Significance
This study demonstrates a distinct mode of RIPK1 activation mediated by a detergent-insoluble, highly ubiquitinated, activated RIPK1 species (iuRIPK1) which functions as a critical intermediate between TNF-receptor-associated complex I and assembly of the cytosolic caspase activation platform complex II in RIPK1-dependent apoptosis (RDA). By conducting a systematic screen for RDA regulators, we reveal the regulation of iuRIPK1 by Parkinson’s disease (PD)-associated LRRK2, E3 ubiquitin ligase c-Cbl, and ALS-associated NEK1. These results point to possible mechanistic links between RIPK1-mediated apoptosis and neurodegenerative diseases such as ALS and PD.
Keywords: RIPK1, necroptosis, apoptosis, TNF, ubiquitination
Abstract
Stimulation of cells with TNFα can promote distinct cell death pathways, including RIPK1-independent apoptosis, necroptosis, and RIPK1-dependent apoptosis (RDA)—the latter of which we still know little about. Here we show that RDA involves the rapid formation of a distinct detergent-insoluble, highly ubiquitinated, and activated RIPK1 pool, termed “iuRIPK1.” iuRIPK1 forms after RIPK1 activation in TNF-receptor-associated complex I, and before cytosolic complex II formation and caspase activation. To identify regulators of iuRIPK1 formation and RIPK1 activation in RDA, we conducted a targeted siRNA screen of 1,288 genes. We found that NEK1, whose loss-of-function mutations have been identified in 3% of ALS patients, binds to activated RIPK1 and restricts RDA by negatively regulating formation of iuRIPK1, while LRRK2, a kinase implicated in Parkinson’s disease, promotes RIPK1 activation and association with complex I in RDA. Further, the E3 ligases APC11 and c-Cbl promote RDA, and c-Cbl is recruited to complex I in RDA, where it promotes prodeath K63-ubiquitination of RIPK1 to lead to iuRIPK1 formation. Finally, we show that two different modes of necroptosis induction by TNFα exist which are differentially regulated by iuRIPK1 formation. Overall, this work reveals a distinct mechanism of RIPK1 activation that mediates the signaling mechanism of RDA as well as a type of necroptosis.
TNFα is an important proinflammatory cytokine involved in mediating a myriad of human inflammatory and neurodegenerative diseases (1–3). Activation of TNFR1 (TNF receptor 1) by TNFα leads to the recruitment of RIPK1, TRADD, and other signaling components to a complex associated with the intracellular death domain of TNFR1 at the cytoplasmic membrane, known as complex I (also called TNF receptor signaling complex, or TNF-RSC) (4). In complex I, ubiquitination of RIPK1 by cIAP1/2 and LUBAC leads to the recruitment of TAK1 and formation of the NEMO/IKK complex, which subsequently promotes the activation of NF-κB and downstream transcription of prosurvival proteins and cytokines (5, 6). Inhibition of this transcriptional response by the protein synthesis inhibitor cycloheximide (CHX) in the presence of TNFα promotes RIPK1-independent apoptosis mediated by complex II, a cytosolic complex composed of RIPK1, FADD, and caspase-8, which drives the activation of caspase-8 (4, 7). However, when caspases are inhibited, cell death can occur by necroptosis. In TNFα-induced necroptosis, necrotic cell death is executed by RIPK1-mediated activation of RIPK3, which in turn mediates the phosphorylation of MLKL to disrupt the integrity of plasma membrane (8).
Alternatively, inhibition or depletion of specific complex I members, such as TAK1, cIAP1/2, or NEMO, promotes RIPK1-dependent apoptosis (RDA) in response to TNFα (9–14). For example, intestinal epithelial cell-specific ablation of NEMO promotes apoptosis of Paneth cells and colonocytes, which can be blocked by inactivation of RIPK1 kinase (13). RDA is distinct from apoptosis induced by TNFα/CHX, as the loss of NF-κB activity alone leads to RIPK1-independent apoptosis (15, 16). Unlike in RIPK1-independent apoptosis, the kinase activity of RIPK1 is required for the formation of complex II in RDA (11, 15, 17, 18). However, unlike in necroptosis, neither MLKL nor the kinase activity of RIPK3 is required for the execution of cell death in RDA, which is instead carried out by caspases (17, 18). Thus, RDA is mechanistically distinct from both RIPK1-independent apoptosis and necroptosis. However, while RIPK1 is activated in both RDA and necroptosis mediated by TNFα, it is unclear if and how the mechanisms that regulate the activation of RIPK1 in RDA and necroptosis differ.
Investigations on the mechanism of RDA have focused on two main lines of inquiry. The first is to understand how known components of complex I suppress the kinase activity of RIPK1 via its ubiquitination and phosphorylation, and thus prevent RDA under normal conditions. K63 and linear ubiquitination of RIPK1 by cIAP1/2 and LUBAC, respectively, allows recruitment of TAK1 and the IKK complex to RIPK1 and inhibits RDA (9, 19, 20). Phosphorylation of RIPK1 by TAK1 and its downstream kinases, including IKK and MK2, suppresses the activation of RIPK1 and formation of complex II (15, 21–23). The second is to explore the role of known apoptotic and necroptotic signaling components in RDA, such as NEMO, CYLD, FADD, and RIPK3 (9, 17, 18). Critically, no systematic screen for regulators of RDA has been performed. Further, the mechanism by which RIPK1 is activated and the manner by which the kinase activity of RIPK1 regulates its binding to FADD and the formation of complex II in RDA are unknown.
In this study, we identify the formation of a highly ubiquitinated, insoluble, activated RIPK1 intermediate which precedes the formation of complex II in RDA. By conducting a targeted siRNA screen, we identify the Parkinson’s disease (PD)-associated kinase LRRK2 and the E3 ligase c-Cbl as promoters of the formation of this insoluble, activated RIPK1 intermediate. Further, we identify NEK1, a kinase associated with ALS, as a suppressor of RIPK1 activation. Finally, we demonstrate two distinct pathways of RIPK1 activation and necroptosis initiation by TNFα, which are differentially regulated by LRRK2 and c-Cbl.
Results
RDA Features Rapid RIPK1 Activation and Cell Death Compared with RIPK1-Independent Apoptosis and Necroptosis.
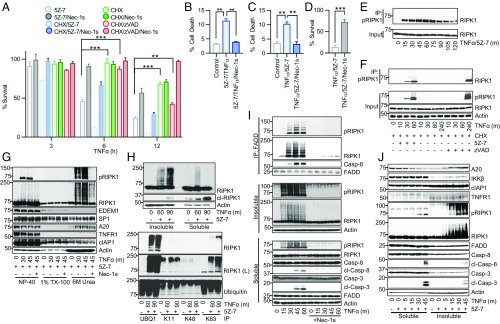
The sensitivity of 661W cells, which express neuronal markers NeuN, MAP2, and β–III tubulin (24), to RDA was first tested. The 661W cells rapidly died in response to TNFα and the TAK1 inhibitor 5Z-7-oxozeanol (5Z-7), which was blocked by the highly specific RIPK1 kinase inhibitor R-7-Cl-O-Necrostatin-1 (Nec-1s) (25) (Fig. 1A). While RDA is known to proceed more rapidly than RIPK1-independent apoptosis, its kinetics compared to necroptosis have not been examined (12, 15, 16, 19). In both 661W cells and mouse embryonic fibroblasts (MEFs), RDA induced by TNFα/5Z-7 occurred more rapidly than both necroptosis induced by TNFα, CHX, and the caspase inhibitor zVAD.fmk (zVAD) and apoptosis induced by TNFα/CHX (Fig. 1A and SI Appendix, Fig. S1A). TNFα/5Z-7 also killed primary CNS-derived cells, including primary neuronal cultures, primary astrocytes, and primary microglia, which was blocked by Nec-1s, suggesting that different cell types in the CNS can undergo RDA (Fig. 1 B–D).
Fig. 1.
Induction of RDA by TNFα leads to rapid and elevated activation of RIPK1 in a detergent-insoluble cellular compartment before formation of complex II. (A) The 661W cells treated with TNFα/5Z-7 to induce RDA, TNFα/CHX to induce RIPK1-independent apoptosis, or TNFα/CHX/zVAD to induce necroptosis in the presence or absence of Nec-1s. Cell viability determined by CellTiter-Glo. (B) Primary neuronal cultures treated with TNFα/5Z-7 to induce RDA for 8 h and cell death measured by Toxilight. (C) Primary astrocytes treated for 5 h and cell death measured by Toxilight. (D) Primary microglia treated for 5 h and cell survival measured by CellTiter-Glo. (E–J) Cells treated as indicated and samples analyzed by Western blotting using indicated antibodies; pRIPK1 is RIPK1-pS166. (E) The 661W cells treated with TNFα/5Z-7 to induce RDA. Lysates immunoprecipitated with RIPK1-pS166 ab. (F) WT MEFs lysates immunoprecipitated with RIPK1-pS166 ab. (G) The 661W cells treated with TNFα/5Z-7 to induce RDA, harvested with Nonidet P-40 buffer, and cleared by centrifugation to yield the Nonidet P-40 fraction. Pellet resuspended in Triton X-100 buffer and cleared by centrifugation to yield the TX-100 fraction. Resulting pellet was resuspended in buffer with 6 M urea and cleared by centrifugation to yield the urea fraction. (H) The 661W cells harvested as in G; Nonidet P-40 fraction marked as detergent “soluble” fraction, 6 M urea fraction marked as detergent “insoluble” fraction (Top). Urea fraction used for immunoprecipitation with anti-linear ubiquitin (UBQ1) or anti-K11, -K48, or -K63 ubiquitin antibodies (Bottom). (I) Tak1−/− MEFs treated with TNFα to induce RDA and harvested as in H. Nonidet P-40 soluble lysate used for complex II immunoprecipitation with anti-FADD (M-19). (J) The 661W cells harvested as in H. Concentrations of reagents: TNFα, 10 ng/mL (A–G, I, and J), 1 ng/mL (H); CHX, 1–2 μg/mL; 5Z-7, 500 nM; Nec-1s, 10 μM; zVAD, 20 μM; SM-164, 500 nM. Same concentrations of these compounds were used in subsequent experiments unless noted. All data shown are mean ± SD of three or more independent experiments. **P ≤ 0.01 and ***P ≤ 0.001 by one-way ANOVA (A–C) or Student’s t test (D).
The kinetics of RIPK1 activation in RDA was characterized using two established antibodies for RIPK1-phospho-Ser166 (pS166), a bona fide marker of RIPK1 kinase activity (25–28). Induction of RDA by TNFα/5Z-7 led to robust RIPK1 activation within 15 min and peak activation at 1 h (Fig. 1E). Rapid activation of RIPK1 in RDA was not due to an off-target effect of 5Z-7, as similar kinetics were observed in response to TNFα alone in Tak1−/− MEFs (SI Appendix, Fig. S1B). Both RIPK1 inhibitor Nec-1s and kinase dead Ripk1D138N MEFs blocked RIPK1-S166 phosphorylation (SI Appendix, Fig. S1 B and C).
In contrast, significant RIPK1 activation was not detected until 4 h following necroptosis induction by TNFα/CHX/zVAD (Fig. 1F). As expected, no activation of RIPK1 occurred in RIPK1-independent apoptosis induced by TNFα/CHX (Fig. 1F), or in response to TNFα alone (SI Appendix, Fig. S1D). Rapid induction of RIPK1 kinase activity was also observed in primary astrocytes in response to TNFα/5Z-7 (SI Appendix, Fig. S1E). Overall, the kinetic difference in RIPK1 activation in RDA vs. necroptosis induced by TNFα/CHX/zVAD suggests that distinct mechanisms may regulate RIPK1 activation in these two cell death pathways.
Formation of an Insoluble Pool of Ubiquitinated, Activated RIPK1 in RDA After Complex I and Before Complex II.
Activated RIPK1 is found in detergent-insoluble fractions in postmortem pathological samples from patients with the neurodegenerative diseases Alzheimer’s disease (AD), ALS, and multiple sclerosis (27, 29–31). To determine if accumulation of activated RIPK1 in the insoluble fraction may be relevant to RDA, we developed a protocol to analyze the distribution of activated RIPK1 in cells undergoing RDA. Briefly, cells were harvested in 0.2% Nonidet P-40 buffer and centrifuged to yield the Nonidet P-40–soluble fraction, which was sufficient to release endoplasmic reticulum protein EDEM1 and partially release nuclear protein SP1. The insoluble pellet was extracted using buffer with 0.2% Nonidet P-40 + 1% Triton X-100 (TX-100) and centrifuged to yield the TX-100–soluble fraction. This vigorous extraction with 1% TX-100 buffer further lysed cellular organelles and further released SP1 (Fig. 1G). Finally, the insoluble pellet was extracted again using buffer with 0.2% Nonidet P-40 + 6 M urea and centrifuged to yield the detergent-insoluble but 6 M urea-soluble fraction.
Upon induction of RDA, a distinct pool of activated, highly modified RIPK1 appeared in the 6 M urea fraction as a distinct ladder as assessed by SDS-PAGE (Fig. 1G). This highly modified form of activated RIPK1, insoluble in Nonidet P-40 or TX-100 buffers, but soluble in 6 M urea, appeared prominently within 30 min following RDA induction.
Due to its ladder-like appearance, we hypothesized that the highly modified RIPK1 found in the urea fraction was ubiquitinated as well as phosphorylated. To determine if activated, insoluble RIPK1 was ubiquitinated and the type of linkages present, ubiquitin chain-specific antibodies were used for immunoprecipitation from the urea fraction following induction of RDA (Fig. 1H). Upon induction of RDA, no increase in K11- or K48-linked ubiquitinated RIPK1 was detected, suggesting that the insoluble RIPK1 ladder was not proteasome-targeted RIPK1. Instead, immunoprecipitation indicated that the insoluble RIPK1 was modified by both linear and K63-linked polyubiquitin chains (Fig. 1H). Therefore, we named this critical signaling intermediate in RDA “iuRIPK1” for “insoluble, ubiquitinated RIPK1.”
Formation of iuRIPK1 was inhibited by Nec-1s, indicating that it was a consequence of rapid RIPK1 activation in RDA (Fig. 1 G and I). TNFα stimulation of Tak1−/− MEFs also led to the formation of iuRIPK1 (Fig. 1I). In addition, we observed induction of RDA in response to very low doses of TNFα (SI Appendix, Fig. S1F), and, correspondingly, robust formation of iuRIPK1 occurred in response to 1 ng/mL TNFα within 60 min (SI Appendix, Fig. S1G).
We probed the 6 M urea fraction to see if other complex I proteins were associated with iuRIPK1. We did not find TNFR1 in the insoluble fraction, nor was there any change in cIAP1 in the insoluble fraction in response to the induction of RDA (Fig. 1G). These data suggest that iuRIPK1 is not associated with the entire TNF-RSC, and thus iuRIPK1 can be distinguished from complex I. However, we found that A20 levels increased in the 6 M urea fraction. Since A20 is an important deubiquitinating enzyme for RIPK1 (2), the association of A20 with the insoluble fraction is consistent with the highly ubiquitinated nature of iuRIPK1.
Formation of iuRIPK1 peaked before the formation of complex II and caspase activation in RDA (Fig. 1I). Specifically, iuRIPK1 formed at 15 min and peaked at 30 min, while complex II began to form at 30 min and peaked at 45 min. We verified that RIPK1 was recruited to complex I before significant iuRIPK1 formation in an extended time-course analysis comparing the formation of complex I, iuRIPK1, and complex II (SI Appendix, Fig. S1H). However, we found complex II components FADD and caspase-8 in the insoluble fraction without TNFα stimulation, which did not substantially change upon induction of RDA (Fig. 1J). Based on these findings, we propose that the iuRIPK1 induced during RDA may represent a distinct intermediate which stabilizes and concentrates activated RIPK1 to facilitate formation of complex II and caspase activation.
Rapid Formation of iuRIPK1 Specifically in RDA.
We hypothesized that if iuRIPK1 was responsible for rapid complex II formation and cell death in RDA, then it would be rapidly induced in RDA but not RIPK1-indepedent apoptosis or necroptosis induced by TNFα/CHX/zVAD. Consistent with this hypothesis, iuRIPK1 did not form in RIPK1-independent apoptosis (Fig. 2A). The activation of caspases also occurred faster and more robustly in RDA than in apoptosis induced by TNFα/CHX (Fig. 2A). We assessed ATP release from dying cells as another biomarker to characterize the speed and strength of caspase activation, as caspase activation in apoptosis mediates the opening of the plasma membrane channel protein pannexin 1, resulting in selective secretion of nucleotides (32). Induction of RDA in 661W cells and Tak1−/− MEFs led to rapid secretion of ATP within 1.5–2 h (SI Appendix, Fig. S2 A and B).
Fig. 2.
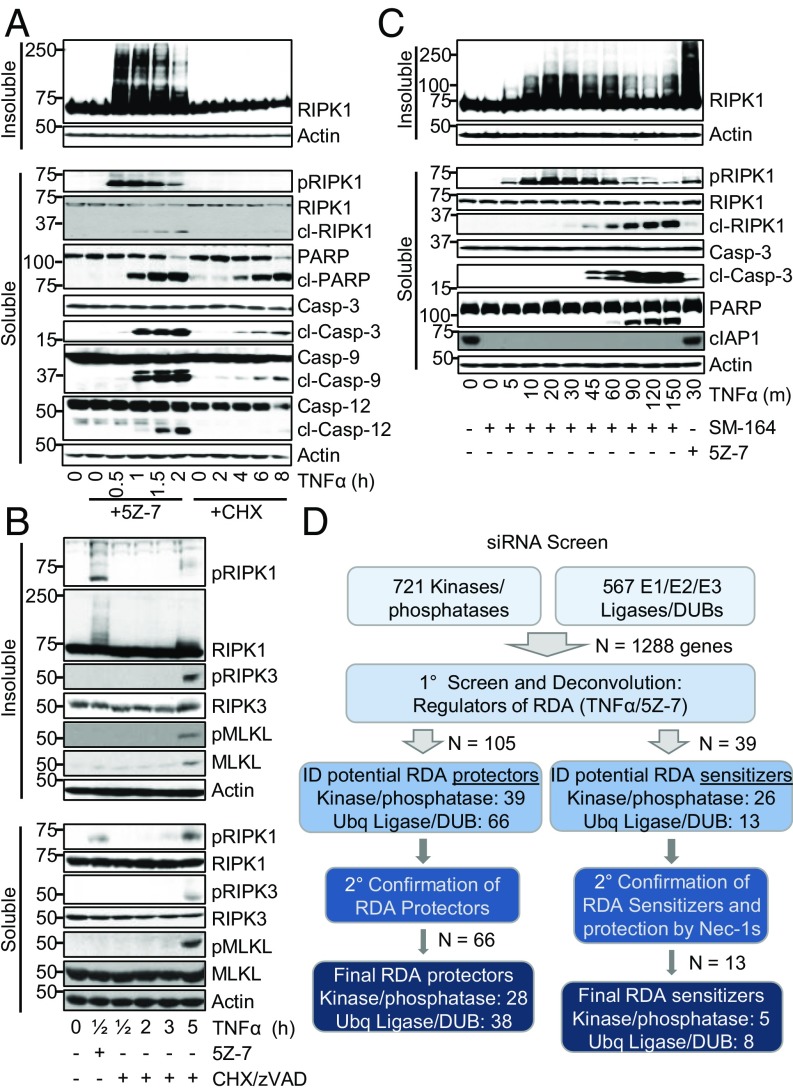
A siRNA screen identifies potential regulators of RDA and iuRIPK1 formation. (A–C) Cells treated as indicated and samples analyzed by Western blotting using indicated antibodies; pRIPK1 is RIPK1-pS166, pRIPK3 is RIPK3-pT231/S232, and pMLKL is MLKL-pS345. (A) WT MEFs treated with TNFα/5Z-7 to induce RDA or TNFα/CHX to induce RIPK1-independent apoptosis and fractionated to yield the detergent-soluble and -insoluble fractions. (B) WT MEFs treated with TNFα/5Z-7 to induce RDA or TNFα/CHX/zVAD to induce necroptosis and analyzed as in A. (C) The 661W cells pretreated with SM-164, RDA induced by TNFα, and cells fractionated to yield the detergent-soluble and -insoluble fractions. (D) Scheme and summary of siRNA screen for regulators of RDA in 661W cells. Evaluation of hits was performed by normalizing values to nontargeting control siRNA and averaging three replicates.
Critically, formation of iuRIPK1 did not occur in necroptosis induced by TNFα/CHX/zVAD in 661W cells or MEFs (Fig. 2B and SI Appendix, Fig. S2C). Consistent with previous reports (33), activated RIPK1 and RIPK3 and phosphorylated MLKL were found in the insoluble fraction 3–5 h after necroptosis induction. High-molecular-weight iuRIPK1 did not form, and a mild increase in RIPK1 in the insoluble fraction in necroptosis occurred only at 5 h, unlike iuRIPK1 formation at 30 min in RDA. Taken together, these results suggest that the formation of iuRIPK1 species is specific for RDA and may link the rapid RIPK1 activation observed in RDA to RIPK1-kinase-dependent formation of complex II and cell death.
iuRIPK1 Forms Independently of cIAP1/2.
Since cIAP1/2 directly mediate K63-linked ubiquitination and indirectly promote LUBAC-mediated linear ubiquitination of RIPK1 in complex I (6), we next examined the role of cIAP1/2 in iuRIPK1 formation. Similar to inhibition of TAK1, depletion of cIAP1/2 with SM-164 is known to induce RDA in combination with TNFα (9). iuRIPK1 was detected in RDA induced by TNFα/SM-164 despite robust depletion of cIAP1 (Fig. 2C). While depletion of cIAP1/2 reduced higher-molecular-weight (>150 kDa) iuRIPK1 species, the levels of modified iuRIPK1 species with lower molecular weights (75–100 kDa) increased, as did the accumulation of activated RIPK1 (Fig. 2C). Thus, consistent with the role of cIAP1/2 in mediating the activation of TAK1 (2), the loss of cIAP1/2 also promoted RIPK1 activation and the formation of iuRIPK1. Further, these results suggest that while cIAP1/2 contribute to the ubiquitination of iuRIPK1, additional, prodeath, E3 ubiquitin ligases may be involved in ubiquitinating RIPK1 to promote iuRIPK1 formation and RIPK1 activation in RDA.
A Targeted siRNA Screen Identifies Regulators of RDA.
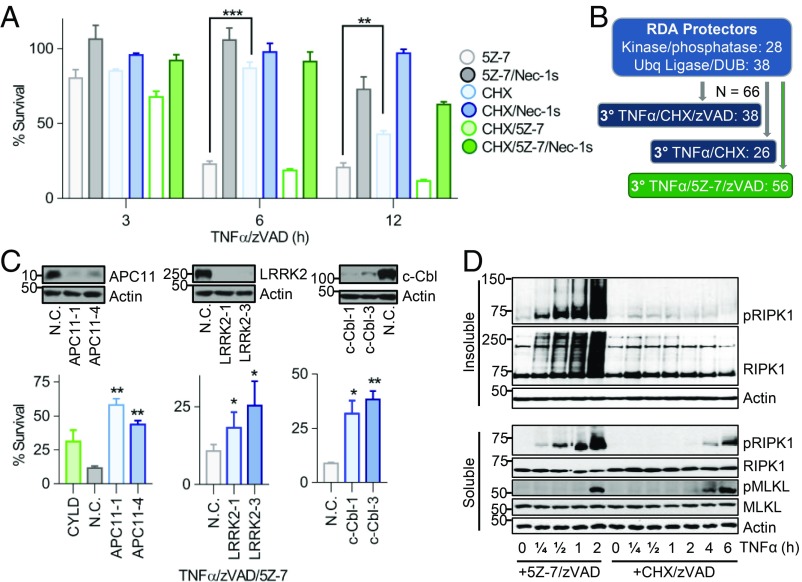
Since we found that rapid RIPK1 activation and iuRIPK1 formation were distinct features of RDA and did not occur in necroptosis induced by TNFα/CHX/zVAD or RIPK1-independent apoptosis induced by TNFα/CHX, we hypothesized that RDA may be regulated by distinct genes not previously known to be involved in TNFα-induced cell death. Since iuRIPK1 is heavily ubiquitinated and phosphorylated, we screened the Dharmacon mouse siRNA libraries targeting 1,288 genes encoding ubiquitin ligases, deubiquitinases, kinases, and phosphatases for their role in RDA (Fig. 2D and SI Appendix, Fig. S3A). Of 1,288 genes screened, we identified 66 genes that when knocked down protected against RDA induced by TNFα/5Z-7, including RIPK1 and CYLD, indicating success of the screen (Fig. 2D and SI Appendix, Fig. S3B). Twenty-two of these genes had a lower P value than RIPK1, and of these CYLD was the top deubiquitinase (DUB) and APC11 the top E3 ligase, with three individual duplexes showing protection against RDA (SI Appendix, Fig. S3 C and D). Overlap with the hits from a previous screen for necroptosis regulators in L929 cells was a minimal seven hits, demonstrating that regulation of RDA is distinct from that of necroptosis (SI Appendix, Table S1). We also identified 13 genes that when knocked down sensitized to RDA, including complex I components A20, XIAP, HOIL-1, and HOIP, further indicating reliability of the screen data (Fig. 2D and SI Appendix, Table S2). The top sensitizer to RDA identified by a combination of both cell death and cell survival assays was NEK1 (SI Appendix, Fig. S3E).
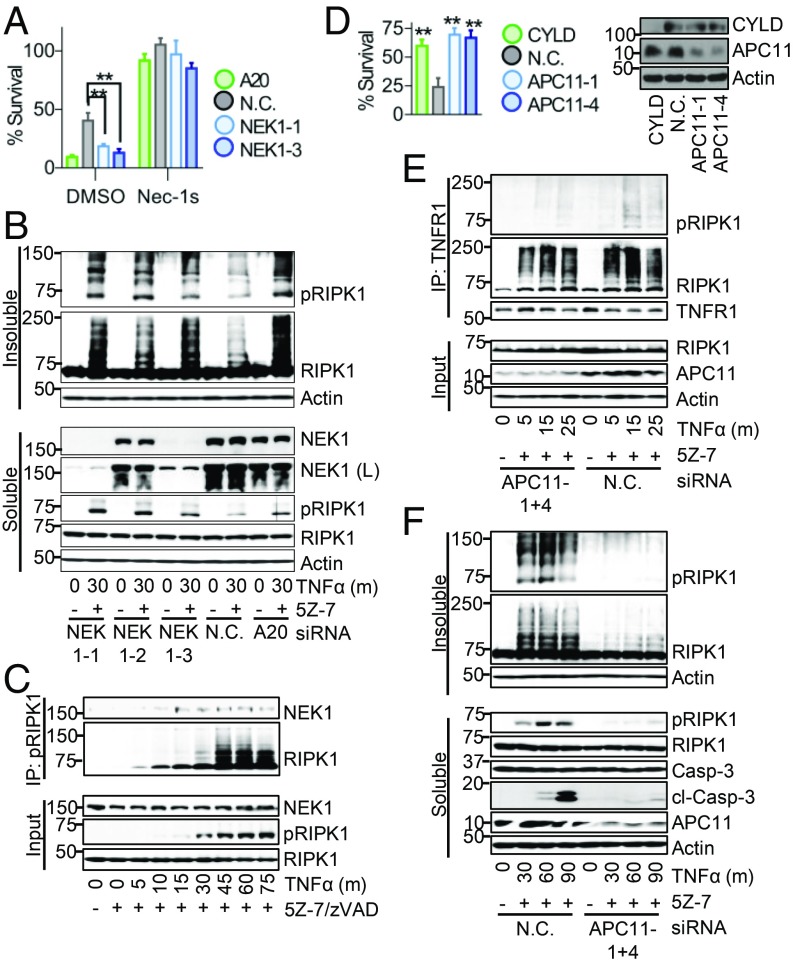
NEK1 Restricts RIPK1 Activation and iuRIPK1 Formation.
Loss-of-function mutations in NEK1 (NIMA-related kinase 1) are implicated in ∼3% of ALS cases (34, 35). Therefore, we characterized the role of the top sensitizer screen hit NEK1 in RDA and investigated its interaction with RIPK1 in RDA. Knockdown of NEK1 by two different siRNAs sensitized cells to RDA induced by TNFα/5Z-7 (Fig. 3A). Similar results were seen by assaying for cell death as well as cell survival in a time course of RDA induction (SI Appendix, Fig. S4A). Knockdown of NEK1 also sensitized cells to cell death induced by TNFα/CHX, but not TNFα alone (SI Appendix, Fig. S4 B and C). Sensitization to RDA by loss of NEK1 was blocked by Nec-1s, demonstrating that NEK1 may serve as a brake on RIPK1 activation under RDA conditions (Fig. 3A and SI Appendix, Fig. S4A).
Fig. 3.
NEK1 restricts while APC11 promotes RIPK1 activation and iuRIPK1 formation. (A) The 661W cells transfected with siRNAs targeting NEK1, A20 (positive control), or nontargeting control (N.C.) siRNA for 72 h and treated with TNFα/5Z-7 for 6 h to induce RDA. Cell survival determined using CellTiter-Glo. Knockdown efficiency shown in B. (B–F) Cells transfected and/or treated as indicated and samples analyzed by Western blotting using indicated antibodies; pRIPK1 is RIPK1-pS166. (B) The 661W cells fractionated using lysis buffer with Nonidet P-40/TX-100 followed by 6 M urea. (C) Activated RIPK1 isolated from 661W cells treated with TNFα/5Z-7/zVAD by immunoprecipitation with RIPK1-pS166 ab. (D–F) The 661W cells transfected with siRNAs against APC11, CYLD (positive control) or N.C. siRNA for 72 h. (D) Cells treated with TNFα/5Z-7 for 8 h to induce RDA. Cell survival determined using CellTiter-Glo. (E) Immunoprecipitation of complex I with anti-TNFR1 from soluble lysates. (F) Cells fractionated using lysis buffer with Nonidet P-40/TX-100 followed by 6 M urea. Concentrations of reagents: TNFα, 1 ng/mL (A, B, and D–F), 10 ng/mL (C). All data shown are mean ± SD of three or more independent experiments. **P ≤ 0.01 by one-way ANOVA.
Knockdown of NEK1 resulted in increased levels of activated RIPK1 30 min following RDA induction and dramatically increased levels of iuRIPK1, which correlated with NEK1 knockdown (Fig. 3B). However, knockdown of NEK1 did not substantially affect levels of activated RIPK1 in complex I or RIPK1 recruitment to complex I (SI Appendix, Fig. S4D), suggesting that NEK1 acts at the level of iuRIPK1 formation. Immunoprecipitation of activated RIPK1 following induction of RDA showed that NEK1 binds to activated RIPK1 starting at 15–30 min, which may serve to destabilized activated RIPK1 and prevent its transition to iuRIPK1 and thus subsequent cell death (Fig. 3C).
The Catalytic Core of the APC/C Complex Promotes RIPK1 Activation and iuRIPK1 Formation in RDA.
To identify positive regulators of RIPK1 activation and iuRIPK1 formation, we examined a top protective hit in our siRNA screen, APC11. APC11 is the catalytic E3 ubiquitin ligase subunit of the APC/C, a large, multisubunit complex which targets its substrates for degradation via K11- and K48-linked polyubiquitination (36). While its most well-known role is in regulating cell cycle, APC/C is active throughout the cell cycle as well as in postmitotic cells such as neurons (37, 38). Knockdown of APC11 by two different siRNAs inhibited RDA to a similar extent as CYLD knockdown, increasing the survival rate from an average of 24% with nontargeting control knockdown to 70% and 67% with APC11 knockdown (Fig. 3D). In addition, approximately twofold protection was observed by cell death assays at multiple time points (SI Appendix, Fig. S5A). Knockdown of APC11 also protected against cell death and caspase-3 activation in RDA in Tak1−/− MEFs induced by TNFα alone (SI Appendix, Fig. S5 B and C). Consistent with the involvement of the catalytic core of the APC/C complex in RDA, knockdown of core APC/C component APC10 also inhibited caspase activation and cell death induced by TNFα/5Z-7 (SI Appendix, Fig. S5 D and E).
Reduction of APC11 resulted in substantially reduced RIPK1 activation in RDA, particularly at 15 min (SI Appendix, Fig. S5F). Therefore, we examined RIPK1 activation in complex I with and without APC11 knockdown. While knockdown of APC11 resulted in only a mild reduction of RIPK1 association with complex I under RDA conditions, it resulted in substantially reduced RIPK1 activation in complex I (Fig. 3E). Consistent with our results above that early RIPK1 activation in RDA results in iuRIPK1 formation, knockdown of APC11 prevented iuRIPK1 formation and sustained RIPK1 activation (Fig. 3F). Overexpression of APC11 alone did not lead to increased RIPK1 ubiquitination in 293T cells, although interaction between APC11 and RIPK1 was observed (SI Appendix, Fig. S5G).
Overall, APC11 promotes robust RIPK1 activation at complex I and the subsequent formation of iuRIPK1 in RDA. Consistent with its role in RIPK1 activation, APC11 knockdown did not protect against RIPK1-independent apoptosis induced by TNFα/CHX (SI Appendix, Fig. S5H). However, APC11 knockdown did provide mild protection against necroptosis induced by TNFα/CHX/zVAD, suggesting that it may play a more general role in promoting RIPK1 activation (SI Appendix, Fig. S5I).
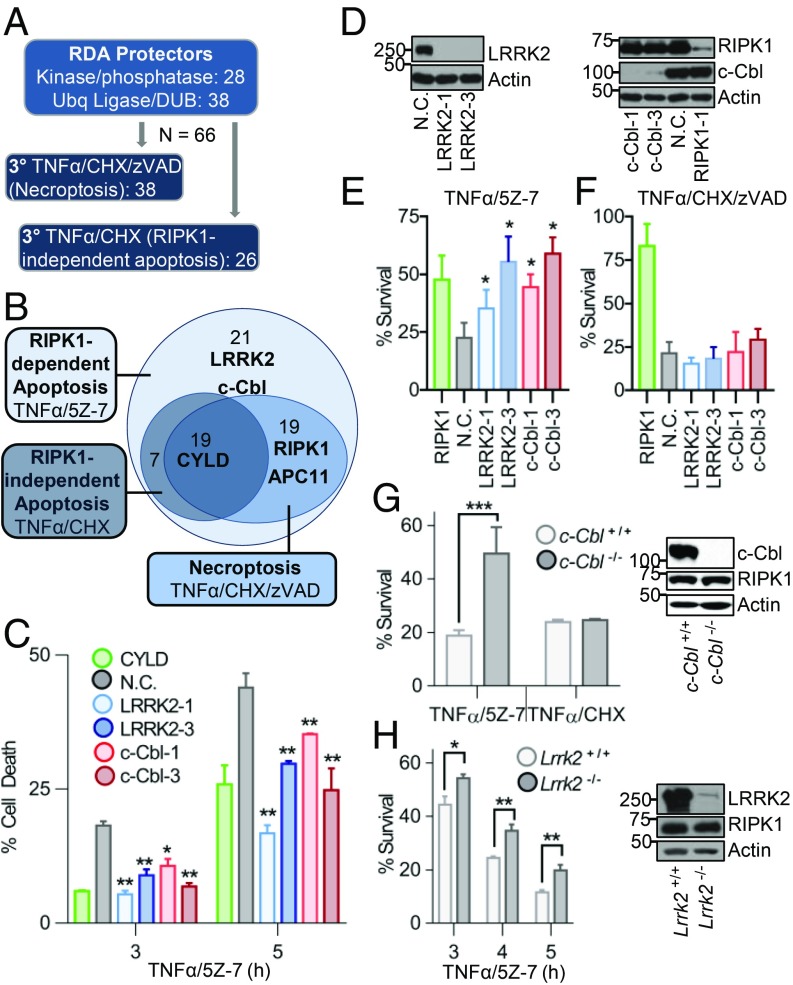
LRRK2 and c-Cbl Are Specific Mediators of RDA.
Next, we sought to identify genes responsible for the RDA-specific rapid RIPK1 activation and iuRIPK1 formation that we have established. To do this, we determined the role of the 66 RDA protectors from our siRNA screen in RIPK1-independent apoptosis induced by TNFα/CHX and necroptosis induced by TNFα/CHX/zVAD to identify genes specifically regulating RDA (Fig. 4A and SI Appendix, Table S3). Knockdown of 19 genes, including CYLD, protected all three modes of cell death and knockdown of 19 additional genes, including RIPK1 and APC11, protected against the two RIPK1-dependent pathways, RDA (TNFα/5Z-7) and necroptosis (TNFα/CHX/zVAD). Knockdown of 21 genes protected specifically against RDA but not the other cell death modes (Fig. 4B). Of these 21 genes, STRING network analysis revealed that the E3 ligase c-Cbl and kinase LRRK2 are connected to TNFR1 and RIPK1 signaling and therefore of particular interest (SI Appendix, Fig. S3B).
Fig. 4.
Tertiary screens identify LRRK2 and c-Cbl as specific positive regulators of RDA. (A) Scheme of tertiary siRNA screens for regulators of RIPK1-independent apoptosis induced by TNFα/CHX and necroptosis induced by TNFα/CHX/zVAD in 661W cells. Evaluation of hits was performed by normalizing values to nontargeting control (N.C.) and averaging three replicates. (B) Euler diagram of screen results. (C–F) The 661W cells transfected with siRNAs against LRRK2, c-Cbl, CYLD, RIPK1 (positive control), or N.C. siRNA for 72 h. (C) Cells treated with TNFα/5Z-7 and cell death determined by Toxilight. (D) Knockdown efficiency for C, E, and F determined by Western blotting of total lysates. (E and F) The 661W cells treated as indicated for 8 h and cell survival determined by CellTiter-Glo. (G and H) Matching (G) c-Cbl WT or KO or (H) Lrrk2 WT or KO MEFs treated as indicated and cell survival determined using CellTiter-Glo. Western blots show loss of protein in mutant MEFs. Concentrations of reagents: TNFα, 1 ng/mL for TNFα/5Z-7; 10 ng/mL for TNFα/CHX and TNFα/CHX/zVAD. All data shown are mean ± SD of three or more independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 by one-way ANOVA.
To confirm these findings, we examined the role of LRRK2 and c-Cbl in RDA by using two different siRNAs for each in both cell death and cell survival assays (Fig. 4 C–E). Each siRNA provided significant, although partial, protection against RDA induced by TNFα/5Z-7. Consistent with the results from the tertiary screen, knockdown of LRRK2 or c-Cbl did not protect against necroptosis induced by TNFα/CHX/zVAD or RIPK1-independent apoptosis induced by TNFα/CHX (Fig. 4F and SI Appendix, Fig. S6A).
To provide further confirmation for the role of LRRK2 and c-Cbl in RDA, we obtained matching WT and KO MEFs for each gene. c-Cbl−/− MEFs were substantially protected from RDA induced by TNFα/5Z-7 but were not protected from apoptosis induced by TNFα/CHX (Fig. 4G). Lrrk2−/− MEFs were partially but significantly protected from RDA induced by TNFα/5Z-7, and this protection was consistent at multiple time points (Fig. 4H). In contrast, Lrrk2−/− MEFs were sensitized to apoptosis induced by TNFα/CHX (SI Appendix, Fig. S6B).
LRRK2 Promotes Increased Association of RIPK1 with Complex I in RDA.
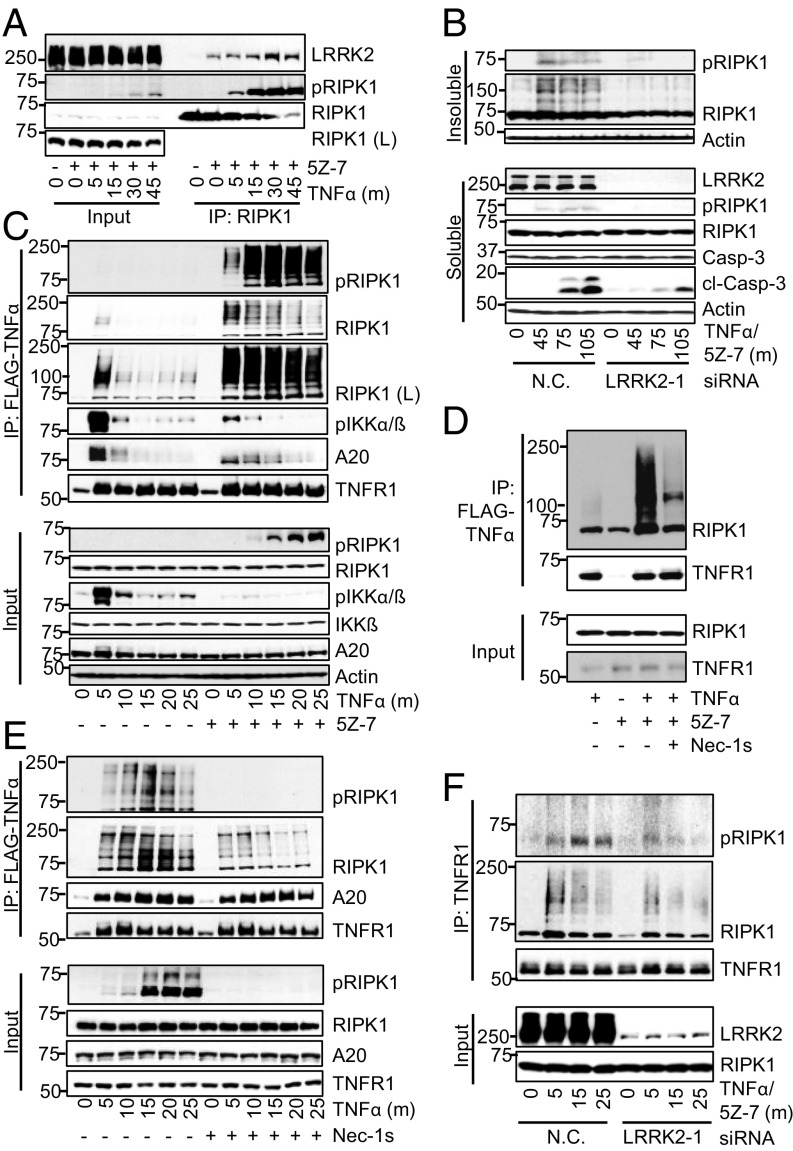
Leucine-rich repeat kinase 2 (LRRK2) is a large, multidomain RIPK1 family member also known as RIPK7. Gain-of-function mutations in LRRK2 are the most common cause of autosomal-dominant PD, and increased kinase activity is thought to be largely responsible for driving pathogenicity (39–41). However, LRRK2 also has a multitude of protein–protein interaction domains, and LRRK2’s armadillo repeat was recently shown to bind to FADD’s death domain and mediate neuronal death (42). LRRK2 kinase inhibitors did not protect against RDA, suggesting that LRRK2’s scaffold function may play a role in RDA (SI Appendix, Fig. S7A). LRRK2 has also been shown to bind to RIPK1 upon overexpression in 293T cells, but interaction of the endogenous proteins has not been shown, nor is it clear what the functional significance of this interaction is (43). Therefore, we first determined if endogenous LRRK2 and RIPK1 interact in RDA. We found that induction of RDA resulted in binding of endogenous RIPK1 to LRRK2 (Fig. 5A). This binding was induced as early as 5 min following RDA induction and peaked at 30 min. Furthermore, the activation of RIPK1 in RDA at both early and late time points, as indicated by the appearance of RIPK1-pS166, was inhibited by LRRK2 knockdown (SI Appendix, Fig. S7B), as was subsequent iuRIPK1 formation and caspase activation (Fig. 5B). Due to the early kinetics of LRRK2 association with RIPK1 in RDA, we next sought to elucidate the regulation of complex I in RDA.
Fig. 5.
LRRK2 promotes RDA-specific up-regulation of RIPK1 activity and association with complex I. Cells transfected and/or treated as indicated and samples analyzed by Western blotting using indicated antibodies; pRIPK1 is RIPK1-pS166. (A) The 661W cells treated with TNFα/5Z-7 to induce RDA. RIPK1 complexes immunoprecipitated using anti-RIPK1. (B) The 661W cells transfected with siRNAs against LRRK2 or nontargeting control (N.C.) siRNA for 72 h and fractionated using lysis buffer with Nonidet P-40/TX-100 followed by 6 M urea. (C) The 661W cells stimulated with FLAG-TNFα. Immunoprecipitation of complex I with FLAG-M2 beads. (D) The 661W cells stimulated with FLAG-TNFα for 15 min and complex I analyzed as in C. (E) Tak1−/− MEFs treated with FLAG-TNFα to induce RDA with and without Nec-1s and complex I analyzed as in C. (F) The 661W cells transfected as in B and complex I analyzed by TNFR1 immunoprecipitation following induction of RDA with TNFα/5Z-7. Concentrations of reagents: TNFα, 1 ng/mL (B and F), 10 ng/mL (A); FLAG- TNFα, 50 ng/mL.
We purified complex I following TNFα stimulation for 0–25 min in cells treated with 5Z-7 or vehicle. RIPK1 association with complex I increased massively upon TAK1 inhibition compared with TNFα stimulation alone, especially at later time points (Fig. 5C). Further, while very little activated RIPK1 was detected in complex I induced by TNFα alone, high levels of modified and activated RIPK1-pS166 were found in complex I after treatment with TNFα/5Z-7. Similar results were obtained with TNFα treatment alone in WT vs. Tak1−/− MEFs (SI Appendix, Fig. S7C). RIPK1 association with complex I in WT MEFs and Tak1−/− MEFs was initially similar at 2–5 min after TNFα stimulation; strikingly, while the levels of RIPK1 in complex I rapidly diminished in WT MEFs after 5 min, the recruitment of RIPK1 to complex I in TAK1-deficient cells continued to increase until 15 min. Further, activated RIPK1 was detected as early as 5 min in complex I in TAK1-deficient cells, before its appearance in the total lysate, suggesting that RIPK1 activation in RDA occurs at complex I (Fig. 5C and SI Appendix, Fig. S7C).
Since the above results were obtained with murine TNFα (mTNFα), we wondered if the activation of TNFR2 might contribute to the activation of RIPK1. Thus, we first used human TNFα (hTNFα), which binds only to TNFR1 in murine cells (44). Stimulation with hTNFα also led to dramatically increased levels of RIPK1 in complex I when TAK1 was inhibited (SI Appendix, Fig. S7D). To directly analyze the contribution of TNFR1, we used an alternate method of complex I purification using anti-TNFR1, and found RIPK1-pS166 in the TNFR1 complex (SI Appendix, Fig. S7 E–G). Further, we found massively increased recruitment of RIPK1 to TNFR1 in RDA induced by TNFα/5Z-7 compared with both TNFα alone and apoptosis induced by TNFα/CHX (SI Appendix, Fig. S7E).
Strikingly, this differential recruitment of RIPK1 to complex I was seen in RDA induced by TNFα/5Z-7 vs. necroptosis induced by TNFα/CHX/zVAD (SI Appendix, Fig. S7F). We found high levels of activated RIPK1 and sustained, increased levels of highly modified RIPK1 in complex I only in RDA. Therefore, TAK1 suppresses RIPK1 accumulation and activation in complex I.
We next asked if the kinase activity of RIPK1 could regulate its accumulation in complex I. Previous studies have not observed a role for RIPK1 kinase activity in regulating complex I but were not carried out in RDA conditions, where we found substantial early RIPK1 activation. Indeed, Nec-1s treatment led to substantially decreased levels of RIPK1 in complex I in 661W cells with TAK1 inhibition (Fig. 5D) and in Tak1−/− MEFs (Fig. 5E). Specifically, Nec-1s led to a mild decrease in RIPK1 levels in complex I at 5 min but prevented sustained RIPK1 accumulation and activation, resulting in substantially decreased association of RIPK1 with complex I at later time points. Similar results were seen following TNFα/5Z-7 treatment in WT vs. RIPK1D138N kinase-dead knock-in MEFs (SI Appendix, Fig. S7G). Nec-1s did not affect RIPK1 levels in complex I during necroptosis induced by TNFα/CHX/zVAD, consistent with previous studies and the lack of substantial RIPK1 activation in complex I under this condition (SI Appendix, Fig. S7H). Overall, our data show that loss of TAK1 activity by pharmacological inhibition or genetic deletion leads to a unique complex I in RDA with sustained levels of highly activated RIPK1, which may act as a prelude to iuRIPK formation.
To determine if LRRK2 promotes iuRIPK1 formation by regulating this unique complex I in RDA, we compared complex I with or without LRRK2 knockdown. Knockdown of LRRK2 reduced RIPK1 association with complex I under RDA conditions (Fig. 5F). While initial RIPK1 activation in complex I at 5 min was not substantially affected by LRRK2 knockdown, this reduction in RIPK1 levels in complex I broke the feed-forward loop driving RIPK1 activation in RDA and resulted in substantially reduced RIPK1 activation in complex I at 15 and 25 min. Overall, these results suggest that the formation of the unique iuRIPK1 intermediate in RDA occurs following a massive increase in RIPK1 levels and activity in complex I specifically in RDA. LRRK2 is involved in recruiting and/or stabilizing activated RIPK1 in complex I in RDA and thus contributes to increased levels of activated RIPK1 in complex I and formation of iuRIPK1.
c-Cbl Promotes K63-Ubiquitination of RIPK1 in Complex I and Formation of iuRIPK1.
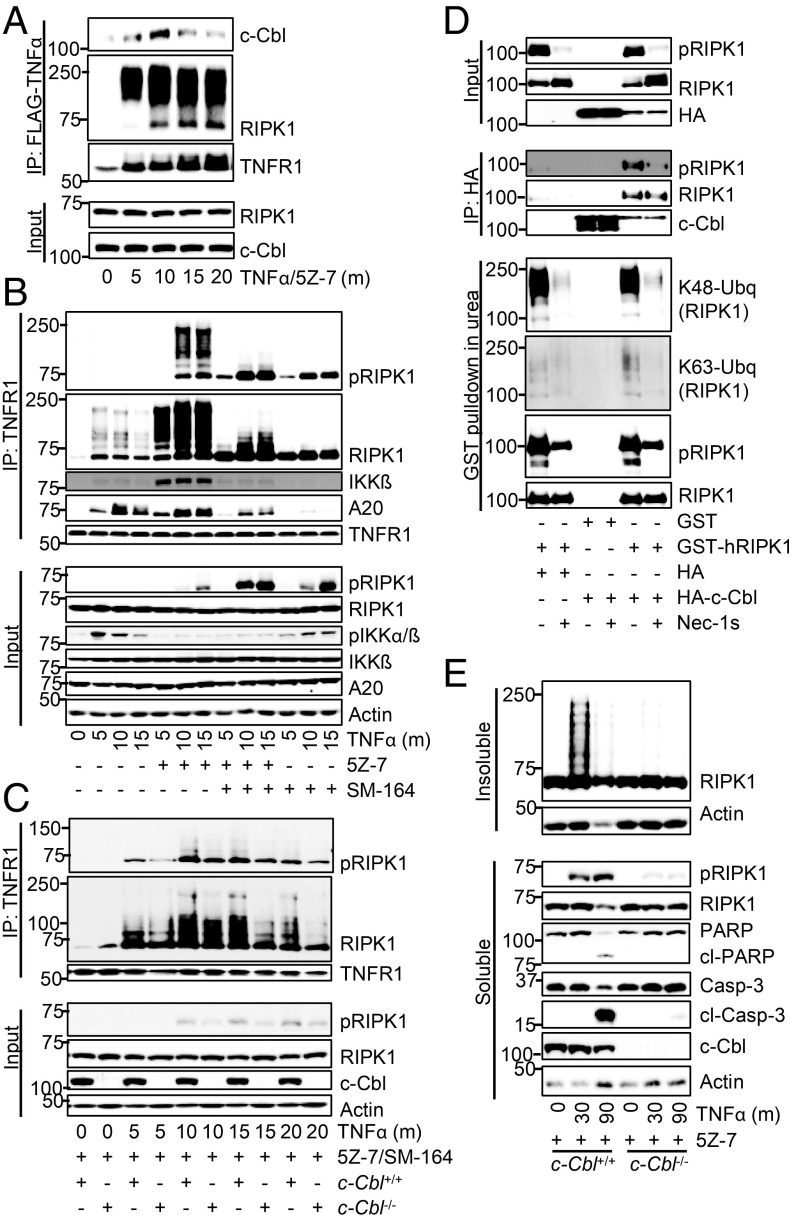
c-Cbl is a RING E3 ligase that can mediate the conjugation of either K48- or K63-linked ubiquitin chains (45). CIN85, a c-Cbl binding partner, can be recruited to complex I upon TNFα stimulation (46). We found that c-Cbl itself was recruited to complex I following ∼10 min TNFα/5Z-7 treatment (Fig. 6A). In addition, knockdown of CIN85 protected cells from RDA (SI Appendix, Fig. S8A). Following induction of RDA by TNFα/5Z-7, RIPK1 activation at 25 min and onward was noticeably reduced in c-Cbl knockdown cells (SI Appendix, Fig. S8B). Therefore, we hypothesized that c-Cbl might be the missing E3 ligase which promotes prodeath K63 ubiquitination of RIPK1 at complex I and iuRIPK1 formation in RDA.
Fig. 6.
c-Cbl promotes K63-ubiquitination of RIPK1 in complex I and subsequent iuRIPK1 formation in RDA. (A–E) Cells transfected and/or treated as indicated and samples analyzed by Western blotting using indicated antibodies; pRIPK1 is RIPK1-pS166. (A) The 661W cells treated with FLAG-TNFα/5Z-7 to induce RDA and complex I immunoprecipitated with FLAG-M2 affinity beads. (B) Immunoprecipitation of complex I from WT MEFs with anti-TNFR1. (C) Immunoprecipitation of complex I from c-Cbl+/+ and c-Cbl−/− MEFs with anti-TNFR1. (D) The 293T cells transfected with indicated plasmids and c-Cbl immunoprecipitated with HA-affinity beads, or RIPK1 purified by GST pulldown in 1 M urea. (E) c-Cbl+/+ and c-Cbl−/− MEFs treated with TNFα/5Z-7 to induce RDA and fractionated using lysis buffer with Nonidet P-40/TX-100 followed by 6 M urea. Concentrations of reagents: TNFα, 1 ng/mL (E), 10 ng/mL (C), 50 ng/mL (B); FLAG- TNFα, 50 ng/mL.
To distinguish the ubiquitination mediated by cIAP1/2 vs. that of c-Cbl, we first examined complex I with cIAP1/2 depletion induced by SM-164, both on its own and in combination with TAK1 inhibition by 5Z-7. As expected, the ubiquitination of RIPK1 was reduced, but not completely blocked, following TNFα/SM-164 treatment (Fig. 6B). Levels of RIPK1 in complex I were increased with TNFα/SM-164 compared with TNFα alone. Further, like TAK1 deficiency, loss of cIAP1/2 promoted the activation of RIPK1 in complex I (Fig. 6B). This suggests that increased association and activation of RIPK1 in complex I are common features of RDA. Importantly, cIAP1/2 depletion before treatment with TNFα/5Z-7 decreased, but did not eliminate, the ubiquitination of RIPK1 in complex I, again suggesting the possible involvement of additional E3 ubiquitin ligases in ubiquitinating RIPK1 under RDA conditions (Fig. 6B). Finally, loss of c-Cbl protected against RDA induced by both TNFα/SM-164 and TNFα/5Z-7/SM-164, demonstrating that cIAP1/2 are not required for c-Cbl to promote RDA (SI Appendix, Fig. S8 C and D).
Therefore, we examined complex I induced by TNFα/5Z-7/SM-164 in c-Cbl+/+ vs. c-Cbl−/− MEFs (Fig. 6C). c-Cbl deficiency resulted in reduced RIPK1 ubiquitination in complex I in RDA at 15–20 min. Similar results were seen with c-Cbl knockdown, particularly at 15 min (SI Appendix, Fig. S8E). Coexpression of c-Cbl and RIPK1 in 293T cells demonstrated that c-Cbl binds to RIPK1 and specifically promotes K63-ubiquitination of RIPK1, which is blocked by Nec-1s (Fig. 6D). Further, purified c-Cbl robustly mediated K63-ubiquitination of purified RIPK1 in an in vitro ubiquitination assay with recombinant E1 and E2 (SI Appendix, Fig. S8F). Finally, iuRIPK1 formation and RIPK1 activation following TNFα/5Z-7 was almost completely blocked in c-Cbl−/− MEFs (Fig. 6E and SI Appendix, Fig. S8G). Subsequent caspase activation and cleavage of caspase substrates were also inhibited (Fig. 6E). Overall, these data suggest that c-Cbl regulates RDA at the level of RIPK1 ubiquitination in complex I. Since the K63 ubiquitination of RIPK1 mediated by c-Cbl can be inhibited by Nec-1s (Fig. 6D), c-Cbl–mediated K63 ubiquitination of RIPK1 may be involved in stabilizing the active conformation of RIPK1 to promote the formation of iuRIPK1.
Two Distinct Pathways of Necroptosis Initiation by TNFα.
Inhibiting caspases in the context of RDA (i.e., TNFα/5Z-7/zVAD treatment) switches the mode of cell death from RDA to necroptosis (18). We wondered how regulation of necroptosis induced by TNFα/5Z-7/zVAD might compare with necroptosis induced by TNFα/CHX/zVAD, and if RDA-specific regulators identified in our screen may differentially regulate the two pathways of necroptosis induction. Necroptosis induced in the context of TAK1 inhibition, by TNFα/5Z-7/zVAD, occurred more rapidly than necroptosis induced by TNFα/CHX/zVAD in both 661W cells and WT MEFs (Fig. 7A and SI Appendix, Fig. S9A). Therefore, the kinetics of cell death induction by TNFα/5Z-7/zVAD is more similar to RDA than necroptosis induced by TNFα/CHX/zVAD. We confirmed that caspase inhibition did in fact change the mode of cell death to necroptosis by confirming dependence on RIPK1 kinase activity, a change in cell death morphology from apoptotic to necrotic by electron microscopy, and dependence on RIPK3 and MLKL in TNFα/5Z-7 + caspase inhibitor-treated cells (Fig. 7A and SI Appendix, Fig. S9 B–D).
Fig. 7.
Two distinct pathways of necroptosis initiation by TNFα. (A) The 661W cells treated with TNFα/zVAD plus either 5Z-7 or CHX to induce necroptosis. Cell viability determined by CellTiter-Glo. (B) Scheme for additional tertiary siRNA screen for regulators of necroptosis induced by TNFα/5Z-7/zVAD. (C) The 661W cells transfected with siRNAs against APC11, LRRK2, or c-Cbl or nontargeting control (N.C.) siRNA for 72 h and treated with TNFα/5Z-7/zVAD for 8 h to induce necroptosis under RDA conditions. Cell survival assayed by CellTiter-Glo. Knockdown efficiency determined by Western blotting. (D) The 661W cells treated with TNFα/5Z-7/zVAD or TNFα/CHX/zVAD to induce necroptosis. Cells fractionated using lysis buffer with Nonidet P-40/TX-100 followed by 6 M urea and analyzed by Western blotting with indicated antibodies; pRIPK1 is RIPK1-pS166, pMLKL is MLKL-pS345. Concentrations of reagents: TNFα, 1 ng/mL (C), 10 ng/mL (A and D). All data shown are mean ± SD of three or more independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 by one-way ANOVA.
Next, we screened the 66 genes identified as RDA protectors in our siRNA screen for protection against necroptosis induced by TNFα/5Z-7/zVAD (Fig. 7B). Consistent with a model of two distinct pathways for necroptosis induction, the majority (85%) of the genes which promoted RDA (TNFα/5Z-7) also promoted TNFα/5Z-7/zVAD, but only a smaller fraction also promoted necroptosis induced by TNFα/CHX/zVAD (SI Appendix, Fig. S9E). To remove any bias based on the arbitrary cutoff used to classify genes as protectors, the fold protection of each siRNA duplex in each mode of cell death vs. its fold protection in RDA was plotted. The correlation between protection in TNFα/5Z-7/zVAD vs. RDA, r = 0.72, was significantly higher than in TNFα/CHX vs. RDA, r = 0.48, or TNFα/CHX/zVAD vs. RDA, r = 0.56 (SI Appendix, Fig. S9F). These data demonstrate that necroptosis induced in the context of TAK1 inhibition is distinctly regulated from necroptosis induced in the context of protein synthesis inhibition in an unbiased manner. Further, these data showed that LRRK2 and c-Cbl, as well as APC11, promote necroptosis induced by TNFα/5Z-7/zVAD, which we confirmed by low-throughput assays (Fig. 7C).
Our data suggest that iuRIPK1 is a critical intermediate bridging complex I and complex II in RDA, and it does not form in RIPK1-independent apoptosis or necroptosis induced by TNFα/CHX/zVAD. The mechanism of iuRIPK1 formation involves sustained and increased levels of activated RIPK1 in complex I, which is regulated by LRRK2, followed by K63 ubiquitination of activated RIPK1 at complex I promoted by c-Cbl. Therefore, if necroptosis induction under TAK1-deficient conditions is distinct from necroptosis induced by TNFα/CHX/zVAD, iuRIPK1 should form only in the former mode of necroptosis. Indeed, we found the formation of iuRIPK1 in necroptosis induced by TNFα/zVAD in Tak1−/− MEFs (SI Appendix, Fig. S9G). In addition, RIPK1 activation and iuRIPK1 formation occurred within 15–30 min, before MLKL phosphorylation at 2 h, in necroptosis induced by TNFα/5Z-7/zVAD but not TNFα/CHX/zVAD, in both 661W cells and MEFs (Fig. 7D and SI Appendix, Fig. S9H).
Discussion
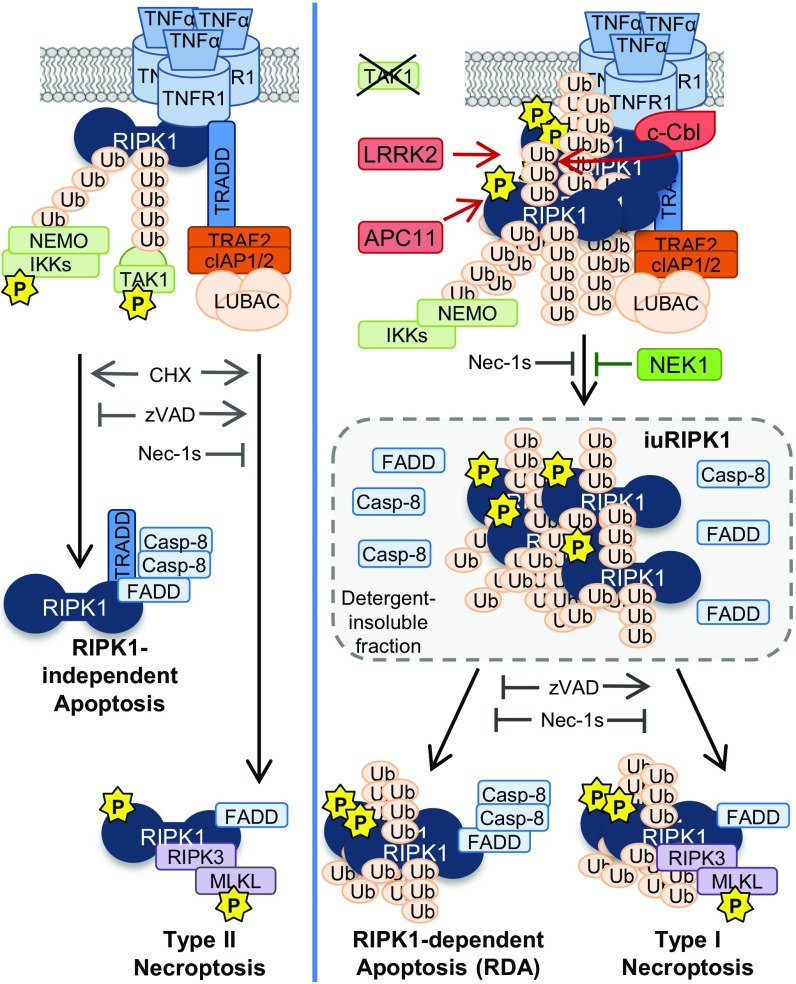
In this paper, we demonstrate that TNFα stimulation of cells combined with the inhibition of TAK1 promotes RDA through the early activation of RIPK1 in complex I and the formation of a distinct pool of highly ubiquitinated and activated RIPK1 in the insoluble compartment, named iuRIPK1. We show that iuRIPK1 may be a critical part of the signaling pathway in RDA by concentrating activated RIPK1, leading to sustained RIPK1 activation and interaction with FADD and caspase-8 to form RIPK1-dependent complex II. The identification of four genes from our siRNA screen, c-Cbl, Apc11, Lrrk2, and Nek1, whose protein products are all involved in regulating both iuRIPK1 formation and caspase activation and cell death in RDA, supports the model that iuRIPK1 is an important platform for subsequent complex II formation (Fig. 8).
Fig. 8.
Model for different modes of TNFα-induced apoptosis and necroptosis. When protein synthesis is blocked, TNFα can lead to RIPK1-independent apoptosis or type II necroptosis, in which active RIPK1 slowly accumulates in the cytosol or in association with complex II. When TAK1 is deficient, TNFα can lead to RDA or type I necroptosis via the accumulation of activated RIPK1 in complex I, which subsequently transitions into an insoluble pool of highly ubiquitinated and activated RIPK1 (iuRIPK1). LRRK2, c-Cbl, and APC11 cooperate to promote the formation of iuRIPK1 to mediate complex II formation, while NEK1 inhibits iuRIPK1 formation.
We found that c-Cbl mediates iuRIPK1 formation by promoting K63 ubiquitination of activated RIPK1 in complex I under RDA conditions. While proteins such as cIAP1/2 promote prosurvival K63 ubiquitination of RIPK1 in complex I, the loss of c-Cbl results in further reduction of K63 ubiquitination of RIPK1 and substantially restricts iuRIPK1 formation in RDA. Thus, c-Cbl mediates prodeath K63 ubiquitination of RIPK1 to promote iuRIPK1 formation and sustained RIPK1 activation independent of cIAP1/2 in RDA. In addition, LRRK2 stabilizes activated RIPK1 associated with complex I and therefore promotes formation of iuRIPK1. NEK1 and APC11 may act by suppressing and cooperating, respectively, with the activation of RIPK1 and formation of iuRIPK1.
Formation of iuRIPK1 occurs in RDA but not in RIPK1-independent apoptosis. It is also relevant to necroptosis. While no iuRIPK1 formation occurs in necroptosis induced by TNFα/CHX/zVAD, iuRIPK1 rapidly forms in necroptosis induced under RDA conditions by TNFα/5Z-7/zVAD. The distinct RIPK1 activation profiles in necroptosis induced by TNFα/5Z-7/zVAD and TNFα/CHX/zVAD suggest that these represent two distinct modes of RIPK1 activation and necroptosis initiation.
We propose explicit naming of the two different necroptosis pathways induced by TNFα “type I” and “type II” necroptosis, as defined by the distinct modes of RIPK1 activation (Fig. 8). RIPK1 activation occurs slowly in the cytosol or associated with complex II in type II necroptosis induced by TNFα/CHX/zVAD, whereas it occurs rapidly in complex I in type I necroptosis induced by TNFα/5Z-7/zVAD. iuRIPK1 forms in RDA and type I necroptosis, but not in type II necroptosis or apoptosis. Therefore, formation of iuRIPK1 may be a useful biomarker for determining if RIPK1 activation observed in vivo is a result of disruption of complex I. Consistent with a differential mode of RIPK1 activation and role for iuRIPK1 in type I vs. type II necroptosis, c-Cbl and LRRK2 specifically promote type I but not type II necroptosis, and APC11 plays a more significant role in type I than in type II necroptosis. Overall, our results demonstrate that the distinct mechanism of RIPK1 activation proceeding from TNFα stimulation when complex I is disrupted by TAK1 inhibition is relevant not just to RDA but to necroptosis as well.
We show that before formation of iuRIPK1, deficiencies in either TAK1 or cIAP1/2 increase RIPK1 levels in complex I and promote its activation in complex I, which defines the first critical regulatory step in the RDA and type I necroptosis pathways. We identify that PD-associated LRRK2, also known as RIPK7, binds to RIPK1 in RDA and plays a role in promoting this first step. Further, while previous studies have focused on the role of RIPK1 kinase activity on complex II formation in RDA (11, 15, 17, 18), we show that RIPK1 kinase activity regulates RIPK1 levels in complex I under RDA conditions, which is possible due to the massive increase in RIPK1 activation in complex I specifically in RDA conditions. The mechanism for increased RIPK1 association and activation in complex I in RDA requires further exploration but may be partially due to direct phosphorylation of RIPK1 in complex I by TAK1 and IKK (15, 21), which may trigger the disassociation of RIPK1 from the TNF-RSC or prevent additional RIPK1 recruitment to complex I. In the absence of TAK1/IKK, an initial increase in RIPK1 levels at complex I occurs, due to increased recruitment and/or decreased disassociation. This may then allow RIPK1 activation to lead to a feed-forward loop to increase the accumulation of activated RIPK1 in complex I and formation of iuRIPK1.
Because of the important role of RIPK1 kinase activity in specifically mediating deleterious outcomes downstream of TNFR1, RIPK1 inhibitors have been advanced into human clinical studies as a therapeutic approach for diseases where anti-TNFα has been efficacious, such as psoriasis, rheumatoid arthritis, and ulcerative colitis (47). RIPK1 inhibitors are being investigated in human clinical trials for neurodegenerative diseases as well, such as ALS and AD, although the precise mechanisms by which RIPK1 kinase activity may contribute to different neurodegenerative conditions remains to be clarified (27, 29, 30). We show that neuronal, astrocytic, and microglial cultures can undergo RDA in response to TNFα and identify four RDA regulators, c-Cbl, Apc11, Lrrk2, and Nek1, which are highly expressed in the CNS (48, 49). Astrocytes in particular express all four genes at high levels, and, interestingly, microglia, which are most sensitive to RDA, are reported to have low expression of RDA suppressor Nek1 (49). Further, while optineurin deficiency in ALS has been shown to drive neuroinflammation and necroptosis and can induce axonal degeneration (29), the potential role of RIPK1 in other genetic variants of ALS, such as NEK1, has not been examined and the mechanism by which NEK1 mutations promote ALS is unclear. Here, we demonstrate a connection between NEK1 and RIPK1-mediated cell death and show that NEK1 binds to activated RIPK1 to suppress RIPK1 activity. Further, LRRK2 mutations are a leading cause of PD, and again the mechanisms by which LRRK2 mutations promote PD are unclear (39, 40). Here, we connect LRRK2 to RIPK1 kinase activation and RDA.
Overall, this work reveals two distinct modes of RIPK1 activation in TNFR1 signaling and provides critical insights on the mechanism of RIPK1 activation and complex II formation in RDA. Furthermore, our study demonstrates possible interesting mechanistic links between RIPK1-mediated apoptosis and the neurodegenerative diseases ALS and PD. Since inhibition of RIPK1 can block both RDA and necroptosis, as well as neuroinflammation mediated by microglia (27, 29, 30), targeting RIPK1 kinase activity presents a unique opportunity to inhibit both cell death pathways and inflammation for the treatment of human diseases, including ALS and possibly PD.
Materials and Methods
Antibodies and Reagents.
All antibodies are commercially available (SI Appendix), except antibodies for ubiquitin immunoprecipitation, a gift from Genentech, and custom-made polyclonal antibodies for RIPK1 (DD), caspase-12, and immunoprecipitation of RIPK1-pS166. FLAG-TNFα was from Enzo. TNFα refers to mTNFα unless specified.
Cell Lines and Tissue Culture.
Cells were provided by D. F. Chen, Harvard Medical School, Boston, MA (611W); H. Band, University of Nebraska Medical Center, Omaha, NE (c-Cbl−/− MEF); and D. Alessi, University of Dundee, Dundee, UK (Lrrk2−/− MEF). Cells were grown at 37 °C with 5% CO2 in high-glucose DMEM with 8.5% FBS supplemented with penicillin/streptomycin.
Animals and Primary Cell Generation.
All animal studies were performed according to ethical guidelines and procedures approved by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School. Primary cells were derived according to standard methods with minor modifications. See SI Appendix.
CellTiter-Glo Assay for Cell Survival.
CellTiter-Glo luminescent cell viability kit was from Promega. Cells were plated in 384-well dishes and cell death was induced 1–3 d later. Cell viability was measured according to the manufacturer’s instructions.
Toxilight Assay for Cell Death.
Toxilight Bioassay Kit was from Lonza. Ten microliters of conditioned cell media was transferred to a 384-well plate and 25 μL of reconstituted Toxilight reagent was added followed by luminescence measurement according to the manufacturer’s instructions.
siRNA Knockdown.
The 661W cells were forward-transfected with 20 nM siRNA using RNAiMax according to the manufacturer’s instructions. For simultaneous transfection of two different siRNA duplexes, 10 nM of each was used for 20 nM total siRNA. MEFs were forward-transfected with 50 nM siRNA using RNAiMax. Cells were plated, transfected, and then treated 72 h later in the same plate.
siRNA Screen.
Dharmacon siGENOME siRNA mouse library subsets were used for the siRNA screen in 661W cells in triplicate. See SI Appendix.
Cell Lysis and Immunoprecipitation.
Cells were lysed and immunoprecipitations done in a standard 20 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol buffer-based buffer with supplementation as described in SI Appendix.
Urea Fractionation.
Soluble fraction was prepared as for immunoprecipitation. After centrifugation of the cell lysate, the pellet was washed with PBS and then resuspended in freshly made 6 M urea + lysis buffer in 20% of the original lysis volume and rotated at 4 °C for at least 1 h before recentrifugation at 21,000 × g at 4 °C for 30 min. The resulting supernatant is termed the “insoluble” fraction. If 0.2% Nonidet P-40 lysis buffer was initially used, the insoluble pellet was first extracted with lysis buffer + 1% Triton X-100 before solubilization with 6 M urea. See SI Appendix for ubiquitin immunoprecipitation from urea fraction.
Protein Purification and in Vitro Ubiquitination Assay.
Human c-Cbl-HA and GST-RIPK1 were expressed and purified from HEK293T cells. In vitro ubiquitination reactions were set up using Boston Biochem Ubiquitin Conjugation Initiation Kit and 1 µM E2 enzyme UbcH5. See SI Appendix.
Western Blotting.
Samples were lysed in the indicated buffer and run on SDS-PAGE gels before transfer to nitrocellulose for Western blotting. Membranes were blocked in 5% milk in Tris-buffered saline + 0.1% Tween 20 (TBST) for 1 h, incubated overnight in primary antibodies in 3% BSA in TBST, and visualized with HRP-linked secondary antibodies and ECL.
Statistical Analysis.
Data are expressed as the mean ± the SD. For cell viability measurements by CellTiter-Glo, the average of 3 or more independent experiments are shown. For cell death measurements by Toxilight, the average of 2–4 independent experiments are shown. Two-tailed Student’s t test was used for comparison of two groups, and one-way ANOVA was used for multiple comparisons with Bonferroni’s post hoc test using GraphPad Prism. In all cases where P values for ANOVA are reported, the global P value was ≤0.05.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Neurological Disorders and Stroke Grant 1R01NS082257, National Institute on Aging (NIA) Grants 1R01AG047231 and RF1AG055521, and NIH Grant T32 GM007306 and NIH/NIA Grant T32 AG000222 (to P.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806973115/-/DCSupplemental.
References
- 1.Aggarwal BB. Signalling pathways of the TNF superfamily: A double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 2.Varfolomeev E, Vucic D. Intracellular regulation of TNF activity in health and disease. Cytokine. 2018;101:26–32. doi: 10.1016/j.cyto.2016.08.035. [DOI] [PubMed] [Google Scholar]
- 3.Vassalli P. The pathophysiology of tumor necrosis factors. Annu Rev Immunol. 1992;10:411–452. doi: 10.1146/annurev.iy.10.040192.002211. [DOI] [PubMed] [Google Scholar]
- 4.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 5.Ting AT, Pimentel-Muiños FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 8.Galluzzi L, Kepp O, Chan FK, Kroemer G. Necroptosis: Mechanisms and relevance to disease. Annu Rev Pathol. 2017;12:103–130. doi: 10.1146/annurev-pathol-052016-100247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 10.Ting AT, Bertrand MJM. More to life than NF-κB in TNFR1 signaling. Trends Immunol. 2016;37:535–545. doi: 10.1016/j.it.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arslan SÇ, Scheidereit C. The prevalence of TNFα-induced necrosis over apoptosis is determined by TAK1-RIP1 interplay. PLoS One. 2011;6:e26069. doi: 10.1371/journal.pone.0026069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donnell MA, Hase H, Legarda D, Ting AT. NEMO inhibits programmed necrosis in an NFκB-independent manner by restraining RIP1. PLoS One. 2012;7:e41238. doi: 10.1371/journal.pone.0041238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlantis K, et al. NEMO prevents RIP kinase 1-mediated epithelial cell death and chronic intestinal inflammation by NF-κB-dependent and -independent functions. Immunity. 2016;44:553–567. doi: 10.1016/j.immuni.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondylis V, et al. NEMO prevents steatohepatitis and hepatocellular carcinoma by inhibiting RIPK1 kinase activity-mediated hepatocyte apoptosis. Cancer Cell. 2015;28:582–598. doi: 10.1016/j.ccell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dondelinger Y, et al. NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol Cell. 2015;60:63–76. doi: 10.1016/j.molcel.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 16.Legarda-Addison D, Hase H, O’Donnell MA, Ting AT. NEMO/IKKgamma regulates an early NF-kappaB-independent cell-death checkpoint during TNF signaling. Cell Death Differ. 2009;16:1279–1288. doi: 10.1038/cdd.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dondelinger Y, et al. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ. 2013;20:1381–1392. doi: 10.1038/cdd.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamothe B, Lai Y, Xie M, Schneider MD, Darnay BG. TAK1 is essential for osteoclast differentiation and is an important modulator of cell death by apoptosis and necroptosis. Mol Cell Biol. 2013;33:582–595. doi: 10.1128/MCB.01225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–424. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Annibaldi A, et al. Ubiquitin-mediated regulation of RIPK1 kinase activity independent of IKK and MK2. Mol Cell. 2018;69:566–580.e5. doi: 10.1016/j.molcel.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng J, et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat Commun. 2017;8:359. doi: 10.1038/s41467-017-00406-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dondelinger Y, et al. MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat Cell Biol. 2017;19:1237–1247. doi: 10.1038/ncb3608. [DOI] [PubMed] [Google Scholar]
- 23.Jaco I, et al. MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol Cell. 2017;66:698–710.e5. doi: 10.1016/j.molcel.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayyad Z, Sirohi K, Radha V, Swarup G. 661W is a retinal ganglion precursor-like cell line in which glaucoma-associated optineurin mutants induce cell death selectively. Sci Rep. 2017;7:16855. doi: 10.1038/s41598-017-17241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger SB, et al. Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol. 2014;192:5476–5480. doi: 10.4049/jimmunol.1400499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ofengeim D, et al. Activation of necroptosis in multiple sclerosis. Cell Rep. 2015;10:1836–1849. doi: 10.1016/j.celrep.2015.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei R, et al. SPATA2 regulates the activation of RIPK1 by modulating linear ubiquitination. Genes Dev. 2017;31:1162–1176. doi: 10.1101/gad.299776.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito Y, et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science. 2016;353:603–608. doi: 10.1126/science.aaf6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ofengeim D, et al. RIPK1 mediates a disease-associated microglial response in Alzheimer’s disease. Proc Natl Acad Sci USA. 2017;114:E8788–E8797. doi: 10.1073/pnas.1714175114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caccamo A, et al. Necroptosis activation in Alzheimer’s disease. Nat Neurosci. 2017;20:1236–1246. doi: 10.1038/nn.4608. [DOI] [PubMed] [Google Scholar]
- 32.Chekeni FB, et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature. 2010;467:863–867. doi: 10.1038/nature09413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenna KP, et al. SLAGEN Consortium NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2016;48:1037–1042. doi: 10.1038/ng.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner D, et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain. 2016;139:e28. doi: 10.1093/brain/aww033. [DOI] [PubMed] [Google Scholar]
- 36.Chang L, Barford D. Insights into the anaphase-promoting complex: A molecular machine that regulates mitosis. Curr Opin Struct Biol. 2014;29:1–9. doi: 10.1016/j.sbi.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konishi Y, Stegmüller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- 39.Martin I, Kim JW, Dawson VL, Dawson TM. LRRK2 pathobiology in Parkinson’s disease. J Neurochem. 2014;131:554–565. doi: 10.1111/jnc.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Chan SL, Tan EK. Targeting LRRK2 in Parkinson’s disease: An update on recent developments. Expert Opin Ther Targets. 2017;21:601–610. doi: 10.1080/14728222.2017.1323881. [DOI] [PubMed] [Google Scholar]
- 42.Antoniou N, et al. A motif within the armadillo repeat of Parkinson’s-linked LRRK2 interacts with FADD to hijack the extrinsic death pathway. Sci Rep. 2018;8:3455. doi: 10.1038/s41598-018-21931-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho CC, Rideout HJ, Ribe E, Troy CM, Dauer WT. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J Neurosci. 2009;29:1011–1016. doi: 10.1523/JNEUROSCI.5175-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis M, et al. Cloning and expression of cDNAs for two distinct murine tumor necrosis factor receptors demonstrate one receptor is species specific. Proc Natl Acad Sci USA. 1991;88:2830–2834. doi: 10.1073/pnas.88.7.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swaminathan G, Tsygankov AY. The Cbl family proteins: Ring leaders in regulation of cell signaling. J Cell Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]
- 46.Narita T, Nishimura T, Yoshizaki K, Taniyama T. CIN85 associates with TNF receptor 1 via Src and modulates TNF-α-induced apoptosis. Exp Cell Res. 2005;304:256–264. doi: 10.1016/j.yexcr.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Harris PA, et al. DNA-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. J Med Chem. 2016;59:2163–2178. doi: 10.1021/acs.jmedchem.5b01898. [DOI] [PubMed] [Google Scholar]
- 48.Dong L, et al. The E3 ubiquitin ligase c-Cbl inhibits microglia-mediated CNS inflammation by regulating PI3K/Akt/NF-κB pathway. CNS Neurosci Ther. 2016;22:661–669. doi: 10.1111/cns.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.