Significance
A peptidoglycan cell wall provides bacteria with protection from environmental stresses, and interfering with assembly of the cell wall is among the most effective strategies for antibiotic development. To build a cell wall, bacteria first synthesize lipid II on the inner leaflet of their membrane and then flip it across to the outer leaflet, where it is used to make peptidoglycan. Here, we report the structure of the lipid II flippase MurJ from Escherichia coli, and we use high-throughput mutagenesis to identify functionally important regions of the protein. Together with evolutionary covariation analysis, these data show that MurJ must exist in at least two discrete conformational states, providing a framework for understanding lipid II flipping.
Keywords: lipid flippase, peptidoglycan, X-ray crystallography
Abstract
The peptidoglycan cell wall provides an essential protective barrier in almost all bacteria, defining cellular morphology and conferring resistance to osmotic stress and other environmental hazards. The precursor to peptidoglycan, lipid II, is assembled on the inner leaflet of the plasma membrane. However, peptidoglycan polymerization occurs on the outer face of the plasma membrane, and lipid II must be flipped across the membrane by the MurJ protein before its use in peptidoglycan synthesis. Due to its central role in cell wall assembly, MurJ is of fundamental importance in microbial cell biology and is a prime target for novel antibiotic development. However, relatively little is known regarding the mechanisms of MurJ function, and structural data for MurJ are available only from the extremophile Thermosipho africanus. Here, we report the crystal structure of substrate-free MurJ from the gram-negative model organism Escherichia coli, revealing an inward-open conformation. Taking advantage of the genetic tractability of E. coli, we performed high-throughput mutagenesis and next-generation sequencing to assess mutational tolerance at every amino acid in the protein, providing a detailed functional and structural map for the enzyme and identifying sites for inhibitor development. Lastly, through the use of sequence coevolution analysis, we identify functionally important interactions in the outward-open state of the protein, supporting a rocker-switch model for lipid II transport.
The peptidoglycan (PG) cell wall is a defining feature of bacterial cell structure, and disrupting its synthesis is among the most effective strategies for treatment of bacterial infections. PG is synthesized from lipid II, which is first assembled on the inner leaflet of the plasma membrane and then flipped across the bilayer. The lipid II headgroup is then polymerized into PG by either class A penicillin binding proteins (PBPs) or SEDS proteins (1, 2). The size and hydrophilicity of the lipid II headgroup preclude spontaneous flipping across the membrane. MurJ is required for lipid II flippase activity in vivo (3). It is a member of the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) family of transporters, which also includes proteins involved in similar biological processes such as the colanic acid flippase WzxC in bacteria and the proposed N-linked glycan flippase Rft1 found in all eukaryotes (4, 5).
Recently, the crystal structure of MurJ from the thermophilic bacterium Thermosipho africanus was reported (6). This high-resolution structure revealed an inward-open conformation not previously observed in any MOP transporter structure and confirmed that MurJ possesses two additional transmembrane (TM) helices at its carboxyl terminus, which are not found in other MOP transporters. MurJ was crystallized in the presence of lipid II, and its structure showed a large central cavity containing electron density consistent with bound substrate, although it was of insufficient clarity to allow modeling of lipid II. While this crystal structure of T. africanus MurJ has been highly informative, this version of the protein is rather divergent from Escherichia coli MurJ (28% sequence identity), which has been the subject of almost all functional studies to date. To better understand MurJ function, we crystallized E. coli MurJ by the lipidic cubic phase method and determined its structure to 3.5-Å resolution.
In addition, we employed two distinct approaches to investigate MurJ function: high-throughput mutagenesis and evolutionary coupling analysis. In the first of these, we sought to exploit the genetic tractability of E. coli through a high-throughput mutagenesis and sequencing (mut-seq) approach (7). This approach entails construction of a large point mutant library, followed by selection for functional MurJ mutants. Next-generation sequencing then provides a comprehensive assessment of almost all possible single-nucleotide substitution mutants, providing a map of functionally important regions of the protein. In a second approach, we used evolutionary coupling analysis (8) as a means of identifying pairs of residues that show evolutionarily conserved interactions, providing insight into other conformational states of the MurJ protein.
Results
Initially, we were able to obtain crystals for full-length E. coli MurJ by the lipidic cupic phase method, but the crystals were small and diffraction was weak. We reasoned that the limited hydrophilic surface area of the protein for crystal packing may have prevented effective crystallization. To address this problem, we adopted a strategy from G protein-coupled receptor crystallography and fused the protein BRIL to the transporter amino terminus (9). The resulting protein crystallized readily, and crystals showed improved diffraction, enabling collection of a dataset to 3.5-Å resolution. The structure was subsequently solved using molecular replacement (MR)-Rosetta phasing with T. africanus MurJ as the search model, and the structure was refined to Rwork/Rfree of 0.28/0.30, with good geometry (SI Appendix, Fig. S1 A and B and Table S1). As expected, the BRIL promotes tight crystal packing through interaction with itself from neighboring molecules along the a axis (SI Appendix, Fig. S1C). Residues 5 to 14 of TM1 appear to be distorted due to the amino-terminal fusion to BRIL (Fig. 1).
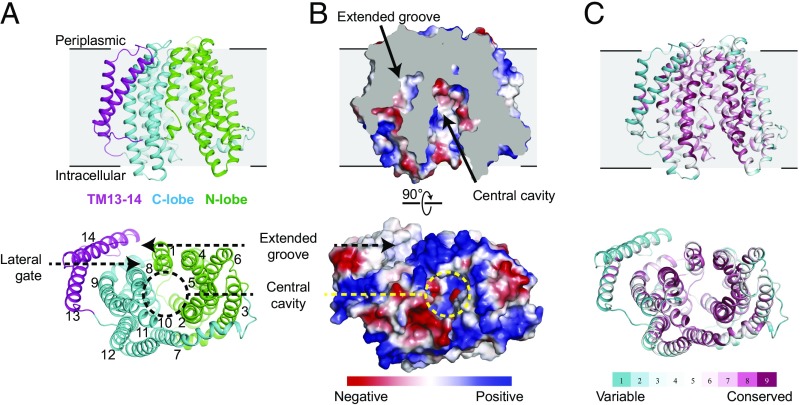
Fig. 1.
Overall structure of MurJ from E. coli. (A) Ribbon representation of MurJ viewed from the side and from the intracellular face, with N-lobe (TM1 to TM6) and C-lobe (TM7 to TM12) colored in green and cyan, respectively, and TM13 and TM14 colored in magenta. (B) Electrostatic potential surface colored from red to blue for negatively to positively charged regions reveals a highly hydrophilic central cavity and hydrophobic extended groove formed by TM13 and TM14. The structure is shown in the same orientation as in A. (C) The structure is colored by sequence conservation using ConSurf server with 483 MurJ sequences from both gram-negative and -positive bacteria, revealing a highly conserved central cavity in contrast to variable peripheral region.
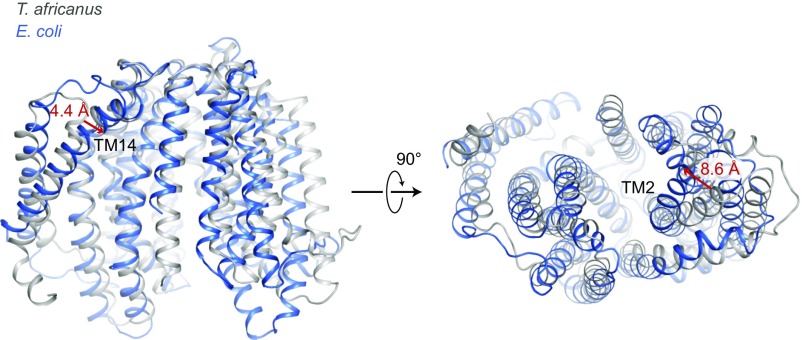
The crystal structure of E. coli MurJ is similar overall to that of T. africanus MurJ, consisting of two homologous six-pass TM bundles, followed by a carboxyl-terminal pair of helices located adjacent to the second bundle (Fig. 1). The two six-helix bundles form two distinct lobes of the protein, which contact each other only on the periplasmic face of the protein, resulting in an overall inward-open conformation similar to the structure of T. africanus MurJ. The MurJ structures from T. africanus and E. coli showed an overall root-mean-square deviation of 2.4 Å over 311 Cα atoms. TM2 in the N-lobe and TM14 in E. coli MurJ were shifted inward by around 8.6 and 4.4 Å, respectively, relative to the C lobe, compared with the equivalent positions in T. africanus MurJ (Fig. 2). This may be a result of the fact that T. africanus MurJ was crystallized in the presence of substrate, while E. coli MurJ is substrate-free, allowing it to adopt a partially occluded conformation. Alternatively, it may reflect intrinsic differences between the MurJ proteins from the two species. A cross-section through the protein shows that the hydrophilic cavity extends almost entirely through the membrane, with a thin hydrophobic barrier where the two lobes of the protein interact on the extracellular face of the membrane (Fig. 2). As in the case of T. africanus MurJ, the E. coli enzyme shows a large, deep groove formed by TM13 and TM14, confirming that this feature is conserved across evolutionarily distant species. In addition, TM13 and TM14 in T. africanus MurJ are shifted outward, perhaps as a result of substrate binding, creating a larger groove compared with E. coli (Fig. 2). This is consistent with its proposed role as the interaction site for the lipid II isoprenoid tail, which must remain in the hydrophobic lipid bilayer throughout the flipping process.
Fig. 2.
Structural overlay of MurJ from T. africanus and E. coli. The two structures were aligned at the C-lobe (left half of the protein as shown). T. africanus MurJ is colored in gray and E. coli in blue. E. coli MurJ reveals a relatively narrow central cavity and extended pocket compared with that in T. africanus. As a reference, the relative shift in orientation for TM14 and TM2 are marked with red arrows.
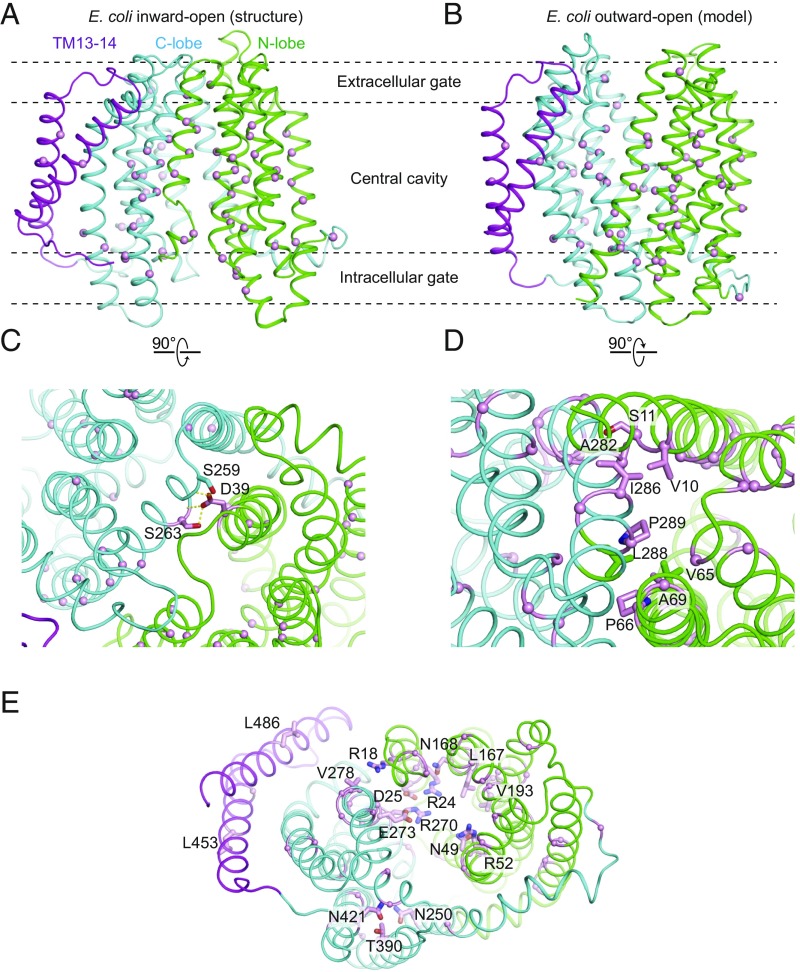
Many functionally important residues have been identified previously in E. coli MurJ, and our structure now provides a framework for their interpretation (10, 11). Of particular note is Asp39, which sits at the extracellular face of the protein and interacts with the positive dipole of TM8 and with Ser263 (Fig. 3). This interaction bridges the two lobes of the protein and may help to stabilize the inward-open conformation. Asp39 is absolutely conserved among MurJ homologs in gram-negative bacteria and is intolerant of substitution (10). Similarly, Ala29 sits at the interface between the two lobes of the protein, where it is almost entirely occluded. Mutation of this residue to Cys is known to be tolerated, but when labeled with a thiol-reactive agent, the transporter exhibits complete loss of function (3). The steric enclosure of Ala29 would prevent labeling of this site in an inward-open state but would be well accommodated in an outward-open conformation. Labeling of the A29C mutant would therefore trap the transporter in single conformation, accounting for the loss of lipid II flipping observed experimentally (3).
Fig. 3.
Mapping of mut-seq data on the MurJ structure. Residues at which mutations show more than a fivefold-frequency decrease between cells grown in arabinose (MurJwt induction) and glucose (MurJwt depletion) are mapped in inward-open (A) and outward-open MurJ (B) structure, shown as pink spheres in each case. (C) Two essential residues (shown as pink sticks) mediate interactions between N-lobe and C-lobe in the extracellular side of inward-open MurJ. Dashed yellow line represents hydrogen bond. (D) Several essential residues compose the intracellular gate of outward-open MurJ. A close-up view of hydrophobic interactions among these residues is shown. (E) Solvent-exposed essential residues located in the central cavity and extended groove (pink sticks).
To explore the functional consequences of MurJ mutagenesis in more detail, we took advantage of the genetic tractability and high transformability of E. coli to perform high-throughput mutagenesis followed by next-generation sequencing (mut-seq; ref. 7). This approach entails creating a large library of murJ mutants and then selecting for their ability to rescue loss of wild-type MurJ in cells. In brief, we constructed an E. coli strain in which wild-type murJ expression is driven by an arabinose promoter and a mutagenized plasmid-borne murJ by the lac promoter. In medium containing arabinose, wild-type MurJ is produced and the cells grow normally, irrespective of whether the plasmid-borne murJ is functional. However, in glucose medium, wild-type murJ expression is repressed, and growth will rely on the expression of mutagenized murJ from the plasmid. Thus, only plasmids that encode functional murJ missense mutants will grow in this condition. By comparing the frequency of any given mutation between cell populations grown in the different media, a fitness measure of murJ mutations can be determined. Those changes that do not affect MurJ function are expected to be present at similar frequencies in the two populations, while those resulting in a loss of function are expected to be depleted from cells grown in glucose-containing medium.
Using this approach, we scored complementation of more than 1,500 individual point mutants in murJ, including substitutions at all 521 codons in the sequence (Dataset S1). Deleterious changes were identified throughout the protein coding sequence, including at known functional sites like Asp39 discussed above, as well as many others. A total of 101 mutations resulted in at least a fivefold depletion in cells grown in glucose medium relative to cells grown in arabinose medium, suggesting a severely deleterious effect consistent with a complete loss of function. Indeed, 40 of these mutations result in the introduction of a premature stop codon. The other 61 mutations can be divided into four major groups (Fig. 3 A and B and SI Appendix, Table S2): (i) mutations of buried hydrophobic residues to charged or bulky residues, likely resulting in a loss of folding; (ii) mutations in residues composing the extracellular gate (e.g., aforementioned D39, S263), located in the contact between two lobes in the extracellular side (Fig. 3C); and (iii) mutations in solvent-exposed residues on the intracellular side, which likely comprise the intracellular gate and may stabilize the outward-open conformation. Mutations belonging to this third group are consistent with a MurJ homology model in the outward-open conformation in which V65, P66, and A69 engage in extensive hydrophobic interactions with L288, P289, and S292 in the pseudosymmetry-related TM8 of the C lobe (Fig. 3D). Mutations that insert large hydrophilic amino acids in these positions would disrupt the interaction between the two lobes, accounting for loss of function in these mutants. (iv) The fourth group of mutations includes those in solvent-exposed residues located in the central cavity and extended groove, which are likely involved directly in substrate binding or play a role in the regulation of transport (Fig. 3E). These also include previously identified charged residues located in the central cavity, which were shown to be essential for PG biogenesis (10, 11).
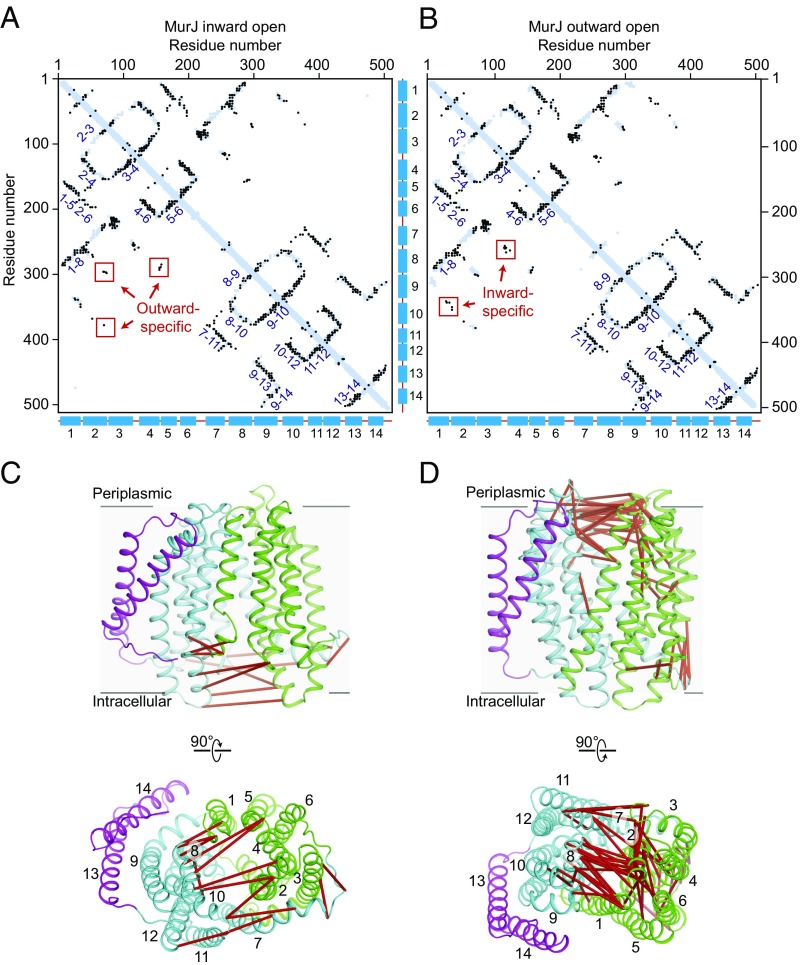
The function of MurJ in transport fundamentally depends on conformational change, but crystallography by nature presents only a static view of the protein. To gain insight into the possible existence of other conformational states, we turned to evolutionary coupling analysis. This technique can identify residues that have coevolved with one another due to spatial proximity in the folded protein structure (8) and can reveal biologically important contacts, irrespective of the structural conformation to which they belong. Using an alignment of 15,972 sequences, we identified a total of 470 evolutionary couplings scoring in the 99th percentile. The majority of coevolving pairs correspond to residues that are located <5 Å from one another in our structure (Fig. 4 A and B). However, a subset of strongly coevolved residues are more distant from one another and do not make any direct contact. These covarying pairs are located almost exclusively at the cytosolic face of the transporter, where they consist of pairs of residues on each of the two lobes of the protein (Fig. 4 C and D). To test whether the coevolution of these pairs can be accounted for by a conformational change to an outward-open state of MurJ, we constructed a outward-open homology model using the template of MATE from Pyrococcus furiosus (PDB ID code 3VVN) and TM13 and TM14 from MurJ. When evolutionary couplings are superimposed on this model, cytosolic couplings are well accounted for, while coevolving residues on the periplasmic face are now distant (Fig. 4 B and D). Together, these data show that both inward-open and outward-open states are under evolutionary selection pressure, providing strong support for a rocker-switch alternating-access model of lipid transport.
Fig. 4.
Evolutionary covariation analysis of MurJ. Residue pairs located less than 5 Å from one another in MurJ inward-open structure (A) and outward-open model (B) are shown as blue dots, while coevolved pairs are shown as overlaid black dots. Strong covariations are observed between helices (labeled by helix number) that are adjacent to one another in the structure. A subset of strongly coevolved residue pairs are distant (>5 Å) in one conformation but close (<5 Å) in the other, indicating that both inward-open and outward-open states have been subjected to evolutionary selection. These violation pairs are mapped on the inward-open structure (C) and outward-open model (D) and red line connected.
Discussion
Flipping of lipid II across the plasma membrane is an essential step in PG biosynthesis and represents a broadly conserved potential therapeutic target. Here, we have reported the crystal structure of E. coli MurJ, the most extensively studied of the broadly conserved MurJ transporters. While it is similar overall to a previously reported structure of T. africanus MurJ, the E. coli enzyme shows a distinct partially closed conformation, which may be representative of the apo-state of the protein. Moreover, the structure confirms that usual features seen in the T. africanus MurJ are conserved across species, attesting to their functional importance. Most notably, this includes a lateral gate between TM1 and TM8, as well as a long hydrophobic groove along TM13 and TM14, which has been proposed as the binding site for the 55-carbon isoprenoid tail of lipid II. Consistent with a functionally important role for this region, high-throughput mutagenesis experiments showed that introduction of premature stop codons between TM12 and TM13 is not tolerated (Dataset S1). Similarly, several mutations in TM13 and TM14 (L453, L459, or L486) are depleted, further supporting a functionally important role for this region of the protein. In addition to TM13/14, high-throughput mutagenesis experiments highlighted other functionally critical regions of the protein, including the inner (cytosolic) gate, the outer (periplasmic) gate, and the central cavity. Among poorly tolerated mutations, substitutions that altered the overall charge of the central cavity were particularly common, indicating the importance of electrostatic interactions for substrate recognition, transport, or both. All these essential residues are extremely conserved among gram-negative bacteria but are divergent in gram-positive bacteria, although MurJ from the two classes can functionally substitute for one another (12). This suggests MurJ from gram-negative and gram-positive bacteria utilize distinct substrate recognition features to catalyze lipid II flipping. Despite their divergence in sequence, some major features are shared between gram-positive and gram-negative MurJ proteins, including an abundance of charged residues in the central cavity and conserved residues at the interface between the amino-terminal and carboxyl-terminal domains of the protein (10).
The identification of MurJ as the lipid II flippase is remarkably recent and has been the subject of some controversy (13). In particular, other work had shown previously that the SEDS-family protein FtsW can flip lipid II in vitro, a result that is at odds with the identification of MurJ as the relevant flippase in vivo (14). However, in vitro flippase assays are notoriously difficult to conduct and interpret, in large part because the presence of improperly folded proteins can result in membrane defects that nonspecifically scramble lipids (15). Recent data from a variety of studies has now solidified the identification of MurJ as the lipid II flippase, including mass spectrometry showing that lipid II binds strongly to MurJ, but not the SEDS protein FtsW (16). Moreover, the SEDS proteins have now been unambiguously shown to catalyze PG polymerization (1, 2), and it is difficult to imagine that the complex and biochemically disparate tasks of lipid II flipping and polymerization are mediated by a single enzyme. While SEDS proteins show sequence (1) and structural (17) similarity to glycosyltransferases, the structures of MurJ from T. africanus and E. coli show clear similarity to transporters. These proteins differ from other MOP-family transporters in exactly the ways expected for a lipid II flippase: they show a larger central cavity suitable to accommodate the bulky lipid II headgroup and they possess a lateral portal to allow the isoprenoid tail to remain in the lipid bilayer throughout flipping. In addition, we have shown that residues on opposite lobes of the cytosolic face of the protein show strong coevolution, indicative of a transport cycle alternating between inward-open and outward-open states. Lastly, the fact that homologous lipid flippases can substitute for MurJ in certain cases (18, 19) further supports its role as the lipid II flippase in cell wall assembly.
With the structure of E. coli MurJ, interpretation of the large body of mutagenesis and other functional data are now straightforward. Together with the previously reported structure of T. africanus MurJ, the structure and saturating mutagenesis of E. coli MurJ provides a framework for the design of future experiments to investigate unresolved aspects of MurJ function. These include identification of the energetic factor(s) driving lipid II flipping and understanding how MurJ and other proteins in the PG synthesis machinery work in concert to assemble the cell wall. Importantly, the combination of structural study, mut-seq, and evolutionary coupling analysis is likely to be generally applicable to other essential proteins involved in bacterial cell wall assembly or other processes. Using a similar approach to that described here may help shed light on the mechanisms of action for other essential cell wall proteins, including those that compose the divisome and elongasome cell wall synthesis complexes. In the long-term, a detailed mechanistic understanding of cell wall assembly will not only be of value for understanding bacterial cell biology but also facilitate discovery of novel antibacterial agents.
Methods
Crystallography.
A fusion protein was constructed consisting of the crystallographic chaperone protein apocytochrome b562RIL (BRIL) fused to the amino terminus of E. coli MurJ (residues 5 to 511), followed by a 3C protease site and a carboxyl-terminal protein C epitope tag (“EDQVDPRLIDGK”). This was cloned into a pET28a expression vector using NcoI and NotI restriction enzymes (New England Biolabs), and the expression vector was transformed into E. coli BL21(DE3) strain. Liquid cultures were inoculated with transformed colonies and grown in LB medium at 37 °C with shaking. The cultures were shifted to 18 °C when the OD600 reached 0.8, and protein expression was induced at this point by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After continued growth for 16 h, cells were harvested by centrifugation and resuspended in lysis buffer containing 25 mM Hepes pH 7.6 and 150 mM NaCl. After lysis by sonication, cell membranes were pelleted by ultracentrifugation at 142,000 x g for 1 h. The pellets were homogenized using a glass dounce tissue grinder in a solubilization buffer containing 20 mM Hepes pH 7.5, 350 mM NaCl, 10% (vol/vol) glycerol, 2 mM CaCl2, 0.5% (wt/vol) lauryl maltose neopentyl glycol [(LMNG) Anatrace], and 0.05% (wt/vol) cholesterol hemisuccinate [(CHS) Steraloids]. The sample was stirred for 1 h at 4 °C and clarified by centrifugation as before for 30 min. The supernatant filtered on a glass microfiber filter was loaded by gravity flow onto anti-protein C antibody affinity resin. The resin was washed extensively with buffer consisting of 20 mM Hepes pH 7.6, 250 mM NaCl, 2 mM CaCl2, 0.01% LMNG, and 0.001% CHS. The protein was eluted in the same buffer without CaCl2, supplemented with 5 mM EDTA and 0.1 mg/mL protein C peptide. The protein C tag was cleaved by 3C protease overnight. The protein was further purified by size exclusion chromatography on Superdex 200 Increase in buffer 20 mM Hepes pH 7.6, 250 mM NaCl, 0.01% LMNG, and 0.001% CHS.
Before crystallization, the protein was concentrated to 45 mg/mL. Protein was reconstituted into lipidic cubic phase by the twin-syringe method (20), using a protein-to-lipid ratio of 1:1.5 (wt/wt). The host lipid was a 10:1 (wt/wt) mixture of monoolein (Hampton Research) and cholesterol (Sigma). After reconstitution, protein samples were dispensed in 35-nL drops on glass sandwich plates using a Gryphon LCP robot (Art Robbins Instruments). Crystals were obtained with a precipitant solution of 100 mM MES pH 6.0, 100 mM potassium phosphate dibasic, 28% PEG 300, and 1% 1,2,3-heptanetriol. Data collection was performed at the General Medical Sciences and Cancer Institutes Structural Biology Facility at Advanced Photon Source (GM/CA @ APS) beamline 23ID-B using a 10-μm beam diameter, a 0.2-s exposure, fivefold attenuation, and a 0.2° oscillation angle. Data from five crystals were integrated, scaled, and merged in HKL2000 (21).
The crystals belonged to space group C2221 and contained one copy of BRIL-MurJ per asymmetric unit. Initial phasing was attempted by MR using Phaser (22), with the recently published MurJ structure from T. africanus (PDB ID code 5T77) as a search model. This was unsuccessful, likely due to low sequence identity and large conformational differences between the template and model. Instead, we were able to solve the phase problem by MR-Rosetta in Phenix (23), with T. africanus MurJ as search model. This approach integrates MR, Rosetta modeling, Autobuild, and density modification and refinement (24). BRIL (PDB ID code 4N6H) was subsequently placed using Phaser. The model was built in COOT and refined with Phenix. The final model incudes BRIL residues 3 to 107, followed by MurJ with residues 5 to 507. Ramachandran analysis showed that 95.05% of residues are in favorable regions and 4.95% are in allowed regions, with no outliers. Data collection and refinement statistics are summarized in SI Appendix, Table S1, and the structure is deposited in the Protein Data Bank (PDB ID code 6CC4).
Mutagenesis.
Mut-seq was performed essentially as described before (1, 7). A plasmid library harboring mutagenized murJ (library size ∼1 million) was transformed into E. coli strain CS7 (Para::murJ) by electroporation and plated on LB with chloramphenicol and 0.2% (wt/vol) arabinose. Cells were scraped and the suspension was diluted to OD600 ∼1. After serial dilution to 10−2, 100 µL of the diluted suspension containing an estimated 106 cells per milliliter was plated on LB-chloramphenicol plates supplemented with 100 µM IPTG, 0.2 (wt/vol) glucose, or 0.2% (wt/vol) arabinose. The plates were incubated overnight at 37 °C, and surviving colonies were harvested and diluted as described above. Plasmids were prepared from cell suspension before plating (input control), and after plating on LB with glucose, arabinose, or IPTG. Under this selection scheme, we did not detect any isolates without murJ insert on plates with glucose or IPTG as judged by colony PCR analysis of 16 isolates using GoTaq polymerase (Promega).
To prepare the sequencing libraries, purified plasmids from cell suspensions were digested with EcoRV and ClaI to release the cloned murJ inserts. The 2-kb fragments released were gel purified (Zymogen), and the amount of DNA recovered was quantitated using Qubit reactions (Invitrogen) according to the manufacturer’s protocol. After diluting the purified DNA samples to 2 ng/µL, 1 µL of the diluted DNA was mixed with 1.25 µL of Tagment DNA Buffer (15027866; Illumina) and 0.25 µL of Tagment DNA Enzyme (15027865; Illumina). The mixture was incubated for 10 min at 55 °C in a thermocycler. Next, 11.2 µL of 2X KAPA Master Mix (KK2612; KAPA Biosystems) and 4.4 µL of primers N705 and S503 (for the control input library), N706 and S503 (for the arabinose library), N701 and S504 (for the glucose library), and N702 and S504 (for the IPTG library) were added. PCR was performed at 72 °C for 3 min, 98 °C for 5 min, 98 °C for 12 cycles of 10 s, 62 °C for 30 s, and 72 °C for 30 s, and a final extension step at 72 °C for 5 min. Quality of the PCR products was judged by running 3.75 µL of PCR product on a 1.5% agarose gel. The resulting products were purified and size selected by AMPure beads (A63881; Agencourt). First, 18.75 µL of the PCR product was mixed with 15 µL of AMPure beads and incubated at room temperature for 5 min. The beads were captured by a magnetic stand, and the supernatant was discarded. The beads were then washed twice with 200 µL of 80% ethanol, air dried, and eluted by incubation with 40 µL of water for 5 min at room temperature. The eluted DNA solution was transferred to a new tube, and the size distribution of the libraries and the amount of DNA was measured by Tapestation (Agilent) and Qubit, respectively. The prepared libraries were sequenced using MiSeq v3 Reagent Kit (150-cycle; MS-102-3001; Illumina) exactly as described in the manufacturer’s protocol. In this case, 7 pM of the DNA was used and typically resulted in a clustering density around 900/mm2.
Data analysis was performed using CLC Workbench (Qiagen). Failed reads were discarded and the sequences were trimmed with a cutoff quality value of 0.005 and a minimal length of 35 bp. The reads obtained were aligned using the global alignment setting to the reference sequence of pCS126, with the length and similarity score set to 1. The unmapped reads from this alignment were collected and further aligned to pCS126, with a similarity score set to 0.98, mismatch cost set to 2, and gap cost set to 3. Lastly, the mutations from the reads were identified by the variant finder tool, using settings of polyploidy equal to 1, minimal frequencies equal to 0.0001, and count equal to 2. The mutations found were then exported to Microsoft Excel for further analysis.
Identifying Evolutionary Couplings in MurJ.
The full-length sequence of MurJ (511 amino acids, MURJ_ECOLI) was used to generate multiple sequence alignments of different depths using five jackhmmer (25) iterations against the April 2017 Uniref100 database (26). To optimize both coverage of the query sequence and the number of sequences included in the alignment, all subsequent calculations were done on the alignment computed at bitscore 0.6 with 97.3% of the sequence including less than 30% gaps in each column and 15,972 total sequences (4,247 effective sequences after downweighting sequences with >80% identity to the query). Evolutionary couplings (ECs) were then computed as described previously (8, 27), and a mixture model was applied to the set of all possible ECs to identify contacts that had a 99% probability of being in the tail of the score distribution (28). This threshold corresponds to 470 amino acid pairings, placing them in the 99.6th percentile of the full space of potential couplings. These high-scoring ECs were then compared with the presented models, with a threshold of 5 Å for identifying true contacts.
MurJ Outward-Open Model Generation.
TM1 to TM12 of the outward-open model was generated using the I-TASSER server as previously described (11). TM13 and TM14 were modeled based on their relative position to the other TM domains, on the assumption that TM7 to TM14 rotate largely as a rigid body during conformational transition.
Supplementary Material
Acknowledgments
We thank the GM/CA @ APS beamline staff for excellent technical assistance with data collection and all members of the T.G.B. and A.C.K. laboratories for helpful discussions and advice. This work was supported by the Center of Excellence for Translational Research (CETR) Grant U19AI109764 (to J.J.M., T.G.B., and A.C.K.) and by the NIH Grant R01GM106303 (to D.S.M.). F.A.R. is supported by the CETR Grant U19AI109764 and NIH Grants R01GM066174 and R01GM076710.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure factors and refined coordinates for Escherichia coli MurJ have been deposited in the Protein Data Bank (PDB ID code 6CC4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802192115/-/DCSupplemental.
References
- 1.Meeske AJ, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emami K, et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol. 2017;2:16253. doi: 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sham LT, et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helenius J, et al. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature. 2002;415:447–450. doi: 10.1038/415447a. [DOI] [PubMed] [Google Scholar]
- 6.Kuk AC, Mashalidis EH, Lee SY. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat Struct Mol Biol. 2017;24:171–176. doi: 10.1038/nsmb.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robins WP, Faruque SM, Mekalanos JJ. Coupling mutagenesis and parallel deep sequencing to probe essential residues in a genome or gene. Proc Natl Acad Sci USA. 2013;110:E848–E857. doi: 10.1073/pnas.1222538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marks DS, et al. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler EK, Tan WB, Joseph H, Ruiz N. Charge requirements of lipid II flippase activity in Escherichia coli. J Bacteriol. 2014;196:4111–4119. doi: 10.1128/JB.02172-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler EK, Davis RM, Bari V, Nicholson PA, Ruiz N. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J Bacteriol. 2013;195:4639–4649. doi: 10.1128/JB.00731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meeske AJ, et al. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci USA. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leclercq S, et al. Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep. 2017;7:43306. doi: 10.1038/srep43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohammadi T, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helenius J, et al. Helenius et al. reply. Nature. 2008;454:E4–E5. [Google Scholar]
- 16.Bolla JR, et al. Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ. Nat Chem. 2018;10:363–371. doi: 10.1038/nchem.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjodt M, et al. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature. 2018;556:118–121. doi: 10.1038/nature25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elhenawy W, et al. The O-antigen flippase Wzk can substitute for MurJ in peptidoglycan synthesis in Helicobacter pylori and Escherichia coli. PLoS One. 2016;11:e0161587. doi: 10.1371/journal.pone.0161587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sham L-T, Zheng S, Kruse AC, Bernhardt T. Evidence for the coupling of substrate recognition with transporter opening in MOP-family flippases. Mol Microbiol, in press. [DOI] [PMC free article] [PubMed]
- 20.Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr F Struct Biol Commun. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiMaio F, et al. Improved molecular replacement by density- and energy-guided protein structure optimization. Nature. 2011;473:540–543. doi: 10.1038/nature09964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH. UniProt Consortium UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopf TA, et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toth-Petroczy A, et al. Structured states of disordered proteins from genomic sequences. Cell. 2016;167:158–170.e12. doi: 10.1016/j.cell.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.