Significance
The aggregation of the amyloid- (A) peptide into amyloid fibrils is associated with Alzheimer’s disease, and several point mutations leading to early-onset disease have been identified in A. By studying the aggregation of five disease-related mutations in vitro, we rationalize their link to familial Alzheimer’s disease. We have determined the effect of mutations on the individual steps of the overall A42 aggregation reaction and find for four of the mutations a significant increase in the rate of self-replication of fibrils, a process that has been linked to the production of toxic oligomeric species. Furthermore, by investigating the nature of the mutation, we determine the importance of the charge and size of specific residues in the aggregation of the wild-type peptide.
Keywords: amyloid, aggregation mechanism, kinetic analysis, self-assembly, driving forces
Abstract
The aggregation of the amyloid-β (Aβ) peptide is linked to the pathogenesis of Alzheimer’s disease (AD). In particular, some point mutations within Aβ are associated with early-onset familial Alzheimer’s disease. Here we set out to explore how the physical properties of the altered side chains, including their sizes and charges, affect the molecular mechanisms of aggregation. We focus on Aβ42 with familial mutations—A21G (Flemish), E22K (Italian), E22G (Arctic), E22Q (Dutch), and D23N (Iowa)—which lead to similar or identical pathology with sporadic AD or severe cerebral amyloid angiopathy. Through global kinetic analysis, we find that for the E22K, E22G, E22Q, and D23N mutations, the acceleration of the overall aggregation originates primarily from the modulation of the nucleation processes, in particular secondary nucleation on the surface of existing fibrils, whereas the elongation process is not significantly affected. Remarkably, the D23 position appears to be responsible for most of the charge effects during nucleation, while the size of the side chain at the E22 position plays a more significant role than its charge. Thus, we have developed a kinetic approach to determine the nature and the magnitude of the contribution of specific residues to the rate of individual steps of the aggregation reaction, through targeted mutations and variations in ionic strength. This strategy can help rationalize the effect of some disease-related mutations as well as yield insights into the mechanism of aggregation and the transition states of the wild-type protein.
In 2017, the number of people suffering from dementia reached 50 million, with an annual increase of nearly 10 million new cases (1). As one of the most common neurodegenerative diseases, Alzheimer’s disease (AD) accounts for 60% to 70% of all dementia cases (1, 2). Several lines of evidence link the aggregation of the amyloid beta (A) peptide to the pathogenesis of AD: A plaques are present in the brain of affected individuals (3), and early-onset AD is observed in people with Down’s syndrome, in which case there are three copies of the A precursor gene (4–6), as well as in people with mutations that lead to increased A production, altered ratio between different A variants, or more aggregation-prone A variants (2, 7–9).
A is cleaved from an amyloid precursor protein (APP). The most common variants have 40 (A40) or 42 (A42) residues, while several other variants that differ in length at the N or C terminus have been discovered. Although A42 is expressed at a much lower level than A40, it shows higher cell toxicity and is found to be the initial and major component in the cerebral senile plaques, which implies that A42 plays a significant role in the pathogenesis of AD (7, 10–12).
In most AD cases, the disease appears to be sporadic, with an average onset age of 65 y, but for a small subset of all affected individuals, a genetic factor linked to the disease has been identified in APP or other proteins (13). Most of these mutations lead to the emergence of AD symptoms before 65 y of age, and the resulting disease is known as early-onset familial AD (FAD) (14).
In this study, we focus on familial mutations that occur inside the A region: A21G (Flemish), E22G (Arctic), E22K (Italian), E22Q (Dutch), and D23N (Iowa) (Fig. 1). Two nearly identical models of A42 WT fibrils have been obtained by solid-state NMR for independently prepared samples, implying a reproducible structure (15, 17). In these models, A21, E22, and D23 are exposed on the surface of the ordered part of the fibril (Fig. 1). A common feature of these mutations is a one- (E22Q, E22G, and D23N) or two- (E22K) unit change in the net charge. The WT peptide is expected to have a net charge between 3 and 4 at neutral pH. The charge leads to electrostatic repulsion between A monomers as well as aggregated species. This electrostatic repulsion plays a central role in attenuating the aggregation behavior of A (18) and is expected to be reduced in these less charged mutants.
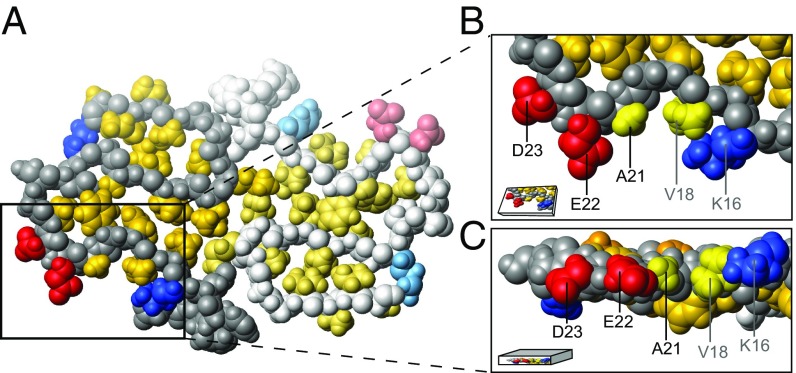
Fig. 1.
Location of residues K16, V18, A21, E22, and D23 in the A42 WT fibril model (5KK3.pdb) resolved by Colvin et al. (15). The structure within the fibril of two monomers within the same plane are shown in A, where the second monomer is displayed in paler color. The hydrophobic patch of one monomer is shown in a zoom-in top view (B) and side view (C). The image was prepared using MOLMOL (16) and shows 70% of the van der Waals radius.
The formation of A fibrils from a supersaturated solution of monomers has been inferred to occur through a double nucleation mechanism (19). While primary nucleation, involving monomers only, is a slow step with a high energy barrier, secondary nucleation involving monomers on the surface of fibrils has a significantly lower energy barrier (20). Thus, secondary nucleation occurs at a much higher rate during a large fraction of the reaction time course (19, 21). While the presence of secondary nucleation in the aggregation of A was originally suggested based on global fitting of nonseeded and seeded aggregation reactions, selective isotope labeling of monomer and fibril in separate experiments clearly identified the monomers as the origin of new aggregates in this fibril catalyzed secondary nucleation process, thus providing further evidence of its existence (19, 22).
The change in total net charge of A due to the E22Q, E22G, D23N, and E22K mutations will reduce the electrostatic repulsion between molecules. Therefore, an increase in aggregation rate, and possibly also a change in mechanism, is expected for these mutations. Many studies have been devoted to investigating the effect of familial mutations on aggregation kinetics. Most of these studies have used synthetic A mutant peptides at one or a few peptide concentrations and have indeed found higher aggregation propensity and cytotoxicity than for synthetic A WT (9, 23–27). To go beyond this expected behavior, here we study the concentration-dependent aggregation kinetics of five familial mutants (A21G, E22Q, E22K, E22G, and D23N) of recombinant A (M1-42) (elsewhere referred to simply as A42). We use nonseeded and seeded samples to investigate the mechanism of aggregation, rate constants of the underlying microscopic steps, and the electrostatic contributions to the observed effects. The aim is to find which microscopic steps in the aggregation process are affected the most by the different mutations and to infer which residues are most relevant for the steric and electrostatic effects during WT aggregation.
Results
Studies of A point mutations known to cause early-onset AD may provide clues as to the molecular driving forces for aggregation and pathology. While several familial mutants, including those in the current study, are reported to have different aggregation propensities than A42 WT, we here seek to understand the molecular basis by focusing on the aggregation mechanism of A42 and its perturbation by five single mutations found in families with increased risk for early development of AD.
Fibril Morphology of A Familial Mutants.
Mature fibrils formed from 10 M monomer [samples collected after reaching the plateau in thioflavin T (ThT) fluorescence; see ThT Fluorescence Versus Time] were observed using cryo-TEM for A42 WT and all five mutants. All six peptides form fibrils of similar width but with apparent differences in twist, length, and fibril–fibril association. Overall, the fibrils are hundreds of nanometers to a few micrometers long with a thickness of 5 to 10 nm with two filaments wound around each other and twisted to a different degree (Fig. 2). A42 WT, E22Q, and E22G have similar morphology, in which the fibrils show a highly twisted structure with a relatively short node-to-node distance. Fibrils of A21G, E22K, and D23N appear to be longer and slightly less twisted with a longer node-to-node distance; these fibrils are less tangled and more evenly distributed over the grids (Fig. 2, wider fields of view are shown in SI Appendix, Figs. S1 and S2). In addition to dispersed fibrils, large dense aggregate clusters that are composed of bundled fibrils were observed. These clusters are found to be surrounded by a network of fibrils, and the diameter of the dense core area is about 1 to 2 m. Aggregate clusters were found for WT, E22Q, E22G, and D23N and most prominently for E22G, where very few isolated fibrils were observed compared with other samples.
Fig. 2.
Cryo-transmission electron microscopy (TEM) images of fibrils formed by A42 WT or familial mutants. The samples initially contained 10 M monomeric A peptide, 6 M of ThT, 20 mM sodium phosphate, 200 M EDTA, and 0.02% NaN3 at pH 8.0 and were imaged after reaching the plateau of ThT fluorescence. (Scale bar, 100 nm.)
Aggregation Kinetics.
The aggregation of A42 WT and five familial mutants—A21G, E22K, E22Q, E22G, and D23N—was followed by recording the ThT fluorescence for nonseeded and seeded samples as a function of time at a range of peptide concentrations (0.4 to 10 M). These experiments start from supersaturated monomer solutions without or with preformed fibrils. Samples are prepared on ice and quickly transferred to 37 °C for incubation and ThT fluorescence measurements.
The aggregation kinetics data at all concentrations of each mutant were analyzed in four stages: (i) plotting of ThT fluorescence versus time, (ii) extraction and comparison of the aggregation half time versus concentration, (iii) fitting of a power function to the half times to estimate the scaling exponent, and (iv) global kinetic analysis—that is, normalization of each aggregation curve and fitting of various models to find the minimal model required to fit the data in a global manner at all peptide concentrations. Stages 1 to 3 represent preliminary analyses that serve to present and discuss findings that can be derived without modeling, while stage 4 constitutes the more stringent analysis to obtain a detailed microscopic mechanism of aggregation.
ThT Fluorescence Versus Time.
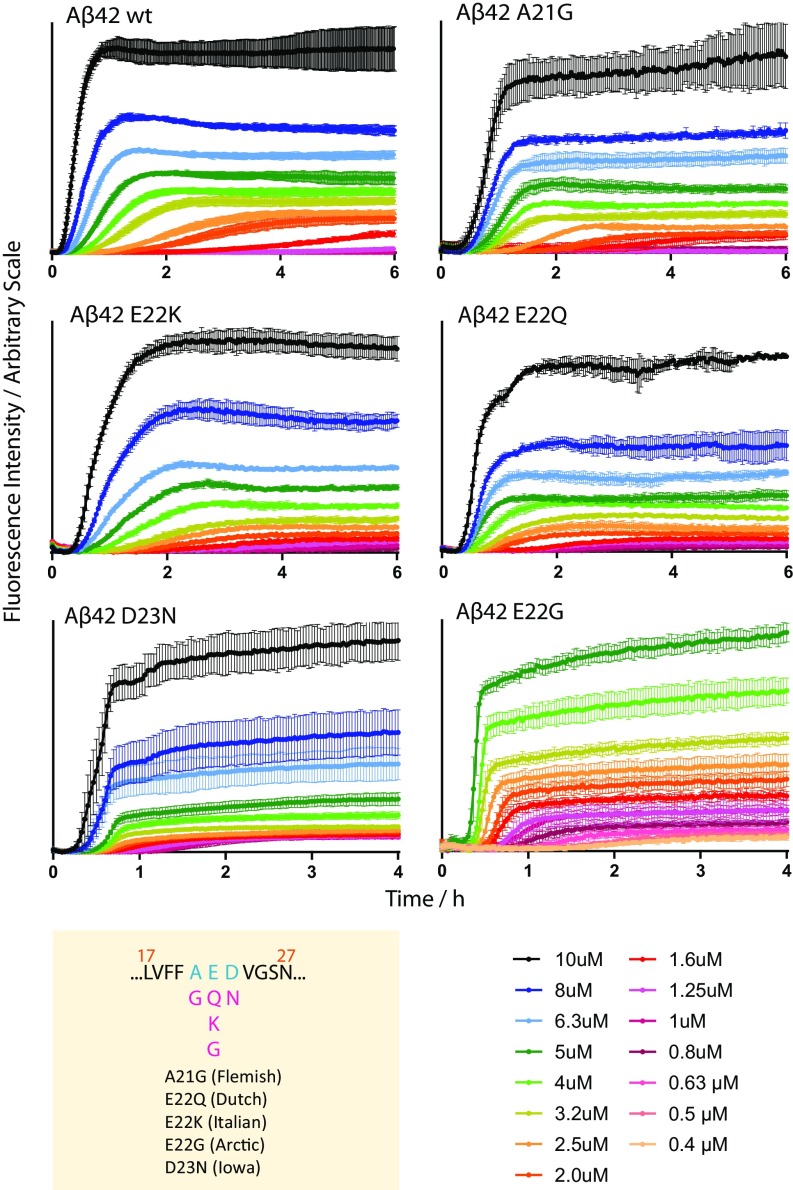
We find that the aggregation curves of the familial mutants are sigmoidal-like, similar to those of A42 WT, but with distinct differences in overall appearance and aggregation rate, depending on the mutation (Fig. 3). Both the aggregation time and the ThT fluorescence intensity depend on the peptide concentration. Moreover, the curve shape is clearly affected; the mutant A21G, E22K, and E22Q appear similar to the WT, whereas E22G and D23N show clear differences and aggregate much faster.
Fig. 3.
Concentration-dependent aggregation kinetics data of A42 WT and five familial mutants—A21G, E22K, E22Q, E22G, and D23N—at 37 °C. ThT fluorescence was monitored as a function of time for initially monomeric samples with peptide concentrations in the range of 0.4 to 10 M. Each color represents the average fluorescence signal intensity of four replicates at the same peptide concentration. All samples contain 6 M ThT, 20 mM sodium phosphate, 200 M EDTA, and 0.02% NaN3, at pH 8.0. The mutation sites of the five familial mutants are shown below the WT sequence (residues 17 to 27) in the yellow panel. Fits of these data are shown in Fig. 5, their half times are shown in SI Appendix, Fig. S4.
Half Time Versus Concentration.
To quantify the aggregation propensity and its monomer concentration dependence, we use the aggregation half time (), which is defined as the time point when the ThT intensity reaches half way between the initial baseline and the final plateau value. A full kinetic model to fit these data are discussed in the next section. The half time is plotted as a function of monomer concentration on double logarithmic scales (Fig. 4). The half time of all A42 familial mutants is dependent on the initial monomer concentration in the form of a power function, Eq. 1, as previously observed for A42 WT.
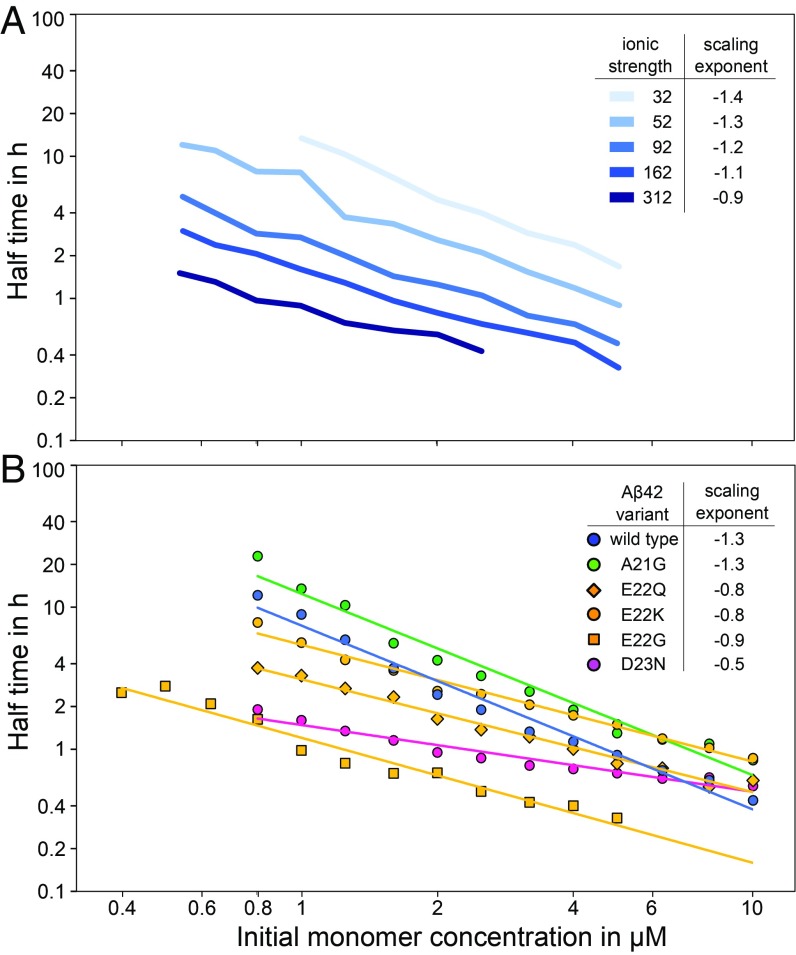
Fig. 4.
Half time of fibril formation as a function of monomer concentration on double logarithmic axes. (A) Previously published data of A42 WT at different ionic strengths from 32 mM (lightest blue) to 312 mM (darkest blue) (28). Monomer dependence decreases with increasing ionic strength. (B) A42 WT and each of the five familial mutants; representative aggregation curves are shown in Fig. 3. Each data point is an average over several repeats of the aggregation experiments, with 3 to 4 replicates at each concentration (resulting error bars are shown in SI Appendix, Fig. S5). The scaling exponent was estimated by fitting a power function to the data for each mutant (Eq. 1). The half time is concentration dependent, and all familial mutants except for A21G show a decreased monomer dependence (higher scaling exponent) compared with A42 WT.
A21G is the least aggregation-prone among all mutants studied, and it behaves similarly to A42 WT. E22K and E22Q aggregate faster than WT in the low concentration range. Even faster aggregation is seen for D23N and E22G. The increased aggregation propensity compared with WT for all mutants, except for A21G, is reflected in a shorter half time than for WT particularly at the lowest peptide concentrations (Fig. 4). The order of aggregation propensity at low peptide concentration of A42 WT and familial mutants observed here agrees with the result of a previous study of synthetic A40 peptides with familial mutations studied at one peptide concentration, 10 M, with agitation (9). However, that system differs due to the effect of C-terminal truncation (29) and the mechanical perturbation caused by agitation.
Scaling Exponent.
The scaling exponent (Eq. 1) was determined for each mutant to quantify the monomer dependence of the half time. In the double logarithmic plot (Fig. 4), the scaling exponent is the slope of the plotted straight line. The slight curvature seen for some of the mutants is a sign of saturation effects, as explained in the full fits below (30). The scaling exponents reported here are thus averages over the whole monomer concentration range sampled. A42 WT and A21G are the most strongly dependent on monomer concentration with the lowest scaling exponent = 1.3, followed by E22K, E22Q, and E22G with a of 0.8 to 0.9 and D23N with the highest scaling exponent, = 0.5, and hence the lowest monomer concentration dependence. Although the E22 mutations all display a similar of around 0.8, thus seen as parallel lines in Fig. 4, E22G is much more aggregation-prone than E22Q and E22K. The half time for E22G is 5 times shorter than for E22Q and 10 times shorter than for E22K over the entire concentration range. The aggregation propensity of D23N lies between that of E22G and E22Q in the concentration range studied, however it has a weaker concentration dependence and aggregates as fast as E22G at the lowest peptide concentration examined, 0.8 M (see Fig. 4). At high peptide concentrations of around 10 M, D23N aggregates with a similar half time as E22Q and WT, whereas below 1 M, D23N aggregates as fast as E22G. Importantly, if extrapolated to the even lower concentrations encountered in a physiological context, the increase in aggregation rate relative to WT of these weakly concentration-dependent mutants—D23N, E22G, and E22Q—is expected to become even more pronounced. This finding underscores the importance of performing experiments over a wide concentration range approaching the one found in vivo.
Global Kinetic Analysis.
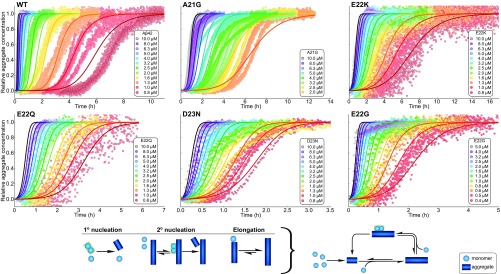
For a full mechanistic analysis, we performed global fits to the ThT data for each mutant using the online fitting platform AmyloFit (18). In this instance, “global” means the kinetic curves at all concentrations were fitted by the same rate constants and reaction orders, giving three free fitting parameters for each mutant. The possible origin of minor deviations between data and fitted curves at high peptide concentrations for D23N, E22G, and E22Q are discussed in SI Appendix, section 2. For all systems, the main source of new aggregates is a surface catalyzed nucleation process, and for none of the six peptides can the data be fitted by a model that includes primary nucleation and elongation only (SI Appendix, Fig. S3). The data for all mutants and the WT are well fit by a model in which the surface catalyzed secondary nucleation process was explicitly treated as a multistep process, akin to Michaelis–Menten-type enzyme kinetics (29). This model explicitly considers the individual events taking place during secondary nucleation, initial attachment of monomers or preformed oligomers, followed by formation and detachment of the nucleus (see mechanism in Fig. 5). At high enough monomer concentrations, the fibril surface will be fully covered, and this secondary nucleation process will be saturated, becoming independent of the solution monomer concentration and limited by the formation and detachment of the newly formed nuclei. The quantity describing the concentration range in which this saturation effect is significant is the Michaelis constant .
Fig. 5.
Global fitting of the data shown in Fig. 3 for A42 WT and five familial mutants—A21G, D23N, E22Q, E22K, and E22G. All data are well reproduced by global fits of models in which the majority of new aggregates is produced by fibril-surface catalyzed formation of nuclei from monomer. Furthermore, this process was found to display saturation, similar to that observed previously for A40 (29), for E22G, E22Q, E22K, and D23N. A schematic of the processes considered in this model and how they fit together in the aggregation reaction network is shown at the bottom. Lower concentrations of A42 WT and A21G are excluded from the fitting due to issues with reproducibility for reactions with such long half times.
Secondary nucleation is half saturated at a monomer concentration of . We find that this value is close to the maximum sampled monomer concentrations for WT and A21G, meaning saturation effects are small in these cases. [Note that in previously published work (19) the aggregation of the WT was fit with a model not explicitly including saturation, as the maximal sampled concentrations there were two-fold lower than in the present work, thus saturation was even less significant.] For D23N, all sampled concentrations are in the fully saturated regime. Thus, we can only conclude that the half saturation concentration, , is below the minimum sampled monomer concentration. To obtain an upper bound during fitting, we increased until a significant worsening of the fits was observed. This upper bound is = 0.3 M, and the other rate constants quoted for D23N are obtained when is set to this upper bound.
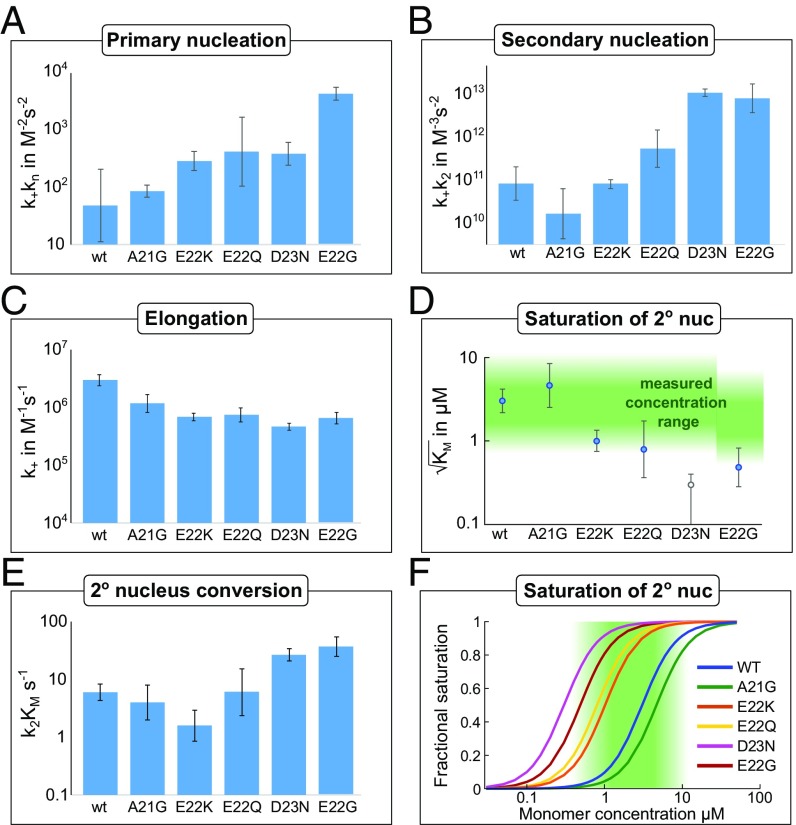
Compared with the WT, we find a substantial (100-fold or more) increase in the combined rate constant for elongation and secondary nucleation, k+k2, for E22G and D23N; a slight increase for E22Q; and comparable rates for E22K and A21G. The combined rate constant for elongation and primary nucleation, k+kn, shows the greatest increase for E22G; the other mutants only show a slight increase or are comparable to the WT. The detailed effects of mutations on each of the microscopic processes of aggregation are described below and summarized in Fig. 6.
Fig. 6.
Results of the global fitting of each mutant. Error bars are standard deviations over replicates and repeats of the experiment. (A) The combined rate constant of primary nucleation and elongation increases significantly, by up to over two orders of magnitude. (B) The combined rate constant of secondary nucleation and elongation also increases significantly, approximately paralleling the combined rate constant of primary nucleation and elongation. This corresponds to the low concentration limit where saturation effects are negligible; see E for the high concentration limit. (C) Estimate of the elongation rate constant from the TEM measurements (errors in that case are standard errors of the mean over the dimensions of the measured fibrils) and fitted rate constants. The values obtained all closely resemble those determined for the WT protein. (D) , the monomer concentration at which secondary nucleation is half saturated. The region of monomer concentrations sampled in this study is marked in green. A42 WT and A21G show little saturation effects, while D23N is fully saturated at all monomer concentrations sampled. Thus, for D23N, we can only obtain an upper bound for , which is displayed as an empty circle in the plot. (E) Rate of conversion of secondary nuclei. This corresponds to the high concentration limit where the system is fully saturated. (F) Illustration of the degree of saturation, using the value of from D. The region of monomer concentrations sampled in this study is marked in green.
Unseeded reactions yield only the combined rate constants, so to estimate the elongation rate constant separately, the average lengths determined from TEM measurements were used (see SI Appendix, section 1). The elongation rate constant was found to vary by less than an order of magnitude from the WT for all mutants (Fig. 6). Therefore, the observed effect of mutations originates mainly from changes to the nucleation rather than the elongation processes.
Seeding Experiment.
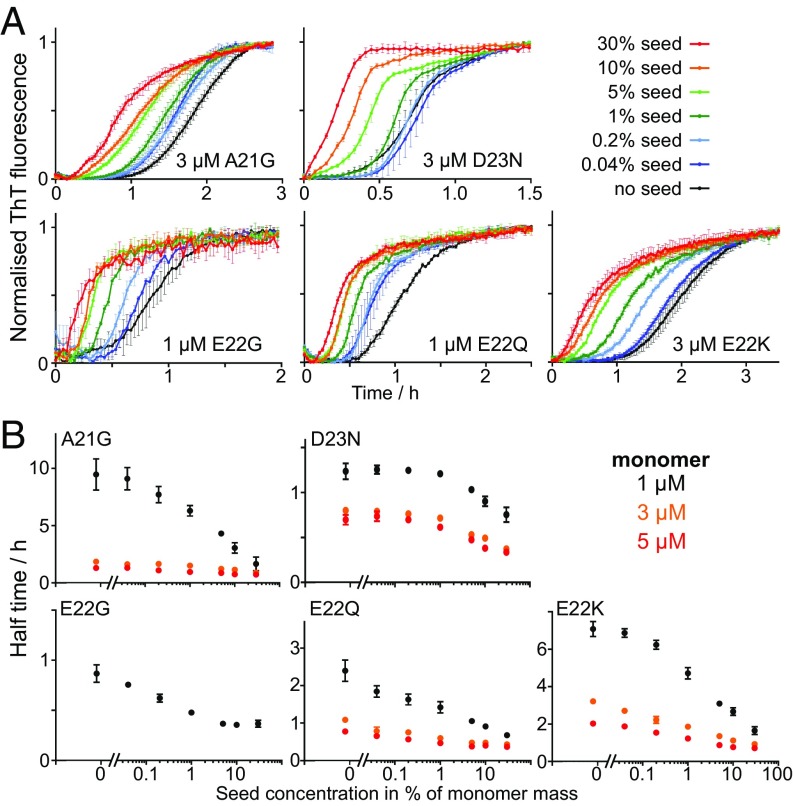
To confirm that secondary mechanisms do indeed dominate the production of new aggregates, as suggested by the global fitting of the data from nonseeded reactions, seeding experiments were performed. In the presence of a secondary nucleation mechanism, addition of a small amount of preformed seeds is expected to lead to a significant shortening of the half time. In this case, initial seeds can be amplified by the secondary process, bypassing a significant portion of the lag phase, whereas in the absence of a secondary process, initial seeds cannot multiply and have little effect on the aggregation kinetics (18).
Aggregation kinetics were thus repeated at monomer concentrations in the range of 1 to 5 M with preformed fibrils at 0%, 0.04%, 0.2%, 1%, 5%, 10%, and 30% in monomer equivalents. We find that the half time is shortened in the presence of seeds at all monomer concentrations studied, and the extent of acceleration is dependent on the seed concentration (see Fig. 7A and SI Appendix, Fig. S6). Most notable, the strong seeding effect at low concentrations of seed (1%) confirms that secondary nucleation is the dominant process of aggregate multiplication. An effect solely due to the direct elongation of seeds, as would be expected in the absence of a secondary process, would not be visible above the noise of the ThT fluorescence signal at such low seed concentrations. Finally, using the rate constants obtained in the fitting of the unseeded data, along with the elongation rate estimated from TEM, the expected curves for the seeded experiments were predicted and were found to be consistent with the data (see SI Appendix, Figs. S7 and S8).
Fig. 7.
Seeding experiments. (A) Normalized ThT fluorescence as a function of time of mutant A21G, E22Q, E22G, E22K, and D23N. Seeds of the same peptide were added at time 0 at concentrations of 0.04%, 0.2%, 1%, 5%, 10%, and 30% monomer equivalents, and the monomer concentration is indicated in each panel. All samples contain 6 M of ThT, 20 mM sodium phosphate, and 200 M of EDTA at pH 8.0. Data at additional monomer concentrations are shown in SI Appendix, Fig. S6. (B) Aggregation half time as a function of the logarithm of the seed concentration for A21G, E22Q, E22G, E22K, and D23N, at one or three different monomer concentrations as indicated in each panel.
Discussion
Reduced Electrostatic Repulsion and Mechanistic Shift.
The detailed kinetic experiments and analyses reveal that the charge substitutions not only affect the rate of aggregation but also cause a change in the underlying mechanism by shifting the relative importance of different microscopic steps. This shift in mechanism is evident in the weaker monomer dependence of for the mutants with reduced absolute charge: All charge substitution mutants aggregate faster than A42 WT in the low concentration range but are more similar to WT at high peptide concentrations.
A mechanism in which the formation of nuclei is dominated by the nucleation of monomers on the surface of existing fibrils, with the potential for saturation of this process, describes the data well for all mutants. In this description, the secondary nucleation process may become saturated and therefore independent of the monomer concentration at high monomer concentrations, when the binding sites on the fibril surface are fully covered in surface-bound species (29). Under such conditions, the conversion to and detachment of species formed after nucleation are rate-limiting at high monomer concentration. We find that secondary nucleation proceeds at a much higher rate for D23N, E22G, E22Q, and E22K compared with WT at low and physiological monomer concentrations such that generation of toxic oligomers from monomers of these mutants at the surface of fibrils may become even more severe than for WT (19, 31–33).
While the saturation effect is not observed to be significant at the A42 WT concentrations sampled here, the fact that it does become significant for the charge mutants, upon reduction of the intermolecular electrostatic repulsion, is in line with our expectations from earlier work (28). In this earlier study, we investigated the effect of electrostatic interactions on the aggregation of A42 by increasing the ionic strength of the solution to screen the electrostatic repulsion between the aggregating species. The overall aggregation propensity increased with increasing ionic strength, and secondary nucleation was found to reach saturation at higher ionic strengths. The half times of aggregation at different ionic strengths up to 312 mM are shown in Fig. 4A; note the flatter curves at higher ionic strengths indicative of saturation. Likewise, in a separate study, we found that a change in pH from 8.0 to 7.4, also accompanied by a decrease in absolute charge of the monomer, leads to a saturation of secondary nucleation (34).
In all these cases, the decreased repulsion between monomers and fibrils, due to the decreased electrostatic interactions, accelerates the microscopic steps that involve several charged species. This leads to an overall increase in aggregation propensity but also makes saturation of secondary nucleation more likely. The monomer-dependent attachment process in secondary nucleation will likely become more favorable when electrostatic repulsion is decreased, but the conversion and detachment process will be less affected, making the latter rate-determining. Therefore, under decreased electrostatic repulsion, secondary nucleation saturates. This means that its rate becomes independent of the monomer concentration and hence the dependence of the half times on the monomer concentration weakens for the charge mutants compared with the WT. However, reduction of charge through mutation is a more localized effect than shielding through increase of ionic strength, and the magnitude of the effect is expected to depend on the specific location of the mutation.
To determine the effect of mutations on the conversion of secondary nuclei—that is, the second step of the secondary nucleation process—we calculated its rate (29) (Fig. 6). Indeed, the rate is only significantly increased for the two fastest mutants, D23N and E22G, and even for those mutants, the effect is less than an order of magnitude. Thus, the origin for the overall increase in rate originates mainly from an increased saturation—that is, a more favorable interaction during the fibril binding step of secondary nucleation.
We will now discuss each of the mutants in turn. In particular, we attempt to determine the importance of electrostatic effects in the observed change in aggregation behavior. To determine the role of a mutated residue during a specific step of the aggregation reaction, it is assumed that the transition state structure of the mutants closely resembles that of the WT.
D23N.
Cryo-TEM images show that D23N forms long and thin fibrils and additional densely packed big fibril clusters (Fig. 2 and SI Appendix, Fig. S1). D23N is a relatively conservative substitution in terms of side-chain volume; however, at pH 8.0, D23N has one unit less negative charge than WT. Both the E22Q and D23N substitutions involve a change from carboxylate to carboxamide, but the enhancement of the aggregation rate is much larger in the case of D23N, suggesting that the charge at D23 is more important in the rate-determining steps of aggregation. The combined rate constant of elongation and primary nucleation (k+kn) shows a slight increase, and the combined rate constant of elongation and secondary nucleation (k+k2) increases drastically, to give the highest among all five mutants studied here (Fig. 6). Two factors contribute to the increased rate of secondary nucleation of D23N: The rate of conversion to secondary nuclei is increased slightly compared with the WT, and the coverage of fibrils by prenuclei is significantly higher. The former effect may be related to particular features of the short hydrophilic Asn– side chain. For example, the higher propensity of Asn– compared with Gln to form hydrogen bonds between the side chain and main chain of the same peptide may provide additional stabilizing interactions during the transition state of nucleus conversion (35, 36). The latter effect, the complete saturation of secondary nucleation, is evident in the weak concentration dependence with a scaling exponent of 0.5 and the low value of . The increased monomer–fibril interactions may be a result of the decreased electrostatic repulsion in D23N.
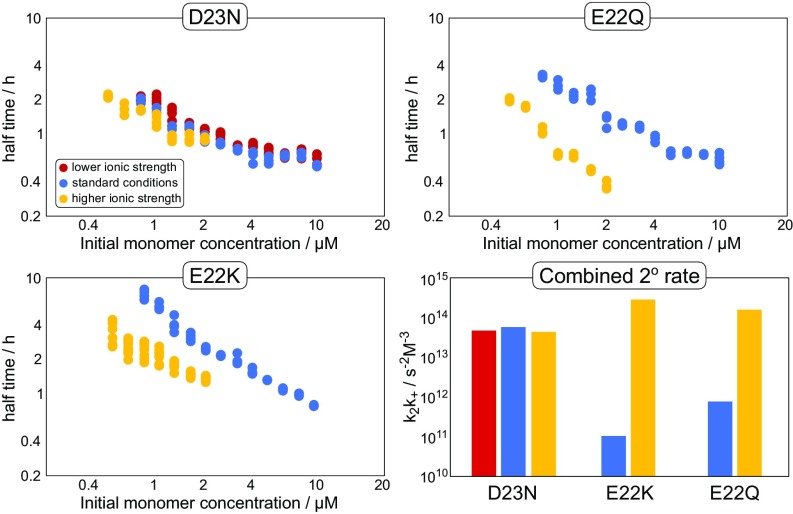
To determine the importance of charge in this observed change of aggregation behavior, we varied the ionic strength of the solution and performed further aggregation experiments of D23N. We find that both an increase of the ionic strength from ca. 60 to 162 mM as well as a decrease to 24 mM has no significant effect on the aggregation kinetics, as evident in the half times in Fig. 8. The fact that a change in ionic strength does not affect the aggregation behavior suggests that electrostatic interactions are not as important for D23N as they are for the WT. This is an indication that a significant part of the electrostatic effects observed in the WT aggregation may be due to the monomer–fibril association step in secondary nucleation, which is saturated and hence no longer kinetically visible in D23N. Furthermore, this finding suggests that D23 is one of the main contributors to electrostatic effects observed for A42 WT and as such is responsible for attenuating the monomer to fibril attachment rate during secondary nucleation. In the model of WT A42 fibrils (Fig. 1), the D23 side chain is exposed on the fibril surface. It is far away from both positive residues K16 and K28; on one side, it is next to the E22 side chain, and on the other side, the V24 side chain docks into the hydrophobic core, leading to complete solvent exposure to that side of D23. Furthermore, D23 is aligned with adjacent layers of monomers, producing bands of negative charge along the length of the fibril, whose removal could conceivably lead to a significant decrease in electrostatic repulsion between the fibril and any adsorbing species.
Fig. 8.
Effect of varying ionic strength on the half times of aggregation for the different mutants. Half times are plotted against initial monomer concentration on a double logarithmic plot. Bottom Right compares the combined rate constant of secondary nucleation and elongation for the different mutants and ionic strengths. Lower, standard, and higher ionic strengths are 24 mM, 60 mM, and 162 mM, respectively. While D23N is not significantly affected by changes in ionic strength, both E22K and E22Q are sped up significantly at higher ionic strengths, suggesting that charge still plays a role during the aggregation of these mutants.
E22Q.
Like A42 WT, E22Q forms short fibrils and also densely packed fibril clusters (Fig. 2 and SI Appendix, Fig. S1). E22Q is a relatively conservative substitution in terms of side-chain volume and hydrogen bonding capacity; however, at pH 8.0, E22Q has one unit less negative charge than WT. This mutation is comparable to D23N in that it replaces a carboxylate with a carboxamide; however, the effect seen here is much weaker than for D23N, both in the overall aggregation propensity as well as concerning the extent of saturation of secondary nucleation. Compared with WT, k+kn and k+k2 increase by about one order of magnitude (Fig. 6), but because of saturation effects, the half times are comparable at higher peptide concentrations (Fig. 4). The fact that the secondary nucleus conversion rate is comparable to that of the WT suggests that no important interactions are formed by the E22 during this conversion step.
To further strengthen our conclusion about D23 being the main group responsible for the electrostatic effects in the aggregation of WT A42, we performed another aggregation experiment with E22Q, increasing the ionic strength from 60 to 162 mM (Fig. 8). In line with our expectations, the overall rate of aggregation is significantly increased upon an increase of ionic strength, indicating that electrostatic repulsion is still an important contribution to the overall kinetics for E22Q. Thus, for A42 WT, the contribution to the electrostatic repulsion is smaller for E22 compared with D23. The fact that two charges in such close proximity have such a different effect implies that one of the residues is involved in specific interactions. One possibility could be the interaction of E22 with the closely located, positively charged, K16 side chain with the possibility for hydrogen bonding and charge–charge interactions leading to local charge compensation. Thus, attenuating the charge due to the E22 group would lead to a less pronounced charge effect compared with the D23.
E22K.
E22K is the least charged peptide in this study, with two units of less negative charge than WT. It pushes the previous mutation of E22Q even further by replacing the negatively charged side chain with a positively charged one, at the same time increasing the side-chain volume. Cryo-TEM images reveal distinct changes in fibril morphology; E22K forms longer fibrils with a longer node-to-node distance than WT and more loosely packed aggregates. The aggregation kinetics are slightly slower than for E22Q and at high peptide concentrations even slower than for the WT peptide. The slow kinetics at high concentrations is in line with the fact that this is the only mutant that shows, relative to the WT, a significantly decreased secondary nucleus conversion rate (Fig. 6), which will determine the overall secondary nucleation rate at high concentrations.
The low aggregation propensity, despite the largest reduction of absolute charge, further strengthens the conclusion that the charge at the E22 position is significantly less important than at the D23 position. As for E22Q, a significant increase in the aggregation propensity upon an increase in ionic strength is observed for E22K also. Furthermore, the lower aggregation propensity of E22K compared with E22Q, and in particular its lower secondary nucleus conversion rate, suggests that the steric constraints of a large lysine side chain at position 22 may affect the stability of the transition states during nucleus conversion. If steric effects are indeed important, then reduction of the size of the group at the 22 position should lead to a significant increase of the aggregation propensity.
E22G.
E22G forms short fibrils, similar to A42 WT, and also densely packed big fibril clusters (Fig. 2 and SI Appendix, Fig. S1). E22G has one unit less negative charge than WT at pH 8.0, similar to E22Q. However, while the E to Q mutation leaves the size of the side chain effectively unchanged, E to G significantly reduces its size. This change in size has a drastic effect on the aggregation propensity, making E22G the most aggregation-prone mutant in the concentration range studied here. Compared with E22Q, the combined rate constants k+kn and k+k2 increase by about two orders of magnitude for E22G (Fig. 6). The aggregation via a saturated secondary nucleation mechanism agrees with the previous result from a study of the synthetic form of E22G (23).
Similar to D23N, E22G shows increased saturation (although not to the same extent as D23N) as well as an increased rate of secondary nucleus conversion. The origin of the latter effect may include the much larger range of allowed dihedral angles for glycine compared with all other amino acids, potentially making lower energy transition states accessible (37). Substitution from glutamate or glutamine to glycine also increases significantly the overall hydrophobicity of monomers as well as fibrils (38). In the A42 WT fibril structure, E22 is located on the fibril surface, gating the hydrophobic patch formed by A21 and V18 (Fig. 1) (15). The glycine substitution disrupts this gating and widens the hydrophobic patch. Overall this increased hydrophobicity may be the reason for the significantly increased fibril affinity during secondary nucleation of E22G compared with both WT and E22Q.
E22G is not only the most aggregation-prone variant among all of the five familial mutants, it is also associated with very aggressive early-onset AD and rapid deposition of plaques in the brain of affected individuals (7). In particular, the fast rate of secondary nucleation might lead to increased oligomer formation at a relatively short time, which is in line with the previous finding of increased protofilament formation (7) and may contribute to the early-onset of AD pathology.
In summary, for the three mutations at position 22, the overall aggregation propensities thus fall in line with the side chain size, with the smallest (Gly) being the fastest and the largest (Lys) being the slowest. The trend does not follow the charge of the side chain, and the kinetics of the mutants are still susceptible to electrostatic shielding. Overall this suggests that the effect of the E22 group on the kinetics of aggregation of the WT is dominated by steric or hydrophobic effects, with electrostatic effects being less important.
A21G.
Unlike the other four mutants studied, the A21G mutation is not a charge substitution mutant. However, it is found in the same region of the peptide as the other mutations and is also linked to early onset FAD. At pH 8.0, both A42 WT and A21G have a net charge between 3 and 4. A21G represents a seemingly small substitution, removal of one methyl group, with severe consequences for early-onset AD or cerebral amyloid angiopathy (14). Cryo-TEM images show a distinct change in fibril morphology; longer and less twisted fibrils are found for A21G compared with WT, and less clumping of the A21G fibrils is observed (Fig. 2 and SI Appendix, Fig. S1). By contrast, the kinetic data reveal that A21G is comparable to the WT, and indeed slightly less aggregation-prone, over the entire concentration range studied, which is in line with the earlier result for A40 A21G relative to A40 WT at one peptide concentration (9). The scaling exponent of the half time versus peptide concentration is unchanged compared with WT. Global fitting reveals that the combined rate constants for A21G show values comparable to A42 WT (Fig. 6). High-resolution models of the A42 fibril structure (15, 17) reveal that A21 forms a hydrophobic patch with V18 and this patch is gated by two hydrophilic amino acids, E22 and K16, that are exposed on the fibril surface (Fig. 1). The mutation of A21 to G21 would not destroy this patch.
Given the lack of an increase in the aggregation propensity, the association of A21G with disease is likely associated to factors other than aggregation propensity. One such factor might then be the increased A42/A40 ratio and an almost doubled expression level (7). A higher resistance to degradation by neprilysin (9), an enzyme that plays a vital role in A catabolism, may also be of relevance. Crucially this finding highlights the complexity of the disease and that the intrinsic aggregation propensity is only one of multiple disease-relevant factors. A kinetic analysis as presented here allows one to quantify, and potentially rule out, the possible effect due to an altered intrinsic aggregation propensity.
Conclusion
While the exact sequence of events causing AD remain to be defined, the aggregation of the A peptide is considered a critical process in its pathology. We argue that, although most cases involve WT A, significant insights can be gained by studying the effect of point mutations causing early-onset AD on individual steps in the aggregation mechanism. The present study represents a level of investigation routed in recent advances of both a highly reproducible experimental setup and a theoretical framework for global analysis of data. In addition to finding strong effects on monomer-dependent secondary nucleation, we dissect the relative influence of side-chain size and charge.
Our results reveal that the aggregation of A42 familial mutants A21G, E22G, E22Q, E22K, and D23N is dominated by secondary nucleation of monomers on fibril surface, and for E22G, E22Q, E22K, and D23N, this process is even more dominant than for WT A42 and partly or fully saturated at the studied monomer concentrations. At peptide concentrations in the sub-M range, E22G, E22Q, D23N, as well as E22K aggregate significantly faster than A42 WT, with the most aggregation-prone mutants being E22G and D23N, while A21G behaves similar to A42 WT. In particular, we find a marked increase in the rate constant for secondary nucleation, a process that was identified as the main generator of A oligomers (19, 31). These oligomers, several studies indicate, are in turn the main species responsible for the toxicity to neuronal cells (19, 31–33). An increased rate of secondary nucleation, which is likely to also result in an increased amount of oligomers, will have implications for the involvement of the mutant A peptides in early-onset AD. By contrast, the lack of effects on the aggregation rate in the case of A21G suggests that interactions involving the A21 side-chain methylene group are not of great importance in the transition states of aggregation. It moreover highlights that factors other than the intrinsic aggregation propensity can be key in the pathology of AD.
A more in-depth analysis of the molecular origin of the effect of the mutations revealed the nature and importance of A21, E22, and D23 side chains in the different steps of aggregation of WT A42. In general, the lack of correlation between effects of mutations on the elongation and the nucleation rates suggests that the transition state for elongation differs clearly from that of nucleation, whereas the transition states of primary and secondary nucleation appear to be affected similarly. This finding also highlights the presence of several rate-determining steps that can be affected differently by mutations (30). In particular, we find that D23 is a main contributor to the electrostatic effects in the nucleation steps. In agreement with earlier work (28), these electrostatic interactions also appear to control the relative rates and thus the saturation of the secondary nucleation process, with the initial fibril attachment step being more affected by electrostatics. By contrast, the contributions of E22 to the nucleation steps are mainly of a hydrophobic and/or steric nature. In general, removal of exposed, charged side chains is likely to facilitate monomer–fibril association and thus speed up secondary nucleation by increasing coverage of fibrils, whereas substitutions promoting or inhibiting conformational rearrangements are likely to affect the conversion steps.
Thus, our analysis of the effect of mutations on the individual steps of aggregation has not only allowed us to rationalize the pathological effect of some disease-related mutants, it also provides insights into the nature of the interactions that govern the different steps of the assembly process.
Materials and Methods
Peptide Expression and Purification.
The genes coding for WT A (M1-42)—that is, with an N-terminal methionine preceding Asp1 (elsewhere referred to simply as A42)—and the five familial mutants A21G, E22G, E22Q, E22K, and D23N were produced using PCR with overlapping oligonucleotide and cloned into the PetSac vector (a Pet3a variant with NdeI and SacI cloning sites) and expressed in Escherichia coli as described before (39). A42 WT, E22K, and D23N were purified in the same way using ion-exchange chromatography (IEC) and size-exclusion chromatography (SEC). Inclusion bodies were isolated by dispersing cell pellet in solution with 10 mM Tris and 1 mM EDTA at pH 7.5, followed by sonication to break the cell wall and centrifugation to collect the insoluble inclusion body. After repeating the same steps three times, the supernatant in each step was discarded and the pellet was dissolved in 10 mM Tris/HCl, 1 M EDTA, pH 8.5 (buffer A) with 8 M urea and then diluted four times with buffer A to reach 2 M urea. Purification was followed by IEC, starting with incubating of diethylaminoethyl cellulose resin with inclusion body solution. The peptide was eluted with decreasing concentration of urea ranging from 1.8 M to 1 M and increasing salt concentration ranging from 50 mM to 200 mM NaCl in buffer A. A peptides were identified by SDS/PAGE for all of the eluting fractions. Eluates with target peptide obtained from IEC were applied on SEC after lyophilization and dissolution in 6 M GuHCl or passed through a 30 kDa molecular mass cutoff filter. After that, solutions of purified monomers were lyophilized and stored at 20 °C as powder until needed. E22G was purified following the same protocol as above with minor modification. For ion exchange, the NaCl concentration range was from 10 mM to 200 mM. The target peptide was eluted with 1 M urea and 100 mM NaCl. After ion exchange, samples were purified by two rounds of SEC. E22Q and A21G purification follows a similar protocol as A42 WT, with the addition of phenyl Sepharose chromatography. The samples were loaded on phenyl Sepharose column in 0.5 M (NH4)2SO4 at pH 8.0. The column was washed with a linear gradient from 0.5 M (NH4)2SO4 to water followed by elution in 8 M urea, pH 8.0. Eluents were applied on SDS/PAGE, and fractions with target peptides were lyophilized for further purification by SEC.
Sample Preparation.
All kinetic experiments start with SEC gel filtration to guarantee the pure monomeric form of A peptide. The lyophilized peptide powder was dissolved in 1.0 mL 6 M GuHCl for 15 to 20 min (to dissolve preexisting aggregates). After that the A GuHCl solution was loaded on a Superdex 75 10/300 GL column (GE Healthcare Life Sciences) using a fast protein liquid chromatography (FPLC) system (Biorad). The A peptide was eluted at 14 mL in phosphate buffer (20 mM NaH2PO4, 0.2 mM EDTA, 0.02% NaN3, pH 8.0). Peptide concentration was calculated from the integrated absorbance of the collected central portion of the monomer peak using Beer–Lambert law with an extinction coefficient = 1,440 M−1cm−1, which was determined by amino acid analysis (BMC).
Aggregation Kinetics Experiments.
ThT, a fluorescent probe that specifically binds to sheet-rich structures and gives enhanced fluorescence intensity, was used here to follow the A aggregation as a function of time. Purified A peptide was diluted to required concentrations with a ThT concentration of 6 M in low-binding tubes (Genuine Axygen Quality) and kept on ice before setting up the experiment. For concentration-dependent aggregation kinetics, samples were prepared at 12 concentrations ranging from 0.08 M to 10 M with logarithmic spacing. Each concentration contained a total volume of 320 L, and the solution was mixed by gently turning the tube upside down to avoid air bubbles. For seeding experiments, fibrils were prepared at a concentration of 10 M; sonicated for 2 min; diluted to 2 0.04%, 0.2%, 1%, 5%, 10%, and 30% of the monomer concentration equivalent; and used later as seeds. Samples were mixed by adding 120 L monomer to 120 L seed solution. Prepared peptide solution was loaded 80 L per well in a 96-well microplate with a PEG-coated surface (Corning) and measured in a Fluostar Omega or Fluostar Optima plate reader (BMG Labtech). Aggregation was monitored at 37 °C in quiescent condition. The ThT fluorescence was recorded with excitation at 440 nm and emission wavelength at 480 nm. Each experiment was repeated at least twice with triplicate or quadruplicate samples at all concentrations.
Cryo-TEM.
The samples used for cryo-TEM were prepared and incubated in the same way as the aggregation kinetics. Samples of 10 M A42 WT or the other familial mutants were incubated at 37 °C. Fresh fibrils were collected when the aggregation curves reach the plateau. A controlled environment vitrification system (CEVS) was used in sample preparation to maintain a stable temperature to avoid evaporation and heat transfer. A 5 L sample was loaded on a lacey carbon-filmed copper grid and blotted from the backside by using filter paper to generate a layer of solution that is less than 300 nm thick. The grid was then plunged into liquid ethane (around 180 °C) to flash freeze all samples “as is” on the grid and also to avoid ice crystals. Well-frozen samples were stored in liquid nitrogen until imaged. The electron microscope (Philips CM120 BioTWIN Cryo) was equipped with a postcolumn energy filter (Gatan GIF100), a cryoholder (Oxford CT3500), and its workstation for transferring samples. The acceleration voltage of the microscope was 120 kV. Images were recorded with a CCD camera under low electron dose conditions.
Data Analysis.
A typical aggregation curve is a sigmoidal curve with a lag phase, a growth phase, and a plateau. Half time, as a means to estimate the overall aggregation rate, is defined as the point where the ThT value is halfway between the initial baseline and final plateau values. Half time () of each concentration was plotted against monomer concentration () on a double logarithmic plot and fitted by a straight line to obtain the scaling exponent :
| [1] |
The differential equations describing the time evolution of aggregate mass concentration, , and aggregate number concentration, , are given by
| [2] |
| [3] |
where is the elongation rate constant; and are the rate constants of primary nucleation and secondary nucleation, respectively; and are the reaction orders of primary nucleation and secondary nucleation, respectively; and is the Michaelis constant of secondary nucleation. As the models are fit to the overall ThT signal, these rate constants represent weighted averages of any conformations that may be present in the fibrillar and monomeric states. An approximate solution to these equations can be obtained as
| [4] |
where the definitions of the parameters are
| [5] |
| [6] |
| [7] |
| [8] |
| [9] |
and
| [10] |
Again is the initial monomer concentration and , and , are the aggregate number and mass concentrations at the beginning of the reaction and at equilibrium—that is, after completion of the aggregation reaction. Note that for unseeded experiments (i.e., experiments starting from monomer alone, without preformed fibrils), the parameters of nucleation and elongation are coupled; that is, only and are constrained, not the rate constants individually.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803539115/-/DCSupplemental.
References
- 1.World Health Organization 2017 Dementia. Available at www.who.int/en/news-room/fact-sheets/detail/dementia. Accessed May 31, 2018.
- 2.Blennow K, de Leon MJ, Zetterberg H. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 3.Roth M, Tomlinson B, Blessed G. Correlation between scores for dementia and counts of ‘senile plaques’ in cerebral grey matter of elderly subjects. Nature. 1966;209:109–110. doi: 10.1038/209109a0. [DOI] [PubMed] [Google Scholar]
- 4.Rumble B, et al. Amyloid A4 protein and its precursor in Down’s syndrome and Alzheimer’s disease. N Engl J Med. 1989;320:1446–1452. doi: 10.1056/NEJM198906013202203. [DOI] [PubMed] [Google Scholar]
- 5.Sleegers K, et al. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–2983. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- 6.Rovelet-Lecrux A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 7.Nilsberth C, et al. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced Abeta protofibril formation. Nat Neurosci. 2001;4:887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 8.Scheuner D, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 9.Betts V, et al. Aggregation and catabolism of disease-associated intra-a mutations: Reduced proteolysis of aa21g by neprilysin. Neurobiol Dis. 2008;31:442–450. doi: 10.1016/j.nbd.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 11.Iwatsubo T, et al. Visualization of A 42(43) and A 40 in senile plaques with end-specific A monoclonals: Evidence that an initially deposited species is A 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 12.Shepherd C, McCann H, Halliday GM. Variations in the neuropathology of familial Alzheimer’s disease. Acta Neuropathol. 2009;118:37–52. doi: 10.1007/s00401-009-0521-4. [DOI] [PubMed] [Google Scholar]
- 13.Weggen S, Beher D. Molecular consequences of amyloid precursor protein and presenilin mutations causing autosomal-dominant Alzheimer’s disease. Alzheimers Res Ther. 2012;4:9. doi: 10.1186/alzrt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L, et al. Early-onset familial Alzheimer’s disease (EOFAD) Can J Neurol Sci. 2012;39:436–445. doi: 10.1017/s0317167100013949. [DOI] [PubMed] [Google Scholar]
- 15.Colvin MT, et al. Atomic resolution structure of monomorphic a42 amyloid fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koradi R, Billeter M, Wüthrich K. Molmol: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 17.Wälti MA, et al. Atomic-resolution structure of a disease-relevant a (1–42) amyloid fibril. Proc Natl Acad Sci USA. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meisl G, et al. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat Protoc. 2016;11:252–272. doi: 10.1038/nprot.2016.010. [DOI] [PubMed] [Google Scholar]
- 19.Cohen SIA, et al. Proliferation of amyloid-beta42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen SIA, et al. Distinct thermodynamic signature of oligomer generation in the aggregation of the amyloid- peptide. Nat Chem. 2018;10:523–531. doi: 10.1038/s41557-018-0023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arosio P, Knowles TPJ, Linse S. On the lag phase in amyloid fibril formation. Phys Chem Chem Phys. 2015;17:7606–7618. doi: 10.1039/c4cp05563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linse S. Monomer-dependent secondary nucleation in amyloid formation. Biophys Rev. 2017;9:329–338. doi: 10.1007/s12551-017-0289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolognesi B, et al. Single point mutations induce a switch in the molecular mechanism of the aggregation of the Alzheimer’s disease associated abeta42 peptide. ACS Chem Biol. 2014;9:378–382. doi: 10.1021/cb400616y. [DOI] [PubMed] [Google Scholar]
- 24.Luheshi LM, et al. Systematic in vivo analysis of the intrinsic determinants of amyloid pathogenicity. PLoS Biol. 2007;5:2493–2500. doi: 10.1371/journal.pbio.0050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peralvarez-Marin A, et al. Influence of residue 22 on the folding, aggregation profile, and toxicity of the Alzheimer’s amyloid beta peptide. Biophys J. 2009;97:277–285. doi: 10.1016/j.bpj.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson AS, et al. Physiochemical characterization of the Alzheimer’s disease-related peptides A 1-42Arctic and A 1-42wt. FEBS J. 2006;273:2618–2630. doi: 10.1111/j.1742-4658.2006.05263.x. [DOI] [PubMed] [Google Scholar]
- 27.Brorsson AC, et al. Intrinsic determinants of neurotoxic aggregate formation by the amyloid peptide. Biophys J. 2010;98:1677–1684. doi: 10.1016/j.bpj.2009.12.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meisl G, Yang X, Dobson CM, Linse S, Knowles TPJ. Modulation of electrostatic interactions to reveal a reaction network unifying the aggregation behaviour of the a[small beta]42 peptide and its variants. Chem Sci. 2017;8:4352–4362. doi: 10.1039/c7sc00215g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meisl G, et al. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the a40 and a42 peptides. Proc Natl Acad Sci USA. 2014;111:9384–9389. doi: 10.1073/pnas.1401564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meisl G, et al. Scaling behaviour and rate-determining steps in filamentous self-assembly. Chem Sci. 2017;8:7087–7097. doi: 10.1039/c7sc01965c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen SIA, et al. The molecular chaperone brichos breaks the catalytic cycle that generates toxic ab oligomers. Nat Struct Mol Biol. 2015;22:207–213. doi: 10.1038/nsmb.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlgren KN, et al. Oligomeric and fibrillar species of amyloid- peptides differentially affect neuronal viability. J Biol Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 33.Hermansson E, et al. The chaperone domain brichos prevents CNS toxicity of amyloid- peptide in drosophila melanogaster. Dis Models Mech. 2014;7:659–665. doi: 10.1242/dmm.014787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meisl G, Yang X, Frohm B, Knowles TPJ, Linse S. Quantitative analysis of intrinsic and extrinsic factors in the aggregation mechanism of Alzheimer-associated a-peptide. Sci Rep. 2016;6:18728. doi: 10.1038/srep18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasudev PG, Banerjee M, Ramakrishnan C, Balaram P. Asparagine and glutamine differ in their propensities to form specific side chain-backbone hydrogen bonded motifs in proteins. Proteins. 2012;80:991–1002. doi: 10.1002/prot.24001. [DOI] [PubMed] [Google Scholar]
- 36.Eswar N, Ramakrishnan C. Deterministic features of side-chain main-chain hydrogen bonds in globular protein structures. Protein Eng. 2000;13:227–238. doi: 10.1093/protein/13.4.227. [DOI] [PubMed] [Google Scholar]
- 37.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 38.Wolfenden R, Andersson L, Cullis PM, Southgate CC. Affinities of amino acid side chains for solvent water. Biochemistry. 1981;20:849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- 39.Walsh DM, et al. A facile method for expression and purification of the Alzheimer’s disease-associated amyloid beta-peptide. FEBS J. 2009;276:1266–1281. doi: 10.1111/j.1742-4658.2008.06862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.