Significance
The transfection of lymphocytes with genetic material is a significant unmet need in immunotherapy and the treatment of many diseases. Current transfection strategies primarily rely on physical methods that must be conducted ex vivo and are often inefficient. We identified mixtures of lipid-varied mRNA delivery vehicles (charge-altering releasable transporters, CARTs) that show enhanced uptake into multiple lymphocyte cell types. The top performing lipid mixtures were rapidly identified using a combinatorial strategy and the resulting information was used to design single-delivery agents incorporating both lipid domains using our two-step organocatalytic ring-opening polymerization. Hybrid-lipid CARTs show >80% mRNA transfection efficiency in vitro and >1.5% lymphocyte transfection efficiency in mice, which is higher than previously reported systems. These materials should enable new immunotherapy strategies and applications.
Keywords: gene therapy, nanoparticle, polyplex, combinatorial, immunotherapy
Abstract
We report a strategy for generating a combinatorial library of oligonucleotide transporters with varied lipid domains and their use in the efficient transfection of lymphocytes with mRNA in vitro and in vivo. This library is based on amphiphilic charge-altering releasable transporters (CARTs) that contain a lipophilic block functionalized with various side-chain lipids and a polycationic α-amino ester mRNA-binding block that undergoes rearrangement to neutral small molecules, resulting in mRNA release. We show that certain binary mixtures of these lipid-varied CARTs provide up to a ninefold enhancement in mRNA translation in lymphocytes in vitro relative to either a single-lipid CART component alone or the commercial reagent Lipofectamine 2000, corresponding to a striking increase in percent transfection from 9–12% to 80%. Informed by the results with binary mixtures, we further show that CARTs consisting of optimized ratios of the two lead lipids incorporated into a single hybrid-lipid transporter molecule maintain the same delivery efficacy as the noncovalent mixture of two CARTs. The lead lipid CART mixtures and hybrid-lipid CARTs show enhanced lymphocyte transfection in primary T cells and in vivo in mice. This combinatorial approach for rapidly screening mRNA delivery vectors has provided lipid-varied CART mixtures and hybrid-lipid CARTs that exhibit significant improvement in mRNA delivery to lymphocytes, a finding of potentially broad value in research and clinical applications.
Emerging DNA and RNA technologies have the potential to revolutionize life science research and medicine. Relying on the delivery of oligonucleotides, these technologies enable a range of applications from protein expression to genome editing and regulation, impacting vaccinology, allergy tolerization, gene therapy, and cancer immunotherapy (1–3). mRNA-based therapies, in particular, have emerged as powerful complements to DNA-based methods due to their decreased risk of mutagenesis, transient effects, and reduced complexity relative to plasmid DNA since mRNA is inherently the minimal genetic vector necessary for protein expression (4–6). However, the effective delivery of oligonucleotides has proven challenging due to the difficulty of delivering large, polar, polyanions across the nonpolar membrane of a cell. To address this challenge, recent delivery methods including cationic polymers (7) and lipid nanoparticles (LNPs) (8) have been employed which circumvent the immunogenicity concerns of viral vectors and the tolerability and efficacy issues of mechanical methods such as electroporation. Notable examples of polymer-based systems include pH-responsive lipid/polyester hybrid materials (9), tunable amine-functionalized polyesters (10), polyamine complexes preassociated with cellular translational machinery (11, 12), and rapidly degrading peptide-like materials (13–15). While nonviral methods for mRNA delivery into cultured cells and in vivo are emerging, there have been very few reports on the safe and effective delivery of mRNA to lymphocytes (16). Typically, limited endocytosis and protein translation, as well as low resulting lymphocyte viability, have been difficult challenges to overcome (17).
Of great significance for further advances in vaccinology and immunology with the advent of advanced cancer immunotherapies using chimeric antigen receptor T cells is the delivery of oligonucleotides, including mRNA, into lymphocytes (18–20). Currently, clinical strategies that rely on lymphocyte transfection using mainly physical methods such as electroporation and nucleofection require ex vivo enrichment of T lymphocytes, which is cost- and efficiency-limited (21, 22). Enhanced chemical methods for ex vivo transfection of lymphocytes, or more ambitiously direct in vivo delivery of mRNA into lymphocytes, would allow for more efficient and time-effective treatments (20, 23, 24). Recent advances in gene delivery to lymphocytes include the use of polycationic polymers, such as lipid substituted polyethylenimines (25) or arginine-rich poly(oxanorbonene)s for the delivery of siRNA (26) as well as poly(β-amino ester)s to deliver DNA (27, 28). While the use of these polycationic nanoparticles has improved gene delivery to B and T cells, new transporters are still needed to enhance transfection and decrease toxicity of the carriers. Previously explored transporters typically only achieve 5–50% gene transfection in vitro (27–31) and less than 1% lymphocyte transfection for mRNA delivery in vivo (16, 32, 33).
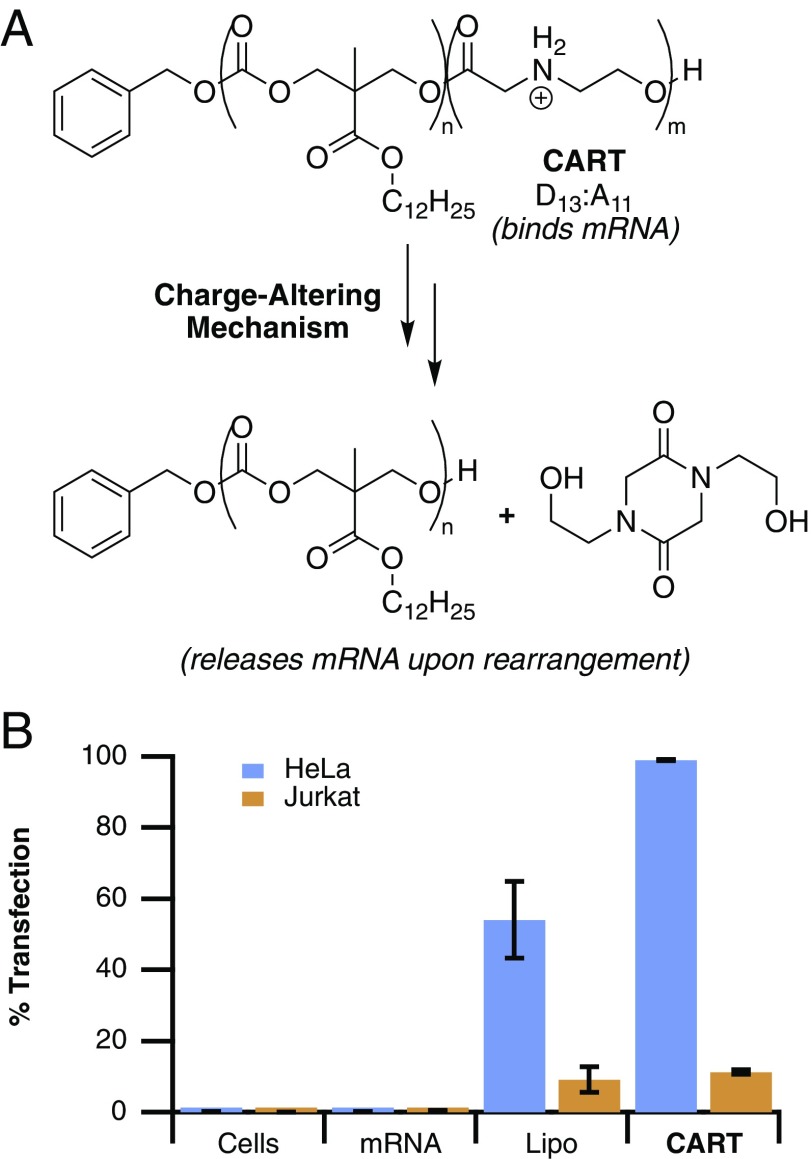
Our group has recently reported the use of poly(carbonate)-b-(α amino ester)s for the delivery of mRNA with >95% efficiency in cultured cells and at high levels in vivo in BALB/c mice (Fig. 1A) (34). Charge-altering releasable transporters, dubbed CARTs, are functionally distinct from most polycation systems in that they are dynamic, undergoing an ester-to-amide rearrangement of the cationic poly(α amino ester) backbone into neutral small molecules (diketopiperizine) over time, thereby providing a mechanism for release of mRNA and avoiding tolerability issues often associated with persistent polycations (35, 36). CARTs are step-economically synthesized and readily formulated with mRNA by simple mixing, rather than the microfluidic methods necessitated by cationic LNP technologies (8, 33, 37, 38). Single-lipid CARTs achieve high transfection of GFP mRNA in many epithelial cell lines such as HeLa (Fig. 1B); however, low delivery efficiencies are observed in lymphatic cells such as Jurkat cells (T lymphocyte), a trend common among many delivery agents (27). Herein we report that systematic tuning of CART lipid domains using a combinatorial method has yielded transporters that significantly outperform previously reported transporters and commercial agents for the transfection of lymphocytes including T cells in culture and in vivo.
Fig. 1.
(A) Structure of previously reported CART D13:A11 and neutral products of the charge-altering mechanism. (B) Transfection efficiencies of EGFP mRNA alone, lipofectamine-formulated EGFP mRNA, and D13:A11-complexed EGFP mRNA into HeLa cells versus Jurkat cells (T lymphocytes). Error bars represent the SD over three experiments in triplicate.
The efficiency of oligonucleotide delivery vehicles is greatly influenced by the type of lipids they contain (16, 37, 39–42). For example, we showed that amphipathic transporters for siRNA perform best with dodecyl-functionalized lipid blocks compared with the analogous hexyl or ethyl materials (43). Langer and coworkers have used high-throughput methods to find optimal lipid compositions for both cationic polymers (44–46) and LNPs (16, 47, 48). Additionally, Tew has shown that the lipid and charge distribution greatly influences siRNA uptake, especially in T lymphocytes (26, 49). Taken together, these studies suggest that the uptake of the newly described CART–mRNA nanoparticles could be influenced by their lipid content, prompting our interest in a rapid method to identify preferred systems.
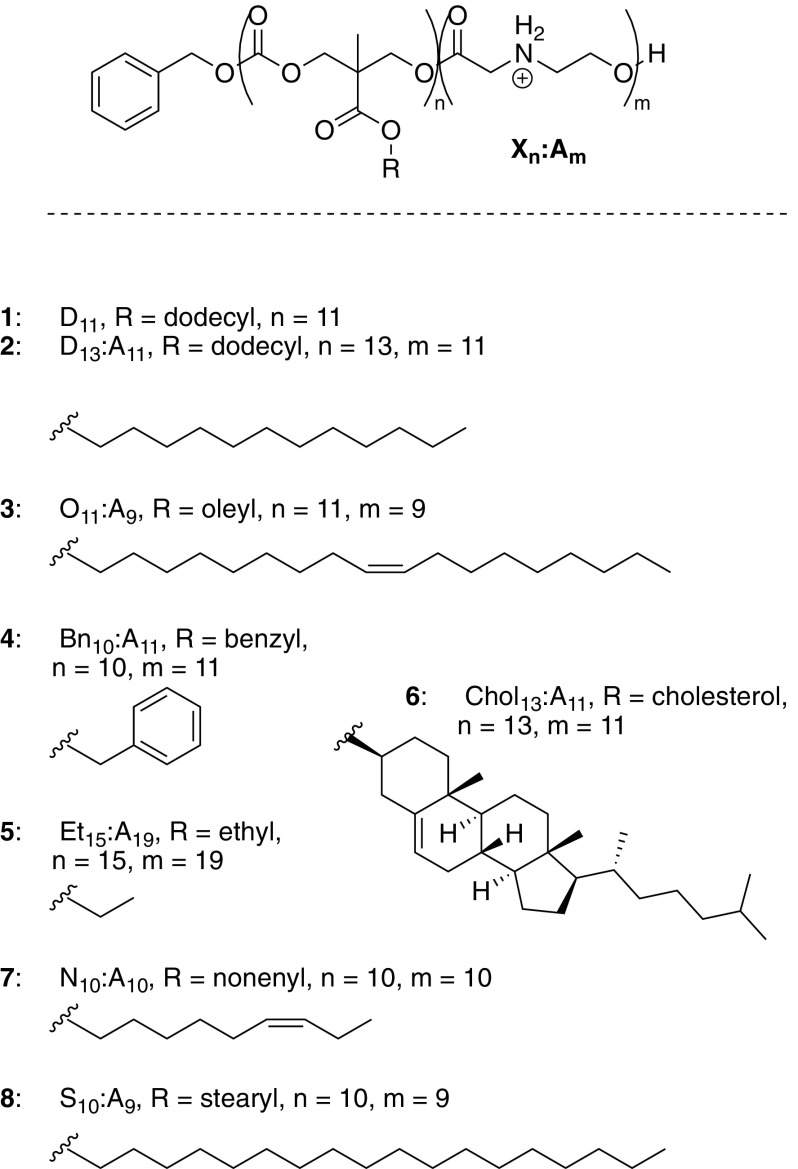
Organocatalytic ring-opening polymerization (OROP) methods (34, 50–52) allow for the rapid (two step) synthesis of CARTs, each with a single but different lipid block. We hypothesized that binary and ternary mixtures of lipid blocks (di- and trilipid blocks) might lead to better-performing CARTs as different compositions would allow for more varied performance at the critical and differing stages of RNA complexation, protection, delivery, and release. However, exploring this idea with even as few as eight different lipids would require the synthesis of 64 different lipid diblocks including the single-lipid diblock repeat (AA, BB, etc.) and mixed-lipid diblocks (AB, BA, BC, CB, etc.). Exploring different lipid diblock ratios would quickly inflate this number and the required synthesis time. In contrast, we reasoned that a high-throughput approach to testing performance of binary (and higher-order) mixtures would be to simply mix and test different monolipid CARTs in pairs. Similar combinatorial approaches by Langer and coworkers (53), Riezman and coworkers (41), and Montenegro and coworkers (54) have been utilized in other contexts for identifying trends in oligonucleotide and protein delivery materials. In this case, exploration of all 64 diblock combinations at a given ratio would require the synthesis of only eight different CARTs and their combinatorial mixing. When an unequal ratio of two CARTs is used, all 64 of those combinations are unique, and by varying the ratio by n-fold one could expand this sampling by n-fold without the need to prepare new materials. Here we report a high-throughput screen of lymphocyte delivery using eight CARTs differing by their lipid domain and representing 64 CART systems (Fig. 2). Using this strategy, we have identified CART mixtures that outperform individual CARTs. Informed by these results, we then designed and prepared hybrid CARTs incorporating both lipid domains of the preferred individual CART mixtures and in the preferred ratios (i.e., a hybrid-lipid CART). We found that these hybrid-lipid CARTs performed as well as the CART mixtures and proved to be highly effective for mRNA delivery to lymphocytes in vitro and in vivo.
Fig. 2.
Structures of CARTs with varying lipid blocks used in this study.
Results and Discussion
Design and Synthesis of Single-Lipid CARTs.
We synthesized eight different CART candidates with variations in their lipid blocks for evaluation in mixtures for mRNA delivery to lymphocytes (Fig. 2). All CARTs were prepared in only two synthetic steps and low dispersity utilizing our previously reported OROP methodology using benzyl alcohol as the initiator (50, 55). To simplify initial screening, overall block lengths were kept constant at ∼12 lipid units and 12 cationic units, in line with previous optimization of α-amino ester CARTs for mRNA delivery (34). A homooligomer of the dodecyl carbonate monomer (D11, 1, Fig. 2) was included to determine if simply increasing the total lipid content relative to the cationic content would be advantageous. The CART panel contained a variety of lipids that have previously shown success for oligonucleotide delivery (43, 56), including saturated (dodecyl 2, stearyl 8), unsaturated (oleyl 3 and nonenyl 7), and aromatic (benzyl 4) lipids, as well as the much more rigid cholesterol (6), which is commonly used in LNPs (57). An ethyl-functionalized CART (5) was also synthesized to determine if partial incorporation of a significantly less hydrophobic CART would be advantageous.
Screening of CART Mixtures for Delivery into Lymphocytes.
A high-throughput assay was developed to evaluate binary combinations of the lipid-containing CARTs for mRNA delivery. For preliminary optimization studies, Jurkat cells were selected as a commonly used model lymphocyte line with low reported transfection efficiencies (26, 27, 58). The experimental design involved pairwise premixing of CARTs 1–8 at specified ratios in a 96-well plate. Mixtures were set up such that each column and row of a 96-well plate corresponded to 66% and 33% of the given mixture (with respect to cationic charges), respectively. Thus, 64 total combinations were prepared for evaluation (each CART by itself, and mixed with the other seven CARTs at both a 2:1 and a 1:2 ratio). For example, column 1, row 2 contains 66% of CART 1 and 33% of CART 2, and column 2, row 2 contains 100% CART 2. By mixing CARTs at 2:1 ratios rather than simply 1:1, redundancy is avoided and 64 unique mixtures can be prepared.
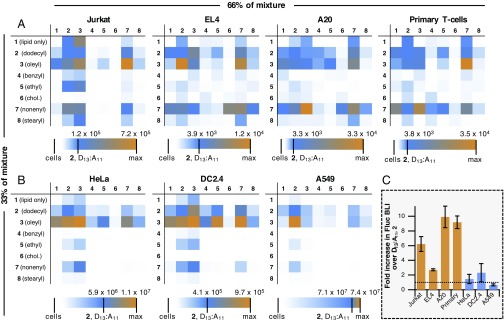
CART mixtures were then added to a second 96-well plate containing firefly luciferase (Fluc) mRNA in PBS at a 10:1 (+/−) charge ratio of total cations on the CARTs to total anions on the mRNA to form electrostatic associated CART–mRNA complexes. Fluc mRNA was selected as a convenient reporter system, because it allows simultaneous measurement of expression levels across an entire 96-well plate in a single image using an IVIS system. After mixing the CART–mRNA for 20 s, these complexes were added to Jurkat cells (50,000 per well) for treatment at a final concentration of 20 ng mRNA per well. Bioluminescence was assessed after 8 h and processed to quantify relative expression of the mixtures compared with the previously reported CART D13:A11 (2) (Fig. 3).
Fig. 3.
High-throughput evaluation of mixtures of amphipathic CARTs for the delivery of luciferase (Fluc) mRNA. For mixture matrices, CARTs in columns and rows correspond to 66% and 33% of the total cationic charge in the mixture, respectively. Each grid image is three-color-weighted to designate the relative expression levels of the given mixture, with white representing no expression, blue representing the expression of CART D13:A11 2, and orange being the highest-performing mixture. The scale bar below each matrix gives the bioluminescence (in photons⋅s−1⋅cm−2⋅sr−1) of each experiment. (A) Expression in nonadherent lymphocytes (Jurkat cells, EL4 cells, A20 cells, and activated primary T cells). (B) Expression in adherent epithelial cells and monocytes (HeLa cells, DC 2.4 cells, and A549 cells). All values represent the average bioluminescence of three separate experiments. (C) Normalized bioluminescence intensity of cells treated with the highest-performing mixture of CART 3 and 7 demonstrating the fold improvement over D13:A11 2 in all cell lines tested. Horizontal dashed line corresponds to expression from unmixed D13:A11 2. Data are the average of three separate experiments, each in triplicate, with error bars representing the SD.
By color-weighting the raw bioluminescence values, it is clear that several CART mixtures significantly outperform D13:A11 (2), the previously reported CART, for mRNA delivery into Jurkat cells. Specifically, a 1:2 mixture of O11:A9 (3) to N10:A10 (7) was the lead performer in Jurkat cells, demonstrating a sixfold enhancement in expression over D13:A11. The 2:1 O11:A9 (3):N10:A10 (7) mixture of these CARTs was also effective, though to a somewhat lesser degree. The mixtures containing the lipid block homooligomer (1) generally did not outperform the analogous CART by itself, suggesting that overall increased lipid content in CART complexes is not by itself advantageous. Notably, mixtures containing the benzyl (4), ethyl (5), and cholesterol (6) CARTs showed very little expression regardless of their lipid CART counterpart. This was somewhat surprising since cationic LNP formulations typically incorporate a cholesterol component of 10–40% (46, 57, 59). However, previously reported systems were monomeric and so result in much lower local concentrations of cholesterol, which, while not studied, could be achieved with CARTs using cholesterol as an initiator (43). Significantly, Jurkat cells treated with all lipid mixtures showed >73% viability, with all high-performing compounds showing >85% viability (SI Appendix, Fig. S1). This can be attributed to their charge-altering degradation to neutral products compared with other reported persistent polycations that exhibit cytotoxicity (60).
We next examined other cells lines and found that the top-performing mixtures of O11:A9 (3) and N10:A10 (7) consistently outperformed the other lipid combinations in all other nonadherent lymphocyte lines tested including EL4 immortalized T cells and A20 immortalized B cells, which showed 3- and 10-fold increases in gene expression, respectively. This trend was further confirmed in activated primary murine T cells (Fig. 3A), which exhibited a ninefold increase in expression relative to the single-lipid CART D13:A11 2. Interestingly, in the adherent monocyte and epithelial cell lines tested (Fig. 3B), including HeLa, DC2.4, and A549 cells, the lipid combinations did not significantly affect luciferase expression, with D13:A11 (2) and O11:A9 (3) performing comparably to or better than the combinations, indicating the enhanced mRNA uptake by mixed nonenyl- and oleyl-based CARTs may be unique to lymphatic cells.
Optimization of Lipid Ratio.
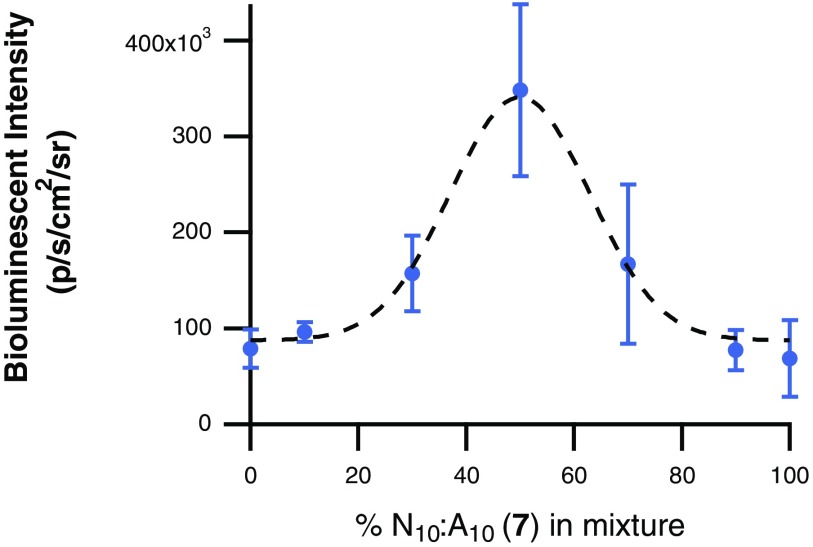
After the high-throughput screen examining 33% and 66% lipid mixtures resulted in increased luciferase expression with the O11:A9 (3) and N10:A10 (7) CART mixtures, we sought to determine the optimal treatment ratio of CARTs 3 and 7 for mRNA delivery into Jurkat cells. For this study, mixtures were prepared as described above but with additional ratios of 3 and 7. Specifically, Fluc mRNA was complexed with 10:0, 9:1, 7:3, 5:5: 3:7, 1:9, or 0:10 ratios of O11:A9 (3):N10:A10 (7) (Fig. 4). Further reflecting this discovery-based approach, the mixtures resulted in a parabolic expression trend, with the 5:5 mixture resulting in the maximum bioluminescence intensity, more than fourfold higher than either CART 3 or 7 alone.
Fig. 4.
Performance of binary mixtures of varying ratios of O11:A9 (3) and N10:A10 (7) for the delivery of Fluc mRNA into Jurkat cells. All values represent the average bioluminescence of three separate experiments with error bars corresponding to the SD.
Synthesis and Evaluation of Hybrid-Lipid CARTs.
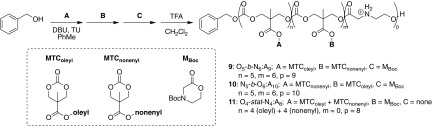
To study whether the enhanced uptake of the mixed CART–mRNA complexes into lymphocytes was due to solely noncovalent CART lipid mixes or whether covalent lipid mixtures would affect activity, we prepared several hybrid CARTs with lipid blocks of both nonenyl and oleyl side chains (Fig. 5). We prepared two different diblock CARTs of varying block order by OROP, with a third block being the self-immolating domain, yielding O5-b-N6:A9 (9) and N5-b-O6:A10 (10) after global TFA deprotection. Additionally, we prepared CART O4-stat-N4:A8 (11) consisting of a first block made up of a 1:1 statistical mixture of oleyl and nonenyl monomers, followed by the α-amino ester cationic block to collectively reflect the properties of the CART mixtures. These hybrid-lipid CARTs were fully characterized by NMR endgroup analysis and gel permeation chromatography (SI Appendix, Fig. S2).
Fig. 5.
Synthesis of CARTs containing mixtures of lipophilic blocks in either block (b) or statistical (stat) architectures. Monomer ratios were selected based on the optimized performance of a 50:50 mixture of O11:A9 (3) and N10:A10 (7).
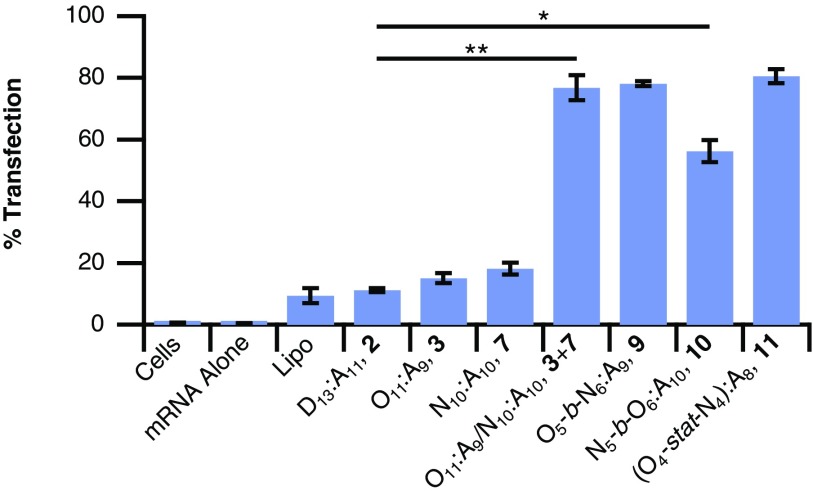
The hybrid-lipid CARTs 9-11 and the lead 1:1 noncovalent mixture of monoblock CARTs 3 and 7 were then assayed for EGFP mRNA delivery into Jurkat cells compared with 2 and commercial reagent Lipofectamine 2000 (Lipo). By flow cytometry, EGFP mRNA alone resulted in no GFP-positive cells and Lipo resulted in only 9% GFP-positive cells (Fig. 6). Both the 1:1 noncovalent mixture of CARTs 3 and 7 as well as the hybrid triblock O5-b-N6:A9 9 and statistical mixture 11 yielded 77–81% GFP-positive Jurkat cells, a striking sevenfold increase over the performance of CART 2 and an ninefold increase over that of Lipo (Fig. 6). Interestingly, CART triblock 10 had slightly lower GFP transfection, 56%, although it still significantly outperformed Lipo (P = 0.0004) and single-lipid CARTs. All hybrid-lipid CART–mRNA complexes tested in Jurkat cells resulted in cells with greater than 80% cell viability relative to untreated cells by a 3-[4,5-dimethylthiazole-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay (SI Appendix, Fig. S3).
Fig. 6.
Performance of noncovalent (3 + 7) and hybrid-lipid CARTs (9, 10, and 11) for the delivery of EGFP mRNA to Jurkat cells, as well as baseline performance of Lipo and single-lipid CARTs (2, 3, and 7). All values represent the average percent of EGFP-positive cells from three separate experiments with error bars corresponding to the SD. Unpaired Student’s t test: *P < 0.0005, **P < 0.0001.
Understanding the Increased Performance of Lipid Mixtures.
We conducted several experiments aimed at exploring possible factors that could contribute to the increased efficacy of the nonenyl and oleyl lipid CART mixtures and hybrid-lipid CARTs. We hypothesized that the increased mRNA expression by these lipid mixed CARTs and hybrid CARTs could be due to (i) increased efficiency in mRNA binding into polyplexes, (ii) increased mRNA uptake possibly due to differences in particle size, or (iii) differences in endosomal escape and/or mRNA release of the mixtures.
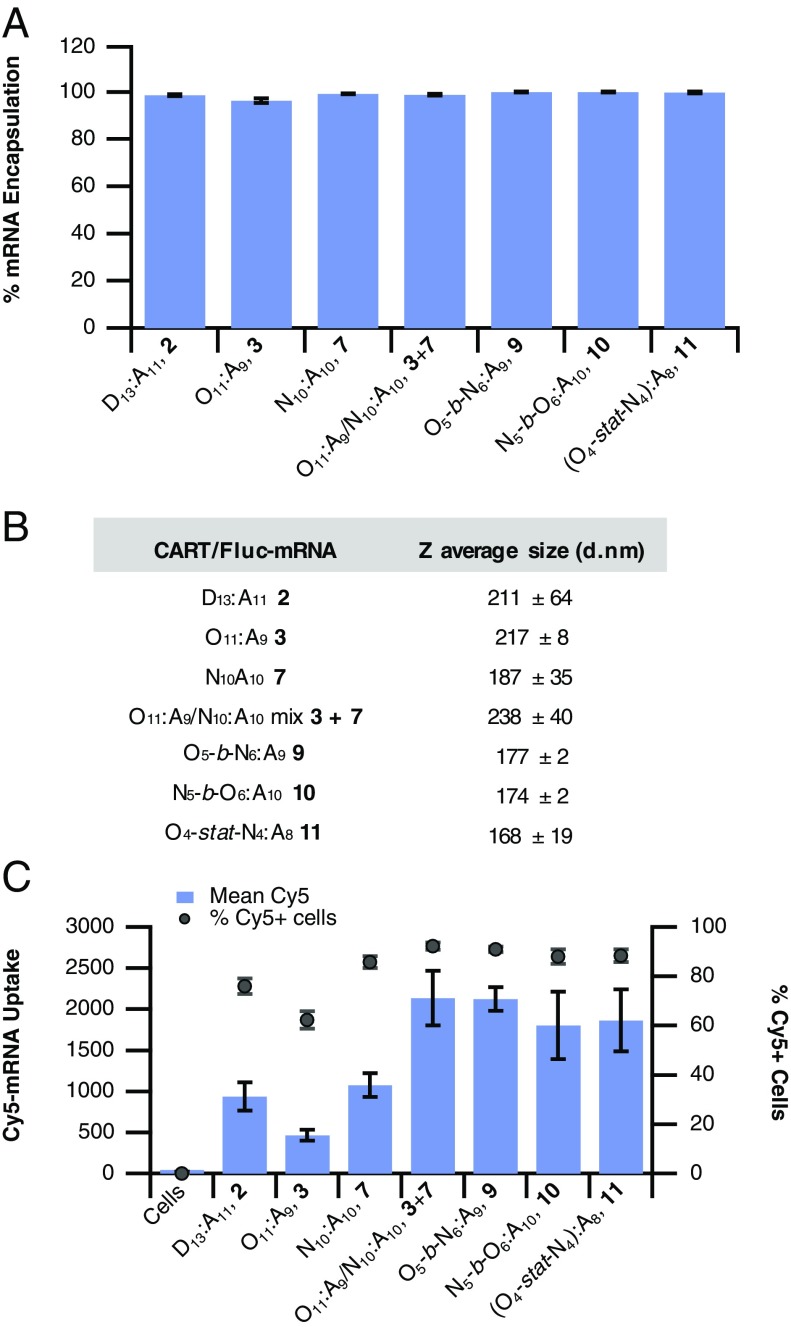
To evaluate the encapsulation efficiency of mRNA into lipid CART mixtures, we used a commercial dye (Qubit-RNA; Life Technologies) that shows significant fluorescence increase upon binding free mRNA but would not show fluorescence to polyplex bound mRNA due to lack of association. Using this probe system, we determined that all CART–mRNA complexes bind 96–99% of mRNA in the formulation (Fig. 7A), which is even higher than the top reported LNP formulations (33, 61, 62). However, this alone likely does not contribute to the increased efficacy of the mixed-lipid CARTs since single-lipid formulations and mixtures show nearly identical encapsulation efficiencies. This is consistent with delivery efficiencies in adherent cells that show similar performance of lipid mixtures and single CARTs (Fig. 3).
Fig. 7.
Evaluating the mechanism of enhanced mRNA delivery by lipid mixtures. (A) Percent encapsulation of free mRNA by CART complexes. Encapsulation is normalized to free mRNA (0%) and Qubit-mRNA dye alone (100%). (B) CART–mRNA complex particle sizes as determined by DLS. (C) Comparison of the uptake of a fluorescently labeled Cy5-mRNA by single-CART oligomer complexes versus lipid CART mixtures and hybrid-lipid CARTs in Jurkat cells. Data are reported as the mean Cy5 fluorescence of all cells measured (bars, left axis) or percentage of Cy5-positive cells (markers, right axis). All values are expressed as the average of three separate experiments with error bars corresponding to the SD.
Next, we used dynamic light scattering (DLS) to determine if the incorporation of multiple lipid domains in the formulation led to differences in particle sizes. Notably, all formulations showed consistent particle sizes from 177 to 238 nm with no obvious trend between delivery efficiency and size within that range, suggesting that particle size alone is likely not a major contributor to the efficacy of the lipid CART mixtures or hybrid-lipid CARTs (Fig. 7B). All particle size distributions were monomodal with low dispersity (SI Appendix, Fig. S4).
Finally, we used a fluorescently labeled mRNA to decouple mRNA uptake from the resulting gene expression. This enables the determination of whether CART mixtures show differences in mRNA uptake which results in their higher efficacy, or if similar amounts of mRNA are delivered compared with unmixed CARTs, suggesting that higher gene expression is a function of endosomal escape and/or mRNA release from CART complexes. To achieve this, we treated Jurkat cells with a panel of CARTs and CART mixtures formulated with Cy5-labeled EGFP mRNA. As expected, due to its limited efficacy for inducing gene expression, the three CARTs with single-lipid domains (dodecyl 2, oleyl 3, and nonenyl 7) showed limited levels of Cy5 mRNA uptake, with somewhat lower percentages of Cy5-positive cells (Fig. 7C). In contrast, both the noncovalent mixture of 3 and 7 and the CARTs containing both oleyl and nonenyl lipids (9–11) showed a nearly twofold increase in the total mRNA delivered, relative to single-lipid CARTs 2, 3, and 7. Taken together with the encapsulation data (Fig. 7A), this suggests that particles formed with the combination of these lipids are primarily more effective at getting into lymphocytes, such as Jurkat cells, rather than enhancing endosomal escape or mRNA release. The mechanism for poly(carbonate)-b-(α-amino ester) CART uptake is primarily endocytotic (34). Therefore, in addition to enhancing cell uptake, the presence of two unsaturated lipids could also lead to increased endosomalytic behavior and mRNA release through hexagonal phase formation, so partial contribution from these factors is also possible (6, 44, 63).
One explanation for the increase in efficiency of uptake for mixed-lipid CARTs is that the presence of two different unsaturated lipids (in this case oleyl and nonenyl) in the same particle significantly increases the fluidity of the particle’s lipid domains. These domains are hypothesized to help enhance membrane fusion of mRNA delivery vehicles and trigger endocytosis (64, 65). This effect may be especially pronounced in T cells, which are known to have low rates of endocytosis and do not fuse with amphipathic polyplexes as well as adherent cells (66).
In Vivo mRNA Delivery Using Hybrid-Lipid CARTs.
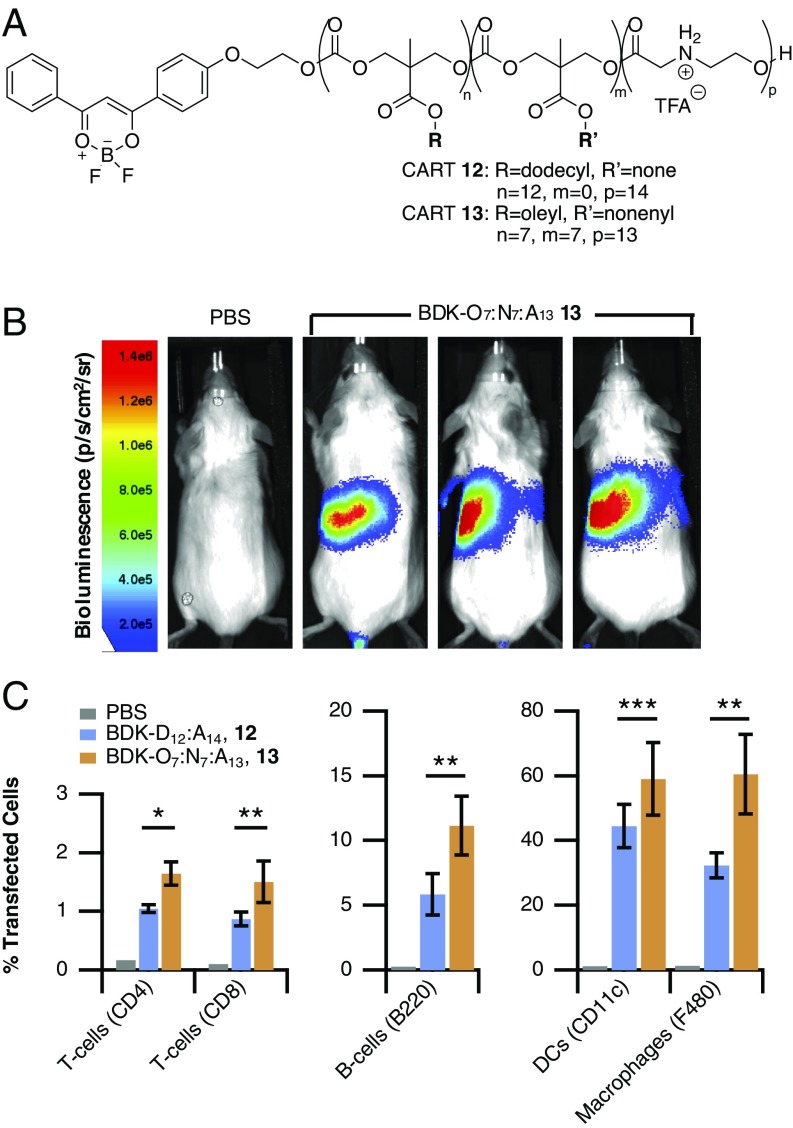
We demonstrated the in vivo efficacy of these mixed-lipid systems for mRNA delivery by delivering luciferase mRNA with fluorescently labeled CARTs and determining both the resulting gene expression by bioluminescence imaging and the transfection efficiency of spleenocytes by flow cytometry. Previous reports in the literature, and our own experience, suggest that the expression of fluorescent reporter genes is difficult to observe when determining cell uptake into specific spleenocyte subpopulations (16, 32, 33). Therefore, we used fluorescently labeled single-lipid CART 12 and hybrid-lipid CART 13 (Fig. 8A). These CARTs are easily prepared using a difluoroboron diketonate (BDK) alcohol (67) as the initiator for ring-opening polymerization. CART stoichiometry results in a nearly 1,000-fold molar excess of CART to mRNA, so the sensitivity of labeling the CART is significantly higher than using a labeled mRNA cargo as in other reports. We have confirmed that both mRNA uptake and resulting gene expression are directly correlated to BDK fluorescence using flow cytometry and that all cells that are positive for BDK-CART fluorescence are also positive for gene expression (SI Appendix, Fig. S5)
Fig. 8.
In vivo transfection of lymphocytes using mixed-lipid CARTs. (A) Structure of fluorescently labeled single-lipid CART 12 and hybrid-lipid CART 13. (B) Representative bioluminescence images following in vivo delivery of luciferase mRNA in BALB/c mice via tail vein injection by hybrid-lipid CART 13. (C) Transfection of lymphocytes and monocytes in the spleen following i.v. injection of CART 12–mRNA and CART 13–mRNA complexes. All values are expressed as the average of separate experiments (CART 12: n = 5; CART 13: n = 6 mice) with error bars corresponding to the SD (*P < 0.001, **P < 0.05, ***P < 0.01, unpaired Student’s t test).
Mice transfected with hybrid-lipid CART 13-mRNA showed high levels of Fluc gene expression localized primarily in the spleen (Fig. 8B), which is advantageous for lymphocyte transfection as those cells reside at significant levels in that organ. Hybrid-lipid CART–mRNA complexes generally showed higher levels of luciferase expression compared with the single-lipid-containing CART 12 (SI Appendix, Fig. S6).
After mice were imaged for luciferase expression they were killed and their spleens analyzed for CART transfection in T cells, B cells, macrophages, and dendritic cells. Higher percentages of transfected cells were observed with mice treated with the mixed-lipid CART relative to the single-lipid system (Fig. 8C and SI Appendix, Fig. S7). A significant increase in T-cell transfection was observed for CD4 T cells from ∼1.0% with CART 12 to 1.6% with CART 13, respectively. A similar trend was also observed in CD8 T cells, with hybrid-lipid CART 13 transfecting 1.5% of CD8 T cells in the spleen. Notably, transfection of T cells at this level has not been observed in vivo for similar mRNA delivery systems, which show well below 1% T-cell transfection (16, 32, 33), presumably due to the vast number of cells in the spleen. The mixed-lipid CART 13 also showed transfection of 11% of B cells, outperforming both CART 12 and previously reported LNP systems which transfect 1–7% of B cells (16, 32). Higher levels of transfection were also observed in monocyte populations of both dendritic cells and macrophages, a desirable characteristic for mRNA-based vaccination approaches.
Overall gene expression levels are higher in mice treated with the mixed-lipid CART, especially in lymphocyte populations, indicating that the optimization of lipid composition in a high-throughput in vitro screen can be used to inform the design of mixed-lipid systems with improved lymphocyte transfection in vivo.
Conclusions
The design and screening of a combinatorial library of 64 noncovalent CART lipid mixtures allowed us to identify a pair of CARTs, oleyl CART 3 and nonenyl CART 7, that are more effective for mRNA delivery than either CART alone for the delivery of mRNA into T and B lymphocytes. Informed by these results, we prepared hybrid-lipid CARTs 9 and 11 incorporating these same oleyl and nonenyl lipid components into a single CART and found them to exhibit similarly high transfection efficiencies (77–81%) in Jurkat cells, sixfold higher than either unmixed CART 3 or 7 alone and ninefold higher than the commercial agent Lipo. Additionally, although many physical transfection methods and cationic transporters lead to decreases in cell viability, cells treated with the CART mixtures maintained high viability along with the high transfection efficiency. Despite the fact that all CART complexes tested were similar in size (177–238 nm) and achieved a similar percent encapsulation, the amount of Cy5-labeled mRNA that entered cells was higher for mixed CARTs (e.g., 3 and 7 together) relative to their individual CART components (3 or 7 alone). These data suggest that both covalent and noncovalent CART mixtures containing both oleyl and nonenyl functionalities are more effective at transfecting lymphocytes than either single-lipid CART. In addition to establishing a high (80%) transfection efficiency in vitro using these mixed-lipid CARTs, we have further shown that these vehicles are effective for in vivo mRNA delivery of a reporter gene, outperforming the single-lipid CART for transfection of CD4 and CD8 T cells in the spleen (>1.5% versus <1%). As there is a great need for improved delivery methods to lymphocytes for both in vivo and ex vivo applications including cancer immunology and vaccinology, the combinatorial strategy reported herein provides a facile method to discover more effective transporter lipid combinations and hybrid-lipid transporters for a potentially wide range of polyanionic cargos. This study has further identified CART combinations and hybrid-lipid CARTs that exhibit high transfection efficiencies and minimal toxicity, providing tools for future biomedical research and therapeutic use.
Materials and Methods
Materials.
Reagents were purchased from Sigma-Aldrich and used as received unless otherwise indicated. The 1-(3,5-bis-trifluoromethyl-phenyl)-3-cyclohexyl-thiourea (51), lipid-functionalized monomers (43, 50), and Boc-morpholinone monomer (55) were all prepared according to literature procedures. Regenerated cellulose dialysis membranes (Spectra/Por 6 Standard RC; molecular weight cutoff 1,000) were purchased from Spectrum Laboratories, Inc. Lipofectamine 2000 was purchased from Life Technologies. MTT was purchased from Fluka.
mRNAs.
EGFP mRNA (5meC, Ψ, L-6101), Fluc mRNA (5meC, Ψ, L-6107), and Cy5-EGFP mRNA (5meC, Ψ, L-6402) were purchased from TriLink BioTechnologies Inc.
Instrumentation.
Particle size was measured by DLS on a Malvern Zetasizer Nano ZS90. Flow cytometry analysis was performed on a BD LSRII.UV FACS Analyzer (Stanford University Shared FACS Facility). Bioluminescence was measured using a CCD camera (IVIS 100; Xenogen Corp.) and analyzed using Living Image Software (PerkinElmer). Fluorescence measurements were taken on a Horiba Jobin-Yvon Spex Fluorolog-3 fluorimeter with a thermoelectrically controlled R928P detector (240–850 nm).
Cell Lines.
Jurkat, A20, and EL4 cells were maintained in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin. Nonadherent cells were treated with the addition of 1% poly(vinyl alcohol) (PVA) to prevent adhesion. For murine primary T cells, single-cell suspensions were prepared from spleens of 6- to 8-wk-old BALB/c mice. From these suspensions, untouched T cells were isolated using the Pan T cell Isolation kit II (Miltenyi Biotec GmbH) according to the manufacturer’s protocol. HeLa, DC2.4, and A549 cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were grown at 37 °C in a 5% CO2 atmosphere. Cells were passaged at ∼80% confluence.
Preparation of CARTs.
Single-lipid and hybrid-lipid CARTs were prepared as previously described (34, 43, 55). Detailed procedures and characterization can be found in SI Appendix.
High-Throughput Screen of Hybrid-Lipid CART–mRNA Complexes.
Nonadherent cells were seeded at 50,000 cells per well in 100 μL of serum-free RPMI media containing 0.1% PVA (∼75 kDa) in black 96-well plates. Adherent cells (seeded at 20,000 cells per well in DMEM) were allowed to adhere overnight while lymphocyte lines were transfected immediately. CART mixtures (in DMSO) were made up in a 96-well V-bottom plate by first diluting from 2 mM (with respect to chains) single-CART stock solutions to equal concentrations with respect to cationic monomers (final cation concentration of 94.4 nM). These were then mixed at 2:1 and 1:2 ratios by taking 3.33 μL of one CART solution and 6.66 μL of the second and mixing to obtain all 64 combinations. In a separate 96-well plate, 13 μL of a 5.15 ng/μL solution of Fluc mRNA in PBS (pH adjusted to 5.5 with HCl) was added. Complexes were formed by taking 2 μL of each CART mixture and mixing with mRNA for 20 s, then 4.46 μL of this solution was added to a black 96-well plate containing the cells to be transfected at a final concentration of 20 ng mRNA per well in 100 μL total volume of media containing 0.35 mg/mL d-luciferin. Cells were incubated with treatment for 8 h at 37 °C, then the resultant luminescence was measured using an IVIS 100 CCD camera. Data represent the average bioluminescence intensity of three experiments, each in triplicate, with error expressed as ±SD.
EGFP and Cy-5 EGFP mRNA Delivery and Expression in Jurkat Cells by Flow Cytometry.
Jurkat cells were seeded at 25,000 cells per well in 100 μL serum-free RPMI media containing 0.1% PVA in 96-well plates. CART mixtures and CART–mRNA polyplexes were prepared as described above adjusted to a total treatment dose of 100 ng EGFP or Cy5-EGFP mRNA. The Lipo control was prepared in OptiMEM per the manufacturer’s instructions. After formulation, 22.5 μL of the CART–mRNA complexes was added to a total volume of 100 μL, all conditions in triplicate, for a final mRNA concentration of 100 ng per well. The cells were incubated for 8 h at 37 °C then resuspended in serum-containing media and centrifuged, the supernatant was removed, and the pelleted cells were redispersed in PBS (125 μL) and transferred to FACS tubes, and read on a flow cytometry analyzer (LSR-II.UV, Stanford University). The data presented are the mean fluorescent signals from 10,000 cells analyzed. For transfection efficiency, untreated cells were gated for no EGFP expression, and the data presented are the percentage of 10,000 cells analyzed with higher EGFP expression than untreated cells. Error is expressed as ±SD.
MTT Viability Assay.
Cellular viability following treatment with CART–mRNA complexes was assayed using a standard MTT/formazan turnover assay. Briefly, Jurkat cells were treated with CART–mRNA complexes at the concentrations used in the combinatorial screen (20 ng mRNA per well) or flow cytometry experiments (100 ng mRNA per well). Cells were grown for 3 d, after which time 2 μL of a 5 mg/mL solution of MTT (Fluka) was added. Cells were incubated for an additional 4 h then lysed and absorbance measured at 570 nm. Viability was determined by dividing the absorbance of treated samples by the average absorbance of untreated cells.
Percent Encapsulation Determination by Qubit Probe.
Encapsulation efficiency was measured by the fluorescence of the RNA-specific Qubit RNA HS dye (Q32852; Invitrogen ). Briefly, mRNA was complexed with a single CART or CART mixtures as described above using 840 ng of EGFP mRNA and enough CART for a net 10:1 (cation:anion) ratio. This was added to 0.75 mL of RNase-free deionized water containing 5 μL of Qubit reagent. The fluorescence of the solution was immediately measuring using an excitation wavelength of 630 (4-nm slit width) and emission wavelength of 680 nm (5-nm slit width). Percent encapsulation was determined by subtracting the fluorescence of the Qubit dye alone (no mRNA) and normalizing to the fluorescence of Qubit with uncomplexed mRNA.
DLS.
CART–mRNA complexes were prepared at a 10:1 (cation:anion) charge ratio as above using 210 ng Fluc mRNA in PBS pH 5.5, mixed for 20 s, then added to 390 μL RNase-free water. The solution was immediately transferred to a disposable clear plastic cuvette and the size measured. The sizes reported are the Z-averages of measurements run in triplicate with error expressed as ±SD.
In Vivo Fluc mRNA Delivery and Flow Cytometry.
BDK-CART–mRNA polyplexes were prepared as described above using 20 μg mRNA per mouse (3 μg Fluc mRNA and 17 μg irrelevant mRNA) in a total volume of 150 μL. Complexes were mixed for 20 s before i.v. injection into tail vain of female BALB/c mice. After 7 h, 150 mg/kg d-luciferin was injected i.p. and luminescence was measured using an AMI Imaging System CCD camera and analyzed using AMI View software. For spleenocyte transfection analysis, animals were killed and single-cell suspensions were prepared by passing the spleens through 70-μm cell strainers followed by lysis of blood cells with ACK Lysing Buffer and washing two times with PBS. Spleenocytes were partitioned into two staining groups and stained fluorescent antibodies for CD8 (APC), CD4 (PE), and B220 (FITC) or F480 (APC), CD11b (FITC), and CD11c (PE). Fluorescence of BDK-CART transfection in subpopulations was detected was detected on LSR-II.UV (BD Biosciences) using the Pacific Blue channel. Experimental protocols were approved by the Stanford Administrative Panel on Laboratory Animal Care.
Supplementary Material
Acknowledgments
We thank Prof. Chris Contag and Prof. Sanjiv Sam Gambhir for materials, tissue culture equipment, and use of the IVIS system; Prof. Richard Zare for the use of the Malvern Zetasizer DLS; Prof. Chaitan Khosla for use of HeLa cells; and Dr. Timothy Blake and Prof. Ronald Levy for thoughtful advice and discussions. This work was supported by Department of Energy Grant DE-SC0018168 and NSF Grant CHE-1607092 (to R.M.W.) and NIH Grants NIH-CA031841 and NIH-CA031845 (to P.A.W.). Support through the Stanford Center for Molecular Analysis and Design (C.J.M.), the Child Health Research Institute, the SPARK program at Stanford University, and the Norwegian Cancer Society (O.A.H.) is acknowledged. Flow cytometry data were collected on an instrument in the Stanford Shared FACS Facility obtained using NIH S10 Shared Instrument Grant S10RR027431-01.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805358115/-/DCSupplemental.
References
- 1.Friedmann T. A brief history of gene therapy. Nat Genet. 1992;2:93–98. doi: 10.1038/ng1092-93. [DOI] [PubMed] [Google Scholar]
- 2.Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012–An update. J Gene Med. 2013;15:65–77. doi: 10.1002/jgm.2698. [DOI] [PubMed] [Google Scholar]
- 3.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 4.Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics–Developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 5.Meng Z, et al. A new developing class of gene delivery: Messenger RNA-based therapeutics. Biomater Sci. 2017;5:2381–2392. doi: 10.1039/c7bm00712d. [DOI] [PubMed] [Google Scholar]
- 6.Guan S, Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 7.Kauffman KJ, Webber MJ, Anderson DG. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J Control Release. 2016;240:227–234. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 8.Reichmuth AM, Oberli MA, Jeklenec A, Langer R, Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X, Fricke J, Kavanagh DG, Irvine DJ. In vitro and in vivo mRNA delivery using lipid-enveloped pH-responsive polymer nanoparticles. Mol Pharm. 2011;8:774–787. doi: 10.1021/mp100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan Y, Xiong H, Zhang X, Cheng Q, Siegwart DJ. Systemic mRNA delivery to the lungs by functional polyester-based carriers. Biomacromolecules. 2017;18:4307–4315. doi: 10.1021/acs.biomac.7b01356. [DOI] [PubMed] [Google Scholar]
- 11.Li J, et al. Structurally programmed assembly of translation initiation nanoplex for superior mRNA delivery. ACS Nano. 2017;11:2531–2544. doi: 10.1021/acsnano.6b08447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, et al. Polyamine-mediated stoichiometric assembly of ribonucleoproteins for enhanced mRNA delivery. Angew Chem Int Ed Engl. 2017;56:13709–13712. doi: 10.1002/anie.201707466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itaka K, Ishii T, Hasegawa Y, Kataoka K. Biodegradable polyamino acid-based polycations as safe and effective gene carrier minimizing cumulative toxicity. Biomaterials. 2010;31:3707–3714. doi: 10.1016/j.biomaterials.2009.11.072. [DOI] [PubMed] [Google Scholar]
- 14.Matsui A, Uchida S, Ishii T, Itaka K, Kataoka K. Messenger RNA-based therapeutics for the treatment of apoptosis-associated diseases. Sci Rep. 2015;5:15810. doi: 10.1038/srep15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchida H, et al. Modulated protonation of side chain aminoethylene repeats in N-substituted polyaspartamides promotes mRNA transfection. J Am Chem Soc. 2014;136:12396–12405. doi: 10.1021/ja506194z. [DOI] [PubMed] [Google Scholar]
- 16.Fenton OS, et al. 2017. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv Mater, 29.
- 17.Ebert O, et al. Lymphocyte apoptosis: Induction by gene transfer techniques. Gene Ther. 1997;4:296–302. doi: 10.1038/sj.gt.3300394. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, Ott PA, Wu CJ. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat Rev Immunol. 2018;18:168–182. doi: 10.1038/nri.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines–A new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fesnak AD, June CH, Levine BL. Engineered T cells: The promise and challenges of cancer immunotherapy. Nat Rev Cancer. 2016;16:566–581. doi: 10.1038/nrc.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, et al. High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol Ther. 2006;13:151–159. doi: 10.1016/j.ymthe.2005.07.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Rivière I. Clinical manufacturing of CAR T cells: Foundation of a promising therapy. Mol Ther Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine BL, Miskin J, Wonnacott K, Keir C. Global manufacturing of CAR T cell therapy. Mol Ther Methods Clin Dev. 2016;4:92–101. doi: 10.1016/j.omtm.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chicaybam L, Sodre AL, Curzio BA, Bonamino MH. An efficient low cost method for gene transfer to T lymphocytes. PLoS One. 2013;8:e60298. doi: 10.1371/journal.pone.0060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landry B, et al. Effective non-viral delivery of siRNA to acute myeloid leukemia cells with lipid-substituted polyethylenimines. PLoS One. 2012;7:e44197. doi: 10.1371/journal.pone.0044197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.deRonde BM, Torres JA, Minter LM, Tew GN. Development of guanidinium-rich protein mimics for efficient siRNA delivery into human T cells. Biomacromolecules. 2015;16:3172–3179. doi: 10.1021/acs.biomac.5b00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao N, et al. Transfecting the hard-to-transfect lymphoma/leukemia cells using a simple cationic polymer nanocomplex. J Control Release. 2012;159:104–110. doi: 10.1016/j.jconrel.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith TT, et al. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat Nanotechnol. 2017;12:813–820. doi: 10.1038/nnano.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tezgel AÖ, et al. Novel protein transduction domain mimics as nonviral delivery vectors for siRNA targeting NOTCH1 in primary human T cells. Mol Ther. 2013;21:201–209. doi: 10.1038/mt.2012.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, et al. T cell-specific siRNA delivery using antibody-conjugated chitosan nanoparticles. Bioconjug Chem. 2012;23:1174–1180. doi: 10.1021/bc2006219. [DOI] [PubMed] [Google Scholar]
- 31.Schallon A, Synatschke CV, Jérôme V, Müller AHE, Freitag R. Nanoparticulate nonviral agent for the effective delivery of pDNA and siRNA to differentiated cells and primary human T lymphocytes. Biomacromolecules. 2012;13:3463–3474. doi: 10.1021/bm3012055. [DOI] [PubMed] [Google Scholar]
- 32.Kranz LM, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 33.Oberli MA, et al. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKinlay CJ, et al. Charge-altering releasable transporters (CARTs) for the delivery and release of mRNA in living animals. Proc Natl Acad Sci USA. 2017;114:E448–E456. doi: 10.1073/pnas.1614193114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballarín-González B, Howard KA. Polycation-based nanoparticle delivery of RNAi therapeutics: Adverse effects and solutions. Adv Drug Deliv Rev. 2012;64:1717–1729. doi: 10.1016/j.addr.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Zylberberg C, Gaskill K, Pasley S, Matosevic S. Engineering liposomal nanoparticles for targeted gene therapy. Gene Ther. 2017;24:441–452. doi: 10.1038/gt.2017.41. [DOI] [PubMed] [Google Scholar]
- 38.Sergeeva OV, Koteliansky VE, Zatsepin TS. mRNA-based therapeutics–Advances and perspectives. Biochemistry (Mosc) 2016;81:709–722. doi: 10.1134/S0006297916070075. [DOI] [PubMed] [Google Scholar]
- 39.Ghanbari Safari M, Hosseinkhani S. Lipid composition of cationic nanoliposomes implicate on transfection efficiency. J Liposome Res. 2013;23:174–186. doi: 10.3109/08982104.2013.779703. [DOI] [PubMed] [Google Scholar]
- 40.Du Z, Munye MM, Tagalakis AD, Manunta MDI, Hart SL. The role of the helper lipid on the DNA transfection efficiency of lipopolyplex formulations. Sci Rep. 2014;4:1–6. doi: 10.1038/srep07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gehin C, et al. Dynamic amphiphile libraries to screen for the “fragrant” delivery of siRNA into HeLa cells and human primary fibroblasts. J Am Chem Soc. 2013;135:9295–9298. doi: 10.1021/ja404153m. [DOI] [PubMed] [Google Scholar]
- 42.Priegue JM, et al. In situ functionalized polymers for siRNA delivery. Angew Chem Int Ed Engl. 2016;55:7492–7495. doi: 10.1002/anie.201601441. [DOI] [PubMed] [Google Scholar]
- 43.Geihe EI, et al. Designed guanidinium-rich amphipathic oligocarbonate molecular transporters complex, deliver and release siRNA in cells. Proc Natl Acad Sci USA. 2012;109:13171–13176. doi: 10.1073/pnas.1211361109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, et al. Effects of local structural transformation of lipid-like compounds on delivery of messenger RNA. Sci Rep. 2016;6:22137. doi: 10.1038/srep22137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love KT, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, et al. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fenton OS, et al. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caffrey LM, deRonde BM, Minter LM, Tew GN. Mapping optimal charge density and length of ROMP-based PTDMs for siRNA internalization. Biomacromolecules. 2016;17:3205–3212. doi: 10.1021/acs.biomac.6b00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pratt RC, Nederberg F, Waymouth RM, Hedrick JL. 2008. Tagging alcohols with cyclic carbonate: A versatile equivalent of (meth)acrylate for ring-opening polymerization. Chem Commun (Camb), 114–116. [DOI] [PubMed]
- 51.Dove AP, Pratt RC, Lohmeijer BGG, Waymouth RM, Hedrick JL. Thiourea-based bifunctional organocatalysis: Supramolecular recognition for living polymerization. J Am Chem Soc. 2005;127:13798–13799. doi: 10.1021/ja0543346. [DOI] [PubMed] [Google Scholar]
- 52.Kiesewetter MK, Shin EJ, Hedrick JL, Waymouth RM. Organocatalysis: Opportunities and challenges for polymer synthesis. Macromolecules. 2010;43:2093–2107. [Google Scholar]
- 53.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lostalé-Seijo I, Louzao I, Juanes M, Montenegro J. Peptide/Cas9 nanostructures for ribonucleoprotein cell membrane transport and gene edition. Chem Sci (Camb) 2017;8:7923–7931. doi: 10.1039/c7sc03918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blake TR, Waymouth RM. Organocatalytic ring-opening polymerization of morpholinones: New strategies to functionalized polyesters. J Am Chem Soc. 2014;136:9252–9255. doi: 10.1021/ja503830c. [DOI] [PubMed] [Google Scholar]
- 56.Benner NL, et al. Functional DNA delivery enabled by lipid-modified charge-altering releasable transporters (CARTs) Biomacromolecules. May 11, 2018 doi: 10.1021/acs.biomac.8b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pozzi D, et al. Transfection efficiency boost of cholesterol-containing lipoplexes. Biochim Biophys Acta. 2012;1818:2335–2343. doi: 10.1016/j.bbamem.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 58.McManus MT, et al. Small interfering RNA-mediated gene silencing in T lymphocytes. J Immunol. 2002;169:5754–5760. doi: 10.4049/jimmunol.169.10.5754. [DOI] [PubMed] [Google Scholar]
- 59.Miller JB, et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv Drug Deliv Rev. 2006;58:1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 61.Patel S, et al. Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano Lett. 2017;17:5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 63.Koltover I, Salditt T, Rädler JO, Safinya CR. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 64.Keller H, et al. Transgene expression, but not gene delivery, is improved by adhesion-assisted lipofection of hematopoietic cells. Gene Ther. 1999;6:931–938. doi: 10.1038/sj.gt.3300896. [DOI] [PubMed] [Google Scholar]
- 65.Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci USA. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Labat-Moleur F, et al. An electron microscopy study into the mechanism of gene transfer with lipopolyamines. Gene Ther. 1996;3:1010–1017. [PubMed] [Google Scholar]
- 67.Zhang G, et al. Multi-emissive difluoroboron dibenzoylmethane polylactide exhibiting intense fluorescence and oxygen-sensitive room-temperature phosphorescence. J Am Chem Soc. 2007;129:8942–8943. doi: 10.1021/ja0720255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.