Significance
The existence of a chemosynthetic subseafloor biosphere was immediately recognized when deep-sea hot springs were discovered in 1977. However, quantifying how much new carbon is fixed in this environment has remained elusive. In this study, we incubated natural subseafloor communities under in situ pressure/temperature and measured their chemosynthetic growth efficiency and metabolic rates. Combining these data with fluid flux and in situ chemical measurements, we derived empirical constraints on chemosynthetic activity in the natural environment. Our study shows subseafloor microorganisms are highly productive (up to 1.4 Tg C produced yearly), fast-growing (turning over every 17–41 hours), and physiologically diverse. These estimates place deep-sea hot springs in a quantitative framework and allow us to assess their importance for global biogeochemical cycles.
Keywords: deep-sea hydrothermal vents, chemosynthesis, Campylobacteria, ecophysiology, NanoSIMS
Abstract
Below the seafloor at deep-sea hot springs, mixing of geothermal fluids with seawater supports a potentially vast microbial ecosystem. Although the identity of subseafloor microorganisms is largely known, their effect on deep-ocean biogeochemical cycles cannot be predicted without quantitative measurements of their metabolic rates and growth efficiency. Here, we report on incubations of subseafloor fluids under in situ conditions that quantitatively constrain subseafloor primary productivity, biomass standing stock, and turnover time. Single-cell-based activity measurements and 16S rRNA-gene analysis showed that Campylobacteria dominated carbon fixation and that oxygen concentration and temperature drove niche partitioning of closely related phylotypes. Our data reveal a very active subseafloor biosphere that fixes carbon at a rate of up to 321 μg C⋅L−1⋅d−1, turns over rapidly within tens of hours, rivals the productivity of chemosynthetic symbioses above the seafloor, and significantly influences deep-ocean biogeochemical cycling.
In 1977, the discovery of deep-sea hot springs revealed unusual ecosystems vastly more productive than other regions in the energy-limited deep sea (1). This productivity is sustained by chemosynthetic microorganisms that harness chemical energy made available when oxidizing seawater and reducing hydrothermal fluid mix. It has long been recognized that the habitat for such organisms may extend far below the seafloor to vast regions of the ocean crust where fluid mixing takes place (1). Fluids exiting this subseafloor biosphere are enriched in microbial biomass relative to surrounding seawater (2) and contain active microorganisms (2–4) that are physiologically and metabolically diverse (5–7). Despite the early realization that subseafloor ecosystems likely contribute significantly to overall chemosynthetic primary productivity (1) and provide nutrition to the surrounding food-limited deep sea, their extent, productivity, and biological dynamics remain poorly constrained (1, 2, 8). It has been generally assumed that above-seafloor production (i.e., microbe-animal symbiotic associations) exceeds production below the seafloor (9). However, this assumption has not been rigorously tested by empirically quantifying subseafloor productivity.
Quantifying subseafloor productivity at submarine hot springs requires knowledge of both the amount of chemical energy that can be supplied by hydrothermal fluids and the efficiency by which microbial communities convert this chemical energy into biomass. Although the chemical composition of hydrothermal fluids is well-described and provides strong indirect evidence for high biological activity in the subseafloor (10, 11), the actual amount of carbon fixed in situ is highly uncertain. Although in situ growth yields can be estimated from pure cultures (12), results from laboratory cultures may not be relevant to complex and largely uncultivated natural communities growing under different physical and chemical conditions. Bulk carbon fixation rates reported for mixed subseafloor microbial communities could likewise be used to constrain subseafloor primary productivity, but only one such measurement has been previously obtained under realistic temperature and pressure conditions (4). Another important consideration is that electron acceptors such as oxygen and nitrate rapidly become limiting during incubation experiments with vent fluids (13, 14), which may lead to carbon fixation rates being greatly underestimated (13). However, it is difficult to ascertain the extent of this bias for existing studies because electron acceptor consumption has not typically been measured alongside carbon fixation. Theoretical estimates of primary productivity have also been derived by combining geochemical measurements with thermodynamic models (15, 16). However, these studies rely on a number of untested assumptions necessary to convert the available energy into biomass.
To overcome these limitations, we used a well-studied low-temperature hot spring known as Crab Spa at the 9°N hydrothermal vent field on the East Pacific Rise (EPR) as a model system to constrain subseafloor chemosynthetic production. Data from incubations conducted under simulated in situ conditions were combined with in situ chemistry and fluid flow rate (14, 17) to arrive at empirical estimates of subseafloor primary productivity in a deep-sea hydrothermal system.
Results and Discussion
Fluids emanating from the subseafloor at Crab Spa are characterized by a temperature of ∼24 °C, microbial abundances substantially elevated relative to bottom seawater, and a chemical composition for most aqueous species that reflects formation by conservative subseafloor mixing of the high-temperature end-member fluid with seawater (14). Dissolved concentrations of redox reactive sulfide, hydrogen, nitrate, and oxygen, however, are substantially depleted relative to values expected for conservative mixing (Table 1), indicating microbial consumption below the seafloor (10, 11, 14). Sixteen independent incubation experiments with Crab Spa fluids were conducted at in situ pressure and temperature (24 °C, ∼246 bar) for ∼24 h, using isobaric gas-tight (IGT) fluid samplers (18); an additional two samples were incubated at in situ pressure and an elevated temperature of 50 °C. For all experiments, natural fluids were amended with NaH13CO3− as a tracer to measure/assess autotrophic carbon fixation. Three experiments were left unmodified as controls, whereas the remaining 15 received additions of nitrate, oxygen, hydrogen, or combinations of these substrates (14). This approach allowed us to quantify the rates and the stoichiometry of redox reactions supporting microbial metabolism and the resulting inorganic carbon fixed into biomass (14). From these measured parameters, we directly calculated the efficiency of new biomass production of the active communities. This value was then combined with measurements of the Crab Spa vent fluid chemistry to constrain the efficiency of energy conversion into biomass in situ. We also explored how variations in environmental parameters influenced primary productivity and microbial community composition.
Table 1.
Predicted versus observed concentrations of potential electron donors and acceptors at Crab Spa
| H2S, µmol/L | H2, µmol/L | CH4, µmol/L | NH4+, µmol/kg | O2, µmol/kg | NO3−, µmol/L | SO42−, mmol/kg | |
| Predicted* | 552 | 29 | 8.1 | 0.2 | 107 | 32 | 25.8 |
| Observed | 184 | <2 | 6.3 | 11.9 | 3.6 | 6.3 | 26.5 |
Units are given either per liter or kilogram of Crab Spa vent fluid.
Values are based on fluid mixing calculations previously described in ref. 14.
Microbial Community Composition and Rate Measurements.
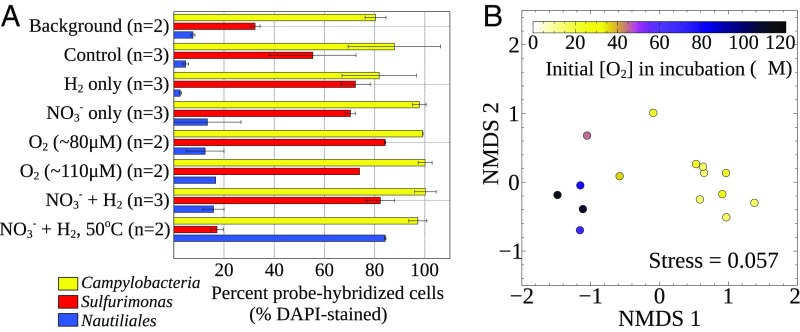
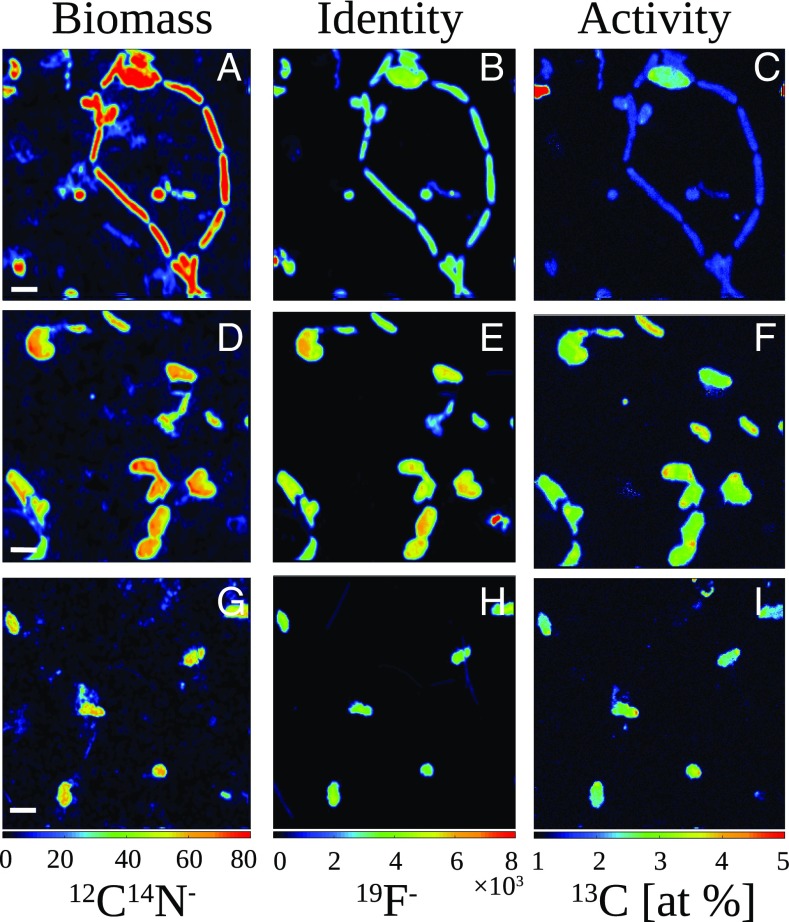
In all incubations, Campylobacteria (7) were the dominant microorganisms, as shown by catalyzed-reporter deposition fluorescence in situ hybridization (CARD-FISH) cell counts [Fig. 1A and SI Appendix, Table S1; 94 ± 11% of total cells (14)] and the proportion of 16S rRNA gene sequences (97 ± 3.7%; SI Appendix, Fig. S1). Identified sequences were related to known chemolithoautotrophs, and a pronounced switch from known mesophilic to thermophilic Campylobacteria occurred in the incubation at 50 °C (Fig. 1A and SI Appendix, Fig. S1 and Table S1). Active carbon fixation was confirmed by specifically measuring H13CO3− tracer incorporation in campylobacterial cells with halogen in situ hybridization-nanoscale secondary ion mass spectrometry [HISH-SIMS (19); Fig. 2]. HISH-SIMS also demonstrated that amendments increased relative CO2 fixation rates, especially for oxygen or a combination of nitrate and hydrogen (Fig. 3). Given that Campylobacteria dominate the natural community in sampled fluids (∼80% of total cells; Fig. 1A and ref. 14) and make up the vast majority of microbes found in the incubations (Fig. 1A and SI Appendix, Fig. S1 and ref. 14), we conclude this group dominates primary productivity in the chronically electron acceptor-limited subseafloor environment at Crab Spa (14), and likely also at other deep-sea vent sites similarly dominated by Campylobacteria (5, 6).
Fig. 1.
Bacterial community composition at the end of incubations. (A) Taxonomic composition inferred from CARD-FISH counts, and (B) Nonmetric multidimensional scaling (NMDS) plot showing the similarity of Sulfurimonas 97% OTU composition between experimental treatments. Each dot represents a different biological replicate for incubations carried out at 24 °C and is colored according to the initial PO2. All CARD-FISH data are averaged by treatment, and errors are presented as SDs (n = 3) or ranges (n = 2) except for the Nautiliales probe in the 110 μM O2 treatment (n = 1). Validation of newly designed probes (Nautiliales = NAUT921 and Sulfurimonas = SFMN287; SI Appendix, Table S2) are described in the Materials and Methods, and specificity tests are shown in SI Appendix, Figs. S5 and S6. Campylobacteria in A corresponds to the combined probes EPSI549 and EPSI914.
Fig. 2.
Metabolic activity of Campylobacteria cells from Crab Spa fluids after short-term incubations at in situ pressure as quantified by HISH-SIMS. Rows represent different experimental treatments as follows: (A–C) control treatment (10% H13CO3−) and (D–I) oxygen amendments (110 μM O2 + 10% H13CO3−). Cells were hybridized with general Campylobacteria probe (A–F) and with a specific Nautiliales probe (G–I), using Fluorine-containing tyramides. Columns display parallel secondary ion images of 12C14N as total biomass indicator (A, D, and G), 19F as a marker for cell identity (B, E, and H) and the 13C enrichment inferred from secondary ions (13C−, 12C−) given as atomic percentage [100 × 13C/(12C + 13C; at %)], as indicator of cell activity. [Scale bar, 2 μm (A–F) and 3 μm (G–I).]
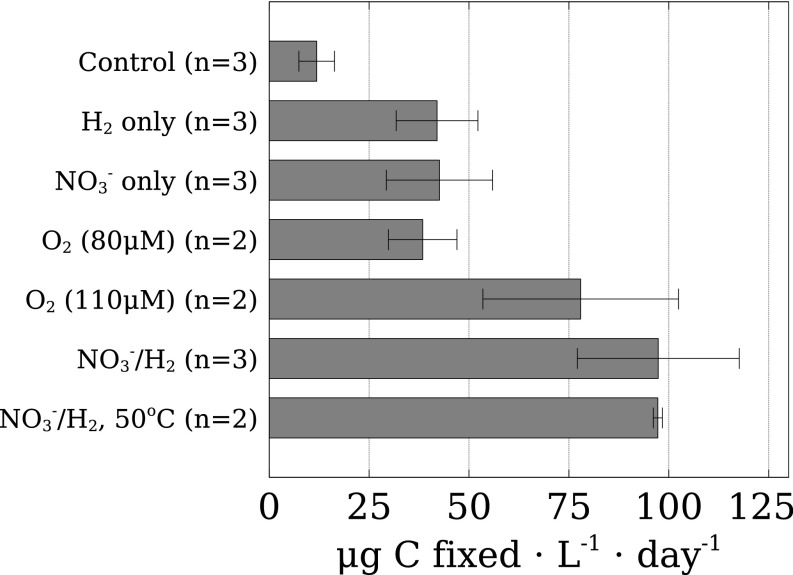
Fig. 3.
Relative estimations of primary productivity in incubations of hydrothermal vent fluids at in situ temperature and pressure determined by HISH-SIMS. Bars represent relative volumetric rates of campylobacterial CO2 assimilation during incubations. Errors are SDs (n = 3) or ranges (n = 2). Values are not corrected for the influence of CARD-FISH procedure (20).
Because CARD-FISH underestimates carbon fixation as a result of tracer loss during sample preparation (20), we also quantified 13CO2 incorporation into bulk microbial biomass by elemental analysis-isotope ratio mass spectrometry (EA-IRMS) (19). Bulk carbon incorporation was, on average, 45% higher than HISH-SIMS, consistent with previous estimates of tracer loss (20). Carbon fixation rates were consistently high (∼17–320 μg C⋅L−1⋅d−1; Table 2 and SI Appendix, Fig. S2), far exceeding values reported from previous microbial incubations of hydrothermal fluids by factors of ∼2–650 (3, 4). This likely reflects the fact that our sampling and incubation approach minimized electron acceptor limitation (13) and changes to the physicochemical environment experienced by microbes (14). The carbon fixation rates reported here are between two and five orders of magnitude higher than carbon production by aerobic ammonia and nitrite oxidation, the two main chemoautotrophic processes in the dark pelagic ocean (21, 22), and are in the range of rates measured for carbon production in the photic zone of the coastal and open ocean (23).
Table 2.
Constraints on subseafloor hydrothermal vent productivity and standing stock from measurements of microbial CGE during incubations at in situ temperature and pressure
| Parameter | Lower bound | Upper bound | Units |
| Absolute carbon fixation rates* | 17.3 | 321.4 | μg C⋅L−1⋅d−1 |
| Chemosynthetic growth efficiency* | 0.06 | 0.13 | Fraction electron equivalents to carbon fixation |
| Estimated in situ carbon fixation† | |||
| Per liter Crab Spa mixed fluid | 104 | 253 | μg C⋅L−1 |
| Per liter Crab Spa end-member fluid | 1.4 × 103 | 3.5 × 103 | |
| Estimated areal animal-microbe symbiotic productivity (area colonized by animals at Crab Spa is ∼1 m2)‡ | 1.25 × 103 | 1.13 × 104 | g C⋅m2⋅y−1 |
| Estimated annual productivity§ | |||
| Crab Spa vent¶ | 6.1 × 103 | 1.5 × 104 | g C⋅y−1 |
| Vent field# | 3.8 × 106 | 9.3 × 106 | |
| Global diffuse-flow vents‖ | 4.5 × 1010 | 1.4 × 1012 | |
| Standing stock**, Crab Spa | 28.6 | NA | g C |
| Biomass residence time††, Crab Spa | 17 | 41 | hours |
| Global standing stock‖ | 1.4 × 109 | 2.7 × 109 | g C |
Derived from incubations at in situ pressure and temperature.
Based on hydrothermal fluid chemical depletions in situ and CGE estimates.
See SI Appendix, SI Text and references cited therein.
Based on fluid flux measurements and estimations of in situ carbon fixation in Crab Spa mixed fluids.
Based on fluid flux of 1.86 L⋅s−1 reported in ref. 17.
Based on fluid flux estimated in ref. 26.
Based on estimates of diffuse-flow fluid flux from ref. 30.
Based on rates of microbial consumption of oxygen and nitrate (14).
Assuming steady state.
Chemosynthetic Growth Efficiency.
Because substrate amendments influenced chemosynthetic activity during incubations (Figs. 2 and 3 and SI Appendix, Fig. S2), we normalized activity to changes in fluid composition (Dataset S1) to determine the amount of carbon fixed per electron acceptor reduced. This parameter, which we term community chemosynthetic growth efficiency (CGE), is the fraction of electron equivalents derived from electron donors used to reduce CO2 into biomass:
| [1] |
where EqCFIX = electron equivalents to carbon fixation and EqDISS = electron equivalents to dissimilatory electron acceptors.
CGE values are a direct empirical constraint on how efficiently hydrothermal vent microbes convert available chemical potential energy into biomass, and also demonstrate which conditions have the largest effect on their ecophysiology. As shown by 16S rRNA gene amplicon data (SI Appendix, Fig. S1) and CARD-FISH counts (Fig. 1A and SI Appendix, Table S1), the majority of organisms present in our incubations were distantly related to the campylobacterial genera Sulfurimonas and Thioreductor (∼93% 16S similarity to cultivars for most Sulfurimonas sequences and 92% for Thioreductor). Within these groups, the 97% operational taxonomic unit (OTU)-level community composition was markedly influenced by temperature (SI Appendix, Fig. S3) and initial oxygen tensions (Fig. 1B). Higher temperatures and oxygen tensions also resulted in an overall lower CGE (SI Appendix, Fig. S4), suggesting these conditions can be tolerated by Campylobacteria, but at a metabolic cost. This tradeoff between tolerance to environmental insult and growth efficiency may be important for driving niche partitioning in situ, where a large number of distinct physicochemical niches exist across steep mixing gradients (24).
Constraints on the Productivity, Turnover, and Standing Stock of the Deep-Sea Vent Subseafloor Biosphere.
With knowledge of chemical depletions and fluid flux, CGE provides a foundation to quantitatively constrain the extent, productivity, and biological dynamics of deep-sea vent subseafloor ecosystems. Using treatment-averaged CGE as upper and lower bounds (0.06–0.13), we estimate that 104–253 μg C is fixed per liter of mixed fluid at our study site. This is equivalent to 1.4–3.5 mg C fixed per liter of end-member hydrothermal fluid (Table 2), which comprises ∼7% of Crab Spa mixed fluids (14). Considering fluid flux (17), annual production at Crab Spa amounts to 6.1–15 kg C (Table 2). This is comparable to the amount of carbon likely fixed in the ∼1 m2 area of Crab Spa colonized by giant tubeworms (Riftia pachyptila), which are the dominant megafaunal species on the East Pacific Rise and the most productive symbiosis described to date (ref. 25 and Table 2 and SI Appendix, SI Text).
Our results can also be used to provide an independent minimum bound on subseafloor biomass standing stock by assuming that cell-specific rates of nitrate and oxygen respiration we reported previously are maximum values for in situ microbial communities [on average, 463 and 70 fmol/cell/d for O2 and NO3−, respectively (14)]. Combining chemical depletions of oxygen/nitrate at Crab Spa (14) with fluid flux (17), we determined total chemical consumption resulting from subseafloor microbial metabolism and the minimum number of cells (1 × 1014 cells or ∼29 g C) necessary to account for these depletions (Table 2 and SI Appendix, SI Text). These values provide empirical constraints on biomass standing stock of the subseafloor biosphere at deep-sea vents and provide insight into its biological dynamics. For example, if we assume that the amount of subseafloor biomass is at steady state, biomass residence time will be short (17–41 h), within the range of doubling times for cultured chemoautotrophic Campylobacteria (refs. 8 and 16 and SI Appendix, SI Text). As both the ambient deep-sea water entering the ocean crust and the endmember hydrothermal vent fluids do not contain any significant numbers of Campylobacteria, yet they constitute the dominant biomass in the fluids exiting the seafloor at Crab Spa (14), their growth must have occurred below the seafloor.
Vent Field and Global Estimates of Productivity and Standing Stock.
On a larger scale, export of biomass into the food-limited deep sea can be assessed by multiplying low-temperature fluid flux of the 9°N EPR vent field (26) with our volumetric primary productivity values (Table 2). Estimated subseafloor chemosynthetic productivity in the vent field ranges from ∼380 to 9,300 g C⋅m2⋅y−1, values that are at least two to four orders of magnitude greater than the amount of photosynthetic biomass reaching this depth [0.4–4 g C⋅m2⋅y−1 (27)]. For the entire 9°N EPR vent field area of 103–104 m2 (26), this corresponds to 3.8–9.3 Mg C y−1. Although we do not know what proportion of newly produced subseafloor biomass reaches the surrounding deep-sea water column, even a small amount would vastly increase the availability of labile carbon for heterotrophic consumers locally, making the deep ocean in the vicinity of vent fields hot spots of microbial activity (28).
Although this study was confined to one hydrothermal vent site, we believe our results are applicable to other subseafloor hydrothermal systems. Although some aspects of fluid chemistry differ between vent fields, there are also some striking similarities. Similar to Crab Spa, most other subseafloor vent fluids are typically enriched in dissolved inorganic carbon and electron donors and contain limited abundances of electron acceptors (9, 11, 15). Under such conditions, it is known that Campylobacteria dominate the in situ microbial community (6, 8, 24, 29). In contrast, the other main group of sulfur-oxidizing chemoautotrophs found at vents, the Gammaproteobacteria, are typically found at interfaces where warm vent fluids and ambient seawater mix turbulently and oxygen concentrations are higher (6, 8, 24, 29). Other potential autotrophic metabolisms could occur in the subseafloor at higher temperatures (such as hydrogenotrophic methanogenesis), but they are likely to be of minor importance quantitatively in basalt-hosted systems compared with aerobic/denitrifying oxidation of sulfide/hydrogen (15, 16). For the example of methanogenesis, fluid composition data for Crab Spa suggest that methane is being consumed in the subseafloor, rather than being produced (Table 1). Collectively, these observations suggest that most moderate-temperature (∼15–60 °C), sulfidic, and oxygen/nitrate-limited subseafloor ecosystems will be dominated by Campylobacteria. Because autotrophic Campylobacteria share fundamental physiological attributes [and therefore core mechanisms of energy conservation (6, 7)], we believe the CGE values derived here for mesophilic and thermophilic communities can be reasonably extrapolated to other systems.
To extend our quantitative estimates of productivity and standing stock to a global perspective, we used low-temperature (5 °C) and high-temperature (350 °C) fluid flux values to calculate the lower and upper bound on global subseafloor productivity, respectively (Table 2) (30). These estimates suggest that subseafloor chemosynthetic productivity at deep-sea hot springs amounts to at most 1.4 Tg C y−1 (Table 2), which is somewhat lower than previous theoretical estimates (refs. 15 and 16 and SI Appendix, SI Text) and representing at most 0.43% of photosynthetic primary productivity reaching depths >2,000 m (27). We also calculate a value for global subseafloor standing stock of 1.4–2.7 Gg C, more than three orders of magnitude lower than previous theoretical estimates (Table 2), which assumed that microbes in the subseafloor are in maintenance mode (16) rather than actively growing as shown here (SI Appendix, SI Text).
Conclusions
Although the paradigm of chemosynthetic microbes transferring geothermal energy to higher trophic levels at deep-sea hot springs has become well-established, the significance of the subseafloor ecosystem to global ocean biogeochemistry is difficult to estimate without reliable quantitative data. Using direct measurements of chemosynthetic growth efficiency and metabolic rates under in situ conditions, our data show that the standing stock of the chemosynthetic subseafloor biosphere is relatively small and turns over rapidly. On the basis of our estimates, subseafloor carbon fixation rivals highly productive animal–microbe symbioses above the seafloor and could therefore constitute a significant source of labile carbon to the otherwise food-limited deep sea. We also identified temperature and oxygen as critical factors driving the niche partitioning of natural communities composed of closely related and physiologically similar taxa, showing how deep-sea hot spring microbes interact with and are shaped by their unique environment.
Moving forward, similar measurements at other vent sites, including additional chemosynthetic processes, and better constraints on the overall fluid flux will be needed to refine our estimates. Although we can now better constrain subseafloor chemosynthetic productivity, how this newly produced organic carbon affects the deep ocean food web and biogeochemistry remains to be determined. Broad application of high-pressure incubations such as those reported here represents a powerful approach to gain quantitative insight into microbially mediated processes in the underexplored deep ocean.
Materials and Methods
Experimental Design.
Fluid samples for all analyses were collected from the Crab Spa vent with the ROV Jason II deployed from the R/V Atlantis during research cruise AT26-10 in January 2014. Crab Spa is located at a depth of 2,506 m at 9°50.3981N, 104°17.4942W. Shipboard incubations of fluids were carried out at in situ pressure in IGT samplers for ∼18–24 h with amendments of H2, NO3−, O2, and H2/NO3− in addition to NaH13CO3 as an isotope tracer for carbon fixation. With the exception of 2 NO3−/H2 incubations carried out at 50 °C, all incubations were conducted at 24 °C, which is the in situ fluid temperature at Crab Spa. During incubations, cell abundance and concentrations of selected chemical species (H2S, H2, NO3−, NH4+, and O2) were measured every ∼6 h. Full details of sampling, incubation procedures, chemical measurements, cell counts, and rate measurements are described in ref. 14.
DNA Analyses.
At the end point of each experiment (∼24 h), the remaining volume of fluid in the IGT (∼8–40 mL) was drawn into a clean, sterile, and DNA-free syringe (Norm-Ject), filtered through a 0.2 μM Sterivex filter cartridge, dried under filtered nitrogen gas, and frozen immediately at –80 °C. DNA was extracted as previously described (31). Amplicons were subsequently sequenced using bacterial primers 27Fmod and 519Rmodbio and 454-pyrosequencing technology (Molecular Research LP).
Pyrotag sequences were analyzed using the QIIME pipeline (32). Sequences were quality filtered with split_libraries.py (-w 50 -r -l,300 -L 1,000 -a 0 -H 6 -b 8 -z truncate_only), then denoised from 454 flowgrams (denoise_wrapper.py). After denoising, chimeras were removed using the script “identify_chimeric_seqs.py,” with USEARCH as the method. This yielded 3,597 ± 1,371 (SD) sequences per sample. Next, 97% OTUs were picked de novo, using the script “pick_otus.py,” with USEARCH as the method and classified with the script “assign_taxonomy.py,” using the SILVA v119 database as a reference. Raw sequences in .sff format are deposited at NCBI under accession number SRP077942.
CARD-FISH and HISH-SIMS 13C Incorporation.
Aliquots of ∼10 mL of fluid were taken from the IGTs at 16 or 24 h after the addition of labels/amendments and preserved with paraformaldehyde (1%, 1 h at room temperature). Cells were then filtered under moderate vacuum onto Au/Pd-sputtered 0.2 μM polycarbonate filters, washed 2× with 10 mL 1× PBS, air-dried, and stored at −20 °C before further analysis.
Filters were embedded in low-melting-point agarose, endogenous peroxidases were inactivated by immersion in 3% H2O2 for 10 min, and cells were permeabilized for 30 min at 37 °C in a 10 mg⋅mL−1 solution of lysozyme in TE buffer. Hybridization and tyramide amplification were conducted at 46 °C for 3 h and 20 min, respectively. Oregon Green 488-X was used for tyramide amplification, which contains two atoms of fluorine per molecule. All newly designed probes and their formamide concentrations are shown in SI Appendix, Table S2. Newly designed probes were tested with positive and negative control cultures across a melting curve to determine both the potential for nonspecific hybridization and the optimum concentration of formamide (SI Appendix, Table S2). Probes were additionally tested to ensure specificity by doing a double hybridization with both the EPSI549-914 combination and the newly designed probes on natural environmental samples (where other organisms aside from Campylobacteria were present). Because cells hybridized with NAUT921 and SFMN287 were also hybridized with the EPSI549-914 probe, this was additional confirmation that these probes are specific to Campylobacteria (SI Appendix, Figs. S5 and S6).
Once hybridized, 5-mm-diameter circular sections were cut out from each filter, and regions of interest were marked with a laser-dissecting fluorescence microscope (Zeiss) with a 63× (NA, 0.75) air objective. The remaining portions of filters were used to count the percentage of DAPI-stained cells hybridized to each specific probe. Seven grids were analyzed per sample, amounting to 400–700 DAPI-stained cells.
Regions of interest or random grids hybridized with the EPSI549-914 probes were analyzed on NanoSIMS 50L Ionprobe from CAMECA (AMETEK), detecting the following secondary ions: 12C, 13C, 12C14N, 13C14N, 19F, Au, 32S and 34P. An average of 49.6 target cells were analyzed per IGT incubation for the EPSI probes, with a range of 22–96. A subset of three samples were also analyzed with the NAUT921 probe (between 14 and 21 cells per sample).
13C Isotope Incorporation into Bulk Biomass.
At the last point during the incubation experiments, a known volume of fluid (∼20 mL) was filtered onto a precombusted GF-75 glass fiber filter (0.3 μM pore size; Advantec), wrapped in combusted aluminum foil, and stored at −80 °C before further analysis.
Filters were subsequently acidified to remove carbonates by exposure to HCl vapor for 3 d at 60–65 °C and then dried for 1 d at the same temperature. Immediately before combustion, dried filters were wrapped with tin foil (Costech part # 041073) and folded into pellets. Samples were combusted in a Carlo Erba/Fisons 1107 Elemental Analyzer “EA” (fitted with a Costech “Zero-Blank” carousel). The EA is attached via Finnigan-MAT Conflo-II interface to a DeltaPlus stable isotope ratio mass spectrometer. Data were acquired using the Isodat (version 2.5) software.
Carbon Fixation Rate Determinations.
For all incubations, 13C-labeled dissolved inorganic carbon (DIC) was supplied as a H13CO3− solution dissolved in filtered bottom seawater and added into low-temperature hot spring fluid (14). The fraction of total DIC as 13C label was determined using measured (DIC) values for background seawater and vent fluid, and was ∼10% in all cases (14). A conversion factor derived from these label percentages was used in both rate determinations below to derive total CO2 fixation rates.
For rate determinations from bulk isotope incorporation measurements, background 13C from an average of background (unincubated) samples was subtracted from detected 13C and normalized as described earlier to determine total CO2 fixed. Rates were determined by dividing total carbon fixed by the time from label addition to when samples were taken.
For HISH-SIMS-derived rates, data were processed with Look@NanoSIMS (33) to demarcate regions of interest for EPSI549/914-hybridized cells based on the 19F signal. Cell biovolume was estimated using the area and length:width ratio parameters for each region of interest, which was then combined with cell carbon density previously reported (34) to estimate carbon content for each cell. The amount of CO2 fixed per cell was then determined by correcting 13C14N/12C14N ratios for background 13C and label concentrations in fluids. This value was then multiplied by EPSI-hybridized cells⋅mL−1 and normalized by time to yield total CO2 fixed per volume per time.
CGE Determinations.
CGE represents the proportion of electrons transferred from energy-yielding oxidation half-reactions (e.g., hydrogen or sulfur oxidation) that are used to reduce CO2 into cell carbon (assuming biomass oxidation state of 0). The inverse proportion, equivalent to the “y” parameter estimated by Klatt and Polerecky (12), is the fraction of electrons transferred to energy-yielding dissimilatory metabolism (e.g., oxygen and nitrate reduction).
Total carbon fixed from bulk isotope measurements was determined as described earlier. The consumption of nitrate and oxygen were also measured, likely the only electron acceptors of importance during incubations (14). The means by which electron equivalents used to reduce these substrates was calculated has been previously described (14). Total electrons oxidized from sulfide and hydrogen were not directly measurable as a result of incomplete oxidation of sulfide (14), so this value was inferred by taking the sum of electron equivalents to carbon fixation and electron equivalents to electron acceptors. CGE was then derived by dividing total carbon fixed by this sum (Dataset S1).
Statistical Analysis.
Correlations of community composition with environmental parameters was carried out with a subset of total sequences and according to statistical analyses that are implemented in scripts of the QIIME pipeline (32). Sulfurimonas 97% OTUs found in 24 °C incubations were first normalized within each sample as the percentages of total Sulfurimonas sequences. Next, beta diversity was calculated using beta_diversity.py with UniFrac as the distance metric. A tree of sequences necessary for the UniFrac metric was generated by aligning sequences using MUSCLE (align_seqs.py) and building a tree using default parameters (make_phylogeny.py). The divergence between these different communities of Sulfurimonas was visualized by using the script nmds.py to generate values for a 2-D Nonmetric Multidimensional Scaling plot. Next, the script compare_categories.py was used with the adonis method to investigate the effect of the following variables (at the beginning of incubations) on final Sulfurimonas OTU composition: pH, [H2], [H2S], [NH3], [NO3−], PO2, [CH4], cell density and time from seafloor until the beginning of incubations. Finally, the script observation_metadata_correlation.py with Pearson correlations was used to look for the effect of PO2 on individual Sulfurimonas OTUs.
Supplementary Material
Acknowledgments
We thank the officers, crew, and pilots of the R/V Atlantis and ROV Jason for their expert help at sea and their outstanding efforts acquiring the samples for this study. We also thank the scientific party for support, with special thanks to Kerry McCulloch, Miriam Sollich, and Xi Wei for invaluable help with shipboard lab experiments. Thanks are also due to Ben van Mooy for lending his laboratory’s oxygen optode system; to Scott Wankel, Carly Buchwald, and Zoe Sandwith for help quantifying nitrate/nitrite; to Virginia Edgcomb for lending an Aanderaa in situ oxygen optode; to Jeremy Rich for providing nitrate measurements for background seawater; to Marc Mußmann and Stefan Dyksma for help designing and testing CARD-FISH probes; to Carl Johnson for the EA-IRMS analyses; and to Lubos Polerecky for answering questions about analyzing NanoSIMS data. We further thank Carl Wirsen for his comments on an earlier version of the manuscript. This research was funded by a grant of the Dimensions of Biodiversity program of the US National Science Foundation (NSF-OCE-1136727 to S.M.S. and J.S.S.). Funding for J.M. was further provided by doctoral fellowships from the Natural Sciences and Engineering Research Council of Canada (PGSD3-430487-2013, PGSM-405117-2011) and the National Aeronautics and Space Administration Earth Systems Science Fellowship (PLANET14F-0075), an award from the Canadian Meteorological and Oceanographic Society, and the WHOI Academic Programs Office. The authors are grateful for using the analytical facilities of the Centre for Chemical Microscopy (ProVIS) at the Helmholtz Centre for Environmental Research, Leipzig, which is supported by European Regional Development Funds (EFRE-Europe funds Saxony) and the Helmholtz Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the NCBI Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra/?term=SRX1898689; accession no. SRX1898689).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804351115/-/DCSupplemental.
References
- 1.Corliss JB, et al. Submarine thermal springs on the Galapagos rift. Science. 1979;203:1073–1083. doi: 10.1126/science.203.4385.1073. [DOI] [PubMed] [Google Scholar]
- 2.Karl DM, Wirsen CO, Jannasch HW. Deep-sea primary production at the Galapagos hydrothermal vents. Science. 1980;207:1345–1347. [Google Scholar]
- 3.Wirsen CO, Jannasch HW, Molyneaux SJ. Chemosynthetic microbial activity at Mid-Atlantic ridge hydrothermal vent sites. J Geophys Res. 1993;98:9693–9703. [Google Scholar]
- 4.Wirsen CO, Tuttle JH, Jannasch HW. Activities of sulfur-oxidizing bacteria at the 21 N East Pacific rise vent site. Mar Biol. 1986;92:449–456. [Google Scholar]
- 5.Huber JA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa S, Takai K. Deep-sea vent chemoautotrophs: Diversity, biochemistry and ecological significance. FEMS Microbiol Ecol. 2008;65:1–14. doi: 10.1111/j.1574-6941.2008.00502.x. [DOI] [PubMed] [Google Scholar]
- 7.Waite DW, et al. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.) Front Microbiol. 2017;8:682, and erratum (2018) 9:772. doi: 10.3389/fmicb.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sievert SM, Vetriani C. Chemoautotrophy at deep-sea vents: Past, present, and future. Oceanography (Wash DC) 2012;25:218–233. [Google Scholar]
- 9.Jannasch HW, Mottl MJ. Geomicrobiology of deep-sea hydrothermal vents. Science. 1985;229:717–725. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- 10.Von Damm KL, Lilley MD. Diffuse flow hydrothermal fluids from 9° 50′ N East Pacific rise: Origin, evolution and biogeochemical controls. In: Wilcock WSD, Delong EF, Kelley DS, Baross JA, Cary SC, editors. The Subseafloor Biosphere at Mid-Ocean Ridges. American Geophysical Union; Washington, DC: 2004. pp. 245–268. [Google Scholar]
- 11.Wankel SD, et al. Influence of subsurface biosphere on geochemical fluxes from diffuse hydrothermal fluids. Nat Geosci. 2011;4:461–468. [Google Scholar]
- 12.Klatt JM, Polerecky L. Assessment of the stoichiometry and efficiency of CO2 fixation coupled to reduced sulfur oxidation. Front Microbiol. 2015;6:484. doi: 10.3389/fmicb.2015.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandernack KW, Tebo BM. In situ sulfide removal and CO2 fixation rates at deep-sea hydrothermal vents and the oxic/anoxic interface in Framvaren Fjord, Norway. Mar Chem. 1999;66:201–213. [Google Scholar]
- 14.McNichol J, et al. Assessing microbial processes in deep-sea hydrothermal systems by incubation at in situ temperature and pressure. Deep Sea Res Part I Oceanogr Res Pap. 2016;115:221–232. [Google Scholar]
- 15.McCollom TM, Shock EL. Geochemical constraints on chemolithoautotrophic metabolism by microorganisms in seafloor hydrothermal systems. Geochim Cosmochim Acta. 1997;61:4375–4391. doi: 10.1016/s0016-7037(97)00241-x. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura K, Takai K. Subseafloor Biosphere Linked to Hydrothermal Systems. Springer; Tokyo: 2015. Geochemical constraints on potential biomass sustained by subseafloor water–rock interactions; pp. 11–30. [Google Scholar]
- 17.Germanovich LN, Hurt RS, Smith JE, Genc G, Lowell RP. Measuring fluid flow and heat output in seafloor hydrothermal environments. J Geophys Res Solid Earth. 2015;120:8031–8055. [Google Scholar]
- 18.Seewald JS, Doherty KW, Hammar TR, Liberatore SP. A new gas-tight isobaric sampler for hydrothermal fluids. Deep Sea Res Part I Oceanogr Res Pap. 2002;49:189–196. [Google Scholar]
- 19.Musat N, et al. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA. 2008;105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musat N, et al. The effect of FISH and CARD-FISH on the isotopic composition of (13)C- and (15)N-labeled Pseudomonas putida cells measured by nanoSIMS. Syst Appl Microbiol. 2014;37:267–276. doi: 10.1016/j.syapm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Pachiadaki MG, et al. Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science. 2017;358:1046–1051. doi: 10.1126/science.aan8260. [DOI] [PubMed] [Google Scholar]
- 22.Reinthaler T, van Aken HM, Herndl GJ. Major contribution of autotrophy to microbial carbon cycling in the deep north Atlantic’s interior. Deep Sea Res Part II Top Stud Oceanogr. 2010;57:1572–1580. [Google Scholar]
- 23.Chavez FP, Messié M, Pennington JT. Marine primary production in relation to climate variability and change. Annu Rev Mar Sci. 2011;3:227–260. doi: 10.1146/annurev.marine.010908.163917. [DOI] [PubMed] [Google Scholar]
- 24.Meier DV, et al. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 2017;11:1545–1558. doi: 10.1038/ismej.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girguis PR, Childress JJ. Metabolite uptake, stoichiometry and chemoautotrophic function of the hydrothermal vent tubeworm Riftia pachyptila: Responses to environmental variations in substrate concentrations and temperature. J Exp Biol. 2006;209:3516–3528. doi: 10.1242/jeb.02404. [DOI] [PubMed] [Google Scholar]
- 26.Lowell R, Farough A, Germanovich L, Hebert L, Horne R. A vent-field-scale model of the east Pacific rise 9°50’N magma-hydrothermal system. Oceanography (Wash DC) 2012;25:158–167. [Google Scholar]
- 27.Lampitt RS, Antia AN. Particle flux in deep seas: Regional characteristics and temporal variability. Deep Sea Res Part I Oceanogr Res Pap. 1997;44:1377–1403. [Google Scholar]
- 28.Meier DV, et al. Heterotrophic Proteobacteria in the vicinity of diffuse hydrothermal venting. Environ Microbiol. 2016;18:4348–4368. doi: 10.1111/1462-2920.13304. [DOI] [PubMed] [Google Scholar]
- 29.Olins HC, et al. Co-registered geochemistry and metatranscriptomics reveal unexpected distributions of microbial activity within a hydrothermal vent field. Front Microbiol. 2017;8:1042. doi: 10.3389/fmicb.2017.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elderfield H, Schultz A. Mid-ocean ridge hydrothermal fluxes and the chemical composition of the ocean. Annu Rev Earth Planet Sci. 1996;24:191–224. [Google Scholar]
- 31.Signori CN, Thomas F, Enrich-Prast A, Pollery RCG, Sievert SM. Microbial diversity and community structure across environmental gradients in Bransfield Strait, Western Antarctic Peninsula. Front Microbiol. 2014;5:647. doi: 10.3389/fmicb.2014.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polerecky L, et al. Look@NanoSIMS–A tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol. 2012;14:1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x. [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Fuhrman JA. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Microbiol. 1987;53:1298–1303. doi: 10.1128/aem.53.6.1298-1303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.