Significance
The onset of chromosomal DNA replication relies on dedicated initiator proteins to chaperone ring-shaped helicases onto DNA. In most eukaryotes, initiators are multisubunit protein complexes that require ATP to bind DNA and to aid helicase recruitment and loading. Although structural studies have recently elucidated high-resolution views of the initiator in isolation or in helicase-containing loading intermediates, how the eukaryotic initiator itself associates with DNA and how these interactions are regulated by conformational changes are not well understood. We use a combination of biochemical and structural studies of the Drosophila initiator origin recognition complex (ORC) to show that conformational alterations in metazoan ORC help regulate its DNA-binding activity, and that ORC, together with its cofactor Cdc6, bends substrate DNA prior to helicase loading.
Keywords: origin recognition complex, DNA replication, initiators, AAA+ ATPase, helicase loading
Abstract
In eukaryotes, the heterohexameric origin recognition complex (ORC) coordinates replication onset by facilitating the recruitment and loading of the minichromosome maintenance 2–7 (Mcm2–7) replicative helicase onto DNA to license origins. Drosophila ORC can adopt an autoinhibited configuration that is predicted to prevent Mcm2–7 loading; how the complex is activated and whether other ORC homologs can assume this state are not known. Using chemical cross-linking and mass spectrometry, biochemical assays, and electron microscopy (EM), we show that the autoinhibited state of Drosophila ORC is populated in solution, and that human ORC can also adopt this form. ATP binding to ORC supports a transition from the autoinhibited state to an active configuration, enabling the nucleotide-dependent association of ORC with both DNA and Cdc6. An unstructured N-terminal region adjacent to the conserved ATPase domain of Orc1 is shown to be required for high-affinity ORC–DNA interactions, but not for activation. ORC optimally binds DNA duplexes longer than the predicted footprint of the ORC ATPases associated with a variety of cellular activities (AAA+) and winged-helix (WH) folds; cryo-EM analysis of Drosophila ORC bound to DNA and Cdc6 indicates that ORC contacts DNA outside of its central core region, bending the DNA away from its central DNA-binding channel. Our findings indicate that ORC autoinhibition may be common to metazoans and that ORC–Cdc6 remodels origin DNA before Mcm2–7 recruitment and loading.
In the three known domains of cellular life, the onset of DNA replication is controlled by dedicated initiator proteins and cofactors, which work in concert to load ring-shaped replicative helicases onto DNA before replisome assembly. In eukaryotes, the six-subunit origin recognition complex (ORC) binds to DNA and, with the help of the Cdc6 and Cdt1 partner proteins, loads the minichromosome maintenance 2–7 (Mcm2–7) helicase onto origins as a head-to-head double hexamer during the G1 phase of the cell cycle (recently reviewed in refs. 1, 2). Upon transitioning into S phase, a subset of available Mcm2–7 complexes are activated to support replisome activity and bidirectional DNA replication.
ORC belongs to the “initiator” clade of the ATPases associated with a variety of cellular activities (AAA+) superfamily (3). AAA+ proteins form homo- or heterooligomeric assemblies that use ATP binding and hydrolysis to regulate interactions with client nucleic acid or protein substrates (4). Five of ORC’s six subunits retain an AAA+ domain, each of which is joined to a single winged-helix (WH) fold (5, 6); these elements of ORC collectively comprise a ring-shaped, pentameric core complex (6). Three of ORC’s AAA+ folds (in Orc1, Orc4, and Orc5) are able to bind ATP (6–9); however, only the ATPase site formed at the interface between Orc1 and Orc4 is essential for ORC function in vivo and has significant ATPase activity in vitro (7, 10–12). Structural studies have suggested that for Drosophila melanogaster ORC (DmORC), the enzymatic activity of the Orc1/4 ATPase site might be controlled by a conformational switch that involves a large rotation of the Orc1 AAA+ domain (6). This movement would allow ORC to transition from an autoinhibited state, in which a central channel in the ORC ring is sterically blocked from binding DNA, to an active form that is competent for DNA binding (6) (Fig. 1A). An autoinhibited conformation of DmORC has not yet been observed in isolated ORC structures from other organisms (9), raising questions as to whether autoinhibition might be a unique feature of the Drosophila complex. How a conformational switch might be triggered to lead to ORC activation in this instance is also unknown.
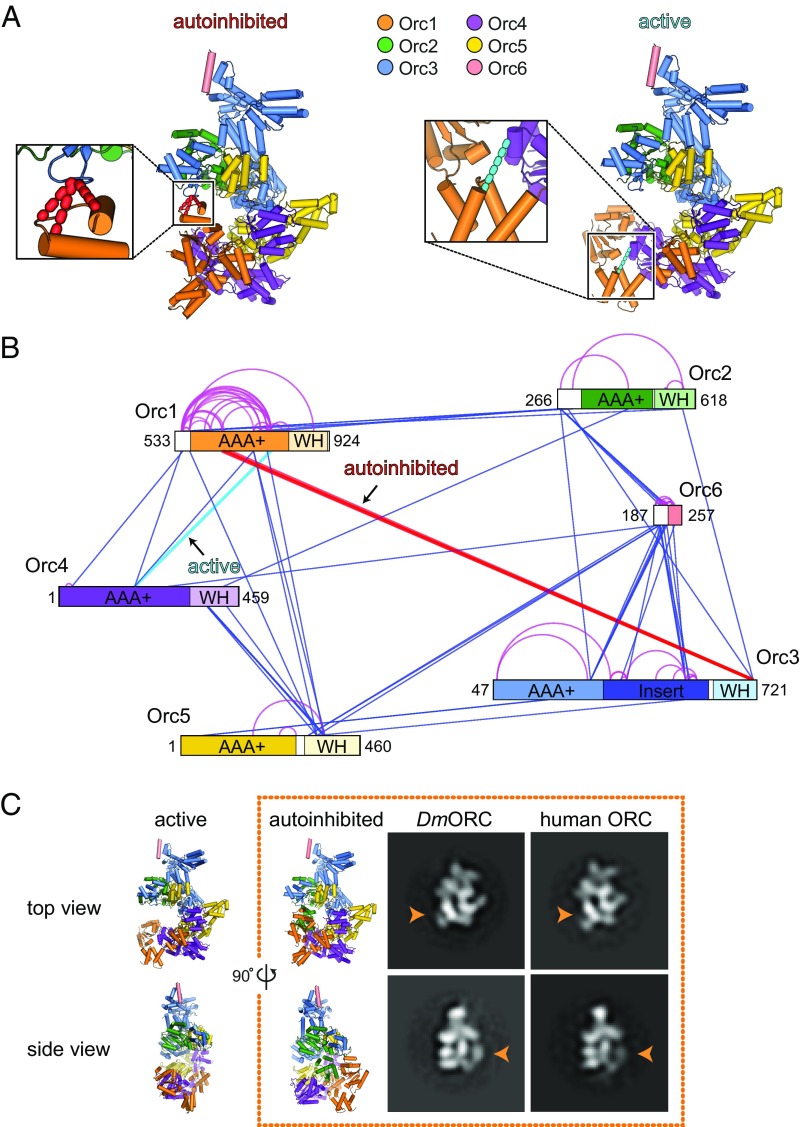
Fig. 1.
The autoinhibited ORC state is a conserved characteristic of ORC and can be detected in solution. (A and B) Cross-linking mass spectrometry of the DmORC core complex reveals specific cross-links expected for the autoinhibited (red dashed lines) and active (cyan dashed line) conformational states. In A, cross-links reporting on the conformational state of ORC are mapped onto the DmORC crystal structure (Left, autoinhibited) and on a model of an “activated” DmORC complex (Right; Materials and Methods). The Orc2 WH domain is omitted for clarity. In B, all DSS-induced cross-links are mapped onto the domain architecture of ORC subunits, with interprotein cross-links depicted as straight blue lines and intraprotein cross-links shown in magenta. The positions of the first and last amino acids of each subunit in the ORC core complex are indicated, with ordered domains and disordered linker regions shown either in color or in white, respectively. Note that one of the cross-links supporting the autoinhibited conformation was detected twice on slightly different peptides due to alternative protease cleavage. (C) Negative-stain EM analysis of DmORC and human ORC1–5 indicates that both complexes can adopt the autoinhibited conformation in the presence of the ATP analog ATPγS. Class averages reflecting the autoinhibited state are shown in two different views, with the characteristic Orc1 density that defines the autoinhibited state highlighted by an orange arrowhead. Top and side views of the crystal structure of Drosophila ORC (6) in the autoinhibited conformation (Right) and an active DmORC model (Left) are displayed for comparison. Subunits are colored as in A.
During Mcm2–7 loading, ORC binds DNA in an ATP-dependent manner (11, 13–15). ORC’s AAA+ and WH domains both contain DNA-binding elements, which line the surface of an interior channel that runs through the center of the Orc1–5 ring (6, 8, 9). In Drosophila ORC, access to these primary binding sites is prevented by the autoinhibited pose of the AAA+ region of Orc1, which folds back onto its own WH domain to seal off a lateral entry gate into ORC’s central channel (6). In metazoans, ORC has been reported to have secondary (and ATP-independent) DNA-binding regions, such as the transcription factor IIB (TFIIB)-like domain found in Orc6 (16, 17). Both yeast and metazoan ORC also have been shown to bend or wrap DNA (18, 19); however, in a DNA-bound structure of ORC in complex with Cdc6, Cdt1, and Mcm2–7 (OCCM), the duplex can be seen to pass through the central channel of both ORC and Mcm2–7 in a relatively straight manner (8). How ORC might accomplish DNA bending, how secondary DNA-binding sites or other ORC regions might modulate this activity, and how DNA deformation might be important for Mcm2–7 recruitment and/or loading are not readily apparent from current structural information.
Here, we seek to better understand the conformational dynamics of ORC and how these states are linked to ATP and DNA binding. Using chemical cross-linking and mass spectrometry, we confirm that DmORC can form an autoinhibited state in solution. Single-particle electron microscopy (EM) reveals that human ORC shares this capability, and that the equilibrium between autoinhibited and active configurations of DmORC is regulated by nucleotide binding, but that ATP alone is unable to drive all DmORC particles into an active configuration. A conserved basic patch in the N terminus of metazoan Orc1 that precedes the subunit’s AAA+ ATPase domain is observed to be critical for the ATP-dependent interactions of ORC with DNA; however, this requirement can be partially overcome by the cobinding of Cdc6, which helps to trap DNA in ORC’s central channel. Interestingly, ORC is found to have a greater affinity for DNA duplexes that are almost twice as long as its central channel. Two-dimensional cryo-EM class averages of an ORC–DNA–Cdc6 complex show that DNA bends away from the central axis of the channel at a particular point, suggesting that the outer surface of the ORC–Cdc6 ring directly contacts DNA to facilitate duplex deformation. Collectively, our findings demonstrate that an ability of ORC to adopt an autoinhibited state is preserved from flies to humans and that ATP binding derepresses this autoinhibition. Our data also show that ORC possesses a distinct element outside the central AAA+/WH domain core that is critical for DNA binding, and that ORC in complex with DNA and Cdc6 actively bends the duplex, possibly as a means to increase the local accessibility of contact sites for recruiting the Mcm2–7 helicase.
Results
The Autoinhibited Conformation of DmORC Is a Genuine Solution State also Adopted by Human ORC.
Previous crystallographic and EM studies identified an autoinhibited state of Drosophila ORC in which the ATPase region of Orc1 did not productively engage catalytic elements on Orc4 but, instead, adopted a conformation sterically incompatible with supporting DNA and Cdc6 binding (6) (Fig. 1A). A subsequent crystal structure of a human ORC1/4/5 subcomplex, together with an 18-Å-resolution cryo-EM structure of a pentameric human ORC1–5 complex, revealed an Orc1/4 conformation that could support both ATP turnover and DNA binding (9). This dichotomy raised questions as to whether an autoinhibited state might be specific to DmORC or was inadvertently induced by experimental conditions.
To address this problem, we used chemical cross-linking and mass spectrometry as an alternative approach to probe the conformational states of DmORC. The recombinant DmORC core complex (which lacks the flexible N-terminal regions of Orc1, Orc2, Orc3, and Orc6), as well as full-length DmORC, was treated with the lysine-reactive reagent disuccinimidyl suberate (DSS; Materials and Methods). Following proteolytic cleavage, cross-linked peptides were fractionated and identified by liquid chromatography (LC) tandem mass spectroscopy (MS/MS) (20). We detected in the range of 50–60 intersubunit cross-links in each sample, with good overlap between the two DmORC complexes (Fig. 1 A and B and SI Appendix, Fig. S1). To identify which cross-links report on the conformational state of DmORC, we measured the distances between the Cα atoms of cross-linked lysine residues in the (autoinhibited) DmORC crystal structure and in an “ATPase-active” model of DmORC, which was generated based on docking the Orc1 AAA+ domain against the Orc4 arginine finger as described by Bleichert et al. (6). Of the observed 57 intersubunit cross-links with the DmORC core complex, 14 could be reliably mapped on these structures using a distance cutoff of 30 Å, while six of 52 cross-links identified with full-length DmORC could be structurally mapped. The remaining intersubunit cross-links resided in regions that were disordered in the DmORC crystal structure or not present in the crystallization construct. Importantly, a subset of mapable cross-links was consistent with the formation of either the autoinhibited state (four cross-links between the Orc1 AAA+ and Orc3 WH domains) or the active conformation (one cross-link between the Orc1 and Orc4 AAA+ domains) (Fig. 1A). For full-length DmORC, only a single cross-link corresponding to the autoinhibited state was observed, whereas no cross-links were found for the active conformation under these experimental conditions; instead, the vast majority of cross-linked species occurred in the N-terminal region of Orc1, which is large and unstructured (SI Appendix, Fig. S1). The appearance of cross-links consistent with the formation of the autoinhibited state seen previously from X-ray diffraction and EM data demonstrates that this conformation reflects a bone fide solution state of the DmORC complex.
To investigate whether only DmORC can adopt the autoinhibited state, we expressed human ORC1–5 in insect cells and analyzed the purified complex by negative-stain EM. Contrary to previous reports (9, 21, 22), the human ORC subunits cofractionated with each other during multiple purification steps in the absence of nucleotides, indicating that apo human ORC1–5 can form a stable pentameric complex on its own (SI Appendix, Fig. S2A). Analysis of recombinant human ORC1–5 by negative-stain EM revealed a relatively monodisperse population of particles (SI Appendix, Fig. S2B). Upon 2D classification, several class averages were observed in which the density attributable to the Orc1 AAA+ domain is clearly disengaged from the neighboring Orc4 subunit (in ∼20% of picked particle images), as would be expected for the autoinhibited ORC conformation (Fig. 1C and SI Appendix, Fig. S2C). Although we also observed human ORC1–5 particles that resemble the activated state (SI Appendix, Fig. S2C), these results nonetheless demonstrate that the ability to adopt the autoinhibited state is not limited to the Drosophila complex.
ATP Binding Licenses Drosophila ORC to Adopt an Active Conformation.
The existence of an autoinhibited ORC state raises questions as to what might activate the complex or shift the equilibrium toward a functional complex. Since activation is expected to be at least partly coupled to the formation of a composite Orc1/4 ATPase site, it would be logical to assume that nucleotide binding to Orc1 might control the conformational switch; however, we previously found that DmORC still adopts the autoinhibited conformation when cocrystallized or analyzed by EM in the presence of the ATP analog adenosine 5′-(3-thiotriphosphate) (ATPγS) (where unhydrolyzed nucleotide could be seen to occupy the ATP-binding pockets of Orc1, Orc4, and Orc5) (6). To investigate whether formation of the active state is simply a rare event for DmORC in the presence of ATPγS, we reexamined ATPγS–DmORC complexes by negative-stain EM, expanding the dataset size (by ∼2.3-fold) to attempt to capture poorly populated states in 2D class averages. A comparison of 2D projections of the autoinhibited DmORC crystal structure and a theoretical, activated DmORC model shows that both states can be readily distinguished in top and side views due to the distinct location of density for the Orc1 AAA+ domain (Fig. 2A). Similar to our previous results, DmORC predominantly adopted the autoinhibited state in 2D class averages when incubated with ATPγS; nonetheless, a very small subset of particles (≤5%) could be found in the larger dataset that resembled the active conformation (Fig. 2A). By contrast, in the presence of ATP, we observed a substantial increase in the number of class averages that represent the activated DmORC state (∼15–20% of particles), although particles reflecting the autoinhibited state were still present, equaling or outnumbering the active ones. Collectively, these data indicate that ATPγS is unable to stabilize Orc1/4 AAA+ domain interactions in DmORC with a similar efficiency as ATP. Our findings also indicate that while ATP-free DmORC predominantly exists in an autoinhibited form, ATP can shift the equilibrium to increase the population of the active state.
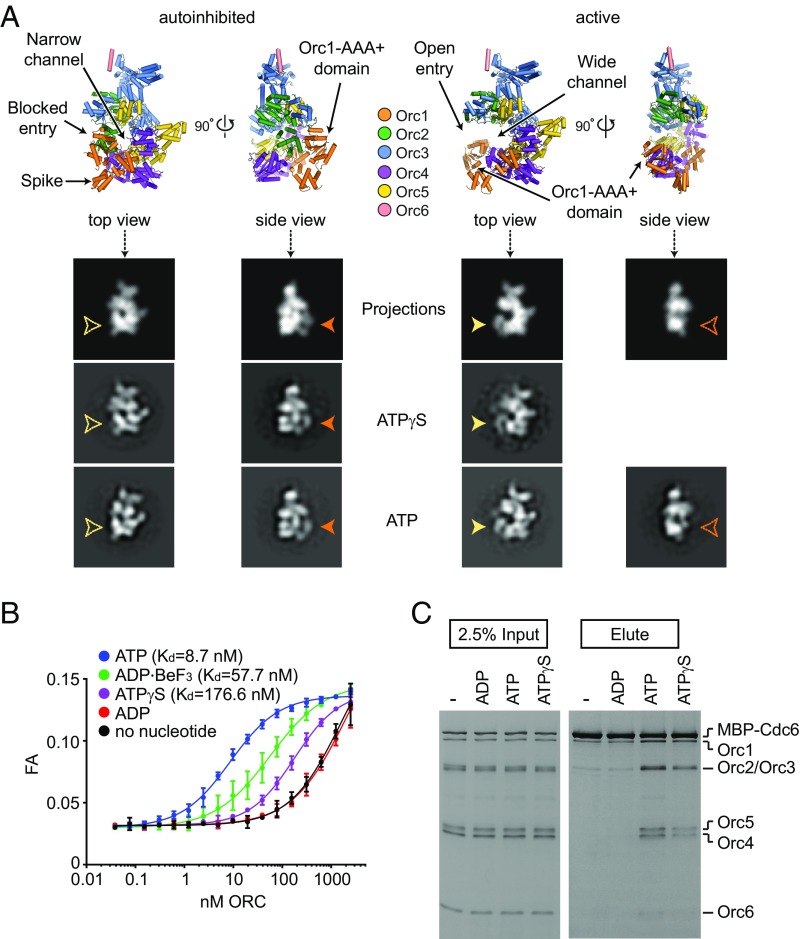
Fig. 2.
ATP and, to a lesser extent, ATPγS, stabilize Drosophila ORC in the active conformation and allow ORC association with both DNA and Cdc6. (A) Top and side views of the DmORC crystal structure (6) (Top Left) and of a model of activated DmORC (Top Right) are shown. Low-pass-filtered 2D projections of both ORC models in top and side views, as well as corresponding class averages of negatively stained DmORC in the presence of ATPγS or ATP, are depicted below. Arrowheads point to the Orc1 AAA+ density that repositions between ATP and ATPγS states (differences in top and side views are indicated in yellow and orange, respectively). Note that no class average depicting the side view of the active state was observed with ATPγS-DmORC. (B) Duplex DNA binding by DmORC was assayed by fluorescence anisotropy (FA) in the absence or presence of different nucleotides at 1 mM concentration. High (low-nanomolar) affinity binding is observed in the presence of ATP, but not ADP or ATPγS. Kds were calculated for the ATP, ADP–BeF3, and ATPγS conditions, but the lack of a plateau in the binding curves obtained with ADP or without nucleotide prevented accurate Kd determination for these conditions. (C) Pull-down assays using MBP-tagged DmCdc6 as bait demonstrate that ATP and, to a lesser extent, ATPγS, can promote the coassociation of ORC with Cdc6. All reactions were performed in the presence of DNA. Input and eluted proteins were separated by SDS/PAGE and visualized by silver staining.
ORC has been shown to bind DNA in an ATP-dependent manner (11, 13–15), an event that likely involves DNA contacts with the ORC central channel (6, 8). Since this DNA-binding site is inaccessible in the autoinhibited ORC state, we asked whether the inefficiency of ATPγS in promoting the formation of a composite Orc1/4 ATPase site dampens the ability of DmORC to bind DNA compared to ATP. Because Cdc6 binds ORC when ATP and DNA are present (23–25), we also probed whether ATPγS and ATP equally support the formation of this ternary complex. To assess the nucleotide-dependent DNA-binding activity of ORC, we measured the affinity of DmORC for a 40-bp duplex DNA under equilibrium conditions using fluorescence anisotropy (Fig. 2B). In the presence of ATP, we observed that DmORC binds DNA with an apparent Kd of ∼9 nM. By contrast, the complex associated much less efficiently with DNA in the absence of nucleotide or with ADP present. Surprisingly, ATPγS reduced the affinity of DmORC for DNA by ∼20-fold compared to ATP, while ADP–BeF3 only showed a mild decrease (approximately sixfold). The decreased ability of ATPγS–DmORC to bind DNA also impaired the coassociation of Cdc6 with DNA-bound DmORC, although the differences between the ATPγS and ATP conditions were less pronounced than what was observed for DNA binding (Fig. 2C), possibly because Cdc6 seals off the pentameric AAA+ ORC ring and prevents duplex escape (8, 26). Taken together, these results demonstrate that although ATPγS can “license” the formation of a competent DNA- and Cdc6-binding ORC state, it is less efficient at doing so than ATP.
A Basic Patch in the N Terminus of Orc1 Stabilizes ATP-Dependent ORC-DNA Contacts.
Structural modeling of an ORC–DNA complex (6), as well as cryo-EM visualization of a helicase-loading intermediate containing DNA and budding yeast OCCM (8), had previously indicated that a core complex of ORC comprising the AAA+ and WH modules of Orc1–5 (but lacking the N-terminal regions that precede the AAA+ folds of Orc1–3) should be sufficient to mediate nucleotide-dependent interactions with DNA (Fig. 3A). However, when we tested such a core complex together with the Orc3-binding portion of Orc6, for ATP-dependent DNA binding by either fluorescence anisotropy or pull-down assays, we unexpectedly found that it did not efficiently engage substrate duplexes (Fig. 3 B and C). To identify regions of ORC that might additionally contribute to DNA affinity, we deleted the N-terminal elements of several DmORC subunits individually and analyzed the ability of these truncated complexes to support DNA binding (Fig. 3 B and C and SI Appendix, Fig. S3A). Removal of the TFIIB-like domain in Orc6, which has previously been shown to bind DNA in metazoans (11, 16, 17), turned out to have only a very small effect on the ATP-dependent ability of DmORC to engage short duplexes. By contrast, deleting the N-terminal region of Orc1 reduced DNA binding to a similar level as the DmORC core complex. When full-length Orc1 was incorporated into the ORC core complex, ATP-dependent DNA binding was restored.
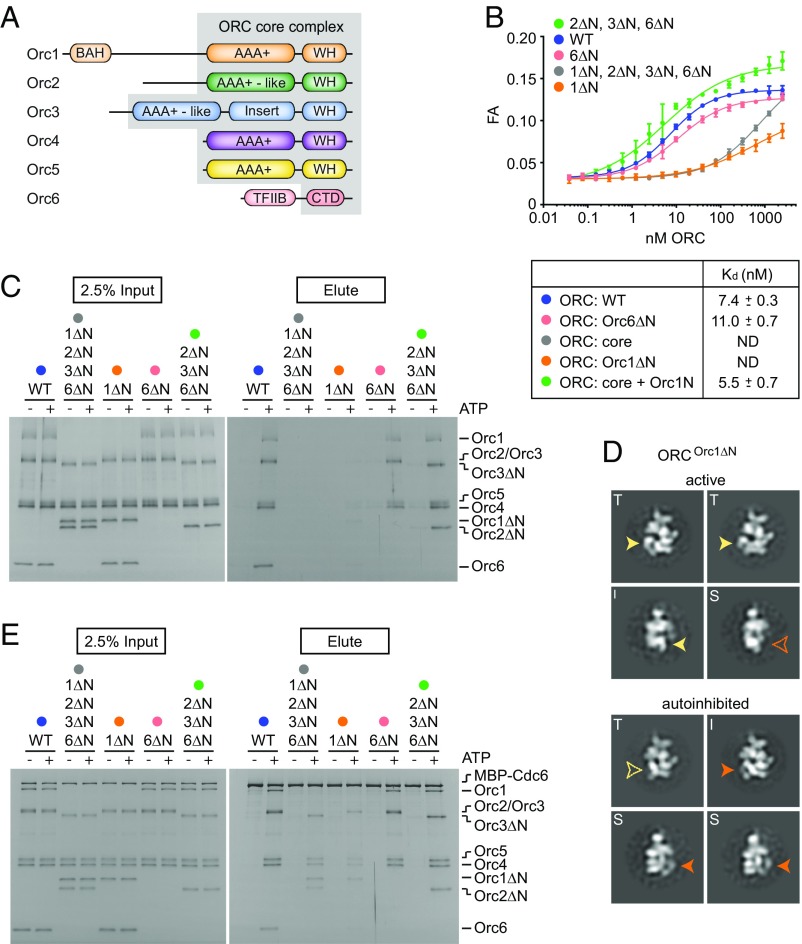
Fig. 3.
The N-terminal region of Orc1 is required for high-affinity, ATP-dependent DNA binding by ORC. (A) Schematic of ORC domain architecture with the ORC core complex lacking the N-terminal regions of Orc1 [including the bromoadjacent homology (BAH) domain], Orc2, Orc3, and Orc6 (including the TFIIB-like domain) bordered in gray. CTD, C-terminal domain; WH, winged-helix domain. ATP-dependent DNA binding by full-length DmORC (WT), the DmORC core (DmORCOrc1ΔN, Orc2ΔN, Orc3ΔN, Orc6ΔN; abbreviated as 1ΔN, 2ΔN, 3ΔN, 6ΔN), or DmORC lacking different combinations of N-terminal regions for subunits Orc1 (1ΔN), Orc2 (2ΔN), Orc3 (3ΔN), and Orc6 (6ΔN) was analyzed by fluorescence anisotropy (FA) (B) and pull-down assays (C). Kds and SEs of the parameter fits for binding curves determined by FA are listed in B. ND, Kds could not be determined because binding curves did not saturate. For the pull-downs in C, a biotinylated DNA duplex was used as bait. Copurifying proteins were eluted by UV cleavage, and input and eluted proteins were analyzed by SDS/PAGE followed by silver staining. (D) Two-dimensional EM analysis of negatively stained DmORC lacking the N-terminal region of Orc1 indicates that the removal of Orc1’s N terminus does not prevent ORC from adopting an active conformation in the presence of ATP. Representative top (T), intermediate (I), and side (S) view class averages of DmORCOrc1ΔN in both the active and autoinhibited states are shown. Arrowheads highlight the repositioning of the Orc1 AAA+ density between autoinhibited and active states (differences in top and intermediate/side views are indicated in yellow and orange, respectively). (E) Pull-down assays using MBP-tagged DmCdc6 as bait were used to assess the ability of full-length DmORC, the DmORC core complex [ORCOrc1ΔN, Orc2ΔN, Orc3ΔN, Orc6ΔN (abbreviated as 1ΔN, 2ΔN, 3ΔN, 6ΔN)], and DmORC lacking different N-terminal regions [ORCOrc1ΔN (abbreviated as 1ΔN), ORCOrc6ΔN (abbreviated as 6ΔN), and ORCOrc2ΔN, Orc3ΔN, Orc6ΔN (abbreviated as 2ΔN, 3ΔN, 6ΔN)] to coassociate with Cdc6 in the presence of DNA. Input and eluted proteins were analyzed by SDS/PAGE followed by silver staining. Note that the Orc6 C-terminal peptide (Orc6ΔN) is not resolved and visible on the gels shown in D and E due to its small size.
The behavior of the DmORC core complex variants suggested to us that the Orc1 N terminus might contain a control element that is essential for high-affinity ORC-DNA contacts. In this regard, we reasoned that the Orc1 N terminus might influence DNA binding by modulating the conformational state of ORC, or it might contain residues that directly contact the DNA duplex. To distinguish between these possibilities, we first analyzed DmORC lacking only the Orc1 N-terminal region (DmORCOrc1ΔN) by EM. Two-dimensional classification of negatively-stained DmORCOrc1ΔN particles demonstrated that this complex does enter the active state in the presence of ATP, although, akin to the observations with full-length DmORC, a substantial portion of particles remained in the autoinhibited conformation (Fig. 3D). These active DmORCOrc1ΔN complexes, as well as the DmORC core complex, could also coassociate with DmCdc6 in the presence of DNA and ATP as seen in pull-down experiments, albeit they did so slightly less efficiently than full-length DmORC (Fig. 3E). The ability of ORC and Cdc6 to copurify in the absence of the Orc1 N terminus is somewhat surprising, given the strong defect of these DmORC assemblies in ATP-mediated DNA binding (Fig. 3 B and C), and suggested that Cdc6 might stabilize ORC on DNA. To test this hypothesis, we compared the ability of full-length DmORC and the DmORC core complex to associate with DNA in the absence or presence of Cdc6 in pull-down assays (SI Appendix, Fig. S4). Our results show that Cdc6 enhances the association of DmORC with DNA, indicating that complexes lacking the Orc1 N terminus engage DNA only weakly or transiently, but that Cdc6 can stabilize these interactions once it binds to ORC and traps DNA in the central channel of the complex.
Since the Orc1 N terminus did not appear to influence the equilibrium distribution of autoinhibited and active ORC states, we hypothesized that this ∼500-aa region might contain an element that binds DNA directly. To narrow down the region responsible for this activity, we truncated Orc1 at different positions N terminal to the AAA+ domain, which did not interfere with the formation of a heterohexameric ORC assembly (SI Appendix, Fig. S3B), and measured the ATP-dependent DNA-binding capabilities of the respective DmORC assemblies by fluorescence anisotropy (Fig. 4 A and B). Deletion of the first 439-aa residues, including deletion of the nucleosome-binding bromoadjacent homology domain, had no impact on high-affinity ORC–DNA interactions. However, removing an additional 75 or 89 residues (ORCOrc1ΔN514, ORCOrc1ΔN528) reduced the affinity of DmORC for DNA by approximately fivefold and >100-fold, respectively.
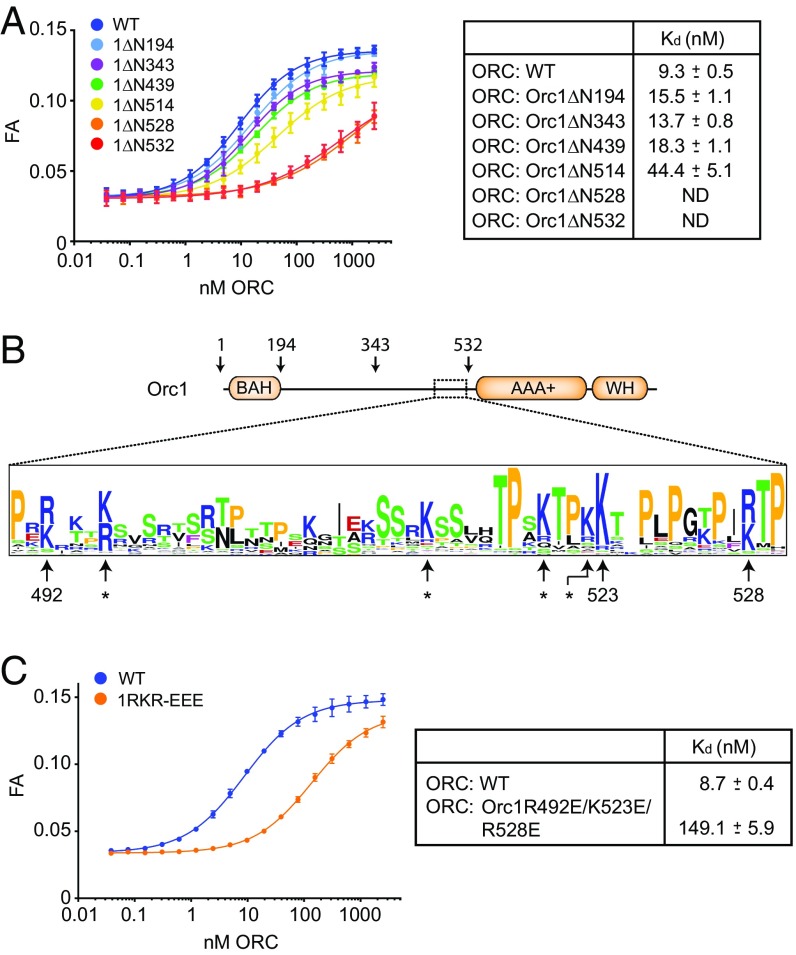
Fig. 4.
A basic patch in the N-terminal region of Orc1 stabilizes ATP-dependent ORC-DNA contacts. (A) ATP-dependent DNA binding of ORC (full-length or containing N-terminally truncated Orc1) to a 40-bp DNA duplex was measured by fluorescence anisotropy (FA). Kds and SEs of the parameter fits are listed for each mutant complex except for ORCOrc1ΔN528 and ORCOrc1ΔN532, for which the lack of a plateau prevented accurate Kd determination. ND, not determined. (B) Schematic of the Orc1 domain architecture depicting the relative positions of conserved bromoadjacent homology (BAH), AAA+ ATPase, and WH domains. Truncations used in A are indicated, as is the region that stimulates ATP-dependent DNA binding (dotted box). A sequence LOGO of this region (corresponding to amino acids 490–530 in DmOrc1), generated using an alignment of metazoan Orc1 protein sequences, reveals several highly conserved basic amino acid residues, including R492, K523, and R528 in DmOrc1. Asterisks denote basic residues that are either missing in several metazoan Orc1 sequences or are not conserved in DmOrc1. (C) Changing conserved basic Orc1 amino acid residues to glutamate decreases the ATP-dependent DNA-binding activity of metazoan ORC. DmORC containing the Orc1R492E/K523E/R528E triple mutant (1RKR-EEE) was purified, and its DNA-binding activity was tested in the presence of ATP by FA as in A.
We next generated multiple sequence alignments of the segment just upstream of the AAA+ domain of metazoan Orc1 proteins. This analysis revealed that the region around amino acids 490–530 of Drosophila Orc1 contains a number of arginines and lysines, several of which are highly conserved (Fig. 4B). To test whether these basic residues are important for ATP-mediated ORC–DNA interactions, we substituted three of these residues (R492, K523, and R528) with glutamate and purified the respective hexameric mutant ORC complex (referred to as DmORCOrc1RKR-EEE; SI Appendix, Fig. S3B). In fluorescence anisotropy experiments, we observed that these charge-reversal mutations impede the ATP-dependent association of DmORC with DNA, increasing the apparent Kd for this interaction by ∼15-fold (from 9 to 149 nM) (Fig. 4C). Collectively, these results further support the notion that N-terminal truncations of ORC subunits do not function to stabilize ORC’s autoinhibited conformation but, instead, directly stabilize high-affinity, ATP-dependent interactions of ORC with DNA, perhaps by directly contacting the duplex.
ORC–Cdc6 Bends DNA.
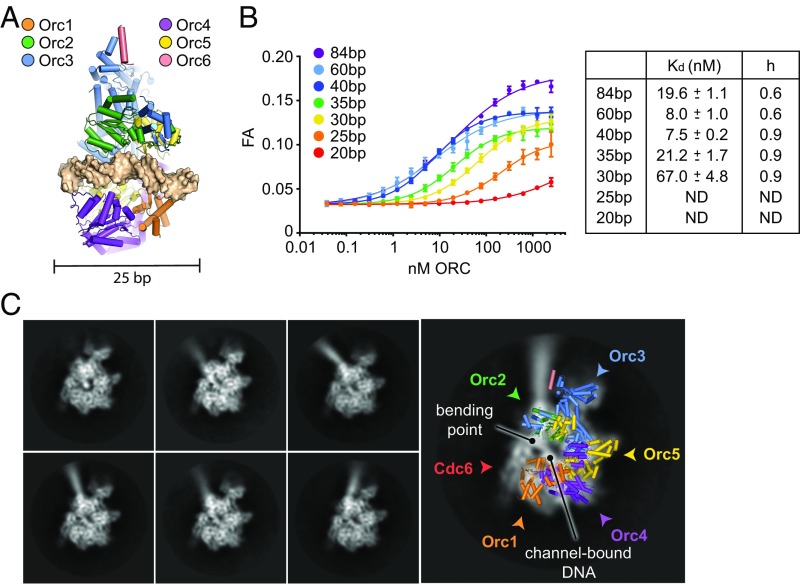
In vivo and in vitro footprinting studies have found that budding yeast ORC footprints 45–50 bp of DNA (13, 25, 27, 28), yet structural studies of ORC in isolation or in complex with other initiation factors suggest that only 20–25 bp of DNA are bound by ORC’s central channel (6, 8) (Fig. 5A). To reconcile this discrepancy, we investigated how DNA duplex length influences the ATP-dependent DNA-binding activity of DmORC by fluorescence anisotropy (Fig. 5B). The resultant data show that the affinity of DmORC for DNA steadily increases from 20 bp up to 40 bp. Extending the length further (e.g., 60 or 84 bp) appears to mildly impede DNA binding, as indicated by a slight increase in the Kd and a decrease in the apparent Hill coefficient; this negative cooperativity and affinity decrease may reflect an attempt by two ORC molecules to associate with DNA that cannot fully accommodate both complexes. These results show that 40 bp is a suitable length of DNA that can be fully engaged by ORC in an ATP-dependent manner. Although competition assays indicate that very long (>3 kb) linear and supercoiled DNAs bind somewhat better (approximately threefold) to the complex than a 40-bp duplex in competition assays, this preference likely is due to the activity of more distal DNA-binding elements such as the Orc6 TFIIB domain (SI Appendix, Fig. S5).
Fig. 5.
ATP-dependent DNA binding by ORC–Cdc6 leads to DNA bending. (A) Docking of a DNA duplex into DmORC’s central channel (based on ref. 6) shows that it can only accommodate 20–25 bp of a DNA duplex. (B) DNA length requirements for ATP-dependent DNA binding by full-length DmORC were assessed by fluorescence anisotropy (FA) using fluorescein-labeled DNA duplexes of indicated lengths in the presence of ATP. Kds and SEs of the parameter fits are listed for DNA duplexes that lead to saturation of DNA binding. ND, Kds could not be determined because binding curves did not saturate. (C) Cryo-EM analysis of DmORC in the presence of DNA and DmCdc6 shows that DNA is bound in ORC’s central channel and is bent toward the domain-swapped Orc2 AAA+/Orc3 WH and Orc3 AAA+/Orc5 WH domains upon exit from one side of the complex. Representative 2D class averages are shown. The locations of ORC subunits and Cdc6 are revealed by superpositioning of a structural model of activated DmORC and highlighted by arrowheads.
Considering that stable ORC–DNA interactions rely on the N-terminal Orc1 basic patch (Figs. 3 and 4) and on DNA duplexes that are almost twice as long as ORC’s central channel (Fig. 5B), we postulated that contacts between DNA and ORC may occur outside the core AAA+/WH domain region. To visualize how ORC might engage DNA more directly, we turned to cryo-EM. Because Cdc6 stabilizes ORC on DNA (Fig. 3E and SI Appendix, Fig. S4), a DmORC–DNA–Cdc6 complex was used for these studies. Two-dimensional classification yielded averages that resembled free ORC (in the usual mix of autoinhibited and active states) or ORC in complex with DNA and Cdc6 (Fig. 5C and SI Appendix, Fig. S6). Interestingly, DNA density was clearly visible in class averages of the ternary complex, and appeared to both emerge from ORC’s central channel and bend toward the domain-swapped Orc2 AAA+/Orc3 WH and Orc3 AAA+/Orc5 WH domains (Fig. 5C). Although the position of the DNA appeared relatively fixed near the ORC–Cdc6 density, the distal DNA end of the 84-bp duplex showed considerable flexibility. Unfortunately, the class averages of the ORC–DNA–Cdc6 complex showed a strong preferred orientation, which prevented us from obtaining sufficiently different views (even from multiple datasets) that would allow us to reconstruct a reliable 3D volume. Nonetheless, our results demonstrate that ORC in complex with Cdc6 (and likely also on its own) engages DNA through a combination of interactions that involve ORC’s central channel and elements outside of this region, and that ORC is capable of bending the duplex through these interactions.
Discussion
Conserved Conformational States in Metazoan ORC.
In the present study, chemical cross-linking and mass spectrometry, as well as EM of negatively-stained human and DmORC assemblies, were used to establish the existence and conservation of an autoinhibited ORC state and to probe for possible mechanisms of activation. Cross-linking/mass spectrometry data show that apo DmORC can exist in an autoinhibited state, providing an independent solution-based confirmation of both our previous crystal and ATPγS-DmORC EM structures (Fig. 1 A and B and SI Appendix, Fig. S1). Further EM analysis of DmORC also demonstrates that Orc1’s autoinhibited conformation is, in part, controlled by the nucleotide-binding status of DmORC and that ATP unfreezes the complex, allowing it to sample the active state (Fig. 2A). Interestingly, however, ATP alone was not sufficient to fully drive the entirety of the DmORC population into an active state (Fig. 2A). It is unclear whether additional factors such as posttranslational modifications or specific ORC-binding proteins might further stimulate or block DmORC activation. The identification of such signals will be an important goal for future studies.
The distinct responses of DmORC to different nucleotide states suggest that these conformational transitions have functional relevance. If this were true, one would also expect the autoinhibited state to be conserved, at least across metazoans. This prediction is borne out by our EM data of human ORC1–5, which show that this complex, like DmORC, can adopt an autoinhibited state under ATPγS conditions (Fig. 1C). The recent crystallographic visualization of an ATP-bound human Orc1/4/5 subcomplex in an active state is consistent with a nucleotide-dependent activation model; however, the same study did not observe autoinhibited human ORC1–5 in the presence of ATPγS using EM (9). This difference could be due to the instability of the human ORC1–5 assembly used in the prior study, which may have limited the ability to detect the autoinhibited state.
The difference between ATP and ATPγS in supporting formation of the active DmORC state is striking and, at first glance, might appear at odds with the known ability of ATPγS to stimulate the formation of ORC-dependent replication initiation intermediates in Drosophila and other systems (8, 11, 13, 30, 31). However, we find ATPγS does stimulate DNA binding compared to nucleotide-free conditions; it just does so less efficiently compared with ATP. This finding is in line with the smaller number of active DmORC particles we observe by EM in the presence of ATPγS versus ATP. These observations suggest that the orientation and position of the γ-phosphate of ATP might be important for stabilizing a closed Orc1/4 ATPase site. DNA binding by ORC is likely further stabilized by ATP-independent, secondary DNA-binding sites in the context of longer DNA duplexes and by the recruitment of Cdc6 (Fig. 2C and SI Appendix, Fig. S4), which closes a gap or “gate” between the Orc1 and Orc2 subunits to prevent the escape of DNA from ORC’s central channel (6, 25, 26). The engagement of substrate DNA by ATP-independent, secondary DNA-binding sites could also explain why prior studies have found that ATP only modestly stimulates the binding of metazoan ORC to long DNA substrates (11, 12, 15), whereas we observe a 100-fold affinity difference between nucleotide-dependent and -independent binding modes using short DNA duplexes in the absence of any competitor DNA (Fig. 2B).
Protein and DNA Requirements for High-Affinity ORC–DNA Interactions.
In addition to providing insights into the activation mechanism of DmORC, the present study advances our understanding of how ORC associates with DNA. Notably, we uncovered a basic patch in metazoan Orc1 that is essential for ATP-dependent ORC–DNA interactions (Figs. 3 and 4). The basic patch resides in the intrinsically disordered amino-terminal region of Orc1, preceding the ATPase domain, and thus is distinct from the DNA-binding elements of ORC’s central channel. This N-terminal Orc1 region likely corresponds to the enhanced eukaryotic origin sensor (EOS) motif described for Saccharomyces cerevisiae Orc1 (32). However, whereas the budding yeast EOS was found to be dispensable for ORC binding to nonorigin DNA, and was therefore proposed to play a role in sequence-specific origin recognition (32), our findings suggest that the basic patch is a more general DNA-binding element that cooperates with AAA+ and WH domains in ORC’s central channel to stabilize ORC on DNA when ATP is present. Interestingly, a helical insert in the Orc4 WH domain, which is specific to the Saccharomycetes group of fungi, was recently identified and appears to confer sequence-specific recognition of budding yeast origins (8), suggesting the S. cerevisiae Orc1-EOS might not be crucial for this activity. Our DNA-binding studies also show that amino acids 1–439 in DmOrc1, which correspond to residues 1–394 in human Orc1, are not required for high-affinity ORC–DNA interactions (Fig. 4A), although this region in human Orc1 has previously been implicated to perform a similar function to the S. cerevisiae Orc1-EOS element (32). This discrepancy can be explained by the poor overall conservation of the Orc1 N terminus, which likely led to a misalignment of the budding yeast EOS with metazoan Orc1 regions in the study of Kawakami et al. (32).
Prior structural studies of ORC, both alone and in complex with other initiation factors, have highlighted DNA-binding elements in ORC’s central channel that mediate ATP-dependent interactions with DNA over a duplex length of ∼20–25 bp (6, 8) (Fig. 5A). However, in testing the length dependencies of the ATP-mediated association of ORC with DNA, we found that ORC not only binds relatively poorly to duplexes of this size but that its affinity for DNA increases as the substrate is lengthened, plateauing at 40 bp (Fig. 5B). This length requirement agrees well with the size of the footprint identified for budding yeast ORC in vivo and in vitro (13, 28), and suggests that ORC-DNA contacts occur outside the AAA+/WH domain core. In agreement with this notion, we observe that DNA is significantly bent in 2D cryo-EM class averages of an ORC–DNA–Cdc6 complex (Fig. 5C). The DNA bend is likely induced by the initiator itself, as DmORC was previously found to wrap DNA and alter DNA linking number (14, 18); similar observations have also been reported using ORC from other species (19, 33). Due to lack of sufficient particle orientations, we cannot conclude with certainty from our EM data that DNA bending occurs as the duplex exits ORC’s channel at the WH face (as opposed to the AAA+ face); however, we favor this interpretation because the WH domains of ORC and Cdc6 serve as a binding interface for Mcm2–7 during helicase loading (6, 8). From this viewpoint, the path of the DNA in these class averages suggests that bending might be mediated by interactions of WH domains of Orc3 and/or Orc5 with DNA, although the exact nature of these contacts remains unclear.
Surprisingly, our biochemical studies did not reveal a role for Orc6 in facilitating the ATP-dependent association of ORC with DNA (Fig. 3). Orc6 contains a TFIIB-like domain that has been shown to bind DNA in studies using the Drosophila and human proteins (16, 17, 34). Drosophila Orc6 was also reported to be essential for ATP-dependent DNA binding by DmORC (11). Our results show that the Orc6 TFIIB-like domain is dispensable for the formation of high-affinity ORC-DNA contacts, suggesting it may fulfill other functions. In addition, prior DNA-binding studies with DmORC were performed with ATPγS and with high competitor DNA concentrations. Thus, it is possible that previous assays measured a relatively incremental change between nonspecific and ATP-dependent DNA binding. Our results do not preclude a function for metazoan Orc6 in targeting ORC to chromosomal origins or in stabilizing ORC on long DNA fragments; however, they do reconcile apparent differences between studies involving budding yeast, human, and Drosophila ORC, and are consistent with reports that Orc6 is not required for ATP-dependent recruitment of ORC or Cdc6 to DNA in biochemical assays in yeast and human systems (11, 12, 19, 35).
Implications for ORC Activation and Mcm2–7 Loading.
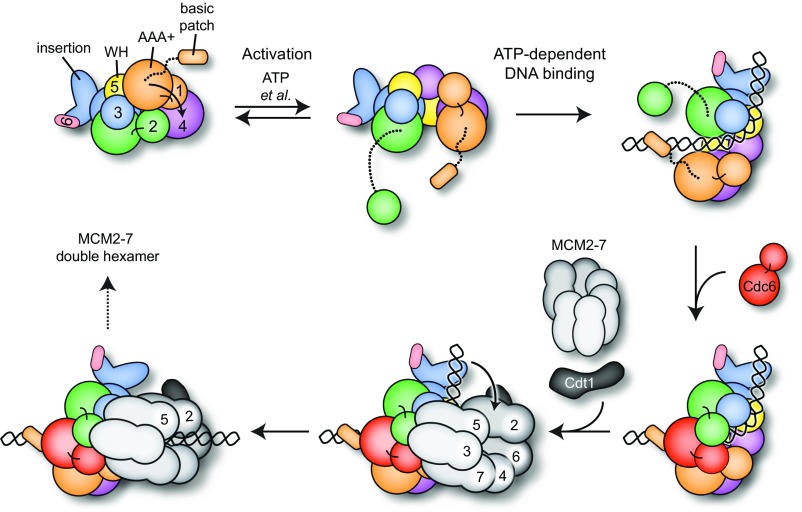
Collectively, the present findings demonstrate that both ATP and a previously uncharacterized basic patch in the Orc1 N terminus that lies proximal to the subunit’s ATPase domain help to regulate the DNA-binding activity of metazoan ORC. ATP permits a structural transition to occur between two different conformational states, one active and one autoinhibited, which we have shown to occur in both Drosophila and human ORC (Fig. 6). Switching back and forth between both conserved conformations may be exploited in vivo to regulate ORC’s capability to productively engage origins, as only the active state sterically permits the binding of DNA in ORC’s central channel; this event also requires the Orc1 basic patch (or EOS) that cooperates with other elements in the channel to establish high-affinity, ATP-dependent ORC-DNA contacts.
Fig. 6.
Model for ORC activation, ATP-dependent DNA binding, and Mcm2–7 loading. ATP binding to the Orc1/4 ATPase site and other as yet unidentified factors allows ORC to more readily sample an active configuration that enables the ATP-dependent binding of DNA to the Orc1 basic patch and to elements in the AAA+/WH domain channel, as well as Cdc6 association. DNA and Cdc6 binding is accompanied by bending of the DNA toward the Orc2 AAA+/Orc3 WH and Orc3 AAA+/Orc5 WH units (the WH elements domain swap with the AAA+ modules of adjacent subunits), which allows an Mcm2–7 hexamer to dock onto the ORC–Cdc6 ring in a manner that aligns the DNA duplex with the Mcm2/5 gate. Conformational changes in ORC likely alleviate DNA-ORC contacts required for bending, allowing a straightened DNA segment to engage the Mcm2–7 pore.
Our observation that Cdc6 can partially overcome the need for the Orc1 basic patch in ATP-stimulated DNA-binding studies indicates that Cdc6 coassociation further stabilizes DNA association with ORC, likely by simply sealing off the Orc1/2 gate that allows duplex entry and exit. As DNA extends beyond the ORC–Cdc6 channel, contacts between the duplex and the WH surface of ORC appear to bend DNA away from the channel axis (Fig. 6). We speculate that DNA bending is required to provide space for the formation of a productive encounter complex between ORC–DNA–Cdc6 and Mcm2–7–Cdt1 before DNA insertion into the pore of an Mcm2–7 heterohexamer, which is likely mediated by interactions between Mcm3 and Cdc6 (30, 35) and does not have to occur at a sharp angle if DNA is bent away. Interestingly, in the structure of the budding yeast OCCM, a DNA entry gate present between the Mcm2 and Mcm5 subunits is positioned near the WH domains of Orc3 and Orc5 (8). This juxtaposition suggests that DNA is bent toward these elements of ORC to help align the DNA duplex with the crack in the Mcm2–7 ring, facilitating helicase loading (Fig. 6). Future efforts aimed at capturing, characterizing, and imaging different pre- and postloading intermediates will be needed to elaborate on the precise mechanism of Mcm2–7 loading and the role of the Orc1 and Cdc6 ATPase in controlling structural transitions during this process.
Materials and Methods
Expression and Purification of ORC.
Drosophila and human ORC assemblies used in this study were expressed and purified from High5 cells as described previously, with minor modifications (SI Appendix, SI Materials and Methods).
DNA and Cdc6 Binding by ORC.
DNA binding to DmORC was measured using a fluorescence anisotropy-based DNA-binding assay using a 40-bp (Table 1) fluorescein-labeled duplex DNA in a Clariostar or a Pherastar FSX plate reader (BMG). Anisotropy averages and the SD of at least three independent experiments were plotted as a function of ORC concentration, and data points were fitted to the Hill equation to obtain Kds. To investigate the DNA length dependence of ORC–DNA interactions, duplex lengths were varied from 20 to 84 bp (Table 1). Competition DNA-binding experiments were performed as described in SI Appendix, SI Materials and Methods. ORC–DNA interactions were also confirmed in pull-down assays using an 84-bp ARS1-derived DNA duplex (Table 1) labeled with a photocleavable 5′-biotin (IDT) as bait.
Table 1.
DNA oligonucleotides used for fluorescence DNA-binding assays
| DNA duplex length/name | DNA duplex sequence |
| 20-bp duplex | |
| Top | 5′-FluorT/AGATCTAAACATAAAATCTG-3′ |
| Bottom | 5′-CAGATTTTATGTTTAGATCT-3′ |
| 25-bp duplex | |
| Top | 5′-FluorT/CATAAAAGATCTAAACATAAAATCT-3′ |
| Bottom | 5′-AGATTTTATGTTTAGATCTTTTATG-3′ |
| 30-bp duplex | |
| Top | 5′-FluorT/GCAAGCATAAAAGATCTAAACATAAAATCT-3′ |
| Bottom | 5′-AGATTTTATGTTTAGATCTTTTATGCTTGC-3′ |
| 35-bp duplex | |
| Top | 5′-FluorT/GAAAAGCAAGCATAAAAGATCTAAACATAAAATCT-3′ |
| Bottom | 5′-AGATTTTATGTTTAGATCTTTTATGCTTGCTTTTC-3′ |
| 40-bp duplex | |
| Top | 5′-FluorT/TTTTGAAAAGCAAGCATAAAAGATCTAAACATAAAATCTG-3′ |
| Bottom | 5′-CAGATTTTATGTTTAGATCTTTTATGCTTGCTTTTCAAAA-3′ |
| 60-bp duplex | |
| Top | 5′-FluorT/CCTGCAGGCCTTTTGAAAAGCAAGCATAAAAGATCTAAACATAAAATCTGTAAAATAACA-3′ |
| Bottom | 5′-TGTTATTTTACAGATTTTATGTTTAGATCTTTTATGCTTGCTTTTCAAAAGGCCTGCAGG-3′ |
| 84-bp duplex | |
| Top | 5′-FluorT/TTTGTGCACTTGCCTGCAGGCCTTTTGAAAAGCAAGCATAAAAGATCTAAACATAAAATCTGTAAAATAACAAGATGTAAAGAT-3′ |
| Bottom | 5′-ATCTTTACATCTTGTTATTTTACAGATTTTATGTTTAGATCTTTTATGCTTGCTTTTCAAAAGGCCTGCAGGCAAGTGCACAAA-3′ |
Fluor, fluorescein.
To investigate the ability of different ORC assemblies to associate with Cdc6, we performed pull-down assays using MBP-tagged DmCdc6 as bait. Full-length, wild-type DmORC or DmORC assemblies lacking specific N-terminal regions (main text) were incubated with 84-bp duplex DNA and DmCdc6, either in the absence or presence of 1 mM nucleotide. DmCdc6 and interacting proteins were purified using amylose beads (New England Biolabs) and analyzed by SDS/PAGE electrophoresis and silver staining.
Chemical Cross-Linking and Mass Spectrometry.
Full-length, wild-type DmORC or the DmORC core assembly was cross-linked with 100 μM DSS at 1 mg/mL of the respective DmORC assembly. After incubation for 45 min at 25 °C, the cross-linking reaction was quenched with ammonium bicarbonate and proteins were digested with endoproteinase Lys-C (Wako) and trypsin protease (Promega; also SI Appendix, SI Materials and Methods). The digested sample was fractionated by size exclusion chromatography, and fractions were analyzed by LC-MS/MS on an Orbitrap Fusion Lumos mass spectrometer as described previously (20, 36). The resulting data were searched against the database containing sequences for full-length or “trimmed” DmORC subunits with xQuest (20, 37).
EM.
For negative-stain EM, full-length Drosophila ORC, DmORC lacking the N-terminal 532-aa residues in Orc1, or human ORC1–5 was spotted onto glow-discharged, continuous-carbon film EM grids and stained with 2% uranyl formate. Grids were imaged either in a Tecnai T12 TWIN transmission electron microscope operated at 100 keV or a Tecnai G2 Spirit BIOTWIN electron microscope operated at 120 kV, both equipped with a LaB6 cathode as an electron source and a FEI Eagle CCD camera.
For cryo-EM, the ORC–DNA–Cdc6 complex was assembled by mixing full-length DmORC, an 84-bp DNA duplex (Table 1), and DmCdc6 at final concentrations of 80 and 100 nM, respectively. Four microliters of sample were applied for 22 s at 22 °C and 100% humidity to a glow-discharged, 400-mesh, C-flat 1.2/1.3 EM grid coated with a thin film of continuous carbon and plunge-frozen in liquid ethane using a Vitrobot. Cryo-EM grids were imaged in a Titan Krios electron microscope (at Janelia Research Campus) operated at 300 kV and equipped with a spherical aberration (Cs) corrector, an energy filter (slit width of 20 eV), and a post-GIF Gatan K2 Summit direct electron detector. Data collection and image processing were performed as described in SI Appendix, SI Materials and Methods. Although high-resolution features were visible in 2D class averages, a strong preferred orientation of the particles on the thin carbon layer unfortunately prevented the reconstruction of any reliable 3D volume.
Supplementary Material
Acknowledgments
We thank Rick Huang and Zhiheng Yu (Janelia Research Campus) for assistance with the cryo-EM data collection. This work was supported by the Novartis Research Foundation (F.B.), the National Cancer Institute (Grant R01-CA030490 to J.M.B. and M.R.B.), and the European Research Council (Grant ERC-2014-AdG 670821 to R.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The crosslinking mass spectrometry data has been deposited to the PRIDE (Proteomics Identifications) data repository under the identifier PXD009526.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806315115/-/DCSupplemental.
References
- 1.Bleichert F, Botchan MR, Berger JM. Mechanisms for initiating cellular DNA replication. Science. 2017;355:eaah6317. doi: 10.1126/science.aah6317. [DOI] [PubMed] [Google Scholar]
- 2.Riera A, et al. From structure to mechanism-understanding initiation of DNA replication. Genes Dev. 2017;31:1073–1088. doi: 10.1101/gad.298232.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 5.Duncker BP, Chesnokov IN, McConkey BJ. The origin recognition complex protein family. Genome Biol. 2009;10:214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleichert F, Botchan MR, Berger JM. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519:321–326. doi: 10.1038/nature14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 8.Yuan Z, et al. Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol. 2017;24:316–324. doi: 10.1038/nsmb.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tocilj A, et al. Structure of the active form of human origin recognition complex and its ATPase motor module. eLife. 2017;6:e20818. doi: 10.7554/eLife.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc Natl Acad Sci USA. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giordano-Coltart J, Ying CY, Gautier J, Hurwitz J. Studies of the properties of human origin recognition complex and its Walker A motif mutants. Proc Natl Acad Sci USA. 2005;102:69–74. doi: 10.1073/pnas.0408690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 14.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vashee S, et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S, et al. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci USA. 2011;108:7373–7378. doi: 10.1073/pnas.1013676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarey MG, Botchan M, Nogales E. Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J Struct Biol. 2008;164:241–249. doi: 10.1016/j.jsb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc. 2014;9:120–137. doi: 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- 21.Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci USA. 2006;103:4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282:32370–32383. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima T, Takahashi N, Stillman B. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- 24.Seki T, Diffley JF. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc Natl Acad Sci USA. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J, et al. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012;20:534–544. doi: 10.1016/j.str.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 28.Diffley JF, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- 29.Bleichert F, et al. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. eLife. 2013;2:e00882. doi: 10.7554/eLife.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J, et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20:944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami H, Ohashi E, Kanamoto S, Tsurimoto T, Katayama T. Specific binding of eukaryotic ORC to DNA replication origins depends on highly conserved basic residues. Sci Rep. 2015;5:14929. doi: 10.1038/srep14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houchens CR, et al. Multiple mechanisms contribute to Schizosaccharomyces pombe origin recognition complex-DNA interactions. J Biol Chem. 2008;283:30216–30224. doi: 10.1074/jbc.M802649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci USA. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eliseev B, et al. Structure of a human cap-dependent 48S translation pre-initiation complex. Nucleic Acids Res. 2018;46:2678–2689. doi: 10.1093/nar/gky054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walzthoeni T, et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9:901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.