Significance
Alternative splicing is a form of genetic regulation that enables the production of multiple proteins from a single gene. This study is one of the first to investigate variation in alternative splicing during a major evolutionary transition. We analyzed RNA from wild and domesticated sunflowers to examine differentiation in splice patterns during domestication. We identified divergent splice forms that may be involved in seed development, a major target of selection during domestication. Genetic mapping revealed that relatively few regulatory switches affecting many proteins have been altered in domesticated sunflowers. Our findings indicate that differences in splicing arose rapidly during a recent evolutionary transition and appear to contribute to adaptation and population divergence.
Keywords: alternative splicing, domestication, RNA-seq, quantitative trait loci
Abstract
Alternative splicing enables organisms to produce the diversity of proteins necessary for multicellular life by using relatively few protein-coding genes. Although differences in splicing have been identified among divergent taxa, the shorter-term evolution of splicing is understudied. The origins of novel splice forms, and the contributions of alternative splicing to major evolutionary transitions, are largely unknown. This study used transcriptomes of wild and domesticated sunflowers to examine splice differentiation and regulation during domestication. We identified substantial splicing divergence between wild and domesticated sunflowers, mainly in the form of intron retention. Transcripts with divergent splicing were enriched for seed-development functions, suggesting that artificial selection impacted splicing patterns. Mapping of quantitative trait loci (QTLs) associated with 144 differential splicing cases revealed primarily trans-acting variation affecting splicing patterns. A large proportion of identified QTLs contain known spliceosome proteins and are associated with splicing variation in multiple genes. Examining a broader set of wild and domesticated sunflower genotypes revealed that most differential splicing patterns in domesticated sunflowers likely arose from standing variation in wild Helianthus annuus and gained frequency during the domestication process. However, several domesticate-associated splicing patterns appear to be introgressed from other Helianthus species. These results suggest that sunflower domestication involved selection on pleiotropic regulatory alleles. More generally, our findings indicate that substantial differences in isoform abundances arose rapidly during a recent evolutionary transition and appear to contribute to adaptation and population divergence.
Alternative splicing is a mechanism that allows organisms to create an assortment of RNA transcripts and proteins by using information from a single gene. This is particularly important during development, when differentiated tissues manufacture proteins with different functions from the same pre-mRNA. For example, different tissues in maize express distinct splice variants, or isoforms (1). Plasticity in pre-mRNA splicing enables organisms to respond to environmental stress including extreme temperatures (2, 3) and drought (1, 4). Proteome diversity is greatly enhanced by splicing, with as many as 70% of genes undergoing alternative splicing in some plant species (5) and sometimes dozens of distinct isoforms produced from a single gene (6). There are many case studies in which alternatively spliced transcripts have been shown to affect important phenotypes; in plants, these include immunity (6), circadian rhythm regulation (7), and flowering time regulation (8). The functional diversity enabled by alternative splicing is therefore vital to the biology of complex organisms.
In addition to differential splicing between tissues, splicing patterns often differ between genotypes. Population genetic variation therefore includes variation in how transcripts are spliced. Luo et al. (9) identified thousands of differential splicing events among three radish samples. Although some studies report a high degree of splicing conservation among taxa (10), others report varying degrees of splicing differentiation among taxa, with more dissimilar groups having greater divergence in splicing (11). For example, there are genotype-dependent splicing differences between tomato and (i) its relative Solanum habrochaites in response to cold stress (2) and (ii) its wild ancestor in response to growth environment (12). Considerable evolution may be mediated by modifications in alternative splicing (11, 13, 14); however, little is known about the process of splice evolution and the role it plays in adaptation and speciation. Is splicing variation frequently targeted by selection, and does it contribute to reproductive isolation? Conversely, is splicing differentiation mainly a kind of near-neutral evolutionary noise that accumulates gradually through time and is largely unlinked to speciation?
Novel isoform variation originates when mutations affect how the splicing machinery, called the spliceosome, interacts with intron/exon boundaries. Sequence variation associated with novel isoforms may be classified into four distinct types. Mutations may convert an intronic sequence into an exon, known as exonization (15, 16). “Exon shuffling” creates novel isoforms via rearrangement or duplication of existing exons. “Transition” describes when local mutations cause an exon to be skipped and effectively transformed into an intron. The literature has emphasized the three aforementioned processes, which all involve changes in or near the gene sequence undergoing splicing, i.e., cis-effects (11, 13, 15, 16).
A fourth class of mutation can generate novel isoform variation via mutations in trans-regulatory loci that affect splice site recognition (15). Trans-regulatory loci that affect alternative splicing may include spliceosome snRNAs or proteins (17), serine/arginine (SR) proteins (18), or other types of regulatory loci. There is potential for a single regulatory allele to interact with other regulatory loci, i.e., epistasis, or affect multiple downstream splicing events, i.e., pleiotropy, resulting in a suite of novel proteins and corresponding phenotypic effects. The relative contribution of cis- and trans-regulatory mutations to the evolution of splicing variation has not been extensively investigated.
Domestication is an extreme evolutionary transition mediated by novel selective pressures, which results in altered demography, physiology, morphology, and potentially splicing patterns. Wild Helianthus annuus, the common sunflower, is distributed across most of North America in diverse habitats. H. annuus was domesticated approximately 5,000 y ago, and genetic and phenotypic variation in the domesticated lineages have been shaped by strong selection for agricultural productivity. Modern breeding, and potentially earlier domestication changes, have involved introgression from wild H. annuus and several other Helianthus species (19). Compared with wild genotypes, domesticated H. annuus shows distinct domestication-related phenotypes, including a larger, single flower head; smaller root system; weaker response to drought; and larger seeds. Domestication also resulted in changes in other seed-related phenotypes, including the losses of shattering, self-incompatibility, and seed dormancy. Ample genomic resources and infrastructure support the study of Helianthus, including seed banks, a reference genome (20), and high-quality genetic maps for several species. Combined, these elements make Helianthus an excellent system in which to investigate alternative splicing differentiation during population divergence.
Here we use seedling transcriptomes and existing genomic resources to examine the genetics of alternative splicing evolution. We find that, through the process of domestication, sunflowers have accumulated differences in the composition of isoforms produced from each gene. Next we use genetic mapping to determine whether splicing differentiation is mainly associated with genetic variation in or near the gene undergoing splicing (cis-effects) or if trans-regulation may play a role. Last, we analyze domestication-specific splicing patterns in diverse wild and early-domesticate sunflower genotypes to study the origins of divergent isoform accumulation.
Results
Divergence in Isoform Composition Between Domesticated and Wild Sunflower Genotypes.
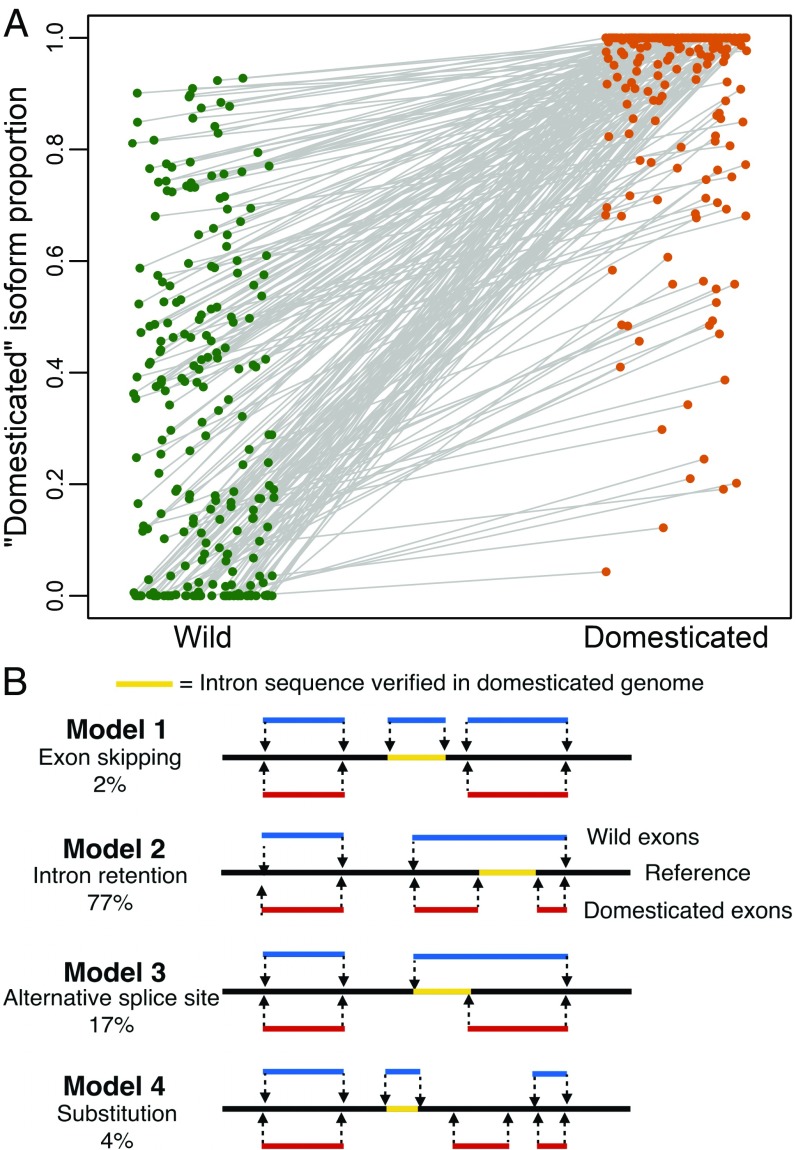
A large number of genes showed consistent differences in RNA isoform accumulation between the domesticated parental sample HA 89 and the extant wild parental sample Ann1238 (Fig. 1A). To examine variation in how transcripts are spliced, we first assembled isoform cDNA sequences de novo and aligned them to the sunflower reference genome (20). We required the aligned transcripts to conform to one of four models illustrated in Fig. 1B to verify splicing. Note that we included only the highest-confidence examples of alternative splicing. Nevertheless, our filtering scheme yielded a conservative list of 564 genes with verified alternative splicing.
Fig. 1.
(A) Parental types have differentiated isoform composition for 226 transcripts. Points show the mean proportion of the domesticated isoform in each parental type, where the domesticated isoform is the primary isoform of HA 89. Gray lines connect each isoform between the samples, with the slope representing the magnitude of differentiation. (B) Basic alignment configurations for validating alternative splicing.
For these loci, we tested for differences in isoform composition, i.e., statistical differences in the relative proportion of each isoform, between the domesticated and wild parental genotypes. However, compositional data violate the assumptions of ordinary statistical tests because components sum to one and no component is independent of the others (21). Therefore, we applied an isometric log-ratio (ILR) transformation (SI Appendix, Fig. S1) to the isoform proportions before testing for a difference in splicing composition. The ILR transformation is commonly used in the analysis of compositional data (22) and transforms the D-dimensional composition (in our case, two isoform proportions) to D-1–independent, transformed values in Euclidean space (one value representing isoform composition). We identified 226 transcripts with significantly different isoform composition between wild and domesticated parental samples after false discovery rate (FDR) correction (P < 0.05; Fig. 1A and SI Appendix, Table S1). In 110 of these cases, one isoform is completely absent in at least one parental type, indicating extreme changes in splicing regulation. The remaining cases represent shifts in isoform composition, ranging from subtle changes to more extreme changes.
In most differential splicing cases we examined (77%), the Ann1238 transcript retained an intron relative to the HA 89 transcript (Fig. 1B, model 2). In 17% of cases, the Ann1238 transcript had an alternate splice site relative to the HA 89 transcript. Substitution-type splicing made up 4% of cases, meaning both accession’s transcripts contained an exon not found in the other accession. Last, in 2% of cases, the HA 89 transcript skipped an exon relative to Ann1238. The relative frequencies of these four splice types found in the 226 differential splicing cases (ratio, 77:17:4:2) is not distinguishable (χ2 test, P = 0.36) from the set of 338 undifferentiated transcripts (ratio, 74:20:2:4).
Population Divergence in Isoform Composition.
Next we evaluated whether each of the 226 cases of differential splicing between HA 89 and Ann1238 are consistently diverged between a broader set of domesticated and wild populations. Isoform composition was compared between five domesticated and five wild H. annuus genotypes (not including HA 89 or Ann1238). By using one-tailed t tests, 19% of cases had divergent isoform composition (P < 0.05; SI Appendix, Fig. S2) between populations, and another 11% differed marginally (0.05 < P < 0.1). The number of important splicing cases may be underestimated as a result of the noisy character of transcript abundance data, which is affected by (i) sampling error, (ii) biological variability among cells and individuals, and (iii) technical factors during RNA extraction (23). Therefore, we suspect that some of the marginally significant transcripts may appear significant if additional samples were included. However, the otherwise flat distribution of P values obtained from this analysis indicates that most cases of differential splicing may not be consistently differentiated between domesticated and wild genotypes, although they are different between HA 89 and Ann1238. Because there is substantial genetically based variation within wild and domesticated groups, our results identify important segregating variation as well as the fixed wild/domesticated differences.
Domestication-Specific Function in Differentially Spliced Transcripts.
Sunflowers are cultivated primarily for their seeds, and breeders have focused on seed-specific characteristics during domestication such as larger, more edible fruit and faster seed development. Notably, wild sunflowers have a seed dormancy period lasting as long as several years, which makes germination unpredictable (24, 25). Seed dormancy has been mostly eliminated from domesticated sunflowers (26, 27), and domesticated seeds are strikingly larger than wild seeds. Therefore, we checked for seed-specific function in the transcripts with differentiated splicing patterns to examine whether splicing divergence has contributed to sunflower domestication. Of the transcripts with significant sequence similarity to annotated Arabidopsis protein sequences (BLASTX e-value <10−20), eight showed homology to proteins with seed-specific functions, e.g., embryo development ending in seed dormancy, which is a larger proportion than among the undifferentiated transcripts (χ2 = 6.0; P = 0.01; SI Appendix, Table S2). Apart from seed development, differentiated transcripts aligned to proteins involved in diverse processes ranging from basic cellular functions to plant-specific functions in the chloroplast. Therefore, splicing differentiation in sunflowers does not solely affect seeds, but seed-specific functions are disproportionately associated with splicing differences. To examine if any other functional categories including those not specific to domestication are overrepresented in the differentially spliced transcripts, we used an enrichment analysis in ErmineJ (28). Although no category showed significant enrichment after ErmineJ’s FDR corrections, the four seed-specific categories did have the strongest statistical support for enrichment among all 618 categories (P < 0.006; SI Appendix, Table S3).
Next we explored whether isoform composition in the differentiated transcripts predicts germination success in HA 89 × Ann1238 recombinant inbred line (RIL) seedlings grown in a common garden experiment. RIL sequences were aligned to the parental transcriptome to determine isoform composition, and principal component analysis was used to reduce the dimensionality of the 226 splicing patterns. The proportion of germinated seeds correlated with splicing principal component 1 (P = 0.02). This supports a functional connection between splicing variation and a domestication-fitness metric. Individual transcripts may have functional connections with germination, e.g., c96929_g1 (P = 0.001) and c57942_g1 (P = 0.003), but no tests met the threshold for statistical significance after FDR corrections.
Regulation of Differential Alternative Splicing.
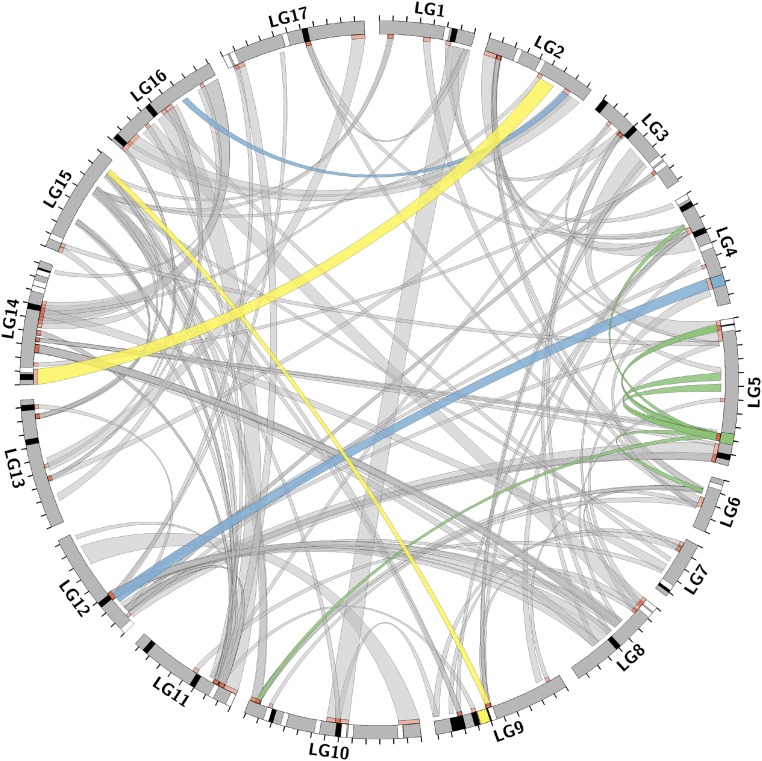
To map regions of the genome associated with splicing events that are differentiated between HA 89 and Ann1238, we treated isoform composition as a phenotype for quantitative trait loci (QTL) analysis. The ILR-transformed proportion of each isoform is unitless and back-transforms to the vector of proportions of the two isoforms, [pi1, 1 − pi1]. The mapping population of 100 RILs were genotyped at 20,000 markers, and we found significant association between isoform accumulation and genetic loci for 144 differential splicing cases (Fig. 2 and SI Appendix, Table S4). These QTLs appear to represent regions that directly or indirectly regulate splicing; however, we have not confirmed any functional relationships.
Fig. 2.
Differential splicing regulation diagram. The 17 linkage groups are depicted circularly, with clockwise megabase-pair tick marks. Links connect QTLs (orange-red) to genes with differential splicing with which they are associated. Link width corresponds to percentage variance explained. White bands are genes with splicing that is entirely cis-regulated. Black bands are QTLs that regulate splicing on the same chromosome but >1 cM away. Band widths do not represent QTL widths. Colored bands and links are referenced in the text. Figure made with Circos (29).
We were able to conclusively determine the position of the gene undergoing splicing relative to regulatory QTLs for 134 transcripts with differential isoform accumulation. Of these transcripts, 39 were only found to be cis-regulated (QTL ≤1 cM away from the gene whose transcript is being spliced), 80 cases were detected as only trans-regulated (QTL >1 cM away from the gene), and 15 cases supported a combination of cis- and trans-regulation. The mean percentage of variance in isoform composition explained by individual QTL was 32.2% (SD = 28.4) for cis-regulated splicing and 9.7% (SD = 16.4) for trans-regulated splicing. For alternative splicing associated with both cis- and trans-QTL, the cis-QTL usually represented most of the variance explained (mean fraction of total percentage variance explained, 0.79, SD = 0.17). Therefore, cis-QTLs generally explained a greater percentage of variance in isoform composition. We verified that the domesticated allele at each QTL was associated with predominance of the domesticated isoform for the majority of cases.
QTLs Contain Spliceosome Genes.
To determine if the QTL we identified contain spliceosome genes, protein sequences from the Solanum lycopersicum spliceosome pathway were obtained from the KEGG pathway database (30) and aligned to the sunflower reference genome (TBLASTN e-value <10−20). Forty-one of 108 QTLs (38%) contained sequences similar to sequences encoding spliceosome components or regulators (SI Appendix, Table S4). To determine the probability of observing this proportion of overlap by chance, we permuted the positions of QTL 1 million times and never observed a comparable proportion of QTLs containing spliceosome genes (P < 0.001). This implies that spliceosome proteins are overrepresented among the detected QTLs associated with differential alternative splicing. We do not claim that any specific spliceosome protein is causing differential alternative splicing, although we see clear statistical evidence that the QTLs associated with differential splicing in general contain some spliceosome function.
Consistent with this finding, a large proportion of QTLs were detected as affecting multiple isoform accumulation phenotypes that differ between HA 89 and Ann1238. The number of differential splicing cases associated with individual QTLs ranged from 1 to 13, with a mean of 2.15 (SD = 1.85). The most pleiotropic QTL (Fig. 2, green) is associated with differences in isoform accumulation for seven genes located within 1 cM of the QTL and another six genes that are more genetically distant or unlinked. In contrast, many spliceform differences have complex genetic bases, with some regulated by as many as seven distinct QTLs.
Evolutionary Origins of Differential Splicing Patterns.
Splicing patterns that differentiate HA 89 from Ann1238 were examined in various H. annuus ecotypes and other Helianthus species to determine the origins of variation in splicing. In most cases, the HA 89 splicing pattern was present in at least 1 of 10 Native American-domesticated landrace samples analyzed, suggesting that most splice patterns diverged early in the domestication process. However, there were two cases, transcripts c95368_g1 and c98741_g1, in which all landrace samples expressed the Ann1238 splicing pattern. This suggests that at least some HA 89 splicing patterns originated more recently, during modern breeding.
Next we examined whether the HA 89 splicing pattern exists among the standing variation in wild H. annuus. In most cases, the HA 89 splicing pattern was present in at least one of five samples from various wild H. annuus populations (not including Ann1238). However, in four cases, no wild H. annuus samples expressed the HA 89 splicing pattern (SI Appendix, Fig. S3). In three of those cases (Fig. 2, yellow) the HA 89 splicing pattern is present in other Helianthus species, including Helianthus argophyllus, Helianthus debilis, Helianthus deserticola, Helianthus petiolaris, and Helianthus praecox. Therefore, most splicing patterns observed in HA 89 were likely derived from standing variation in the ancestral populations, but we do see smaller contributions that likely result from introgression from other species. Furthermore, two QTLs from our mapping analysis overlapped with narrow regions of known genomic introgression from H. argophyllus (19). One QTL, on LG2, is associated with splicing of gene c110619_g2 on LG 16 (Fig. 2, blue). The other QTL is on LG4 and regulates splicing of two genes, c106588_g1 on LG 4 and c110699_g2 on LG12.
Discussion
Alternative Splicing Divergence.
Heritable differences that distinguish HA 89, a domesticated sunflower genotype, from Ann1238, a wild sunflower genotype, include changes in the composition of isoforms produced by alternative splicing. We discovered 226 cases of splicing differentiation and hypothesize that these differences have contributed to domestication. Studies that compare transcriptome-wide splicing differentiation between closely related genotypes are limited (but see refs. 3 and 31). Our study is unique because we examined the role and origins of alternative splicing variation during a recent and rapid evolutionary transition.
Human-mediated selection for agricultural productivity has shaped genetic and phenotypic variation in domesticated H. annuus, and has led to divergence from wild H. annuus (19, 20). Multiple lines of evidence support the putative relationship between some of the observed splicing differences between HA 89 and Ann1238 and selection pressures during the domestication process. Despite our small sample size, some differential splicing cases differed consistently between tested domesticated and wild sunflower lines. The five domesticated lines (and HA 89) were bred specifically for traits that are beneficial to commercial agriculture. Indeed, seed-development functions appear to be enriched in the differentiated transcripts. Likewise, germination success in the mapping population was correlated with splicing variation in the differentiated transcripts.
After conservatively filtering thousands of candidate transcripts down to several hundred clear examples of alternative splicing, 40% showed differential isoform composition between parental genotypes. Thatcher et al. (31) found ∼3% of splicing cases to be differentiated between two maize genotypes. Jakšić and Schlötterer (3) found <1% differential splicing between two Drosophila melanogaster genotypes and 10% when raised at extreme temperatures. The large degree of splicing differentiation we observed may be attributable to three factors. First, we found evidence for spliceosome differentiation, which may affect a large proportion of, or all, downstream splicing. Second, there was little variation among biological replicates for each parental sample, which led to small-magnitude differences appearing statistically significant. Last, some of the observed differences in isoform proportions might result from slight differences in the accumulation of plant tissues between wild and domestic sunflower seedlings.
Regulation of Differential Splicing.
Heritable differences in isoform composition indicate genetic variation at loci involved in regulating splicing. The differential splicing events examined in our study had an excess of trans-regulation, which contradicts current theory regarding novel splicing variation (15, 16). Results from other studies show 90% cis-acting QTLs (31), suggesting that splicing differentiation is caused primarily by sequence changes in or near the gene being spliced. In contrast, there is a deficit of experimental evidence for trans-regulatory splicing factors in alternative splicing evolution (15). Trans-acting loci may include spliceosome snRNAs or proteins (17), SR proteins (18), or other types of regulatory loci. We speculate that trans-regulated splicing may allow conservation of critical exonic sequences, while enabling their reordering to increase phenotypic diversity and adaptive potential. Regulation of splicing discussed here includes only alternative splicing that is differentiated between genotypes, and does not address variation in splicing among tissues or developmental stages within the same plant. Studies in other taxa are required to verify the generality of this result.
Regulation of alternative splicing in sunflowers is complex. Splicing patterns for each transcript are often associated with multiple QTLs, including combinations of cis- and trans-QTLs. The inverse is also true: a large proportion of the QTLs we identified are associated with altered splice patterns for more than one transcript, with some QTLs associated with dozens of differential splicing cases. Our findings show that QTLs associated with differential splicing are highly pleiotropic and suggest that splicing differentiation involves rewiring existing regulatory networks. Consistent with the highly pleiotropic trans-regulation identified via QTL mapping, we found that a disproportionate number of QTLs contain spliceosome protein coding genes that likely affect a wide range of downstream splicing activities. It may be an efficient evolutionary mechanism to allow a single regulatory mutation to alter many transcripts, producing a suite of novel proteins. This is consistent with the theory that alternative splicing complexity is associated with rate of evolution and speciation (11). Note, however, that a pleiotropic regulatory allele may confer some combination of advantageous, deleterious, or neutral effects for an individual’s phenotype. For example, a theoretical mutation might be subject to strong positive selection because it alters the structure of the spliceosome in a way that indirectly increases the mass of seed tissue, while simultaneously affecting the arrangement of leaf nodes or other “nontarget” phenotypes.
Framework for Alternative Splicing Evolution.
Domestication of H. annuus began approximately 5,000 y ago, and the genome of domesticated H. annuus primarily reflects wild H. annuus (19, 20). Consistent with the domestication history of sunflowers, we found that most differentiated HA 89 splicing patterns occur at low to intermediate frequencies in wild H. annuus. Most of these splice types are also expressed in the sunflower landraces (early domesticates), making it likely that splicing differentiation occurred early during the domestication process. However, modern introgression events from other Helianthus species during the past few hundred years have also shaped the domesticated sunflower genome (19, 20). Introgression can be a powerful tool for achieving desired phenotypes during crop domestication and improvement (32). Not unexpectedly, several splicing patterns and QTLs appear to be introgressed from other Helianthus species. Two introgressed splicing patterns are also clearly absent in the landraces, implying that they evolved relatively recently. Unexplained splicing variation may have come from novel mutations, introgression from species not examined in this study, or insufficient sampling in the examined populations.
A large proportion of differential splicing events between HA 89 and Ann1238 were of the intron retention splice type, which is the most commonly observed type of alternative splicing in plants (2, 5, 12). Note that the type of splicing discussed here refers specifically to splicing that differs between genotypes, which does not represent the full range of sunflower alternative splicing. However, similar proportions of each splice type were found in the full set of transcripts, including transcripts with undifferentiated splice patterns. This suggests that the type of splicing selected from standing variation during domestication appears to be unimportant and/or utilizes the same splicing machinery as general alternative splicing in sunflowers.
The results from this study lead us to propose the following framework for the role of alternative splicing in evolution. We show that frequency changes of existing alleles are sufficient to considerably alter how RNA is spliced. This suggests that alternative splicing is not rigidly conserved, and contributes to phenotypic variation within and between populations. In fact, some alternative splicing differentiation appears to be functionally important, underlying important domestication traits in sunflowers. Divergence at splicing regulatory loci may alter many downstream transcripts and phenotypic traits simultaneously. Although this may potentially facilitate rapid phenotypic evolution, such phenotypic change may not be universally beneficial and probably includes a haphazard mix of beneficial, neutral, and deleterious effects. The results from this study appear partially consistent with the regulatory hypothesis (33), which holds that morphological changes evolve mainly by altering expression of conserved proteins. Our study builds on this theoretical framework by demonstrating the importance of trans-regulated alternative splicing. We speculate that the degree of alternative splicing differentiation between natural populations will be less dramatic than that observed in domesticated sunflowers, which are the product of strong artificial selection. Additional research is required to determine whether alternative splicing divergence resulting from domestication is similar to alternative splicing divergence in nature, and whether allopatric vs. ecological divergence leads to different patterns of splicing differentiation.
Materials and Methods
Plant Material and RNA Sequencing.
HA 89 is an inbred line frequently used in research/plant breeding (US Department of Agriculture Ames 3963). Ann1238 was derived from material collected at Cedar Point Biological Station, Keith County, Nebraska. Three seedlings from each parental accession were grown in greenhouse conditions (34) to address stochastic variation in expression. Above-ground tissue for parentals and sixth-generation RILs was frozen in liquid nitrogen, and total RNA was extracted following standard protocols (35) (SI Appendix). Nonnormalized Illumina RNA-sequencing (RNA-seq) library preparation and sequencing on a HiSEq2000 system were performed at the Michael Smith Genome Sciences Centre in Vancouver, BC, Canada. All other samples were prepared for other studies (SI Appendix, Table S5).
Identification of Alternative Splicing.
Trimmomatic v0.32 was used to trim read ends with quality below 10 and remove reads shorter than 90 bp in the parental samples and RILs (70 bp in other samples). We used Trinity (2014-07-17) (36) to assemble a transcriptome containing alternative splicing information and align_and_estimate_abundance.pl (36) to estimate isoform abundances. This resulted in 159,764 transcripts with substantial expression [transcripts per million (TPM) > 1] from 104,732 hypothetical genes. The number of transcripts that were alternatively spliced was 25,893, representing the majority of known genes, with as many as 23 isoforms per transcript and a mean of 3.13 isoforms (SD = 1.92). To examine splicing variation, we first applied basic filters to each set of estimated isoforms: (i) sufficient total expression (TPM > 1) to retain biologically meaningful transcripts, (ii) exactly two isoforms, and (iii) all six parental samples show expression of at least one isoform. After basic filtering, half of the transcripts in our dataset had differentiated isoform composition (P < 0.05) between HA 89 and Ann1238. It is possible at this point that many alternative splicing examples represent paralogs, allelic variation of intron-sized indels, or other assembly errors. Therefore, we next applied the following conservative filters to our dataset; (i) the entire length of the isoform aligns (BLASTN ID > 90%) to a single region in the HA412-HO genome; (ii) the alignment conforms to one of the basic splicing models presented (Fig. 1B), as opposed to more complex or biologically unlikely alignment configurations; and (iii) the retained intronic sequence is present in the reference and in the Ann1238 isoform. Because there is relatively little genomic differentiation between inbred HA 89 samples and the HA412-HO reference, and we require the intron sequence retained by Ann1238 to be present in the HA412-HO reference, it is unlikely that a genomic insertion or deletion polymorphism is the origin of the two different transcripts in our dataset. Because we do not have an Ann1238 genome, it is possible that the few cases representing model 4 (Fig. 1B) are actually not substitution-type alternative splicing but genomic indels in Ann1238. As a post hoc check that our transcripts were assembled correctly, we aligned isoforms with verified splicing to Sanger-sequence datasets and found that our transcripts were reasonably represented (SI Appendix).
Differential Splicing.
To avoid proportions with values of zero, we first added the TPM of one read to each isoform expression value. The t.test function in R was used to test for a difference in ILR-transformed isoform composition, followed by adjustment for FDRs in R by using the package fdrtool (37). Genotypes analyzed in the population-level comparison are as follows: HA369, HA384, RHA274, sunrise, VNIIMK8931 (domesticated), Academy2, Academy7, LEW1, NEW, and TEW (wild). We chose these genotypes because RNA from each sample was sequenced by using Illumina, and their domestication history is unambiguous, e.g., genotypes marked as “weedy” were not included because the category name implies hybrid origins. To maximize statistical power, transcripts with less than 0.25 TPM for any sample were not analyzed. The expression values from each isoform were converted to proportions after adding 0.01 TPM to avoid zero proportions and then ILR-transformed. We used the one-tailed distribution in t tests to test specifically whether the splicing patterns in the domesticated and wild lines followed that of HA 89 and Ann1238, respectively.
Phenotype Associations.
We aligned transcripts with verified splicing to the TAIR10 and Araport11 protein BLAST tables from The Arabidopsis Information Resource (TAIR; www.arabidopsis.org/), retaining the best hit for each gene. The 226 transcripts with differentiated splicing patterns between Ann1238 and HA 89 aligned to 134 different protein sequences in the combined BLAST set (SI Appendix, Table S1). Transcripts that did not align well (e-value >10−20) were excluded. The 338 transcripts with undifferentiated splicing aligned to 224 different Arabidopsis proteins. Annotation for the Arabidopsis proteins were obtained from the “ATH_GO_GOSLIM” annotation table. To check for enrichment in other functional categories, we first aligned our sunflower transcripts to Arabidopsis thaliana amino acid sequences available from TAIR by using BLASTX (38). We filtered for BLAST hits with e-values <10−4, keeping the single best hit for each sunflower transcript. As such, multiple sunflower genes were assigned to the same A. thaliana gene. We then adopted the Gene Ontology categories for A. thaliana genes (available from TAIR) to their sunflower transcript homologs. P values from testing for splicing differentiation (shown here earlier) were used as input to an overrepresentation analysis using the “gene score analysis” method in ErmineJ. For the germination experiment, seeds from all RILs were scarified and placed on wet filter paper to germinate in the dark as in a previous work (39) at the University of British Columbia in Spring 2010 (data in SI Appendix, Table S6).
QTL Mapping.
SNPs were identified by aligning RNA-seq reads to the HA412-HO transcriptome (40) by using bwa mem v0.7.15. SNPs were called by using SAMtools v1.4.1. The number of SNPs that showed fixed differences between wild and domesticated parental samples is 62,000, distributed over 13,000 contigs. We obtained genetic map positions for each fixed parent SNP by aligning transcriptome contigs to the HA412-HO genome by using BLAST (e value ≤10−20; ID ≥90%) and linearly interpolating cM positions from an existing genetic map (20). Six thousand contigs had excellent hits on single chromosomes, producing base pair and genetic map positions for 26,700 SNPs, all of which were placed onto the genetic map between previously mapped markers. These SNPs were examined in the RILs, and filtered by heterozygosity (≤0.2), proportion of wild ancestry (>0.05), and missing data (<0.5), resulting in 20,000 filtered SNPs.
We used the R package “qtl” following recommendations in the R/qtl manual (41). We used 1,000 permutations to estimate a significance threshold (α = 0.05) for each phenotype (ILR-transformed isoform composition). We applied single-QTL analyses to the ILR-transformed data for each differentially spliced transcript. Nearby QTL peaks on the same chromosome were treated as distinct peaks if (i) they were separated by at least 10 cM and (ii) logarithm of the odds ratio (LOD) fell below the significance threshold between the peaks. Final QTL regions were established as the range within 15% of the LOD peak. Subsequently, we used the maximum LOD position of each QTL as the QTL position in a multiple-QTL model to partition the variance explained by individual QTLs. QTLs explaining less than 1% of variance were removed from further analysis. Last, we consolidated overlapping QTLs.
Comparative Analysis.
RNA-seq datasets from various sunflower species and ecotypes obtained from the sequence read archive (SRA) were trimmed and aligned to the HA 89+Ann1238 Trinity assembly. A splicing pattern threshold was calculated as the midpoint between the least extreme HA 89 and Ann1238 samples. Each sample was assigned as having the HA 89 splicing pattern if its proportion of the HA 89 isoform was on the HA 89 side of the threshold. If an individual had less than 0.25 total expression of the two isoforms, they were excluded from the analysis. A transcript was considered to be represented in the landraces if at least 9 of the 10 samples had nonmissing data and represented in wild H. annuus if all five samples had nonmissing data. Analyses were accomplished by using the aforementioned software, and custom python (v2.7.12) and R scripts (v3.3.3) are available at https://github.com/c70smith/SunflowerAlternativeSplicing.
Supplementary Material
Acknowledgments
This work was funded by Genome Canada and Genome BC (LSARP2014-223SUN), and the NSF Plant Genome Program (DBI-0820451 and DBI-1444522).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data reported in this paper have been deposited in the Sequence Read Archive (Bioproject no. PRJNA417714).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803361115/-/DCSupplemental.
References
- 1.Mei W, et al. A comprehensive analysis of alternative splicing in paleopolyploid maize. Front Plant Sci. 2017;8:694. doi: 10.3389/fpls.2017.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H, et al. A comparison of the low temperature transcriptomes of two tomato genotypes that differ in freezing tolerance: Solanum lycopersicum and Solanum habrochaites. BMC Plant Biol. 2015;15:132. doi: 10.1186/s12870-015-0521-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jakšić AM, Schlötterer C. The interplay of temperature and genotype on patterns of alternative splicing in Drosophila melanogaster. Genetics. 2016;204:315–325. doi: 10.1534/genetics.116.192310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thatcher SR, et al. Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 2016;170:586–599. doi: 10.1104/pp.15.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chamala S, Feng G, Chavarro C, Barbazuk WB. Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front Bioeng Biotechnol. 2015;3:33. doi: 10.3389/fbioe.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzo S, Jia Y. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci. 2009;177:468–478. [Google Scholar]
- 7.Filichkin SA, Mockler TC. Unproductive alternative splicing and nonsense mRNAs: A widespread phenomenon among plant circadian clock genes. Biol Direct. 2012;7:20. doi: 10.1186/1745-6150-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macknight R, et al. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell. 2002;14:877–888. doi: 10.1105/tpc.010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo X, et al. Comparative transcriptomics uncovers alternative splicing and molecular marker development in radish (Raphanus sativus L.) BMC Genomics. 2017;18:505. doi: 10.1186/s12864-017-3874-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iñiguez LP, Ramírez M, Barbazuk WB, Hernández G. Identification and analysis of alternative splicing events in Phaseolus vulgaris and Glycine max. BMC Genomics. 2017;18:650. doi: 10.1186/s12864-017-4054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 12.Wang G, Weng L, Li M, Xiao H. Response of gene expression and alternative splicing to distinct growth environments in tomato. Int J Mol Sci. 2017;18:E475. doi: 10.3390/ijms18030475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondrashov FA, Koonin EV. Evolution of alternative splicing: Deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 2003;19:115–119. doi: 10.1016/S0168-9525(02)00029-X. [DOI] [PubMed] [Google Scholar]
- 14.Gamazon ER, Stranger BE. Genomics of alternative splicing: Evolution, development and pathophysiology. Hum Genet. 2014;133:679–687. doi: 10.1007/s00439-013-1411-3. [DOI] [PubMed] [Google Scholar]
- 15.Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 16.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 17.Erkelenz S, et al. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19:96–102. doi: 10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch KW, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 19.Baute GJ, Kane NC, Grassa CJ, Lai Z, Rieseberg LH. Genome scans reveal candidate domestication and improvement genes in cultivated sunflower, as well as post-domestication introgression with wild relatives. New Phytol. 2015;206:830–838. doi: 10.1111/nph.13255. [DOI] [PubMed] [Google Scholar]
- 20.Badouin H, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546:148–152. doi: 10.1038/nature22380. [DOI] [PubMed] [Google Scholar]
- 21.Filzmoser P, Hron K, Reimann C. Univariate statistical analysis of environmental (compositional) data: Problems and possibilities. Sci Total Environ. 2009;407:6100–6108. doi: 10.1016/j.scitotenv.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Egozcue JJ, Pawlowsky-Glahn V, Mateu-Figueras G, Barcelo-Vidal C. Isometric logratio transformations for compositional data analysis. Math Geol. 2003;35:279–300. [Google Scholar]
- 23.McIntyre LM, et al. RNA-seq: Technical variability and sampling. BMC Genomics. 2011;12:293. doi: 10.1186/1471-2164-12-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heiser CB, Smith DM, Clevenger SB, Martin WC. The North American sunflowers (Helianthus) Mem Torrey Bot Club. 1969;22:1–218. [Google Scholar]
- 25.Brunick RL. Seed dormancy in domesticated and wild sunflowers (Helianthus annuus L.): Types, longevity and QTL discovery. PhD Thesis. Oregon State University; Corvallis, OR: 2007. [Google Scholar]
- 26.Snow A, Moran-Palma P, Rieseberg L, Wszelaki A, Seiler G. Fecundity, phenology, and seed dormancy of F1 wild-crop hybrids in sunflower (Helianthus annuus, Asteraceae) Am J Bot. 1998;85:794. [PubMed] [Google Scholar]
- 27.Gandhi SD, et al. The self-incompatibility locus (S) and quantitative trait loci for self-pollination and seed dormancy in sunflower. Theor Appl Genet. 2005;111:619–629. doi: 10.1007/s00122-005-1934-7. [DOI] [PubMed] [Google Scholar]
- 28.Gillis J, Mistry M, Pavlidis P. Gene function analysis in complex data sets using ErmineJ. Nat Protoc. 2010;5:1148–1159. doi: 10.1038/nprot.2010.78. [DOI] [PubMed] [Google Scholar]
- 29.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thatcher SR, et al. Genome-wide analysis of alternative splicing in Zea mays: Landscape and genetic regulation. Plant Cell. 2014;26:3472–3487. doi: 10.1105/tpc.114.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali ML, Sanchez PL, Yu SB, Lorieux M, Eizenga GC. Chromosome segment substitution lines: A powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa) Rice. 2010;3:218–234. [Google Scholar]
- 33.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 34.Burke JM, Tang S, Knapp SJ, Rieseberg LH. Genetic analysis of sunflower domestication. Genetics. 2002;161:1257–1267. doi: 10.1093/genetics/161.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawlowski K, Kunze R, De Vries S, Bisseling T. Plant Molecular Biology Manual. Springer; Dordrecht, The Netherlands: 1994. Isolation of total, poly (A) and polysomal RNA from plant tissues; pp. 231–243. [Google Scholar]
- 36.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strimmer K. fdrtool: A versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 38.McGinnis S, Madden TL. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seiler GJ. Germination and viability of wild sunflower species achenes stored at room temperature for 20 years. Seed Sci Technol. 2010;38:786–791. [Google Scholar]
- 40.Renaut S, et al. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat Commun. 2013;4:1827. doi: 10.1038/ncomms2833. [DOI] [PubMed] [Google Scholar]
- 41.Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. Vol 46 Springer; New York: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.