BASEQ >30, trimming 4 bp at both ends, and keeping one allele at random. Admixture estimations were performed using ADMIXTURE software (22). FST distances were calculated using smartpca (39). Identity-by-descent proportions were estimated using PLINK (40), and heterozygosity estimations were produced using a newly developed method for low-coverage genomes (SI Appendix, Supplementary Note 8). Estimates of f statistics were calculated using admixtools software (24). All plots were prepared using R software (41). Detailed information about methods is included in SI Appendix.
Supplementary Material
Acknowledgments
C.D.B. and R.F. were funded by a grant from the National Science Foundation (1201234); R.F. was funded by a Fundación Canaria Dr. Manuel Morales fellowship; M.D.C.-M. and D.M.-S. were funded by Spanish Ministry of Economy and Competitiveness Grant HAR2016-78197-P; A.E.R.S. was funded by a Ciencia sem Fronteiras fellowship from the Brazilian Federal Agency for Support and Evaluation of Graduate Education; and B.S. and J.K. were funded by a grant from the Gordon and Betty Moore Foundation (GBMF-3804).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data are available through the European Nucleotide Archive (PRJEB22699). Consensus mtDNA sequences are available at the National Center of Biotechnology Information (accession nos. MF991431–MF991448).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800851115/-/DCSupplemental.
References
- 1.Nielsen R, et al. Tracing the peopling of the world through genomics. Nature. 2017;541:302–310. doi: 10.1038/nature21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazaridis I, et al. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linstädter J, Medved I, Solich M, Weniger GC. Neolithisation process within the Alboran territory: Models and possible African impact. Quat Int. 2012;274:219–232. [Google Scholar]
- 4.Zilhão J. Early prehistoric navigation in the Western Mediterranean: Implications for the Neolithic transition in Iberia and the Maghreb. Eurasian Prehistory. 2014;11:185–200. [Google Scholar]
- 5.Martínez-Sánchez RM, Vera-Rodríguez JC, Pérez-Jordà G, Peña-Chocarro L, Bokbot Y. The beginning of the Neolithic in northwestern Morocco. Quat Int. 2018;470:485–496. [Google Scholar]
- 6.Mulazzani S, et al. The emergence of the Neolithic in North Africa: A new model for the eastern Maghreb. Quat Int. 2016;410:123–143. [Google Scholar]
- 7.Barton N, et al. Human burial evidence from Hattab II Cave and the question of continuity in late Pleistocene–Holocene mortuary practices in Northwest Africa. Camb Archaeol J. 2008;18:195–214. [Google Scholar]
- 8.Lubell D, Sheppard P, Jackes M. Continuity in the Epipalaeolithic of northern Africa with emphasis on the Maghreb. In: Wendorf F, Close A, editors. Advances in World Archaeology. Academic; New York: 1984. pp. 146–191. [Google Scholar]
- 9.Linstädter J. Epipalaeolithic-Neolithic-transition in the Mediterranean region of Northwest Africa. Quartär. 2008;55:41–62. [Google Scholar]
- 10.de Groote I, Humphrey LT. Characterizing evulsion in the Later Stone Age Maghreb: Age, sex and effects on mastication. Quat Int. 2016;413:50–61. [Google Scholar]
- 11.Henn BM, et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arauna LR, et al. Recent historical migrations have shaped the gene pool of Arabs and Berbers in North Africa. Mol Biol Evol. 2017;34:318–329. doi: 10.1093/molbev/msw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fadhlaoui-Zid K, et al. Genome-wide and paternal diversity reveal a recent origin of human populations in North Africa. PLoS One. 2013;8:e80293. doi: 10.1371/journal.pone.0080293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallego Llorente M, et al. Ancient Ethiopian genome reveals extensive Eurasian admixture throughout the African continent. Science. 2015;350:820–822. doi: 10.1126/science.aad2879. [DOI] [PubMed] [Google Scholar]
- 15.Schuenemann VJ, et al. Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat Commun. 2017;8:15694. doi: 10.1038/ncomms15694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Varela R, et al. Genomic analyses of Pre-European Conquest human remains from the Canary Islands reveal close affinity to modern North Africans. Curr Biol. 2017;27:3396–3402.e3395. doi: 10.1016/j.cub.2017.09.059. [DOI] [PubMed] [Google Scholar]
- 17.van de Loosdrecht M, et al. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science. 2018;360:548–552. doi: 10.1126/science.aar8380. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Borja P, Aura-Tortosa JE, Bernabeu-Auban J, Jorda-Pardo JF. Nuevas perspectivas sobre la neolitizacion en la cueva de Nerja (Malaga-España): La ceramica de la sala del vestibulo. Zephyrus. 2010;LXVI:109–132. [Google Scholar]
- 19.Pennarun E, et al. Divorcing the Late Upper Palaeolithic demographic histories of mtDNA haplogroups M1 and U6 in Africa. BMC Evol Biol. 2012;12:234. doi: 10.1186/1471-2148-12-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Secher B, et al. The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents. BMC Evol Biol. 2014;14:109. doi: 10.1186/1471-2148-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haak W, et al. Members of the Genographic Consortium Ancient DNA from European Early Neolithic farmers reveals their Near Eastern affinities. PLoS Biol. 2010;8:e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maca-Meyer N, et al. Ancient mtDNA analysis and the origin of the Guanches. Eur J Hum Genet. 2004;12:155–162. doi: 10.1038/sj.ejhg.5201075. [DOI] [PubMed] [Google Scholar]
- 24.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sánchez-Quinto F, et al. North African populations carry the signature of admixture with Neandertals. PLoS One. 2012;7:e47765. doi: 10.1371/journal.pone.0047765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prufer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortes Sanchez M, et al. The Mesolithic-Neolithic transition in southern Iberia. Quat Res. 2012;77:221–234. [Google Scholar]
- 30.Carpenter ML, et al. Pulling out the 1%: Whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am J Hum Genet. 2013;93:852–864. doi: 10.1016/j.ajhg.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindgreen S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res Notes. 2012;5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ginolhac A, Rasmussen M, Gilbert MT, Willerslev E, Orlando L. mapDamage: Testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27:2153–2155. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- 35.Skoglund P, Stora J, Gotherstrom A, Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Archaeol Sci. 2013;40:4477–4482. [Google Scholar]
- 36.Kloss-Brandstätter A, et al. HaploGrep: A fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 2011;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 37.Schroeder H, et al. Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean. Proc Natl Acad Sci USA. 2015;112:3669–3673. doi: 10.1073/pnas.1421784112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C, et al. FUSION Study Ancestry estimation and control of population stratification for sequence-based association studies. Nat Genet. 2014;46:409–415. doi: 10.1038/ng.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 40.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team 2008 R: A language and environment for statistical computing, ISBN 3-900051-07-0 (R Foundation for Statistical Computing, Vienna)., Available at www.R-project.org. Accessed March, 6, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.