Significance
Mechanisms underlying androgen receptor (AR) splice variant 7 (AR-V7) oncogenic function at the genomic level remain poorly defined. Studies here found that AR-V7 cistromes are cell-context–dependent in castration-resistant prostate cancer (CRPC) cells and tissues, resulting in tremendous diversity in AR-V7–regulated transcriptomes across CRPC patients. Thus, few downstream targets of AR-V7 can universally account for CRPC progression, leaving us without adequate, common, viable therapeutic targets for this heterogeneous disease in which AR-V7 itself is not druggable by antiandrogens. Remarkably, we discovered that HoxB13 governs the diverse AR-V7 cistromes among CRPC, thus shifting focus from the previously characterized role of HoxB13 in androgen-dependent prostate cancer to a distinct role in CRPC. These findings will significantly impact therapeutic strategies for AR-V7–driven CRPC, for which there is no approved therapy.
Keywords: AR-V7, castration-resistant prostate cancer, HoxB13, motif-resolution cistromes
Abstract
The constitutively active androgen receptor (AR) splice variant 7 (AR-V7) plays an important role in the progression of castration-resistant prostate cancer (CRPC). Although biomarker studies established the role of AR-V7 in resistance to AR-targeting therapies, how AR-V7 mediates genomic functions in CRPC remains largely unknown. Using a ChIP-exo approach, we show AR-V7 binds to distinct genomic regions and recognizes a full-length androgen-responsive element in CRPC cells and patient tissues. Remarkably, we find dramatic differences in AR-V7 cistromes across diverse CRPC cells and patient tissues, regulating different target gene sets involved in CRPC progression. Surprisingly, we discover that HoxB13 is universally required for and colocalizes with AR-V7 binding to open chromatin across CRPC genomes. HoxB13 pioneers AR-V7 binding through direct physical interaction, and collaborates with AR-V7 to up-regulate target oncogenes. Transcriptional coregulation by HoxB13 and AR-V7 was further supported by their coexpression in tumors and circulating tumor cells from CRPC patients. Importantly, HoxB13 silencing significantly decreases CRPC growth through inhibition of AR-V7 oncogenic function. These results identify HoxB13 as a pivotal upstream regulator of AR-V7–driven transcriptomes that are often cell context-dependent in CRPC, suggesting that HoxB13 may serve as a therapeutic target for AR-V7–driven prostate tumors.
In the lethal castration-resistant prostate cancer (CRPC), androgen receptor (AR) is still expressed and functional in most cases (1). Thus, targeting AR is a major strategy to treat CRPC. Indeed, novel hormonal therapy for CRPC employs inhibitors of intratumor and adrenal androgen synthesis (e.g., abiraterone acetate) or more potent AR antagonists (e.g., enzalutamide). However, these agents only provide a temporary response and modest increase in survival, indicating a rapid evolution of resistance and reactivation of AR function (2). Therefore, understanding the mechanisms for AR reactivation and developing novel therapeutic strategies for CRPC remain urgent needs.
AR is a ligand-dependent transcription factor (TF) containing an N-terminal domain, a DNA-binding domain, and a C-terminal ligand-binding domain (LBD) (3). The LBD is the major drug-target site for AR inhibition in CRPC. Unfortunately, aberrant AR mRNA splicing is able to generate AR splice variants (AR-Vs) with short, variant-specific peptides to replace the AR LBD, which has been postulated as an important mechanism contributing to sustained AR signaling and resistance to AR-targeting therapy in CRPC (4, 5). Among more than 20 AR splice variants, AR-V7, which retains the first three exons of the AR gene followed by a variant-specific cryptic exon CE3, is the most frequently expressed variant with the greatest clinical and functional relevance (4, 6–9). Indeed, clinical studies have revealed that AR-V7 expression is associated with poor prognosis of CRPC patients and resistance to AR LBD inhibitors (8–10), and preclinical studies have demonstrated that AR-V7 promotes CRPC cell growth in vitro and in vivo (6, 11, 12). However, it remains poorly understood whether AR-V7, a TF lacking the LBD, is able to bind to CRPC genomes and exert its oncogenic function. Addressing this question is of clinical importance as it may lead to the identification of novel therapeutic targets for AR-V7–driven CRPC in which AR-V7 itself is not druggable by AR LBD inhibitors.
In this study, we have defined motif-resolution AR-V7 cistromes in human CRPC cell models and CRPC patient tissues by using a ChIP-exonuclease sequencing (ChIP-exo) approach (13, 14) with a specific antibody targeting endogenous AR-V7. We found that AR-V7 cistromes are highly heterogeneous among CRPC. Importantly, the cell-context–dependent AR-V7 cistromes and their controlled diverse oncogene sets involved in CRPC are governed by a key common upstream regulator, HoxB13. Thus, HoxB13 may serve as a general therapeutic target for the heterogeneous, AR-V7–driven CRPC.
Results
AR-V7–Regulated Transcriptomes and AR-V7 Cistromes Are Heterogeneous Among CRPC Cells.
To define AR-V7 genomic function, we first examined AR-V7 expression in two CRPC cell lines, CWR22RV1 (22RV1) and LNCaP95 (LN95) (SI Appendix, Fig. S1A), which model different CRPC patients with aberrant AR splicing (11, 15, 16). Interestingly, while AR-V7 and full-length AR (AR-FL) were coexpressed in CRPC cells, AR-V7 did not form a protein complex with AR-FL within the cell nucleus in the absence of hormone (SI Appendix, Fig. S1 B–E), suggesting that AR-V7 and AR-FL may have distinct roles in CRPC. Indeed, silencing of AR-V7 but not AR-FL dramatically decreased castration-resistant cell growth of 22RV1 (SI Appendix, Fig. S2A) and LN95 (SI Appendix, Fig. S2B). To identify AR-V7–regulated genes contributing to CRPC progression, we performed an RNA-seq assay in hormone-depleted 22RV1 and LN95 cells transfected with an siRNA targeting AR-V7 and a control siRNA (Fig. 1A and SI Appendix, Fig. S3 A and B). We found that AR-V7 up-regulated but not down-regulated genes were involved in several cancer-related processes and in prostate cancer recurrence (SI Appendix, Fig. S3C and Dataset S1). Remarkably, while AR-V7 controls similar biologic and disease pathways in 22RV1 and LN95 cells (SI Appendix, Fig. S3C), it regulates distinct sets of genes in these two CRPC cell models reflecting the heterogeneity of AR-V7–driven CRPC (Fig. 1B and Dataset S1).
Fig. 1.
AR-V7 cistromes and their regulated transcriptomes are heterogeneous in AR-V7–driven CRPC. (A) A heatmap of differentially expressed genes [false-discovery rate (FDR) < 0.05, fold-change > 2] after AR-V7 silencing. The gene expression [reads per kilobase per million mapped reads (RPKM)] values for each gene were normalized to the standard normal distribution to generate z-scores. The scale bar is shown with the minimum expression value for each gene in blue and the maximum value in red. (B) Venn diagrams show AR-V7 up-regulated and down-regulated genes in 22RV1 and LN95 cells, respectively. (C) Overlap of AR-V7 binding sites between 22RV1 and LN95 cells. (D and E) ChIP-exo raw tags distribution (1-bp resolution) over AREs on the forward (blue, Left) and reverse (red, Right) strands, respectively in 22RV1 cells (D) and LN95 cells (E). The Center panels represent the bound ARE sequences ordered as in the Left and Right.
To investigate the underlying regulatory mechanisms for AR-V7–regulated gene expression in CRPC cells, we defined high-resolution AR-V7 cistromes in hormone-depleted 22RV1 and LN95 cells using our ChIP-exo approach (13, 14). AR-V7 ChIP-exo was performed using an AR-V7–specific antibody (SI Appendix, Figs. S1 B–E and S2). As a control, AR-FL ChIP-exo was conducted in the same population of CRPC cells using an antibody recognizing the AR LBD (SI Appendix, Fig. S1 B and C). Consistent with the distinct AR-V7 transcriptomes, AR-V7 cistromes were also heterogeneous between 22RV1 and LN95 cells (Fig. 1C and SI Appendix, Fig. S3D). In addition to AR-V7 binding locations that overlapped with AR-FL binding regions, AR-V7 preferred binding sites (63.2% for 22RV1 and 41.6% for LN95) were identified (SI Appendix, Fig. S3E). Close inspection of several examples of AR-FL and AR-V7 binding in CRPC cells further confirmed this binding preference (SI Appendix, Fig. S3 F and G and Dataset S2). Our ChIP-exo motif analysis (13, 14) showed that AR-V7 bound to the FL androgen responsive element (ARE) with clear protected signals (Fig. 1 D and E). Interestingly, ChIP-exo–based screening of collaborating TF motifs within the AR-V7 binding locations discovered a Homeobox motif that significantly co-occurred with AR-V7–bound AREs (SI Appendix, Fig. S4A). These data suggest that the FL ARE mediates diverse AR-V7 genomic binding and that TFs recognizing Homeobox motifs may play important collaborative roles in AR-V7 genomic function.
HoxB13 Universally Interacts with AR-V7 on Open Chromatin in the CRPC Genomes.
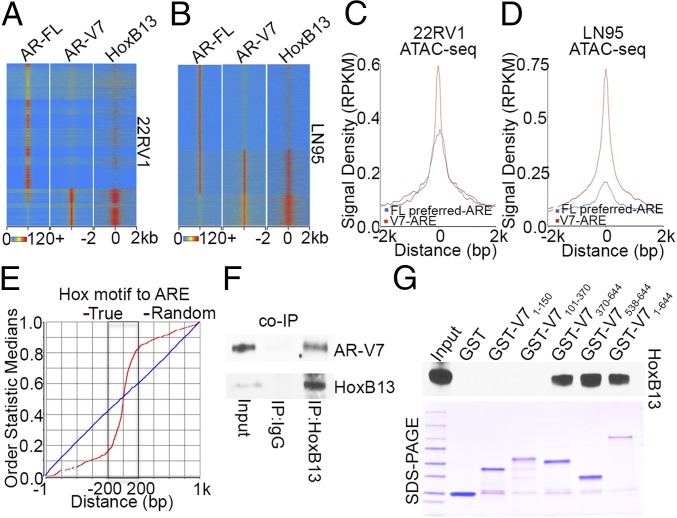
Given that our RNA-seq analysis found that HoxB13 is the most abundant transcript among Homeobox genes expressed in 22RV1 and LN95 cells (Dataset S3), we defined high-resolution HoxB13 cistromes by performing HoxB13 ChIP-exo (SI Appendix, Fig. S4B). Remarkably, integrative analysis of HoxB13, AR-V7, and AR-FL cistromes revealed a precise colocalization between HoxB13 and AR-V7 genomic binding in both cell lines, whereas the HoxB13 cistrome did not overlap with the AR-FL preferred cistrome (Fig. 2 A and B). Global correlation analysis demonstrated that AR-V7 binding signal densities were positively correlated with those of HoxB13 (SI Appendix, Fig. S4C). Examples of AR-V7 and HoxB13 colocalization in 22RV1 and LN95 cells are shown in SI Appendix, Fig. S4D. Because chromatin structures affect AR binding (13), we next performed the assay for transposase-accessible chromatin by sequencing (ATAC-seq) (17) using hormone-depleted 22RV1 and LN95 cells. Integrative analysis of ATAC-seq data with AR-V7/AR-FL ChIP-exo data found that chromatin accessibility was markedly higher on AREs in HoxB13 and AR-V7 binding regions than on AREs in the AR-FL preferred cistrome (Fig. 2 C and D and SI Appendix, Fig. S4E). These data suggest that chromatin accessibility may facilitate preferred genomic colocalization between HoxB13 and AR-V7 vs. AR-FL. Remarkably, when we calculated the genomic distance between precisely defined HoxB13 binding motifs (SI Appendix, Fig. S4 F and G) and AREs bound by AR-V7 (Fig. 1 D and E), we found that the vast majority of these motif instances were separated by just 150 bp (Fig. 2E and SI Appendix, Fig. S4H). Together, these findings indicate an intimate genomic interaction between AR-V7 and HoxB13 on open chromatin in CRPC. To investigate the functional relationship between HoxB13 and AR-V7 genomic binding, we performed AR-V7 ChIP in HoxB13 silenced cells as well as HoxB13 ChIP in AR-V7 silenced cells. We found that HoxB13 silencing significantly decreased AR-V7 binding without affecting AR-V7 protein expression (SI Appendix, Fig. S5 A, B, E, and F and Dataset S2), while silencing of AR-V7 had no effect on HoxB13 binding (SI Appendix, Fig. S5 C, D, G, and H). These data suggest that HoxB13 acts upstream of AR-V7 recruitment to the genome. To explore the physical association of AR-V7 and HoxB13, we first performed coimmunoprecipitation of the endogenous proteins. As shown in Fig. 2F, HoxB13 interacted with AR-V7 in vivo. We next expressed the FL AR-V7 and four regions of AR-V7 as GST fusion proteins (Dataset S2) and tested their ability to interact with in vitro-translated HoxB13. AR-V7 showed a strong interaction with HoxB13 protein via its DNA binding domain (Fig. 2G and SI Appendix, Fig. S4I). These data support a direct protein–protein interaction between AR-V7 and HoxB13.
Fig. 2.
AR-V7 and HoxB13 interact genomically and physically. (A and B) Heatmaps show the ChIP-exo signal intensity of AR-FL, AR-V7, and HoxB13 binding in 22RV1 cells (A) and LN95 cells (B). (C and D) Chromatin accessibility in 22RV1 (C) and LN95 (D) cells was determined by ATAC-seq. Normalized averaged ATAC-seq signal tag distribution over AR-V7 ARE regions and AR-FL preferred-ARE regions is shown. The window indicates ±2-kb regions of AREs. (E) Cumulative genomic position distributions of precisely defined HoxB13 motifs relative to AR-V7–bound AREs in AR-V7/HoxB13 binding locations in 22RV1 and LN95 cells. (F) Whole-cell lysates from hormone-depleted 22RV1 cells were immunoprecipitated with HoxB13 antibodies, and Western blots were performed with antibodies against AR-V7 and HoxB13. (G) GST pull-down assays were performed by incubating in vitro translated HoxB13 with GST ∼ AR-V7 fusion proteins. Western blots were performed using an anti-HoxB13 antibody.
HoxB13 and AR-V7 Coup-Regulate Diverse Target Oncogenes Across CRPC Cells and Tissues.
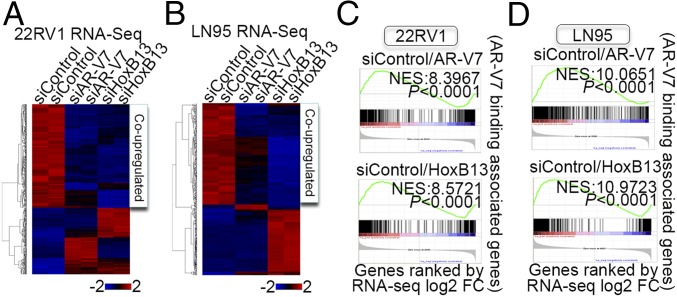
Having established a strong genomic and physical interaction between AR-V7 and HoxB13, we next asked whether AR-V7 and HoxB13 coregulate target genes in CRPC cells. RNA-seq analysis was performed in HoxB13 silenced 22RV1 and LN95 cells, and an unsupervised hierarchical clustering of gene-expression data from AR-V7 silenced, HoxB13 silenced, and control cells was conducted. Strikingly, we found that AR-V7 and HoxB13 up-regulated a common set of genes in both 22RV1 and LN95 cells (Fig. 3 A and B, SI Appendix, Fig. S6A, and Dataset S2), although the coup-regulated genes were largely different between the two cell lines (SI Appendix, Fig. S6B and Dataset S1). In contrast, no codown-regulated gene set was found, consistent with our finding that AR-V7 down-regulated genes were not involved in cancer-related processes (Fig. 3 A and B and SI Appendix, Fig. S3C). To correlate AR-V7 and HoxB13 binding with their regulated genes, we used an integrated approach (14) combining the GREAT algorithm (18) and our RNA-seq data. AR-V7 and HoxB13 showed similar distal binding to regulated genes (SI Appendix, Fig. S6C). These distal AR-V7 and HoxB13 binding sites directly up-regulated genes involved in prostatic neoplasms and metastasis (SI Appendix, Fig. S6D and Dataset S1). Importantly, gene set enrichment analysis (GSEA) (19) found that genes associated with AR-V7 binding sites were significantly enriched not only in AR-V7 up-regulated genes but also in HoxB13 up-regulated genes (Fig. 3 C and D and SI Appendix, Fig. S6 E and F). A similarly pronounced enrichment of genes associated with HoxB13 binding was also observed for both HoxB13 and AR-V7 up-regulated genes (SI Appendix, Fig. S6 G and H). These data reveal a strong direct coup-regulation of target genes by AR-V7 and HoxB13 in CRPC cells.
Fig. 3.
HoxB13 and AR-V7 coup-regulate diverse target oncogenes in CRPC. (A and B) Heatmaps of regulated genes (FDR < 0.05, fold-change > 2) after AR-V7 or HoxB13 silencing in 22RV1 (A) and LN95 cells (B). (C and D) GSEA analyses compare associated genes within ±50 kb of AR-V7 binding locations with AR-V7 or HoxB13 regulated genes determined by RNA-seq analysis in 22RV1 (C) and LN95 (D) cells.
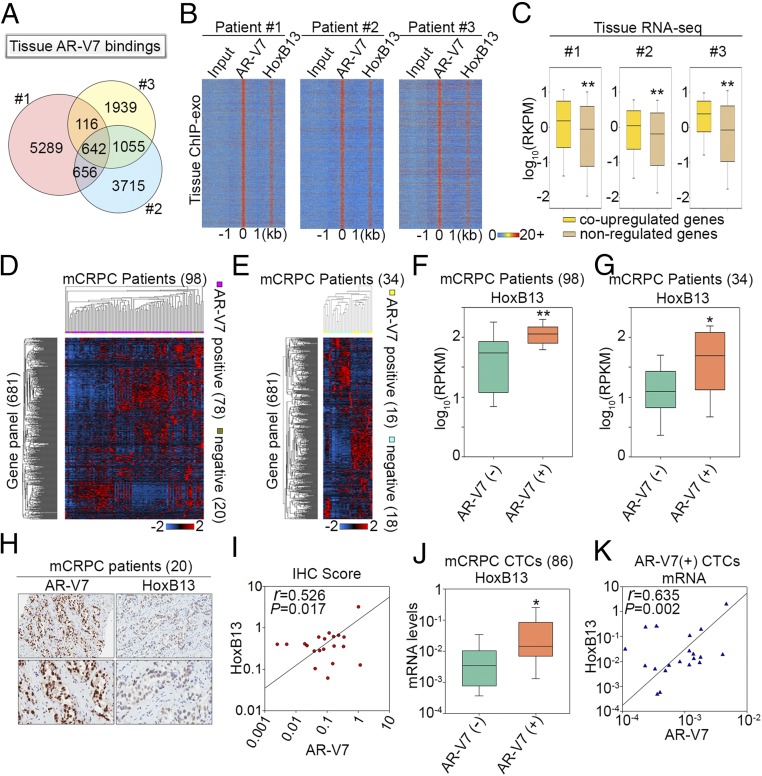
To examine the clinical relevance of transcriptional coregulation by AR-V7 and HoxB13 in CRPC cells, we performed AR-V7 and HoxB13 ChIP-exo as well as RNA-seq in tumors from three AR-V7–expressing CRPC patients (9) (SI Appendix, Fig. S7A). Although AR-V7 binding locations were largely different between the three CRPC patients (Fig. 4A), genomic colocalization between AR-V7 and HoxB13 was observed for all patients (Fig. 4B). Consistent with the heterogeneous nature of AR-V7 cistromes between CRPC patients, most genes associated with AR-V7 were different between the three patients (SI Appendix, Fig. S7 B and C). Integrative analysis of tissue ChIP-exo/tissue RNA-seq data (Fig. 4 A and B and SI Appendix, Fig. S7) with 22RV1/LN95 ChIP-exo/RNA-seq data (Figs. 2 and 3 and SI Appendix, Fig. S6) revealed significantly higher gene expression in CRPC tissues of AR-V7/HoxB13 coup-regulated genes compared with AR-V7/HoxB13-nonregulated genes (Fig. 4C and SI Appendix, Fig. S7B). Importantly, the putative AR-V7/HoxB13 coup-regulated genes (681) identified in CRPC tissues were associated with cancer progression and metastasis (SI Appendix, Fig. S7B and Dataset S1) and were highly expressed in CRPC circulating tumor cells (CTCs) (SI Appendix, Fig. S7D) and independent CRPC tissues (SI Appendix, Fig. S7E). Taken together, these data suggest that AR-V7 and HoxB13 cobinding sites in CRPC patients are functional in up-regulating diverse oncogenic target genes involved in CRPC progression.
Fig. 4.
HoxB13 and AR-V7 coregulate transcriptomic diversity among CRPC patients. (A) A Venn diagram shows AR-V7 binding site diversity between the three CRPC patients. (B) Heatmaps show the ChIP-exo signal intensity of AR-V7 and HoxB13 binding in three CRPC patients. (C) Box plots compare expression of genes associated with tissue AR-V7/HoxB13 binding sites. Coup-regulated genes refer to genes that were also coup-regulated by AR-V7/HoxB13 in CRPC cells. (D and E) Heatmaps show expression of the 681-gene panel among the 98 metastatic CRPC (mCRPC) patients in the Robinson et al. (20) cohort (D), and 34 mCRPC cases in the Beltran et al. (21) cohort (E). (F and G) Box plots compare HoxB13 gene expression between AR-V7+ and AR-V7− patients in the Robinson et al. (20) cohort (F) and in the Beltran et al. (21) cohort (G), respectively. The significance was determined by Mann–Whitney rank sum test. *P < 0.01, **P < 0.001. (H) Representative AR-V7 and HoxB13 immunoreactivity in mCRPC tissues (Upper 20×, Lower 40× original magnification). (I) Correlation between AR-V7 and HoxB13 staining in 20 CRPC cases. Slides were scanned using an Aperio Digital Pathology Slide Scanner (Leica Biosystems) at 40× magnification and staining quantified using the Aperio Image Scope (v11). Data represent the ratio of positive pixels per total pixels from areas of tissue containing mCRPC. (J) A box plot compares HoxB13 mRNA levels in 86 CTCs from mCRPC patients. The significance was determined by one-tailed t test. *P < 0.05. (K) Correlation of mRNA expression between HoxB13 and AR-V7 in AR-V7+ CTCs.
To examine AR-V7 and HoxB13 transcriptional coregulation in large CRPC patient cohorts, we reanalyzed RNA-seq data from 98 CRPC cases in the Robinson et al. (20) cohort and 34 CRPC cases in the Beltran et al. (21) cohort. Based on the presence or absence of exon 3/CE3 splicing junctions (SI Appendix, Fig. S7A), we divided the CRPC patients from these two cohorts into AR-V7+ and AR-V7− groups (SI Appendix, Fig. S8A). Unsupervised hierarchical clustering of gene-expression data from 135 patients (20, 21), including the 3 AR-V7+ patients from this study, failed to identify genes showing similar expression patterns across individuals in each group (SI Appendix, Fig. S8A). Importantly, focused analysis of the 681 genes identified as putative targets of AR-V7/HoxB13 in CRPC tissues (SI Appendix, Fig. S7B) also failed to discover genes showing uniformly high or low expression across AR-V7+ or AR-V7− cases (Fig. 4 D and E). Nonetheless, HoxB13 and AR mRNA expression levels were significantly higher in the AR-V7+ group compared with the AR-V7− group (Fig. 4 F and G and SI Appendix, Fig. S8 B–E). These data reveal AR-V7/HoxB13-regulated transcriptomic diversity among CRPC cases, and suggest that AR-V7 and HoxB13 might be coexpressed in CRPC cases. We thus performed immunocytochemistry (IHC) analysis of AR-V7 and HoxB13 using tissues from 20 CRPC patients. This revealed a high concordance between AR-V7 and HoxB13 protein expression (Fig. 4 H and I). We also conducted an analysis of AR-V7 and HoxB13 gene expression in 86 CTCs isolated from CRPC patients. HoxB13 mRNA expression was significantly higher in AR-V7+ CTCs versus AR-V7− CTCs, and the expression of HoxB13 mRNA and AR-V7 mRNA was positively correlated (Fig. 4 J and K and Dataset S2). These results support the clinical relevance of AR-V7/HoxB13 transcriptional coregulation.
Silencing of HoxB13 Decreases AR-V7–Driven CRPC Growth.
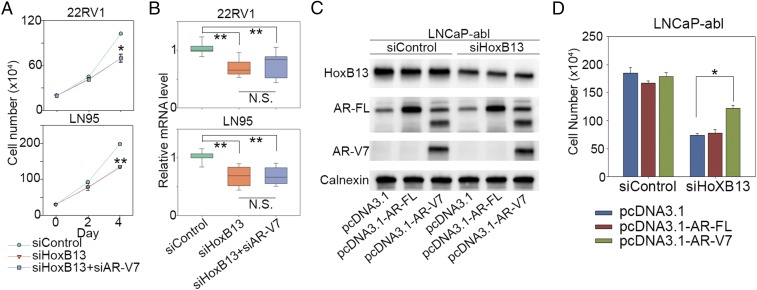
Finally, given that AR-V7 drives CRPC growth in vitro and in vivo (6, 11, 12), we examined whether silencing HoxB13 decreases AR-V7–driven CRPC growth in vitro and in vivo. Silencing of HoxB13 significantly decreased in vitro cell growth of both 22RV1 and LN95 cells (Fig. 5A and SI Appendix, Fig. S9A). Interestingly, simultaneous silencing of both HoxB13 and AR-V7 did not further inhibit cell growth (Fig. 5A) and expression of AR-V7/HoxB13 coup-regulated genes (Fig. 5B), suggesting that the biological function of AR-V7 is mainly determined by HoxB13. To address whether the growth inhibition conferred by HoxB13 knockdown is mediated through AR-V7 loss, we performed cell growth rescue experiments in an AR-V7− CRPC model, LNCaP-abl, by overexpression of AR-V7 or AR-FL in cells with partial HoxB13 silencing or control silencing (Fig. 5C). In the hormone-depleted condition, overexpression of AR-V7 but not AR-FL significantly rescued LNCaP-abl cells from the growth inhibitory effects of HoxB13 protein reduction (Fig. 5D and SI Appendix, Fig. S9B), suggesting that AR-V7 is an important mediator of HoxB13 function in CRPC. To examine the effect of HoxB13 silencing in vivo, 22RV1 and LN95 cells infected with lentivirus encoding HoxB13 shRNA or a control shRNA were injected into castrated mice. Tumor weights and tumor volumes were significantly decreased in shHoxB13-treated groups compared with control groups (Fig. 6 A–F and SI Appendix, Fig. S10A), whereas no mouse body weight decrease was observed in shHoxB13-treated groups versus control groups (SI Appendix, Fig. S10B). Importantly, ChIP analysis of engrafted tumor tissue showed that HoxB13 silencing significantly decreased AR-V7 binding regardless of whether an accompanying decrease in AR-V7 expression was observed (LN95) or not (22RV1) (Fig. 6 G and H, SI Appendix, Fig. S10 C and D, and Dataset S2). These data suggest that HoxB13 governs AR-V7–expressing CRPC growth in vivo through pioneering AR-V7. To further examine whether HoxB13 determines AR-V7 oncogenic function in vivo, 22RV1 and LN95 cells infected with lentiviral shRNA targeting HoxB13 only, both HoxB13 and AR-V7 in combination, or control sequences (SI Appendix, Fig. S10E) were inoculated into castrated mice. Tumor weights were significantly decreased in shHoxB13 only and combined shHoxB13/shAR-V7–treated groups versus shControl groups (SI Appendix, Fig. S10 F and G). Remarkably, there was no significant difference in tumor weights between the shHoxB13 only and the combined shHoxB13/shAR-V7 groups (SI Appendix, Fig. S10 F and G), suggesting that the oncogenic function of AR-V7 in vivo is largely determined by HoxB13.
Fig. 5.
AR-V7 is an important mediator of HoxB13 function in CRPC. (A and B) 22RV1 cells and LN95 cells were transfected with control siRNAs, siRNAs targeting HoxB13, and combined siRNAs targeting HoxB13 and AR-V7. Cell proliferation was measured by direct cell count assays (A), and expression of HoxB13 and AR-V7 coregulated genes AAK1, CROT, NEDD4L, and GRIN3A was analyzed by RT-PCR (B). The results for cell proliferation (A) are shown as mean ± SD (n = 2). The significance for cell proliferation (A) and RT-PCR (B) was determined by one-way ANOVA. *P < 0.01, **P < 0.001. (C and D) LNCaP-abl cells were cotransfected with siRNAs combined with pcDNA3.1 vector, pcDNA3.1–AR-FL (32), or pcDNA3.1–AR-V7 (11). Western blots were performed using the indicated antibodies (C), and cell proliferation on day 5 was measured by direct cell count assays (D). The significance was determined by one-way ANOVA. *P < 0.01.
Fig. 6.
HoxB13 silencing inhibits AR-V7-driven CRPC growth in vivo. (A and D) HoxB13 protein expression in 22RV1 cells (A) and LN95 cells (D) infected with lentivirus encoding HoxB13 shRNA or a control shRNA after 48 h. Western blots were performed using the same cells used for injection into castrated mice. (B and E) Representative picture of shControl 22RV1 tumors and HoxB13-silenced 22RV1 tumors at time of collection (B, day 24), and shControl LN95 tumors and HoxB13-silenced LN95 tumors at time of collection (E, day 34). (C and F) Average 22RV1 tumor weight for control (C, n = 24) and HoxB13 silenced groups (C, n = 23), and average LN95 tumor weight for control (F, n = 24) and HoxB13 silenced groups (F, n = 22). The results are shown as mean ± SE. The significance was determined by one-tailed t test. *P < 0.01, **P < 0.001. (G and H) AR-V7 and HoxB13 tissue ChIP analysis using engrafted 22RV1 (G, tumors n = 12, regions n = 8) and LN95 (H, tumors n = 20, regions n = 8) tumor tissues. The significance was determined by Mann–Whitney rank sum test. **P < 0.001.
Discussion
Despite the emerging clinical importance of AR-V7, the molecular and genomic mechanisms underlying the cancer-promoting function of AR-V7 in CRPC are largely unknown. Foremost, it is unknown whether AR-V7, a TF lacking the LBD, is able to globally bind to chromatin to exert its genomic function. In fact, recent ChIP-seq studies have attempted to address whether AR-Vs can globally bind to the genome in CRPC cell lines (22, 23). Unfortunately, the lack of an AR-V–specific antibody in these ChIP-seq studies made it difficult to distinguish AR-V7 signal from that of AR-FL and other AR-Vs within the same population of CRPC cells. Moreover, although AR-V7 is overexpressed at the protein level in CRPC patients (6, 7, 24), it is unknown whether AR-V7 binds to CRPC patients’ genomes. In this study, we have defined high-resolution AR-V7 cistromes in AR-V7–expressing CRPC cells and patient tissues using our ChIP-exo approach with an AR-V7–specific antibody. By integrating ChIP-exo data with ATAC-seq and RNA-seq analysis, we find that AR-V7 clearly binds to open chromatin in the CRPC genomes through recognizing the FL ARE, and directly regulates diverse target oncogenes in similar biologic and disease pathways contributing to CRPC (Figs. 1–4). Our data support AR-V7 as a major driving AR-V in CRPC progression.
Consistent with the heterogeneous nature of CRPC (25), we found that the AR-V7 cistromes are cell-context–dependent in both CRPC cell lines and clinical specimens, resulting in tremendous diversity in AR-V7–regulated transcriptomes across CRPC patients (Figs. 1–4). Remarkably, a homogeneous feature of CRPC is the colocalization of HoxB13 that accompanies and supports the heterogeneous binding of AR-V7, and these two TFs coup-regulate diverse target genes involved in CRPC progression (Figs. 2–4, SI Appendix, Fig. S7, and Dataset S1). Although controversies exist on the role of HoxB13 in promoting or inhibiting AR-FL function in androgen-dependent prostate cancer (26–28), our results demonstrate that HoxB13 governs heterogeneous CRPC growth mechanisms by directing AR-V7 oncogenomic function (Figs. 2–6). Indeed, in AR-V7–expressing CRPC, the oncogenic function of AR-V7 is mainly determined by HoxB13, and AR-V7 is a critical mediator of HoxB13 function. Interestingly, our RNA-seq analysis failed to identify the rare prostate cancer predisposing G84E mutation in HoxB13 (29) in any CRPC samples examined in this study or in 132 CRPC patients from two cohorts (20, 21), suggesting that this mutation does not contribute to evolution of the novel genomic function of HoxB13 in CRPC. It is possible that chromatin accessibility (Fig. 2 C and D) or other genetic/epigenetic factors may reprogram HoxB13 function in CRPC genomes. Importantly, because few downstream target genes of AR-V7 can universally account for progression of heterogeneous CRPC in which AR-V7 itself is not druggable by AR antagonists, future translational research on AR-V7-driven CRPC should focus on targeting the pivotal upstream genomic regulator HoxB13. Finally, as silencing of HoxB13 and/or AR-V7 significantly decreases but does not completely inhibits CRPC growth, future studies should also consider targeting HoxB13 together with other key growth-promoting mechanisms for CRPC.
Materials and Methods
Cell Lines.
CWR22RV1 cells were obtained from the American Type Culture Collection (ATCC). LNCaP95 cells were provided by J.L., and LNCaP-abl cells were kindly provided by Zoran Culig, Innsbruck Medical University, Innsbruck, Austria. Further details are provided in the SI Appendix, SI Materials and Methods.
Human Tissues.
CRPC tissues utilized in ChIP-exo and RNA-seq analysis were rapid autopsy specimens described in a previous study (7). CRPC tissues for IHC analysis were obtained from Duke University. Samples were obtained from CRPC patients who signed written informed consent. All experimental procedures were approved by the Johns Hopkins University and Duke University Institutional Review Boards.
RNA Interference.
All transient transfections of siRNA into cell lines followed the standard protocol for Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher). Further details are provided in the SI Appendix, SI Materials and Methods.
Cell Proliferation Assays.
Cell proliferation was measured by a direct viable cell count assay, as described previously (13).
Western Blots and Coimmunoprecipitation Assays.
Western blots and coimmunoprecipitation were performed as previously described (30). Antibodies used were anti–AR-V7 (AG10008) from Precision Antibody, and anti-AR (N20) and anti-HoxB13 (H-80) from Santa Cruz Biotechnology.
RNA-Seq and Data Analysis.
RNA-seq analysis was performed and analyzed as previously described (14). Further details are provided in the SI Appendix, SI Materials and Methods.
ChIP-Exo and Data Analysis.
ChIP-exo was performed and analyzed as previously described (13, 14). Further details are provided in the SI Appendix, SI Materials and Methods.
Standard ChIP and RT-PCR Assays.
ChIP and RT-PCR assays were performed as described previously (13).
ATAC-Seq Assays.
ATAC-seq library preparation and data analysis were performed as described previously (17). Further details are provided in the SI Appendix, SI Materials and Methods.
GST Pull-Down Assay.
GST-Pull down assay was performed as previously described (30) with modifications. Further details are provided in the SI Appendix, SI Materials and Methods.
IHC Analysis.
IHC was performed as previously described (11) with modifications. Further details are provided in the SI Appendix, SI Materials and Methods.
Analysis of AR-V7 and HoxB13 mRNA Expression in CTCs from CRPC Patients.
Patients with metastatic CRPC were enrolled in a prospective study evaluating clinical correlates of CTC AR biomarkers, as described previously (10). The study protocol was approved by the Institutional Review Board at the Johns Hopkins University (IRB-6). Written informed consent was obtained from all enrolled patients. CTC analysis was performed as described previously (9). Further details are provided in the SI Appendix, SI Materials and Methods.
Mouse Xenograft Studies.
Mouse xenograft studies were performed as described previously (31). All experiments were conducted in accordance with the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International and The Ohio State University Institutional Animal Care and Use Committee-approved protocol. Further details are provided in the SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants U54 CA217297 (to Q.W., V.X.J., and T.H.-M.H.), R01 GM120221 (to Q.W.), R01 CA200853 (to Q.W., J.H., and S.K.C.), R01 CA172603, R01 CA205001, and R01 CA212403 (to J.H.); Department of Defense Grant W81XWH-16-1-0291 (to Q.W., S.K.C., and J.H.); and the Prostate Cancer Foundation Joyce and Larry Stupski Prostate Cancer Precision Oncology Special Challenge Award (to J.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The ChIP-exo, ATAC-seq, and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE99378).
See Commentary on page 6528.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718811115/-/DCSupplemental.
References
- 1.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 2.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, et al. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maughan BL, Antonarakis ES. Clinical relevance of androgen receptor splice variants in castration-resistant prostate cancer. Curr Treat Options Oncol. 2015;16:57. doi: 10.1007/s11864-015-0375-z. [DOI] [PubMed] [Google Scholar]
- 5.Antonarakis ES, Armstrong AJ, Dehm SM, Luo J. Androgen receptor variant-driven prostate cancer: Clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis. 2016;19:231–241. doi: 10.1038/pcan.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Z, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu R, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hörnberg E, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonarakis ES, et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol. 2017;35:2149–2156. doi: 10.1200/JCO.2016.70.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu R, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, et al. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, et al. Agonist and antagonist switch DNA motifs recognized by human androgen receptor in prostate cancer. EMBO J. 2015;34:502–516. doi: 10.15252/embj.201490306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z, et al. Ligand-dependent genomic function of glucocorticoid receptor in triple-negative breast cancer. Nat Commun. 2015;6:8323. doi: 10.1038/ncomms9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, et al. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–2117. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato M, et al. Cotargeting androgen receptor splice variants and mTOR signaling pathway for the treatment of castration-resistant prostate cancer. Clin Cancer Res. 2016;22:2744–2754. doi: 10.1158/1078-0432.CCR-15-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corces MR, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beltran H, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, et al. The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells. J Urol. 2015;193:690–698. doi: 10.1016/j.juro.2014.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan SC, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015;43:5880–5897. doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu Y, et al. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep. 2015;5:7654. doi: 10.1038/srep07654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar A, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norris JD, et al. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36:405–416. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pomerantz MM, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet. 2015;47:1346–1351. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung C, Kim RS, Zhang HJ, Lee SJ, Jeng MH. HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 2004;64:9185–9192. doi: 10.1158/0008-5472.CAN-04-1330. [DOI] [PubMed] [Google Scholar]
- 29.Ewing CM, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J Biol Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, et al. CCI-779 inhibits cell-cycle G2-M progression and invasion of castration-resistant prostate cancer via attenuation of UBE2C transcription and mRNA stability. Cancer Res. 2011;71:4866–4876. doi: 10.1158/0008-5472.CAN-10-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Udayakumar TS, Vasaitis TS, Brodie AM, Fondell JD. Mechanistic relationship between androgen receptor polyglutamine tract truncation and androgen-dependent transcriptional hyperactivity in prostate cancer cells. J Biol Chem. 2004;279:17319–17328. doi: 10.1074/jbc.M400970200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.