Significance
Type I interferons (IFNs) are cytokines that are essential for host defense against virus infection, but when they are inappropriately produced, they can cause severe autoimmune disease in humans. We have found that the sumoylation pathway of protein modification is essential for preventing an ectopic IFN response. Specifically, we have identified SUMO2 and SUMO3 as the two SUMO proteins that redundantly prevent IFN production. Remarkably, the potent IFN response caused by loss of SUMO2/3 is independent of all known inducers of the antiviral response, revealing a distinct mechanism of IFN production that has implications for our understanding of antiviral immunity.
Keywords: innate immunity, type I interferons, sumoylation
Abstract
Detection of nucleic acids by innate immune sensors triggers the production of type I interferons (IFNs). While IFNs are essential for host defense against viral infection, dysregulated production of IFNs underlies numerous autoinflammatory diseases. We have found that the loss of sumoylation results in a potent, spontaneous IFN response. Vertebrates possess three small ubiquitin-like modifiers (SUMOs) that can be conjugated onto target proteins and alter protein function in diverse but still poorly characterized ways. We demonstrate that regulation of IFN by sumoylation is redundantly mediated by both SUMO2 and SUMO3, but not SUMO1, revealing a previously unknown function of SUMO2/3. Remarkably, this IFN response is independent of all known IFN-inducing pathways and does not require either of the canonical IFN-associated transcription factors IRF3 or IRF7. Taken together, our findings demonstrate that SUMO2 and SUMO3 are specific and essential negative regulators of a noncanonical mechanism of IFN induction.
Detection of foreign nucleic acids by innate pattern recognition receptors triggers an antiviral response through the production of type I interferons (IFNs). These potent antiviral cytokines signal in both an autocrine and paracrine fashion through the IFN alpha receptor (IFNAR) to induce expression of hundreds of antiviral genes known as IFN-stimulated genes (ISGs). There are four principal ways of triggering the IFN response in vertebrates. First, cGMP-AMP synthase (cGAS) detects double-stranded intracellular DNA and signals through the adaptor protein STING to activate the kinase TBK1 and the transcription factor IFN regulatory factor 3 (IRF3), leading to the transcriptional activation of the IFNB1 gene (1). Second, the RIG-I–like receptors (RLRs) detect intracellular viral RNA and bind to the adaptor protein MAVS to trigger a similar TBK1-IRF3 response (2). Third, toll-like receptors (TLRs) 3 and 4 use the adaptor protein TRIF to activate TBK1-IRF3 (3). Finally, TLRs 7 and 9 signal through MyD88 and IRF7, specifically in plasmacytoid dendritic cells, to activate a potent IFN response (4). Importantly, all of the known IFN-inducing pathways require IRF3 and/or IRF7 (4), revealing a conserved module that mediates the canonical IFN response.
These nucleic acid sensing pathways are essential for antiviral defense, but they must be tightly regulated to avoid inappropriate activation by endogenous RNA and DNA. One key mechanism that sets thresholds for activation of the intracellular nucleic acid sensors involves the activity of enzymes that modify or metabolize self nucleic acids. One example of this form of regulation is three-prime repair exonuclease 1 (TREX1), a DNA exonuclease that prevents the activation of cGAS by endogenous DNA (5, 6). In humans, loss of function mutations in the TREX1 gene cause Aicardi–Goutieres Syndrome (AGS), a rare and severe autoimmune disease that presents with symptoms similar to those of a congenital viral infection (7). Several additional AGS genes have been identified, all of which encode proteins important for nucleic acid metabolism or sensing (8). Moreover, numerous other monogenic autoinflammatory diseases are characterized by a type I IFN signature that can be identified in peripheral blood cells, collectively referred to as “interferonopathies” (8). The underlying causes of these rare interferonopathies can be classified into three categories: those that cause excessive activation of nucleic acid sensors, those that impact type I IFN receptor signaling, and those that exert their effects through currently unknown mechanisms (8–10). Importantly, the genetic definition of these rare diseases and their underlying mechanisms has provided insights into a number of more common human autoimmune disorders that share a type I IFN signature as a defining feature, including systemic lupus erythematosus, systemic sclerosis, and Sjogren’s syndrome. To understand the dysregulation underlying the autoinflammatory and autoimmune diseases associated with a type I IFN signature, it is important to understand the full breadth of mechanisms that regulate type I IFN responses.
We have found that loss of sumoylation triggers a potent, spontaneous type I IFN response in the absence of exogenous stimuli. Conjugation of small ubiquitin-like modifiers (SUMOs) onto lysines of target proteins can affect protein function in diverse but still poorly understood ways. Of the three vertebrate SUMOs, we have determined that monomeric SUMO2 and SUMO3 are essential for preventing this spontaneous IFN response, while SUMO1 is not involved. To our surprise, the IFN response driven by loss of sumoylation is independent of STING and MAVS, the type I IFN receptor, the kinases TBK1/IKKε/IKKα/IKKβ, and the canonical IFN-β transcription factors IRF3 and IRF7. Our findings reveal that loss of sumoylation triggers a potent IFN response through the activation of an unanticipated, noncanonical mechanism that is independent of all known IFN-inducing pathways. This regulatory mechanism may be relevant for IFN-associated autoimmune disorders.
Results
Loss of Sumoylation Triggers a Spontaneous Type I IFN Response.
In a yeast 2-hybrid screen for proteins that interact with Trex1, we identified the E1 SUMO ligase SAE2. We therefore hypothesized that sumoylation of Trex1 might influence its function as a negative regulator of the type I IFN response. Sumoylation requires the heterodimeric E1 ligase comprised of SAE1 and SAE2 and the monomeric E2 ligase UBC9. To determine whether sumoylation limits type I IFN responses, we employed a lentiCRISPR approach to target the human genes that encode the E1 and E2 SUMO ligases: SAE1, UBA2 (SAE2), and UBE2I (UBC9). We transduced human THP-1 monocytes with lentiviral constructs encoding the endonuclease Cas9 and a guide RNA (gRNA) specific for each gene. As controls, we transduced THP-1s with constructs encoding a nontargeting gRNA and a gRNA specific for TREX1, a known negative regulator of the cytosolic DNA sensing pathway. Loss of each SUMO ligase was verified by Western blot (Fig. 1A). Interestingly, we found that disruption of SAE1 resulted in decreased expression of SAE2 protein and vice versa, demonstrating that each component of the E1 SUMO ligase is essential for the stability of its partner. After differentiation of THP-1 cells with PMA, we used quantitative RT-PCR to evaluate expression of IFNB1 and the IFN stimulated genes (ISGs) IFI27 and ISG15 (Fig. 1A). Disruption of SAE1, UBA2 (SAE2), or UBE2I (UBC9) resulted in potently elevated expression of IFNB1, IFI27, and ISG15 mRNA compared with the nontargeting control (Fig. 1 B and C). Moreover, we found that SUMO ligase-targeted cells had dramatically elevated IFNB1 expression in comparison with TREX1-targeted cells (Fig. 1B). Therefore, disruption of SUMO ligases in THP-1 cells drives an aberrant and potent type I IFN response in the absence of exogenous stimulation. These data corroborate the recent report of enhanced IFN and ISG responses caused by inducible deletion of Ube2i (UBC9) in mouse cells (11) and reveal that each of the essential components of the sumoylation pathway are required to prevent a spontaneous IFN response.
Fig. 1.
Loss of sumoylation triggers a spontaneous IFN response in THP-1 cells. THP-1 monocytes were targeted by lentiCRISPR endoding the indicated gRNAs. (A) Western blot evaluation of the indicated proteins. (B and C) Evaluation of IFNB1, IFI27, and ISG15 mRNA expression is shown in PMA-differentiated THP-1s by quantitative RT-PCR. (D) THP-1 monocytes were transduced with lentiviruses encoding GFP, wild-type SAE2 (WT), and a catalytic mutant of SAE2 (C173S), and then transduced with a lentiCRISPR lentivirus encoding Cas9 and the indicated gRNA. Western blot evaluation of the indicated proteins. (E) Evaluation of IFNB1, IFI27, and ISG15 mRNA expression in PMA-differentiated THP-1s by quantitative RT-PCR. Statistical analysis in B was performed using a one-way ANOVA and comparing control cells to targeted cells. In D, a two-way ANOVA was used, comparing GFP expressing THP-1s to WT and C173S reconstituted cells. Both tests corrected for multiple comparisons using the Holm–Sidak method. n = 3–5 where n is the number of unique polyclonal cell lines. n.s., not significant; *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001.
To confirm that the elevated IFN response in SUMO ligase-deficient cells is due to loss of sumoylation and not due to an unanticipated function of these enzymes, we tested whether the catalytic activity of SAE2 was required to prevent the IFN response. Cysteine 173 of SAE2 forms a thioester bond between SAE2 and SUMO, allowing the subsequent transfer of SUMO to a lysine residue on a target protein (12). We generated constructs encoding HA-tagged wild-type or C173S SAE2, rendered CRISPR resistant by silent mutations in the UBA2 (SAE2) gRNA targeting site. We transduced THP-1 cells with lentiviral constructs encoding a GFP control, wild-type SAE2 (WT), or C173S SAE2, and then subsequently disrupted the endogenous UBA2 (SAE2) gene. Successful reconstitution and subsequent targeting after puromycin selection was verified by Western blot (Fig. 1D). Expression of wild-type SAE2, but not the C173S catalytic mutant, prevented the expression of IFNB1, IFI27, and ISG15 in SAE2-targeted cells (Fig. 1E). Thus, the catalytic activity of SAE2 is essential for the regulation of type I IFN by SUMO ligases.
The E1 and E2 SUMO ligases frequently act in concert with one of several E3 SUMO ligases. However, and unlike the E1 and E2 ligases, the E3 ligases are not essential for SUMO conjugation onto all substrates, but are instead thought to enhance sumoylation of specific substrates (12). Previous research indicates that the E3 ligase protein inhibitor of activated STAT1 (PIAS1) dampens inflammatory and IFN responses to exogenous ligands through regulation of both IRF3 and STAT1 (13, 14). To determine whether PIAS1 or other well characterized E3 SUMO ligases in the PIAS family prevent a spontaneous IFN response, we used lentiCRISPR to disrupt the genes encoding PIAS1, PIAS2, PIAS3, and PIAS4 (SI Appendix, Fig. S1A). We found that disruption of each PIAS gene did not result in increased expression of IFNB1, IFI27, or ISG15 compared with the nontargeting control (SI Appendix, Fig. S1B), indicating that no individual PIAS gene is essential for the regulation of IFN by sumoylation.
SUMO2 and SUMO3 Redundantly Inhibit a Spontaneous IFN Response.
How might SUMOylation prevent a spontaneous IFN response? Like ubiquitination, conjugation of proteins with SUMO can modify their functions in diverse ways. However, the functional consequences of SUMOylation are less understood than those of ubiquitination. This is in part because there are three SUMO genes that encode three SUMO proteins. SUMO2 and SUMO3 are 97% identical to each other, whereas SUMO1 is only 50% identical to SUMO2/3. An individual SUMOylation site in a specific target protein can be modified by more than one SUMO protein (15), suggesting redundancy of SUMO conjugation that complicates efforts to assign specific functions to each SUMO protein. However, Sumo2−/− mice are embryonic lethal at 10.5 d of gestation, whereas Sumo1−/− mice and Sumo3−/− mice are viable. At the mRNA level, SUMO2 is the most abundant, accounting for ∼80% of total SUMO mRNA in both embryonic and adult murine tissues (16).
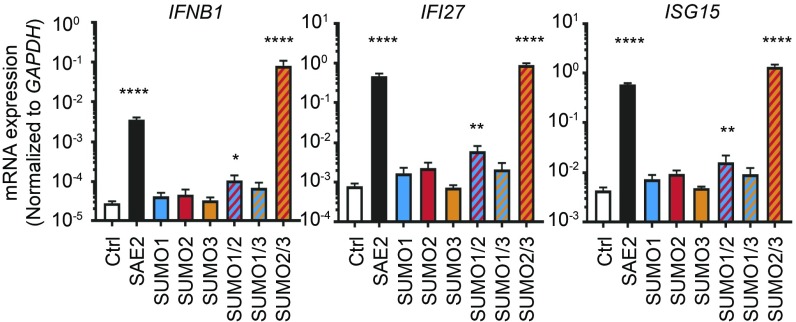
The potent IFN induction caused by loss of all SUMOylation allowed us to test whether a particular SUMO protein was required for preventing this antiviral response. We used lentiCRISPR to target each of the three SUMO genes, alone or in pairwise combinations. We confirmed targeting of SUMO proteins by Western blot (SI Appendix, Fig. S2). To our surprise, deletion of any individual SUMO gene did not result in a spontaneous IFN response. However, the combined disruption of SUMO2 and SUMO3 yielded an IFN response that was even more potent than what we observed in SAE2-targeted cells (Fig. 2). We noted a small increase in ISGs in cells doubly targeted for SUMO1 and SUMO2, likely reflecting the fact that SUMO2 is the most abundant SUMO protein in cells. Our findings demonstrate that SUMO2 and SUMO3, but not SUMO1, are potent negative regulators of the type I IFN response.
Fig. 2.
The combined loss of SUMO2 and SUMO3 triggers a spontaneous IFN response. THP-1 monocytes were transduced with lentiCRISPR lentiviral constructs encoding Cas9 and the indicated gRNAs. Evaluation of IFNB1, IFI27, and ISG15 mRNA expression is shown in PMA-differentiated THP-1s by quantitative RT-PCR. Statistical analysis was performed using a one-way ANOVA and comparing all cell lines to the nontargeting control, correcting for multiple comparisons using the Holm–Sidak method. n = 3 where n is the number of unique polyclonal cell lines. *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001.
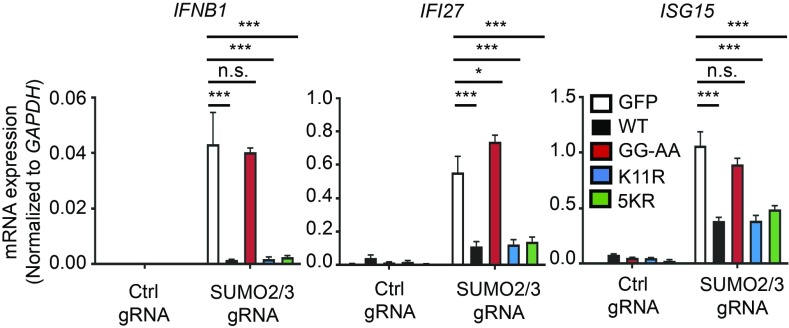
SUMO2 and SUMO3 each contain a canonical sumoylation motif at lysine 11 to which additional SUMOs can be conjugated, resulting in the formation of poly-SUMO chains. Additionally, recent proteomics studies have revealed additional SUMO conjugation sites on four other lysines: 5, 7, 21, and 33 (17). These polymeric SUMO chains are known to play important roles in the recruitment of SUMO-targeted Ubiquitin ligases (STUbLs), which mediate the polyubiquitination and proteasomal degradation of sumoylated proteins (18). To further explore how SUMO2/3 regulates type I IFN responses, and to test whether polymeric SUMO chains are required for this regulation, we generated lentiviral expression vectors encoding N terminus HA-tagged versions of wild-type SUMO2 (WT), an unconjugatable SUMO2 with a mutated diglycine motif (GG-AA), SUMO2 with the canonical polysumoylation site mutated (K11R), and SUMO2 in which all five known conjugatable lysines were mutated to arginines (5KR). Silent mutations were introduced into the gRNA target site of each SUMO2 expression vector to allow for CRISPR targeting of the endogenous gene only. THP-1 cells were transduced with lentiviral constructs encoding these SUMO2 proteins and then subsequently transduced with lentiCRISPR constructs targeting SUMO3 and SUMO2. Successful preconstitution and targeting were verified by Western blot (SI Appendix, Fig. S3). As expected based on our previous findings with the SAE2 catalytic mutant (Fig. 1), expression of GFP or the GG-AA mutant failed to rescue the type I IFN response in SUMO2/3-targeted cells. However, preconstitution of SUMO2/3-targeted cells with WT SUMO2 or either of the polysumoylation-deficient SUMO2 mutants dramatically reduced the expression of IFNB1, IFI27, and ISG15 (Fig. 3). These data corroborate the redundant functions of SUMO2/3 in IFN regulation (Fig. 2) and confirm that SUMO conjugation is essential for this regulation. Moreover, our findings implicate monosumoylation, not polysumoylation, as the key mechanism that regulates the IFN response.
Fig. 3.
The formation of polymeric SUMO2 chains is not essential for the regulation of IFN by sumoylation. THP-1 monocytes were transduced with lentiviral constructs encoding GFP, wild-type SUMO2 (WT), an unconjugatable diglycine mutant (GG-AA), a K11R mutant, or a SUMO2 mutant with all known conjugatable lysines mutated to arginines (5KR). Each cell line was then transduced with the indicated lentiCRISPR constructs. Evaluation of IFNB1, IFI27, and ISG15 mRNA expression by quantitative RT-PCR is shown in PMA-differentiated THP-1 cells. Statistical analysis was performed using a two-way ANOVA and comparing WT, GG-AA, K11R, and 5KR transduced cells to GFP-transduced cells, correcting for multiple comparisons using the Holm–Sidak method. n = 3 where n is the number of unique polyclonal cell lines. n.s., not significant; *P ≤ 0.05; ***P ≤ 0.001.
Sumoylation can target proteins for proteasomal degradation through the recruitment of SUMO-targeted ubiquitin ligases (STUbLs). Multiple components of IFN signaling pathways are degraded by the proteasome after their activation, and loss of function mutations in proteasome subunits in humans cause proteasome-associated autoinflammatory syndrome, which is associated with a chronic IFN signature (10). To determine whether SUMO-targeted ubiquitin ligases could link the type I IFN response we observe in sumoylation-deficient cells to the proteasome, we used lentiCRISPR constructs to target the two known vertebrate STUbLs, RNF4 and RNF111 (18, 19). Successful targeting was verified using a restriction fragment length polymorphism (RFLP) assay as described previously (SI Appendix, Fig. S4A). In both RNF4 and RNF111-targeted THP-1 cells, we observed no increase in IFNB1, IFI27, or ISG15 expression compared with the nontargeting control (SI Appendix, Fig. S4B). While this does not rule out a role for the proteasome in the spontaneous IFN response caused by loss of sumoylation, it does indicate that neither of the two known STUbLs are individually important for this process.
Sumoylation Prevents a Noncanonical Type I IFN Response.
We sought to identify the pathway(s) responsible for triggering the IFN response in sumoylation-deficient cells. Since the majority of known Mendelian interferonopathies are caused by mutations in regulators of nucleic acid sensing and metabolism, we first tested whether the cGAS-STING or RLR-MAVS pathways were responsible for driving the spontaneous type I IFN response in sumoylation-deficient cells. We generated lentiCRISPR constructs targeting either TMEM173 (STING) or MAVS. These cells were then transduced with nontargeting control, TREX1-targeting, or UBA2-targeting gRNAs. Successful targeting was verified by Western blot (Fig. 4A), and functional disruption of STING and MAVS was evaluated by transfection of specific nucleic acid ligands. In response to calf thymus DNA, a cGAS ligand, TMEM173 (STING)-targeted THP-1 cells had decreased expression of IFNB1 compared with both nontargeted and MAVS-targeted THP-1 cells (Fig. 4B). Similarly, MAVS-targeted cells had a reduced IFNB1 response to transfection with a triphosphate RNA ligand that specifically activates RIG-I (Fig. 4B and ref. 20). As expected, TREX1-targeted THP-1 cells, which had a milder ISG signature than SAE2-targeted cells, showed a decrease in ISG expression in TMEM173 (STING)-targeted cells, but not in MAVS-targeted cells (Fig. 4C). However, neither targeting of STING nor MAVS had any impact on the potent expression of IFNB1, IFI27, or ISG15 in SAE2-targeted THP-1 cells (Fig. 4C).
Fig. 4.
The spontaneous IFN response in sumoylation-deficient cells is not dependent on either STING or MAVS. THP-1 monocytes were transduced with lentiCRISPR lentiviral constructs encoding Cas9 and the indicated gRNAs. (A) Western blot evaluation of the indicated proteins. (B) PMA-differentiated THP-1 cells targeted with the indicated gRNAs were transfected with either 1 μg of CT-DNA or 1 μg of RIG-I ligand. Four hours after transfection, RNA was harvested and expression of IFNB1 was evaluated quantitative RT-PCR. The data are representative of two experiments. (C) Evaluation of IFNB1, IFI27, and ISG15 mRNA expression by quantitative RT-PCR in PMA-differentiated THP-1 cells. Statistical analysis was performed using a two-way ANOVA and comparing STING and MAVS-targeted cells to the nontargeted control, correcting for multiple comparisons using the Holm–Sidak method. n = 3 where n is the number of unique polyclonal cell lines. n.s., not significant; *P ≤ 0.05; ****P ≤ 0.0001.
We investigated whether induction of type I IFNs in sumoylation-deficient cells was due to enhanced signaling through the type I IFN receptor instead of through a primary ligand-activated pathway. We made a lentiCRISPR construct targeting IFNAR1, generated a population of robustly targeted cells verified by RFLP (SI Appendix, Fig. S5), and then transduced these cells with nontargeting control, UBA2 (SAE2), or UBE2I (UBC9) lentiCRISPR constructs. Functional disruption of IFNAR1 was evaluated by treatment with recombinant human IFN-β and by transfection of calf thymus DNA (Fig. 5A). In response to IFN-β treatment, we observed robust IFI27 and ISG15 that was absent in IFNAR1-targeted cells. In response to calf-thymus DNA, we observed intact but decreased inducible expression of IFNB1 in IFNAR1-targeted cells, consistent with the known enhancement of IFN production by IFNAR signaling. Importantly, the DNA-activated expression of IFI27 and ISG15 was severely impaired in IFNAR1-targeted cells (Fig. 5A), verifying functional disruption of IFNAR signaling. We then disrupted UBA2 (SAE2) and UBE2I (UBC9) in the IFNAR1-targeted cells and evaluated the expression of the primary response gene IFNB1 and the ISGs IFI27 and ISG15. We found that the potent IFNB1 response caused by loss of sumoylation was intact in IFNAR1-targeted cells, but the ISG response was severely impaired (Fig. 5B). Together, these data demonstrate that sumoylation prevents a primary, IFNAR-independent type I IFN response that is independent of both STING and MAVS.
Fig. 5.
The spontaneous IFN response in sumoylation-deficient cells is not dependent on IFNAR. THP-1 monocytes were transduced with lentiCRISPR lentiviruses encoding Cas9 the indicated gRNAs. (A) Evaluation of IFNB1, IFI27, and ISG15 mRNA expression in PMA-differentiated THP-1s treated with either 50 U/mL recombinant IFN-β or transfected with 1 μg CT-DNA for 6 h is shown. (B) Control and IFNAR1-targeted THP-1 cells were transduced with lentiCRISPR lentiviruses encoding Cas9 and the indicated gRNA. Evaluation of IFNB1, IFI27, and ISG15 mRNA expression in PMA-differentiated THP-1s by quantitative RT-PCR is shown. Statistical analysis was performed using a two-way ANOVA and comparing IFNAR1-targeted cells to the control line, correcting for multiple comparisons using the Holm–Sideak method. n = 3 where n is the number of unique polyclonal cell lines. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
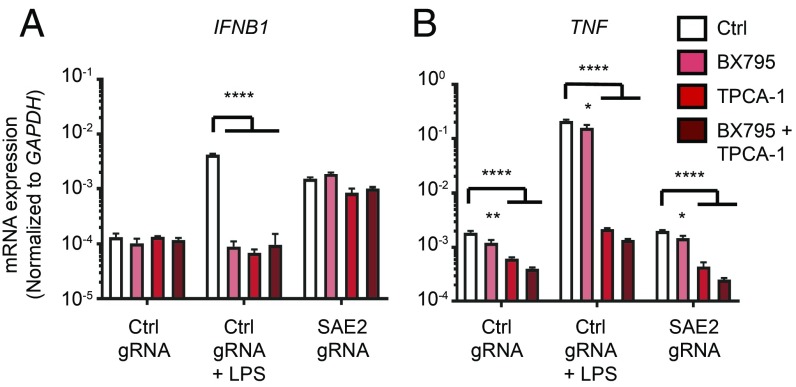
Based on the findings in Figs. 4 and 5, we explored the possibility that sumoylation may regulate type I IFN responses through shared signaling components utilized in all canonical IFN-inducing pathways. Downstream of both STING and MAVS, the related kinases TBK1 and IKKε phosphorylate and activate the transcription factors IRF3 and IRF7 (21). The Toll-like receptors TLR3 and TLR4 signal through TRIF to activate TBK1-dependent IRF3 phosphorylation (3). Finally, in plasmacytoid dendritic cells, TLR7/8/9 signal through the adaptor protein MyD88, which recruits the kinase IKKα to phosphorylate and activate IRF7 (22). Thus, the kinases TBK1/IKKε and IKKα/IKKβ represent the essential signaling nexus required for all canonical IFN-inducing pathways. To test whether these kinases contribute to the IFN response in sumoylation-deficient cells, we took advantage of the chemical inhibitors BX795 and TPCA-1, which inhibit TBK1/IKKε and IKKα/IKKβ, respectively. Nontargeted and SAE2-targeted, differentiated THP-1 cells were treated for 48 h with BX795, TPCA-1, or both of these inhibitors in combination. To verify the efficacy of these inhibitors over the complete course of treatment, nontargeted cells were treated with lipopolysaccharide (LPS) for the final 4 h before RNA harvest. LPS signals through TLR4 to activate both a type I IFN response dependent on TBK1/IKKε and an inflammatory response dependent on the activation of NF-κB by IKKα/IKKβ. We found that BX795 blocked the IFN response to LPS, whereas TPCA-1 blocked both IFN and the proinflammatory TNF response (Fig. 6). However, the IFN response in SAE2-targeted cells was unaffected by either inhibitor, alone or in combination (Fig. 6). These data suggest that none of the four TBK1-related kinases are essential for the IFN response caused by loss of sumoylation.
Fig. 6.
Neither TBK1/IKKε nor IKKα/IKKβ are required for the spontaneous IFN response in sumoylation-deficient cells. THP-1 monocytes were transduced with lentiCRISPR lentiviruses encoding Cas9 and the indicated gRNAs. PMA-differentiated THP-1s were treated for 48 h with DMSO, 1 μM BX795, 20 μM TPCA-1, or 1 μM BX795 and 20 μM TPCA-1. Four hours before harvest, 1 μg of LPS was added to nontargeted THP-1 cells. (A and B) Evaluation of IFNB1 (A) and TNF (B) mRNA expression by quantitative RT-PCR. Statistical analysis was performed using a two-way ANOVA comparing BX795 and TPCA-1 treated cells to the DMSO treated control, correcting for multiple comparisons using the Holm–Sidak method. n = 3 where n is the number of treated cell lines. Data are representative of four separate experiments. *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001.
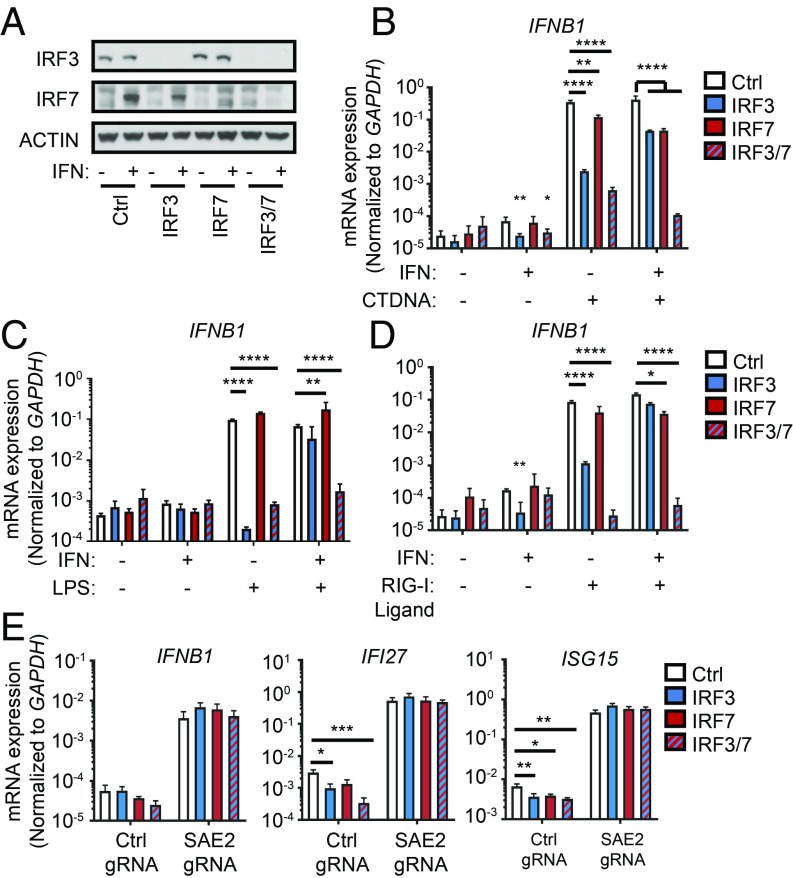
We next investigated the role of the canonical IFN-inducing transcription factors IRF3 and IRF7. IRF3 is essential for STING-, MAVS- and TRIF-dependent IFN production in resting cells. However, IRF7 can functionally compensate for IRF3 in cells that have been pretreated with IFN because IRF7 is itself a potent ISG. Together, these two transcription factors are essential for all canonical IFN-inducing pathways (4). To test whether these transcription factors contribute to the spontaneous IFN response in sumoylation-deficient THP-1 cells, we generated lentiCRISPR constructs targeting IRF3 and IRF7 in vectors with distinct selection markers. We transduced THP-1 cells with constructs targeting IRF3, IRF7, or both, together with corresponding nontargeting controls. To avoid the complication of incomplete targeting, we derived clonal lines of IRF3-, IRF7-, and IRF3/7-targeted cells. Successful targeting was verified by Western blot (Fig. 7A). To validate functional disruption of IRF3 and IRF7, we transfected each clonal line with either calf thymus DNA (Fig. 7B) or RIG-I Ligand (Fig. 7C), or treated cells with LPS (Fig. 7D). IRF3 KO cells failed to mount an IFNB1 response to DNA, RNA, or LPS, but pretreatment of these cells with recombinant IFN-β largely restored the IFNB1 response (Fig. 7 B–D). This restoration after IFN pretreatment was entirely dependent on IRF7, because IRF3/7 double KO cells were completely unable to respond to any of the stimuli, even after pretreatment with recombinant IFN-β. Thus, our IRF3/7 double knockout human cells are functional knockouts for the STING-, MAVS-, and TRIF-IFN pathways. Importantly, these data also demonstrate that no other transcription factor in these cells can functionally compensate for the combined loss of IRF3 and IRF7. We did not test MyD88-dependent IFN signaling in these cells because the IFN response activated by TLR7/9 is specifically restricted to plasmacytoid dendritic cells, and THP-1 cells do not make IFN in response to TLR7/9 ligands. We transduced these targeted cells with a nontargeting or SAE2-targeting gRNA and evaluated the IFN response. Remarkably, the expression of IFNB1, IFI27, and ISG15 caused by loss of SAE2 was completely intact in clonal THP-1 cells lacking IRF3, IRF7, or both (Fig. 7E). Thus, the potent IFN response caused by loss of sumoylation is independent of all canonical IFN-inducing pathways, revealing a distinct mechanism of IFN regulation.
Fig. 7.
The spontaneous IFN response in sumoylation-deficient cells is not dependent on IRF3 or IRF7. THP-1 monocytes were transduced with lentiCRISPR lentiviruses encoding Cas9 the indicated gRNAs. Transduced THP-1s were selected and then single cell cloned. (A) Western blot evaluation of the indicated proteins. (B and C) Evaluation of IFNB1 mRNA expression in PMA-differentiated THP-1s treated with 50 U/mL recombinant IFN-β overnight and then either transfected with 1 μg of CT-DNA (B) or 1 μg of RIG-I ligand for 4 h (C), or treated with 100 ng/mL LPS for 4 h (D). Statistical analysis was performed using a two-way ANOVA and comparing each clonal line to the control, correcting for multiple comparisons using the Holm–Sidak method. n = 3 where n is the number of treated cell lines. (E) Evaluation of IFNB1, IFI27, and ISG15 mRNA expression in PMA-differentiated THP-1s by quantitative RT-PCR. Statistical analysis was performed using a two-way ANOVA and comparing IRF3, IRF7, and IRF3/7-targeted cells to the control line, correcting for multiple comparisons using the Holm–Sidak method. n = 3 where n is the number of unique polyclonal cell lines. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Discussion
Dysregulation of type I IFN production is associated with both monogenic and complex autoimmune diseases. A deeper understanding of the pathways that drive IFN production and the mechanisms that set thresholds for activation of the IFN response will allow for the mechanistic definition of type I interferonopathies, with important implications for the development of new treatments for these diseases (8, 9). In this study, we identify an unanticipated mechanism of IFN regulation and a noncanonical pathway that potently activates the type I IFN response. We show that SUMO2 and SUMO3 are redundant and essential negative regulators of a spontaneous IFN response. Surprisingly, we find that the potent IFN response caused by loss of SUMO2/3 is mediated by a pathway that is completely independent of all known IFN-inducing sensors, kinases, and transcription factors.
We began to study sumoylation because we identified SAE2 as a specific Trex1-interacting protein in a yeast 2-hybrid screen. This led us to hypothesize that sumoylation might influence the ability of Trex1 to regulate the cGAS-STING pathway of DNA sensing. Importantly, the DeJean group recently reported that mouse dendritic cells with conditional deletion of the Ube2i gene that encodes the UBC9 E2 SUMO ligase had increased basal IFN responses and dramatically enhanced inflammatory responses to innate immune stimuli (11). They identified an enhancer region near the mouse Ifnb1 locus that is bound by a SUMO1-conjugated protein under basal conditions. Upon LPS stimulation, SUMO1 disappears from this enhancer, leading to transcription of an enhancer-derived RNA that may influence Ifnb1 transcription. While our data corroborate the role of sumoylation in suppressing the IFN response, we identify SUMO2/3, not SUMO1, as the essential SUMOs for IFN regulation. We therefore suggest that while this enhancer RNA may play an important role in influencing Ifnb transcription, its potential repression by SUMO1 is insufficient to explain the elevated IFN response we observe in sumoylation-deficient cells. Importantly, our understanding of specific functions for individual SUMO proteins is not well developed, in part because of the three distinct SUMO proteins and the fact that specific sumoylation sites can be conjugated by multiple SUMO proteins. Our data reveal an essential function mediated by SUMO2 and SUMO3, but not by SUMO1, which will enable more mechanistic studies of these isoform-specific SUMO functions. Additionally, it is possible that the contributions of specific SUMO proteins to repression of the IFN response may differ among cell types.
We unexpectedly found that neither the cGAS-STING pathway nor any other canonical IFN-inducing pathway is responsible for the potent IFN response caused by loss of sumoylation. Moreover, the transcription factors IRF3 and IRF7, which are essential for all canonical type I IFN responses, are completely dispensable for this ectopic IFN response. How, if not through IRF3 or IRF7, does sumoylation regulate type I IFN production? Two major possibilities remain. One possibility is that alternative transcription factors are regulated by sumoylation, and in sumoylation-deficient cells, they are able to trigger an IFN response. For example, IRF1 was originally identified as a transcription factor that drives expression of IFN-β, although later experiments demonstrated that IFN production by cytosolic nucleic acid sensing pathways and TLRs are unaffected by the loss of IRF1 (4). However, isolated examples persist in the literature of noncanonical IFN responses driven by IRF1 activation (23, 24). Alternatively, IRF5 has been shown to be important for IFN response to some RNA viruses (25). Whether these or other related factors are activated by loss of sumoylation will require further work, but we emphasize that neither IRF1 nor IRF5 are able to compensate for the combined loss of IRF3 and IRF7 in all known canonical IFN-inducing pathways (Fig. 7). Thus, their contributions to IFN production must be through a currently uncharacterized mechanism. A second possibility is that sumoylation modifies the IFN-β locus itself, potentially altering the chromatin structure, and that increased accessibility allows for the activation of IFNB transcription by noncanonical transcription factors. The identification of relevant targets of SUMO2/3 monosumoylation may reveal new components of the IFN response that will help distinguish between these two possibilities.
In summary, we have found that SUMO2 and SUMO3 are specific and essential negative regulators of a noncanonical mechanism of type I IFN induction that cannot be placed within the known IFN-inducing pathways. We propose that further definition of this pathway will provide insights into the protective and pathological functions of type I IFNs.
Experimental Procedures
Cell Lines and Tissue Culture.
HEK 293T cells were grown in DMEM supplemented with 10% FCS, l-glutamine, penicillin/streptomycin, sodium pyruvate, and Hepes. THP-1 cells were cultured in RPMI-1640 supplemented as above. Where indicated, THP-1 cells were differentiated by culturing in 100 nM PMA for 24 h and then culturing for an additional 24 h in fresh medium without PMA before treatment.
Lentiviral Transduction.
VSV-G pseudotyped, self-inactivating lentivirus was prepared by transfecting a nearly confluent 10-cm plate of 293T cells with 1.5 μg of pVSV-G, 3 μg of psPAX-2, and 6 μg of the pRRL lentiCRISPR vector for 24 h and then aspirating and replacing the media with fresh media. Between 2 and 4 × 106 THP-1 cells were transduced with sterile filtered lentiviral supernatant. Twenty-four hours after transduction, the lentiviral supernatants were replaced with fresh media, and 48 h after transduction, the cells were placed under selection with either 5 μg/mL puromycin (Thermo Fisher), 10 μg/mL blasticidin (Thermo Fisher), or 80 μg/mL hygromycin B (Thermo Fisher) for at least 6 d.
LentiCRISPR/Cas9 Gene Targeting.
For CRISPR/Cas9 gene targeting, we generated pRRL lentiviral vectors in which a U6 promoter drives expression of a gRNA, and an MND promoter drives expression of Cas9, a T2A peptide, and either a puromycin, blasticidin, or hygromycin resistance cassette. gRNAs were designed in Benchling. The sequences of the gRNA target sites are listed in the SI Appendix. Where indicated, THP-1 cells were transduced with pSYG lentiviral vectors encoding an HA epitope tag, the protein of interest, an internal ribosome entry site, and a blasticidin resistance cassette. After selection, these cells were transduced with lentiCRISPR vectors.
Western Blot.
Where indicated, targeting of genes by lentiCRISPR was confirmed at least 10 d after transduction by immunoblot analysis of whole cell extracts. Cells were harvested in lysis buffer (20 mM Hepes pH 7.4, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 1 mM DTT) supplemented with complete protease inhibitor mixture (Thermo Fisher), incubated on ice for 15 min, and cleared of insoluble material by centrifugation. Cleared lysates were separated using a 4–12% Bis-Tris SDS/PAGE (Life Technologies) and transferred to Immobilon-P PVDF membrane (Millipore). Antibodies are listed in the SI Appendix.
Restriction Fragment Length Polymorphism Assay.
Targeting of genes by lentiCRISPR was confirmed by a restriction fragment length polymorphism assay where indicated. PCR products were amplified with Phusion High-Fidelity DNA polymerase (Thermo Fisher) or EmeraldAmp (Clontech). Primers and restriction enzymes used for RFLP are listed in SI Appendix.
Cell Treatments.
For transfections, calf-thymus DNA (Thermo Fisher) or RIG-I ligand (20) were complexed with lipofectamine 2000 (Thermo Fisher) at a ratio of 1 μg of nucleic acid to 1 μL of lipid. For LPS stimulations, LPS (Sigma) was added directly to PMA differentiated THP-1 cells. For treatment with chemical inhibitors, BX795 (Selleckchem) and TPCA-1 (Sigma) were diluted to 1 μM or 20 μM, respectively, in fresh media and added to cells for 48 h.
Quantitative RT-PCR.
For quantitative RT-PCR, cells were harvested in RNA-STAT60 (Amsbio) or TRIzol (Thermo Fisher). RNA was reverse transcribed into cDNA with SuperScript III (Thermo Fisher) or EcoDry Premix (Clontech). Quantitative RT-PCR was performed with EVA Green reagents (Bio-Rad Laboratories) on a Bio-Rad CFX96 Real-Time system. Primer sequences are listed in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Elizabeth Gray for assistance with LentiCRISPR targeting and to all the members of the D.B.S. laboratory for helpful discussions. This research was supported by funding from National Institutes of Health Grant R01 AI072945. D.B.S. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease and a Howard Hughes Medical Institute Faculty Scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802114115/-/DCSupplemental.
References
- 1.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 4.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360, and erratum (2006) 25:849. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Gao D, et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci USA. 2015;112:E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutières syndrome. J Immunol. 2015;195:1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crow YJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 8.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 9.Crowl JT, Gray EE, Pestal K, Volkman HE, Stetson DB. Intracellular nucleic acid detection in autoimmunity. Annu Rev Immunol. 2017;35:313–336. doi: 10.1146/annurev-immunol-051116-052331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H, Sanchez GA, Goldbach-Mansky R. Insights from Mendelian interferonopathies: Comparison of CANDLE, SAVI with AGS, monogenic lupus. J Mol Med (Berl) 2016;94:1111–1127. doi: 10.1007/s00109-016-1465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decque A, et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol. 2016;17:140–149. doi: 10.1038/ni.3342. [DOI] [PubMed] [Google Scholar]
- 12.Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 13.Li R, Pan Y, Shi DD, Zhang Y, Zhang J. PIAS1 negatively modulates virus triggered type I IFN signaling by blocking the DNA binding activity of IRF3. Antiviral Res. 2013;100:546–554. doi: 10.1016/j.antiviral.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Liu B, et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatham MH, et al. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat Struct Mol Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, et al. SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep. 2014;15:878–885. doi: 10.15252/embr.201438534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 18.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 19.Poulsen SL, et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol. 2013;201:797–807. doi: 10.1083/jcb.201212075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 22.Hoshino K, et al. IkappaB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature. 2006;440:949–953. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- 23.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz F, et al. Interferon-regulatory-factor 1 controls Toll-like receptor 9-mediated IFN-beta production in myeloid dendritic cells. Eur J Immunol. 2007;37:315–327. doi: 10.1002/eji.200636767. [DOI] [PubMed] [Google Scholar]
- 25.Lazear HM, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9:e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.