Prostate cancer remains a leading cause of cancer death in the United States (1). In 2017, it was estimated that over 160,000 prostate cancer cases were diagnosed in the United States, and ∼26,000 men died of prostate cancer (2). Because tumor growth in prostate cancer is predominantly dependent on the androgen receptor (AR), a homodimer-forming transcription factor, the disease is commonly treated via therapies targeting the AR-axis. While this approach has proven to be initially effective, a majority of prostate cancer patients eventually develop resistance to AR-targeting therapies, leading to castration-resistant prostate cancer (CRPC), the lethal stage of prostate cancer. Measuring prostate-specific antigen levels in patient serum is regularly used for evaluating the efficacy of prostate cancer treatment. The serum prostate-specific antigen value, however, does not always correlate with the malignant state of CRPC (3). Therefore, it is of high priority to discover therapeutic targets and biomarkers that can accurately assess the efficacy of treatment in patients who have become resistant to therapies targeting the AR-axis.
Various mechanisms of resistance to AR targeting have been identified, such as AR bypass signaling, AR alternative splicing, and AR gene amplification (4). Notably, aberrant AR splicing seems to be heavily associated with resistance to AR-targeting therapy in CRPC patients. AR-V7 is one of the splice variants that is most strongly associated with CRPC, and its expression is a predictor of poor survival in CRPC patients (5). In PNAS, Chen et al. (5) aim to uncover the mechanism by which AR-V7 promotes CRPC growth, with the goal of revealing therapeutic targets and biomarkers for CRPC patients. Specifically, the authors investigate the target genes of AR-V7 and the putative mediators of AR-V7 binding to these genes.
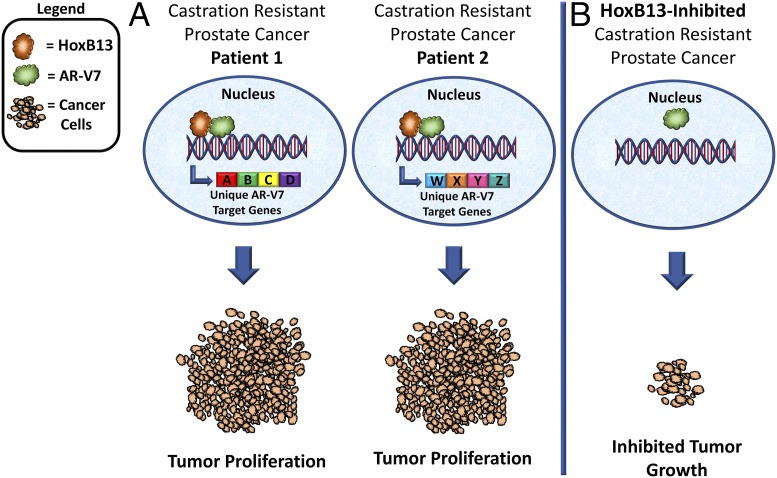
Investigators have previously been unsuccessful in uncovering the oncogenic mechanism driven by AR-V7 in CRPC because of the lack of an AR-V7–specific antibody. Chen et al. (5) developed an AR-V7–specific antibody that was used for ChIP-exonuclease sequencing (ChIP-Exo), a method that determines the gene binding locations of transcription factors at single-base pair resolution, to map where AR-V7 binds within CRPC tumor genomes. Importantly, Chen et al. were also able to validate their findings using two independent patient-derived CRPC tissue libraries, in addition to an in vivo study in which xenograft CRPC cell line-derived tumors were analyzed. The results of this study reveal that AR-V7 target genes are heterogeneous both within independent CRPC cell lines as well as between individual CRPC patient tissues. Results also demonstrate that oncogenic AR-V7 genomic binding is dependent on HoxB13, and targeting HoxB13 is sufficient to inhibit CRPC growth in cell lines and in vivo (Fig. 1). Finally, the role of HoxB13 in AR-V7 genomic binding is reflected in CRPC patient samples and circulating tumor cells (CTCs), suggesting that HoxB13 may be exploited as a therapeutic target for CRPC.
Fig. 1.
Model of HoxB13-mediated AR-V7 oncogenic function. (A) AR-V7 genomic binding is dependent on HoxB13, and genes up-regulated by AR-V7 are heterogeneous between distinct CRPC patients, ultimately leading to tumor cell proliferation. (B) HoxB13 targeting is sufficient to inhibit CRPC tumor growth.
Chen et al. (5) demonstrate that HoxB13 and AR-V7 are coexpressed within the same CRPC patient tissues and CTCs. While the methodology indicates that HoxB13 and AR-V7 are coexpressed within individual CTCs, it has not been determined if they are expressed within the same cell in patient tissues. Furthermore, key experiments demonstrating HoxB13 knockdown-mediated growth inhibition of CRPC cell lines and xenograft tumors only result in a minimal inhibition of cancer cell growth. This could perhaps be a result of a weak HoxB13 knockdown in the xenografted CRPC cell lines. Additionally, while HoxB13 was found to be required for AR-V7 function in both cell lines, the HoxB13 knockdown-mediated antiproliferative effect varies drastically between CRPC xenograft tumors in vivo. This suggests that the effectiveness of HoxB13 therapeutic targeting may be context-dependent. Chen et al. also find that AR-V7 does not dimerize with full-length AR (AR-FL) in vitro in the absence of hormone, suggesting that AR-V7 and AR-FL may have distinct roles within CRPC tumors. This is consistent with studies demonstrating that AR splice variants maintain an AR transcriptional program and promote CRPC growth independent of AR-FL (6).
Moving forward, emphasis should be placed on understanding what mediates the up-regulation of HoxB13 in CRPC. Does AR-V7 enhance HoxB13 expression in CRPC? Alternatively, does HoxB13 promote the AR-V7 splicing variant as cancer cells become resistant to AR-targeting therapies? Kholi et al. (7) have also determined that AR-V9 promotes androgen independent growth and was frequently coexpressed with AR-V7 within CRPC cell lines, patient tumor tissues, as well as CTCs. AR-V9 was also associated with disease progression during AR-targeting therapy. It would be impactful to clarify whether the oncogenic function driven by AR-V9 is similarly dependent on HoxB13. Chen et al. (5) indicate that HoxB13-mediated AR-V7 target genes are largely heterogeneous within independent CRPC cell lines, as well as between patient tissues. These data suggest that the levels of any one gene or set of conserved genes regulated by AR-V7 may serve poorly as a common biomarker for the efficacy of CRPC treatment. Determining if AR-V9 target genes are similarly heterogeneous—or whether they share a common set of target genes—could lead to the identification of biomarkers for measuring CRPC treatment effectiveness. Perhaps elucidating exactly what mediates where HoxB13 directs AR-V7 genomic binding in these different contexts could also help uncover the cause of this overwhelming heterogeneity in AR-V7 gene regulation in CRPC patients, ultimately leading to improved CRPC therapies in the future.
Acknowledgments
The authors are funded by American Cancer Society Grant RSG-17-068-01-TBG, Department of Defense Grant W81XWH-13-1-0470, the Margaret E. Early Medical Research Trust, NIH/National Cancer Institute Grant P50CA092131/University of California, Los Angeles Specialized Program of Research Excellence in Prostate Cancer, and support from the University of California, Los Angeles’s Jonsson Comprehensive Cancer Center, Broad Stem Cell Research Center, Clinical and Translational Science Institute, and Institute of Urologic Oncology.
Footnotes
The authors declare no conflict of interest.
See companion article on page 6810.
References
- 1.Mirzaei M, Mirzadeh M, Mirzaei M. Mortality rate and years of life lost due to prostate cancer in Yazd Province, Iran: A 10-year study. Sultan Qaboos Univ Med J. 2017;17:e424–e429. doi: 10.18295/squmj.2017.17.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eastham J. Prostate cancer screening. Investig Clin Urol. 2017;58:217–219. doi: 10.4111/icu.2017.58.4.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizokami A, et al. Understanding prostate-specific antigen dynamics in monitoring metastatic castration-resistant prostate cancer: Implications for clinical practice. Asian J Androl. 2017;19:143–148. doi: 10.4103/1008-682X.179159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastos DA, Antonarakis ES. CTC-derived AR-V7 detection as a prognostic and predictive biomarker in advanced prostate cancer. Expert Rev Mol Diagn. 2018;18:155–163. doi: 10.1080/14737159.2018.1427068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, et al. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc Natl Acad Sci USA. 2018;115:6810–6815. doi: 10.1073/pnas.1718811115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nyquist MD, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci USA. 2013;110:17492–17497. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohli M, et al. Androgen receptor variant AR-V9 is coexpressed with AR-V7 in prostate cancer metastases and predicts abiraterone resistance. Clin Cancer Res. 2017;23:4704–4715. doi: 10.1158/1078-0432.CCR-17-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]