Significance
Birth defects are the leading cause of infant mortality in the United States and Europe, with cardiac defects being the most prevalent. Here we define the requirement and mechanism of action of CHD4, the catalytic core component of the nucleosome remodeling and deacetylase (NuRD) complex, in embryonic heart development. CHD4 is essential from fly to human and mutations in CHD4 are causative to congenital heart disease, including atrial and ventricular septal defects. By generating a cardiac conditional null allele of CHD4, temporal transcriptional profiling, and systems-level analysis of CHD4 target genes and in utero echocardiography, we define molecular, biochemical, anatomical, and physiological mechanisms for CHD4 and the NuRD complex in repressing inappropriate expression of the skeletal and smooth muscle programs in the developing heart.
Keywords: heart, nucleosome remodeling and deacetylase complex, sarcomere, chromatin, congenital heart disease
Abstract
Cardiac development relies on proper cardiomyocyte differentiation, including expression and assembly of cell-type-specific actomyosin subunits into a functional cardiac sarcomere. Control of this process involves not only promoting expression of cardiac sarcomere subunits but also repressing expression of noncardiac myofibril paralogs. This level of transcriptional control requires broadly expressed multiprotein machines that modify and remodel the chromatin landscape to restrict transcription machinery access. Prominent among these is the nucleosome remodeling and deacetylase (NuRD) complex, which includes the catalytic core subunit CHD4. Here, we demonstrate that direct CHD4-mediated repression of skeletal and smooth muscle myofibril isoforms is required for normal cardiac sarcomere formation, function, and embryonic survival early in gestation. Through transcriptomic and genome-wide analyses of CHD4 localization, we identified unique CHD4 binding sites in smooth muscle myosin heavy chain, fast skeletal α-actin, and the fast skeletal troponin complex genes. We further demonstrate that in the absence of CHD4, cardiomyocytes in the developing heart form a hybrid muscle cell that contains cardiac, skeletal, and smooth muscle myofibril components. These misexpressed paralogs intercalate into the nascent cardiac sarcomere to disrupt sarcomere formation and cause impaired cardiac function in utero. These results demonstrate the genomic and physiological requirements for CHD4 in mammalian cardiac development.
Congenital heart disease remains the most common congenital malformation, and as such, attaining a mechanistic understanding of cardiomyocyte formation is crucial for improving outcomes to structural heart disease (1, 2). Although much emphasis in the last few years has been placed on transcription factor networks that control cardiomyocyte differentiation, these studies have largely focused on transcriptional activation. However, there is growing recognition that alterations in transcriptional repression also lead to congenital heart disease (3–5). Transcriptional repression involves not only cardiac transcription factors but also broadly expressed multiprotein machines that modify and remodel chromatin. Prominent among these is the nucleosome remodeling and deacetylase (NuRD) complex.
The NuRD complex is one of the major transcriptional complexes that function to repress gene expression. The NuRD complex has been reported in most instances to function as a chromatin-modifying complex and demonstrated to act by combining histone deacetylase activity with an ATP-dependent chromatin remodeling helicase to modulate chromatin states at target genes (6–9). The NuRD complex is essential for numerous developmental events, including ensuring proper timing of the switch from stem cell lineages to differentiated cell types, maintaining cell differentiation, and activating DNA damage response pathways (10–16).
The components of the NuRD complex can vary, but it is typically composed of the ATP-dependent chromodomain helicase DNA-binding protein (CHD) 3/4, histone deacetylase (HDAC) 1/2, metastasis-associated protein (MTA) 1/2/3, retinoblastoma binding protein (RBBP) 4/7 (also known as RbAp48/46), GATAD2A/B, and the mCpG-binding domain protein (MBD) 2/3 (7–9, 17, 18). NuRD complex target specificity can be conferred by the association of components of the NuRD complex with tissue-specific cofactors that target the complex to a defined set of loci. Factors include three proteins associated with congenital heart disease: FOG-2, TBX5, and TBX20 (3, 19–32). Consistently, mutations in CHD4 have been found to be causative to congenital heart disease, including atrial and ventricular septal defects (4). Although the functions of the NuRD complex have been studied in a limited context in vivo, its role in cardiac development has yet to be defined and no studies to date have directly addressed the requirement or mechanism for CHD4 in cardiac tissue.
Here we report CHD4 is essential for cardiac development as mice cardiac conditionally null for Chd4 die during midgestation. By performing a systems-level analysis of CHD4 target genes combined with temporal transcriptional profiling, we provide evidence CHD4 directly binds proximal-promoter and distal gene regulatory elements to directly repress many fast skeletal and smooth muscle myofibril genes. Moreover, we find misexpression of fast skeletal and smooth muscle myofibril genes in Chd4 null embryos is direct and not associated with a reactivation of the embryonic skeletal or embryonic smooth muscle program. We report skeletal and smooth muscle proteins are incorporated into cardiomyocytes, forming a hybrid of all three muscle types. Using a noninvasive in utero embryonic echocardiography technique, we show expression of all three muscle types impairs cardiomyocyte function, leading to a decrease in blood flow and ultimately embryonic lethality. Collectively these studies define molecular, biochemical, anatomical, and physiological mechanisms for CHD4 and the NuRD complex in repressing the inappropriate expression of the skeletal and smooth muscle programs in the developing heart.
Results
CHD4 Is Required for Cardiac Development and Myocardial Growth.
To determine the requirement for CHD4 in heart development, we generated Chd4 cardiac conditional null mice, Chd4Δflox/Δflox, by mating Chd4flox/flox female mice to Chd4flox/+; Nkx2-5Cre/+ male mice (14, 33). Heterozygote (Chd4Δflox/+) mice were viable and fertile and displayed no obvious phenotypic abnormalities. By contrast, no mice homozygous for Chd4 ablation were recovered postnatally. Analysis of timed intercrosses to generate Chd4Δflox/Δflox mice failed to identify viable homozygous Chd4Δflox/Δflox embryos subsequent to embryonic day (E)12.5 (SI Appendix, Table S1).
At E11.5 we observed Chd4Δflox/Δflox mice were viable but displayed pericardial edema, pericardial hemorrhage, and stunted growth compared with Chd4flox/flox (no Cre recombinase) littermate controls (SI Appendix, Fig. S1 A–D). Ultrastructural analysis showed Chd4flox/flox and Chd4Δflox/Δflox hearts initiate cardiac chamber formation at E10.5 (Fig. 1 A and B). However, by E11.5, Chd4Δflox/Δflox hearts exhibited hallmarks of cardiac failure, including enlarged left and right atria, a reduced right ventricle, and an enlarged left ventricle (Fig. 1 C and D) (34). Histological analysis on Chd4Δflox/Δflox and Chd4flox/flox hearts at E10.5 revealed a decrease in complexity of the trabecular layer of the right and left ventricles and a significant decrease in the thickness of the compact layer by E10.5 (SI Appendix, Fig. S1 E–I). This myocardial growth defect was concurrent with a decrease in the mitotic index (SI Appendix, Fig. S1J). However, there was no increase in the levels of apoptosis (SI Appendix, Fig. S1K) or any change in the number of endocardial cells relative to cardiomyocytes (SI Appendix, Fig. S2). Since we confirmed Chd4Δflox/Δflox mice lack any cardiac CHD4 protein at E9.5 (SI Appendix, Fig. S3), these data imply CHD4 is required for cardiac development at or before E10.5.
Fig. 1.
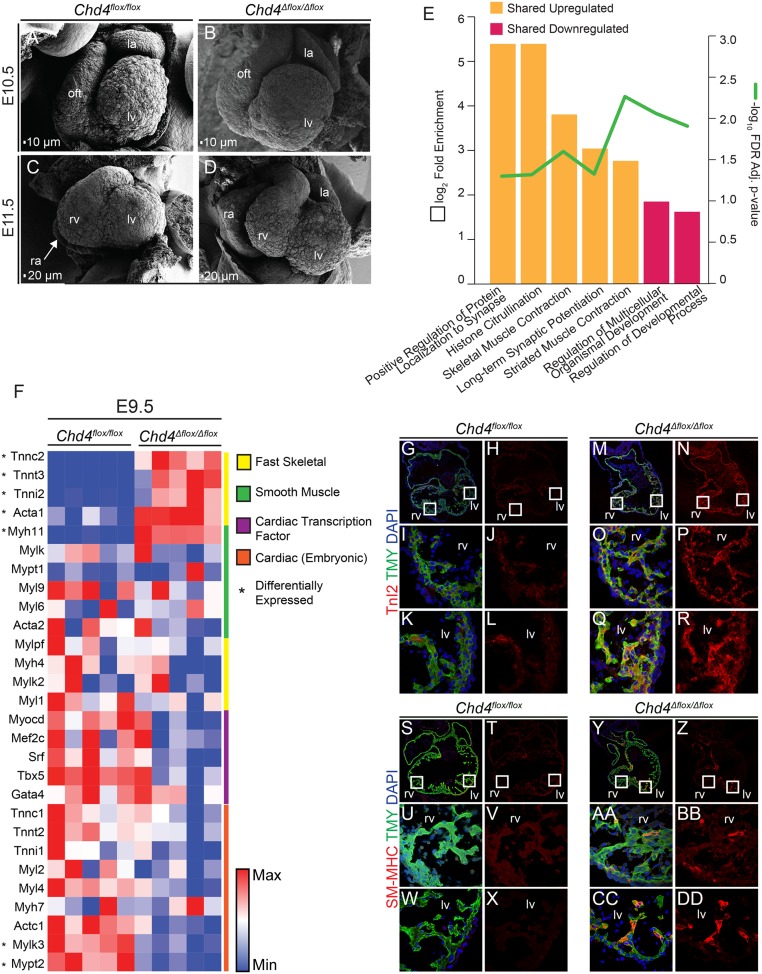
CHD4 is required for transcriptional repression of noncardiac myofibril genes during cardiac development. (A–D) Scanning electron micrographs of Chd4Δflox/Δflox hearts at E10.5 compared with Chd4flox/flox controls demonstrate normal initiation of chamber formation (A and B). By E11.5, Chd4Δflox/Δflox hearts exhibit enlarged atria, a smaller right ventricle, and an enlarged left ventricle compared with Chd4flox/flox hearts (C and D). (E) PANTHER gene ontology (GO) overrepresentation test in biological processes terms for genes up-regulated (orange columns) or down-regulated (pink columns) in Chd4Δflox/Δflox hearts at E9.5 and E10.5. Column height represents log2(fold enrichment) of genes associated with each GO term and green line represents −log10[false discovery rate (FDR)-adjusted P value] of each GO term. (F) Heatmap of fast skeletal, smooth muscle, and cardiac gene expression in Chd4Δflox/Δflox and Chd4flox/flox hearts at E9.5 row scaled to show relative expression reveals lack of transcriptional repression of a set of fast skeletal and smooth muscle myofibril paralogs in the absence of CHD4. (G–R) Fast skeletal Troponin I2 (TnI2) misexpression in cardiomyocytes costained for tropomyosin (TMY) in E10.5 Chd4Δflox/Δflox hearts compared with Chd4flox/flox controls in the right ventricle (I and J compared with O and P) and left ventricle (K and L compared with Q and R). (S–DD) Smooth muscle myosin heavy chain (SM-MHC; Myh11 gene) misexpression in cardiomyocytes costained for TMY in E10.5 Chd4Δflox/Δflox hearts compared with Chd4flox/flox controls in the right ventricle (U and V compared with AA and BB) and left ventricle (W and X compared with CC and DD). Boxed regions denote area of higher magnification. la, left atria; lv, left ventricle; oft, outflow tract; ra, right atria; rv, right ventricle.
CHD4 Regulates Transcription of the Skeletal- and Smooth Muscle-Specific Programs in the Developing Heart.
To explore the molecular mechanism by which CHD4 functions, we performed transcriptomic analysis (RNA-seq) on E9.5 and E10.5 hearts to reflect states before and at the early stage of the observed cardiac defects in Chd4 null hearts (35). Comparing transcript abundances in the presence (Chd4flox/flox) or absence (Chd4Δflox/Δflox) of CHD4 revealed 1,285 differentially expressed genes at E9.5 and 1,318 differentially expressed genes at E10.5 [adjusted P value <0.05, log2(fold change) ≥ ±0.5] (SI Appendix, Fig. S4 A and B). In agreement with the primary role of the NuRD complex as a transcriptional repressor, nearly three times as many genes were up-regulated in the absence of CHD4 relative to down-regulated genes (913 up-regulated versus 372 down-regulated at E9.5; 920 up-regulated versus 398 down-regulated at E10.5) (SI Appendix, Fig. S4 C and D). Of these genes, 327 were coordinately up-regulated, or shared between E9.5 and E10.5, while 65 were coordinately down-regulated at both E9.5 and E10.5 (SI Appendix, Fig. S4 E and F). These genes tended to be those with the highest degree of change in either dataset. Gene ontology (GO) analyses were performed to investigate the roles and pathways of differentially expressed genes in Chd4 null hearts. Surprisingly, the most significant over-represented biological process associated with genes transcriptionally regulated by CHD4 was striated muscle contraction (Fig. 1E) (36).
In cardiomyocytes at early embryonic stages (E8.5–E9.5) myofibril subunits become organized and function as a contractile apparatus that will ultimately develop into mature cardiac sarcomeres. These sarcomeres bear contractile stress to drive the heartbeat and thus circulate nutrients and oxygen throughout the growing embryo (37). Contractility is achieved by cardiomyocyte-specific myofibril subunits, including α-actin (cardiac Actc1 and to a lesser degree skeletal Acta1), β-myosin heavy chain (β-MHC, Myh7), and the troponin complex proteins: cardiac/slow skeletal troponin C1 (Tnnc1), cardiac troponin TnT2 (Tnnt2), and slow skeletal troponin TnI1 (Tnni1 and to a low degree TnI2, Tnni2) (38–42). We found Chd4 null hearts deviate from this normal developmental gene expression pattern by up-regulating the noncardiac paralogs for all of these genes, including smooth muscle Myh11 and fast skeletal Acta1, Tnnc2, Tnnt3, and Tnni2 (Fig. 1F). We confirmed misexpression of noncardiac myofibril isoforms by quantitative PCR (RT-qPCR) (SI Appendix, Fig. S5). Interestingly, aberrant expression of these fast skeletal and smooth muscle paralogs is not accompanied by any significant misexpression of skeletal or smooth muscle transcriptional regulators (Fig. 1F). CHD4 ablation did not significantly alter expression of α-smooth muscle actin (Acta2) or the majority of cardiac myofibril subunits (Fig. 1F and SI Appendix, Fig. S6) (43). We further confirmed increased fast skeletal TnI2 and smooth muscle myosin heavy chain protein levels by immunohistochemistry (Fig. 1 G–DD) (antibody specificity demonstrated in SI Appendix, Fig. S7).
To address whether the misexpression of noncardiac myofibril isoforms was due to CHD4-mediated repression in cardiomyocytes, or a stress response to hemodynamic forces, we conditionally ablated Chd4 with Tnnt2-cre. Results phenocopied that of Nkx2-5Cre/+ in the misexpression of TnI2 and smooth muscle myosin heavy chain (Myh11) (SI Appendix, Fig. S8). Taken together, these studies imply in the absence of CHD4, cardiomyocytes form a hybrid of cardiac, skeletal, and smooth muscle.
CHD4 Binds Proximal Gene Elements to Regulate Myofibril Assembly.
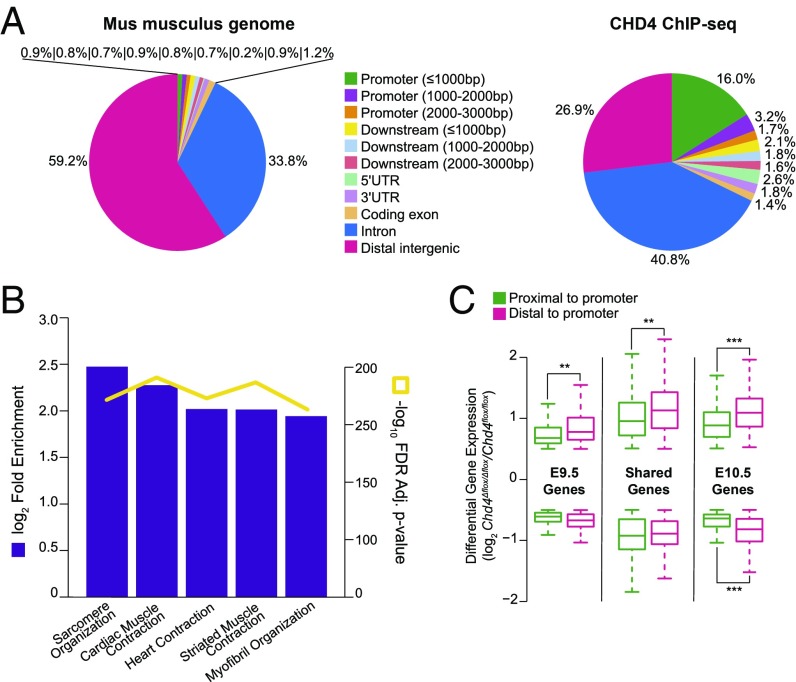
To identify genes directly regulated by CHD4, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) for CHD4 using embryonic hearts collected at E10.0. This is a distinctive report of CHD4 ChIP-seq using noncultured in vivo tissue. We identified 43,818 regions of signal enrichment and found that CHD4 preferentially localizes to proximal-promoter regions (16.0% over 0.9% baseline genome composition) and introns (40.8% over 33.8% baseline genome composition) (Fig. 2A, Right compared with baseline genome composition at Left).
Fig. 2.
CHD4 regulates sarcomere assembly through direct binding to gene regulatory regions in the developing heart. (A, Left) Composition of Mus musculus genome by distance to nearest gene transcription start site (TSS) based on mm10 genome build. (A, Right) Composition of genomic regions bound by CHD4 in wild-type E10.0 hearts by ChIP-seq demonstrates high enrichment at promoter and intronic regions. (B) GO Biological Processes terms for regions bound by CHD4 ranked by log2(fold enrichment) (purple column) and −log10 (FDR-adjusted P value) (yellow line) indicate CHD4 binds genes required for sarcomere organization and myofibril assembly. (C) Comparing the magnitude of gene expression change in Chd4Δflox/Δflox hearts between genes containing proximal promoter (1,500 bp upstream ≥ TSS ≤ 500 bp downstream) or distal intergenic CHD4 ChIP peaks demonstrates a higher degree of change in genes associated with distal regulatory peaks in E9.5, E10.5, and shared up-regulated genes and E10.5 down-regulated genes. ***P value ≤ 0.001, **P value ≤ 0.01.
To elucidate the relationship between CHD4 binding and the previously identified transcriptional changes, CHD4 ChIP-seq peaks were assigned genes by computationally predicted association (44). GO analysis of these genes revealed striking concordance with processes predicted to be transcriptionally regulated by CHD4, specifically sarcomere organization, striated muscle contraction, and myofibril assembly (Fig. 2B). This prompted us to compare genes transcriptionally misregulated in Chd4Δflox/Δflox hearts with genes predicted to contain CHD4 ChIP-seq peaks. Suggestive of direct CHD4-mediated transcriptional regulation, we find a significant enrichment for genes predicted to be directly bound by CHD4 in genes up- or down-regulated in the absence of CHD4 (P < 0.001) (SI Appendix, Fig. S9).
For the E9.5 dataset, 71.4% (652 of 913) of up-regulated genes and 86.3% (321 of 372) of down-regulated genes associated with at least one CHD4 peak; 81.1% (746 of 920) up-regulated and 83.4% (333 of 398) down-regulated genes were associated with at least one CHD4 peak at E10.5. Genes coordinately up- or down-regulated (shared) across E9.5 and E10.5 were also significantly enriched for genes predicted to be directly bound by CHD4; 74.3% (243 of 327) and 84.6% (55 of 65) of genes associating with at least one CHD4 peak, respectively (SI Appendix, Fig. S9). For example, CHD4 peaks are present at regions predicted to associate with the cardiac Mylk3 gene, one of two cardiac sarcomere subunits down-regulated in the absence of CHD4 (Fig. 1F and SI Appendix, Fig. S10). These data are in agreement with studies that have shown CHD4 and the NuRD complex can function in both transcriptional activation and repression (12, 14, 45–48).
As CHD4 appears to directly influence gene regulation, we queried whether there was a difference in the magnitude of regulation based on the location of CHD4 binding, specifically between genes with at least one proximal-promoter site [within 1,500 base pairs (bp) upstream and 500 bp downstream of transcriptional start sites (TSSs)] compared with genes with only distal sites. We observed genes associated with only distal binding sites demonstrated greater differential misregulation in the absence of CHD4 relative to genes with at least one proximal-promoter binding site (Fig. 2C). This unexpected finding may be due to CHD4 localizing at distal active enhancers, as has been previously reported for the NuRD complex (45). Collectively, these data demonstrate CHD4 directly transcriptionally regulates cardiac muscle cell development when cardiac function becomes indispensable for continued embryonic growth.
CHD4 Coordinates a Transcriptional Network to Repress Noncardiac Myofibril Gene Expression.
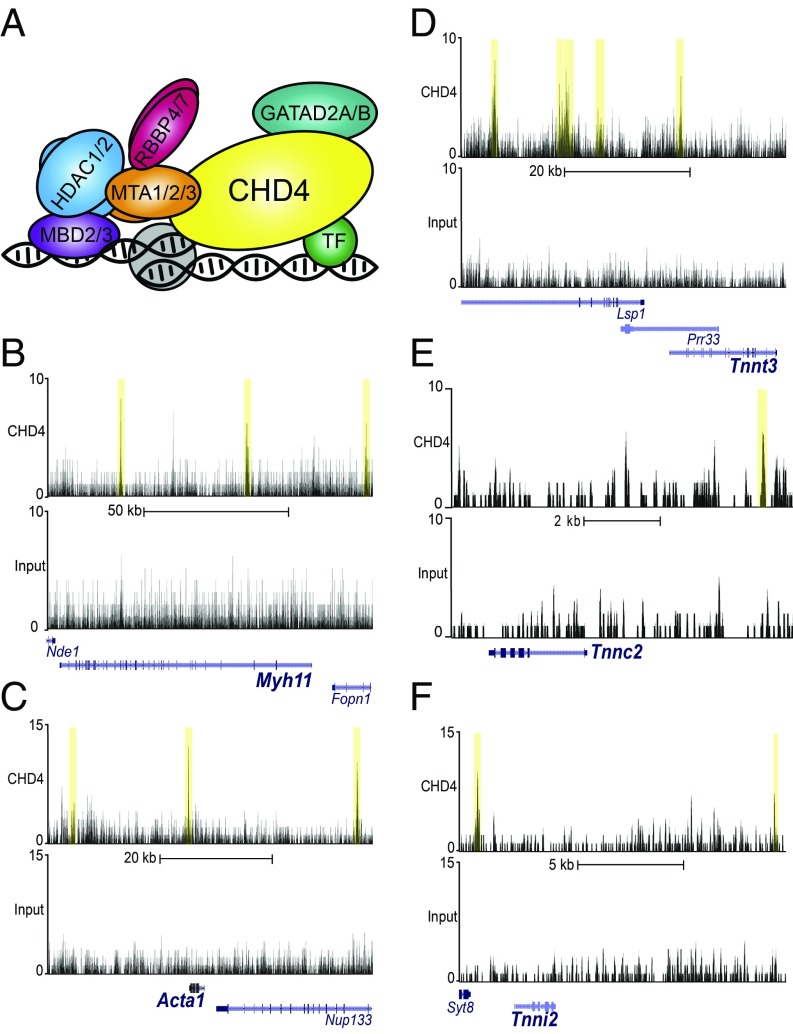
Gene annotation of CHD4-bound regions and visual inspection of browser tracks identified CHD4 as directly bound to regulatory regions for fast skeletal and smooth muscle myofibril paralogs Myh11, Acta1, Tnnt3, Tnnc2, and Tnni2 (Fig. 3). Combined with the previous transcriptomic analyses, these data suggest loss of CHD4 causes derepression of the skeletal and smooth muscle program in cardiomyocytes.
Fig. 3.
CHD4 binds genomic regions linked to Myh11, Acta1, Tnnc2, Tnnt3, and Tnni2. (A) Diagram of CHD4 ChIP-seq approach representing methodology. TF, transcription factor. (B–F) CHD4 binding sites identified by ChIP-seq reads enriched over input DNA at noncardiac myofibril paralog genes are highlighted in yellow.
Misexpression of Noncardiac Myofibril Paralogs Leads to Sarcomere Disarray and Impaired Cardiac Function.
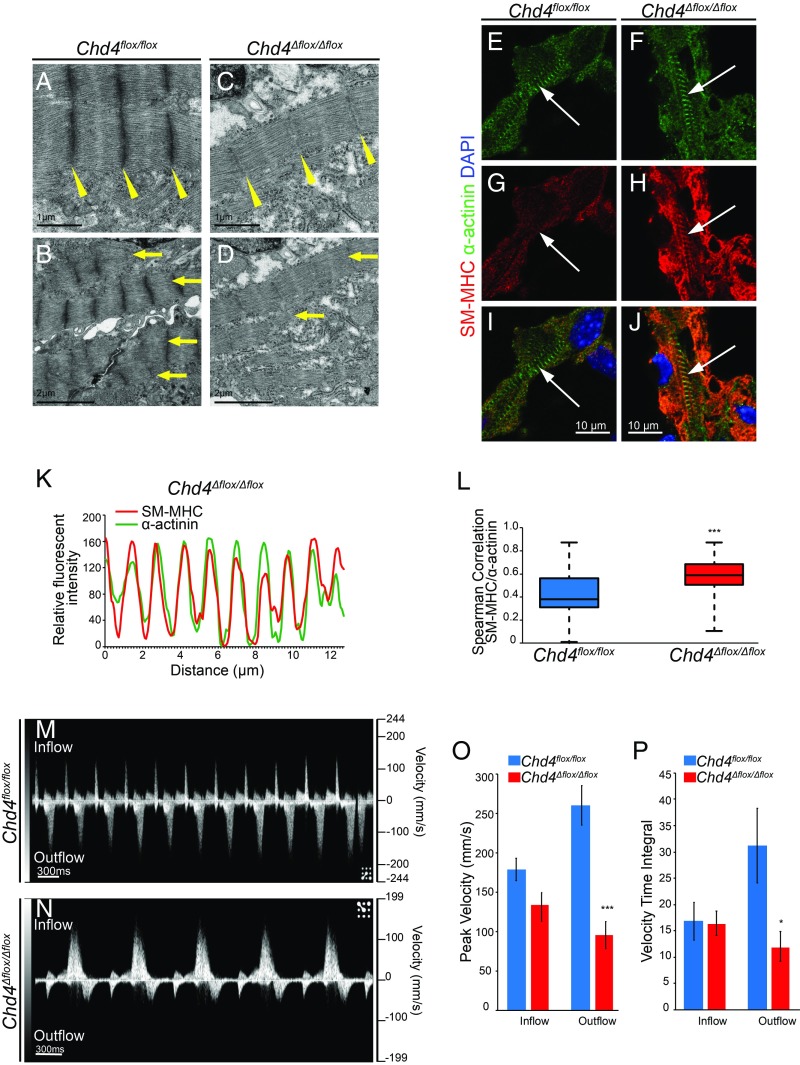
The observation that CHD4 suppresses noncardiac myofibril paralogs led us to hypothesize that misexpression of fast skeletal and smooth muscle myofibril paralogs affects sarcomere organization and ultimately cardiac function in the developing embryo. To test this hypothesis, we analyzed sarcomere formation in the absence of CHD4-mediated transcriptional regulation by transmission electron microscopy. By E10.5 control ventricular cardiomyocytes organize myofibril subunits into discrete sarcomere units in series that are anchored to each other at the Z disc (Fig. 4A, wedge arrows). To provide contractile force, units are arranged in parallel in a higher order structure observed by the alignment of the Z discs across the muscle (Fig. 4B, arrows). In contrast, Chd4Δflox/Δflox hearts show a severe reduction in sarcomere organization and formation of Z discs. This coincides with a decrease in the parallel arrangement of sarcomeres (Fig. 4 C and D). These defects are further associated with a significant decrease in sarcomere organization in Chd4Δflox/Δflox hearts as quantified by the degree of Z-disc and A-band alignment (SI Appendix, Fig. S11).
Fig. 4.
Misexpression of noncardiac myofibril paralogs in the absence of CHD4 leads to sarcomere malformation and altered cardiac function during development. (A–D) Transmission electron microscopy reveals weak, deficient Z-disc formation (yellow wedges) and decreased sarcomere formation and alignment (yellow arrows) in Chd4Δflox/Δflox hearts. (E–J) Costaining for α-actinin (E and F) and smooth muscle myosin heavy chain (SM-MHC) (G and H) demonstrates organization of SM-MHC into striated sarcomere structures and integration into the cardiac sarcomere in Chd4Δflox/Δflox cardiomyocytes compared with Chd4flox/flox controls (I and J). (K) Relative fluorescent signal of SM-MHC and α-actinin plotted against distance in E10.5 Chd4Δflox/Δflox cardiomyocytes reveals intercalation of SM-MHC into the nascent sarcomere. (L) Spearman correlation between SM-MHC/α-actinin signal in Chd4Δflox/Δflox cardiomyocytes is significant compared with Chd4flox/flox controls by Student’s t test, n = 27 vectors per genotype. (M and N) Noninvasive in utero embryonic echocardiography by pulsed-wave (PW) Doppler on E10.5 embryos shows Chd4Δflox/Δflox embryos have pronounced decreases in ventricular (outflow) velocity (N) compared with Chd4flox/flox controls (M). (O and P) Quantification of blood flow velocity from PW Doppler shows significant decrease in ventricular cardiac function in Chd4Δflox/Δflox hearts by peak velocity (O) and velocity time integral (VTI) (P) by Student’s t test, n = 5 embryos per genotype, SEM ±14.35, 18.12, 25.12, and 16.93 (O) and 3.57, 2.30, 7.11, and 2.85 (P). *P value < 0.05, ***P value < 0.001.
To determine whether the skeletal and smooth muscle proteins are incorporated into the developing cardiac muscle, we analyzed colocalization of a cardiac sarcomere Z-disc protein, α-actinin, with misexpressed smooth muscle myosin heavy chain. Fluorescence intensity was measured and quantified over the length of individual sarcomeres. Analysis revealed smooth muscle myosin heavy chain protein organizes into nascent sarcomeres in Chd4Δflox/Δflox cardiomyocytes (Fig. 4 E–L). Taken together, our data imply that by intercalating into the nascent cardiac sarcomere, noncardiac myofibril isoforms displace normal cardiac sarcomere proteins during myofibril assembly.
Coexpression of Cardiac, Smooth Muscle, and Skeletal Muscle Paralogs Compromises Cardiac Contractility and Function.
To determine the physiological consequences of sarcomere disorganization as a result of fast skeletal and smooth muscle myofibril protein misexpression on cardiac function, we developed noninvasive in utero embryonic echocardiography methodologies. We performed ultrasound pulsed-wave (PW) Doppler on E10.5 littermates in utero without surgical manipulation of the dam or embryos. This approach enabled us to measure the effect of sarcomere malformation on cardiac function in the context of the developing heart in situ while avoiding artificial manipulation of cardiac fluid dynamics or maternal stress. PW Doppler recordings of E10.5 control hearts demonstrate consistently strong atrial and ventricular contractions (Fig. 4M and Movie S1). In stark contrast, Chd4Δflox/Δflox hearts with significant cardiac sarcomere disarray show severely reduced ventricular contractions with significant decreases in ventricular outflow peak velocity and velocity time integral (Fig. 4 N–P and Movie S2). Collectively, these data demonstrate CHD4 is required to repress expression of noncardiac myofibril paralogs in the developing heart. In the absence of CHD4, misexpressed fast skeletal and smooth muscle myofibril paralogs intercalate into the cardiac sarcomere, resulting in impaired cardiac function at the point at which cardiac function becomes indispensable for continued embryonic growth.
Discussion
Here we demonstrate CHD4 and the NuRD complex repress both smooth muscle and fast skeletal myofibril paralogs in the developing heart. We find the consequences of activating and incorporating skeletal or smooth muscle myofibrils in cardiomyocytes is a gross disorganization of the sarcomere, a failure of proper contraction and ultimately death of the embryo. There are multiple functional differences between these cell types. For example, cardiac and skeletal muscle have differences in calcium cycling and sensitivity of the troponin complex, as well as cooperativity and activation of the thin filament (49). In contrast to cardiac muscle, smooth muscle cells do not primarily rely on a troponin complex-mediated system of contraction and instead utilize ATP-mediated phosphorylation of the myosin head to induce contraction. Furthermore, unlike cardiac and skeletal muscle, smooth muscle cell myofibrils do not arrange into a striated sarcomere structure and instead organize into an oblique actomyosin cytoarchitecture (50). In striated muscle cells, such as developing cardiomyocytes, myosin heavy chain intercalation is particularly crucial for formation of the thick filament and the associated M-line structure, which is then anchored in alignment between structural Z discs by titin. Substitution of smooth muscle myosin heavy chain into the nascent cardiac sarcomere may therefore be causing the sarcomere formation defects observed in CHD4-null cardiomyocytes, while incorporation of the fast skeletal sarcomere paralogs may further explain the impaired cardiac function in the absence of CHD4-mediated transcriptional regulation.
While some studies have reported misexpression of skeletal or smooth muscle myofibril components in the developing heart at the transcript level, their presence at the protein level had not yet been confirmed (51, 52). Furthermore, the mechanism by which misexpression leads to cardiac developmental defects had not yet been explored. In this report, we have demonstrated that in the absence of CHD4-mediated repression, misexpression of smooth muscle myosin heavy chain, fast skeletal α-actin, and the fast skeletal troponin complex leads to sarcomere disarray and impaired cardiac function. Collectively, this study begins to answer how misexpression of skeletal and smooth muscle myofibril subunits during cardiac development negatively affects sarcomere formation and cardiac function.
We further identified the genomic loci targeted by CHD4 to repress noncardiac myofibril paralogs through an unbiased approach. While some genomic regions have been documented as contributing to repression of these paralogs, these studies have not been performed in a comprehensive fashion using endogenous tissue at the relevant developmental stages of cardiac sarcomere formation (51, 53). Also, the importance of cis-regulatory regions in regulating cardiac development and disease has only recently been emphasized (54). The regulatory regions discovered here thus represent a significant step forward in identifying sites that are crucial for transcriptional regulation and normal heart development.
There is misexpression of many genes in the absence of CHD4-mediated repression in the developing heart. However, at this stage of cardiac development in mouse, continued embryonic growth is dependent on the formation of functional cardiac sarcomere units to initiate systolic function. Our data support a model in which CHD4 loss impedes this process during early heart development. This poses an intriguing question whether in humans, impaired cardiac systolic function associated with certain cardiomyopathies or cardiac failure, in the absence of mutations in cardiac sarcomere subunits, may be due to improper expression and intercalation of noncardiac myofibril paralogs in the cardiac sarcomere. Screening for misexpression of noncardiac myofibril paralogs in cardiac tissue, or for mutations in these putative regulatory elements that may impair CHD4 recruitment or activity, would address this hypothesis and suggest interventions for these patients.
Materials and Methods
A detailed description of materials and methods is provided in SI Appendix, SI Materials and Methods. Briefly, we conditionally ablated Chd4 in the developing murine heart and used transcriptomic, phenotypic, and echocardiography methods to assess differences in cardiac development. We assayed CHD4 genomic localization in the wild-type developmental context via ChIP-seq and bioinformatics analysis. All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee at University of North Carolina Chapel Hill.
Supplementary Material
Acknowledgments
We are grateful to Jeremy Simon for statistical analysis assistance and to the Microscopy Services Laboratory at the University of North Carolina (UNC) for microscopy assistance. The Microscopy Services Laboratory is supported by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. The CH1 monoclonal antibody developed by Dr. Jim Lin was obtained from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242. Sequencing for ChIP-seq experiments was supported by the Epigenomics Core Facility, National Institute of Environmental Health Sciences (NIEHS). This work was funded by Grants R01 HL112618 and R01 HL127640 (to F.L.C.); 5T32 HL069768 and 1F31 HL136100 (to C.M.W.); and the Intramural Research Program of the NIH, NIEHS (ES101965 to P.A.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE109012).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722219115/-/DCSupplemental.
References
- 1.Heron M, et al. Deaths: Final data for 2006. Natl Vital Stat Rep. 2009;57:1–134. [PubMed] [Google Scholar]
- 2.Dolk H, Loane M, Garne E. The prevalence of congenital anomalies in Europe. Adv Exp Med Biol. 2010;686:349–364. doi: 10.1007/978-90-481-9485-8_20. [DOI] [PubMed] [Google Scholar]
- 3.Waldron L, et al. The cardiac TBX5 interactome reveals a chromatin remodeling network essential for cardiac septation. Dev Cell. 2016;36:262–275. doi: 10.1016/j.devcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homsy J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade PA, et al. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 8.Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 9.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y. Biology of the Mi-2/NuRD complex in SLAC (stemness, longevity/ageing, and cancer) Gene Regul Syst Bio. 2011;5:1–26. doi: 10.4137/GRSB.S6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Shaughnessy-Kirwan A, Signolet J, Costello I, Gharbi S, Hendrich B. Constraint of gene expression by the chromatin remodelling protein CHD4 facilitates lineage specification. Development. 2015;142:2586–2597. doi: 10.1242/dev.125450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung H, Kohnken R, Svaren J. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci. 2012;32:1517–1527. doi: 10.1523/JNEUROSCI.2895-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashiwagi M, Morgan BA, Georgopoulos K. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development. 2007;134:1571–1582. doi: 10.1242/dev.001750. [DOI] [PubMed] [Google Scholar]
- 14.Williams CJ, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida T, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo M, et al. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells. 2013;31:1278–1286. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- 17.Marhold J, Kramer K, Kremmer E, Lyko F. The Drosophila MBD2/3 protein mediates interactions between the MI-2 chromatin complex and CpT/A-methylated DNA. Development. 2004;131:6033–6039. doi: 10.1242/dev.01531. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 19.Garnatz AS, et al. FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev Biol. 2014;395:50–61. doi: 10.1016/j.ydbio.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roche AE, et al. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol. 2008;44:352–360. doi: 10.1016/j.yjmcc.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguayo-Gómez A, et al. Identification of copy number variations in isolated tetralogy of Fallot. Pediatr Cardiol. 2015;36:1642–1646. doi: 10.1007/s00246-015-1210-9. [DOI] [PubMed] [Google Scholar]
- 22.Basson CT, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 23.Bruneau BG, et al. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 24.Bruneau BG, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 25.Kirk EP, et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li QY, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 27.Mori AD, Bruneau BG. TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr Opin Cardiol. 2004;19:211–215. doi: 10.1097/00001573-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida A, et al. Genetic mutation analysis in Japanese patients with non-syndromic congenital heart disease. J Hum Genet. 2016;61:157–162. doi: 10.1038/jhg.2015.126. [DOI] [PubMed] [Google Scholar]
- 29.Brown DD, et al. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132:553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaltenbrun E, et al. A Gro/TLE-NuRD corepressor complex facilitates Tbx20-dependent transcriptional repression. J Proteome Res. 2013;12:5395–5409. doi: 10.1021/pr400818c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, et al. Three novel TBX5 mutations in Chinese patients with Holt-Oram syndrome. Am J Med Genet. 2000;92:237–240. doi: 10.1002/(sici)1096-8628(20000605)92:4<237::aid-ajmg2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 32.Svensson EC, et al. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat Genet. 2000;25:353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- 33.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 34.Conway SJ, Kruzynska-Frejtag A, Kneer PL, Machnicki M, Koushik SV. What cardiovascular defect does my prenatal mouse mutant have, and why? Genesis. 2003;35:1–21. doi: 10.1002/gene.10152. [DOI] [PubMed] [Google Scholar]
- 35.Slagle CE, Conlon FL. Emerging field of cardiomics: High-throughput investigations into transcriptional regulation of cardiovascular development and disease. Trends Genet. 2016;32:707–716. doi: 10.1016/j.tig.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi H, et al. PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirschy A, Schatzmann F, Ehler E, Perriard JC. Establishment of cardiac cytoarchitecture in the developing mouse heart. Dev Biol. 2006;289:430–441. doi: 10.1016/j.ydbio.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 38.Nishii K, et al. Targeted disruption of the cardiac troponin T gene causes sarcomere disassembly and defects in heartbeat within the early mouse embryo. Dev Biol. 2008;322:65–73. doi: 10.1016/j.ydbio.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, Reiter RS, Huang QQ, Jin JP, Lin JJ. Comparative studies on the expression patterns of three troponin T genes during mouse development. Anat Rec. 2001;263:72–84. doi: 10.1002/ar.1078. [DOI] [PubMed] [Google Scholar]
- 40.England J, Loughna S. Heavy and light roles: Myosin in the morphogenesis of the heart. Cell Mol Life Sci. 2013;70:1221–1239. doi: 10.1007/s00018-012-1131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saggin L, Gorza L, Ausoni S, Schiaffino S. Troponin I switching in the developing heart. J Biol Chem. 1989;264:16299–16302. [PubMed] [Google Scholar]
- 42.Ilkovski B, Clement S, Sewry C, North KN, Cooper ST. Defining alpha-skeletal and alpha-cardiac actin expression in human heart and skeletal muscle explains the absence of cardiac involvement in ACTA1 nemaline myopathy. Neuromuscul Disord. 2005;15:829–835. doi: 10.1016/j.nmd.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Clément S, et al. Expression and function of alpha-smooth muscle actin during embryonic-stem-cell-derived cardiomyocyte differentiation. J Cell Sci. 2007;120:229–238. doi: 10.1242/jcs.03340. [DOI] [PubMed] [Google Scholar]
- 44.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimbo T, et al. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet. 2013;9:e1004028. doi: 10.1371/journal.pgen.1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada T, et al. Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron. 2014;83:122–134. doi: 10.1016/j.neuron.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miccio A, et al. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010;29:442–456. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen HF, Wade PA, Kutateladze TG. The NuRD architecture. Cell Mol Life Sci. 2013;70:3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z, Yamazaki M, Shen QW, Swartz DR. Differences between cardiac and skeletal troponin interaction with the thin filament probed by troponin exchange in skeletal myofibrils. Biophys J. 2009;97:183–194. doi: 10.1016/j.bpj.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Gómez-Del Arco P, et al. The chromatin remodeling complex Chd4/NuRD controls striated muscle identity and metabolic homeostasis. Cell Metab. 2016;23:881–892. doi: 10.1016/j.cmet.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Heidersbach A, et al. MicroRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. eLife. 2013;2:e01323. doi: 10.7554/eLife.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Postma AV, Bezzina CR, Christoffels VM. Genetics of congenital heart disease: The contribution of the noncoding regulatory genome. J Hum Genet. 2016;61:13–19. doi: 10.1038/jhg.2015.98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.