Abstract
Background
High throughput sequencing (HTS) boosted the discovery of novel viruses and new variants of known viruses. Here we investigated the presence of viruses in 12 pools of sand flies captured in three climatic periods in RAPELD grids at Rio Claro, Chapada dos Guimarães and at Pirizal, North Pantanal, Mato Grosso State, Midwestern Brazil by HTS, viral isolation of a putative Phlebovirus positive pool in Vero cells, RT-PCR and transmission electron microscopy (TEM).
Results
One pool containing three Lutzomyia (Lutzomyia) longipalpis sand flies captured in the transitional climatic period in North Pantanal showed a tripartite genomic sequence of a putative novel Phlebovirus belonging to the phlebotomus fever serogroup. Phylogenetic analysis revealed this virus is closely related and share a common ancestor with phleboviruses included in the same clade: Chagres, Urucuri and Uriurana virus. RNA-dependent RNA polymerase (RdRP) presented 60%, 59% and 58% of amino-acid (aa) similarity with these phleboviruses, respectively. Similarity of Nucleoprotein and NSs protein codified by ambissense strategy of segment S was of 49% and 37%, respectively, with the proteins of the closest phlebovirus, Uriurana virus. Glycoproteins (G1, G2) and NSm protein presented 49% and 48% aa similarity with Chagres and Uriurana virus, respectively. Uriurana virus was isolated from sand flies in Brazilian Amazon and Urucuri from rodents in Utinga forest, Pará State. Chagres virus is an arbovirus responsible for outbreaks of febrile illness in Panama. This phlebovirus was isolated in Vero cells, confirmed by TEM and RT-PCR for the L segment of the virus, and named Viola phlebovirus.
Conclusions
HTS, viral isolation, RT-PCR and TEM showed the presence of one virus in sand flies from North Pantanal with identity to a putative novel Phlebovirus from phlebotomus fever serogroup, named Viola phlebovirus.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2985-3) contains supplementary material, which is available to authorized users.
Keywords: High throughput sequencing, Phylogeny, Viral isolation, RNA virus, Viola phlebovirus, Bunyavirales, Phenuiviridae
Background
Metagenomic studies based on high-throughput sequencing (HTS) have increased the discovery of new viral species and novel variants of known viruses, as well as contributed to viral ecology and evolution studies [1, 2].
The importance of arbovirus transmitted diseases dramatically increased in the past few decades, since emerging arboviruses lead to the occurrence of large viral epidemics in tropical developing regions of the world, including Brazil. Although arboviruses present worldwide distribution, viral species distribution varies among geographical regions. Notably, higher incidence has been observed in tropical areas around the globe [3, 4].
Phlebotomines (Diptera: Psychodidae, Phlebotominae) are medically important insects popularly known as sand flies, involved in the transmission of bacteria, protozoan and arboviruses to humans and animals [5, 6]. At least 530 species of sand flies classified in 22 genera were already reported in the Americas [7]. Species belonging to genus Lutzomyia are highly anthropophilic, occur in a wide geographical distribution and constitute important vectors of human infections in the Americas.
Currently sand flies are associated with transmission of arboviruses belonging to the families Rhabdoviridae (genus Vesiculovirus), Phenuiviridae (genus Phlebovirus) and Reoviridae (genus Orbivirus). Sporadic reports of human cases indicate these infections are incidental in South America, associated mainly with recreational or occupational incursions into tropical sylvatic forests. These viruses are relatively common in Brazil, since at least 20 phleboviruses were discovered in the Brazilian Amazon region between 1954 and 1994 [5]. The lack of differential diagnostic methods and the common febrile symptoms may contribute to the underreporting of these infections.
Sand flies are commonly reported in Mato Grosso (MT) State, including in sylvatic areas of the Pantanal and Cerrado biomes [6, 8] and no viral diversity studies using metagenomics have been performed so far. For this reason, this study aimed to identify the presence of viruses in sand flies captured in North Pantanal and Chapada dos Guimarães National Park of MT.
Methods
Sampling of sand flies
Mato Grosso represents 35% of the Pantanal, the largest tropical humid territory worldwide, with warm and humid climate and mean annual precipitation between 800 and 1400 mm, 80% occur between November and March [9]. Chapada dos Guimarães National Park (CGNP) is a protected area of Cerrado presenting a dry winter from May to September and a rainy summer from October to April, with mean annual precipitation between 1800 and 2000 mm [10].
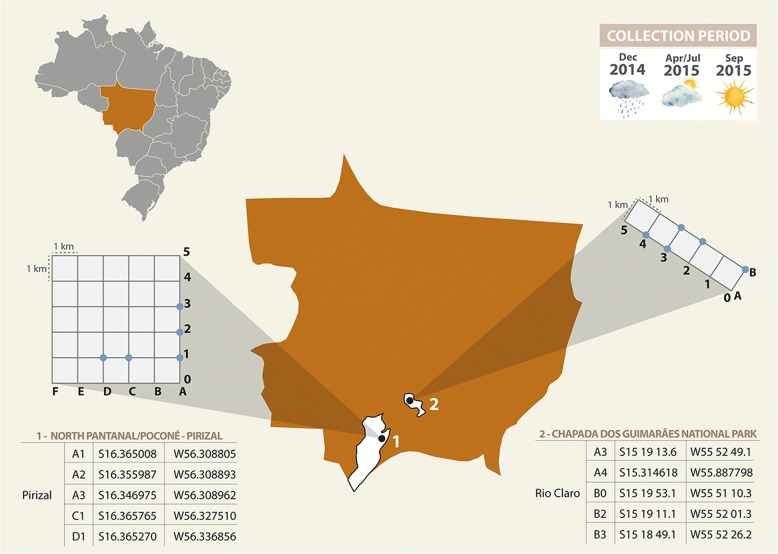
Sand flies were captured in RAPELD grids (Rapid Assessment Program Long Term Ecological Research) at Pirizal, North Pantanal (16°14'06"S, 56°22'70"W) and at Rio Claro, CGNP (15°19'10"S, 55°52'13"W) (Fig. 1).
Fig. 1.
Location of RAPELD systems used in the study for sand flies collections in three climatic periods in North Pantanal (1) and Chapada dos Guimarães National Park (2), Mato Grosso State, Midwestern Brazil. Collection plots are marked with blue dots in each RAPELD system grade
Five plots were selected in each location according to animal presence, proximity to water collections, access to vehicles, presence of riparian vegetation and habitat diversity.
Sand flies were captured with five CDC light traps placed along a 250 m transect at 50 m intervals between 18:00 h and 6:00 h at each sampling plot for two consecutive days in each climatic period. These specimens were immobilized for 1 min at -20 °C, identified according to dichotomous keys [11] and named [7]. Specimens were transported in liquid nitrogen to the virology laboratory. This study was approved by the System of Biodiversity Authorization and Information (SiSBio; ICMBio; IBAMA) under the number 43909-1.
RNA extraction, random PCR and high throughput sequencing (HTS)
Pools of sand flies were macerated in phosphate saline buffer and centrifuged at 4 °C, 5,000× g for 4 min. The supernatant (0.2 ml) was subjected to RNA extraction with High Pure Viral RNA kit (Roche, Basel, Switzerland), without RNA carrier. RNA was then quantified with quantifluor RNA system (Quantus fluorometer, Promega, Madison, Wisconsin, USA).
Reverse transcription was performed with a mean of 152 ng of RNA, followed by cDNA double-stranded (dscDNA) synthesis and random PCR reactions in quintuplicate. PCR products were purified with 20% polyethylene glycol 8000 (20% PEG) and quantified with quantifluor one dsDNA system [12–14].
The library was prepared with TruSeq RNA Sample Prep v2 Kit (Illumina, San Diego, California, USA) (zx ≥ 100 ng of DNA product) followed by 2 × 100 paired-end sequencing in the Illumina HiSeq 2500 (Illumina), using two lanes and generating 60 GB.
Sequence analysis and phylogeny
Sequences were analyzed with FastQC and trimmed (Trimmomatic 0.36) for the removal of primers sequences, adapters, and low-quality fragments (≤ 60 bp reads) removal (parameters: ILLUMINACLIP: TruSeq3-PE.fa:2:20:10, LEADING: 3, TRAILING: 3, SLIDINGWINDOW: 4:30, MINLEN: 60).
The assembled contigs, generated with different kmer (25, 40, 60 and 90) (Velvet v2.1.10), were compared with a viral RefSeq database from the National Center for Biotechnology Information (NCBI) using Blastx. Sequences with e-values > 1e-3 were compared to the entire nr database to exclude non-viral sequences.
The viral related contigs were manually inspected and annotated using Geneious R10. The length of some viral related contigs could be increased when the assembled reads extended beyond the contig ends. Therefore, reads were assembled back to the extended contig until the sequence could be extended no further.
The on-line open access software TMHMM (v2.0) (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict transmembrane domains.
Viral sequences were deposited in the GenBank database under the accession numbers MF289182, MF289183 and MH119632. These sequences were aligned with other viral sequences using MAFFT (v7.221). The best evolutionary model for each dataset was determined with ProtTest (v2.4). Evolutionary history was inferred by maximum likelihood method and Jones-Taylor-Thornton (JTT) model with Gamma distribution with invariant sites (G+I). Phylogenetic trees were generated with MEGA7 and edited with FigTree (v1.4.3).
Viral isolation in Vero cell culture, RT-PCR and transmission electron microscopy
A dilution (1:10 in 1 ml of inoculum) in RPMI medium of the pool supernatant positive for a phlebovirus was inoculated in a Vero cells (ATCC CCL-81) monolayer cultivated in T25 flasks for 2 h at 37 °C under constant agitation. After this period, RPMI culture medium with 5% fetal bovine serum was added and cells monitored daily for 5–7 days. Four passages were performed.
Supernatant was harvested for each passage and stored at -80 °C; monolayers were subjected to total RNA extraction (Trizol, Invitrogen, Carlsbad, Califórnia, USA) followed by a RT-PCR for a region of the L segment of Viola virus [primers SegLF (5'-AAG AGA CTC ATG GAC TCT GCT A-3') and SegLR (5'-CCG GGA AGT ATT TTC ATG GC-3')].
After reverse transcription (8 μl of RNA, primers at 2.5 μM) with 100 U of GoScript (Promega, Madison, Wisconsin, USA), a PCR containing the same primers at 1 μM, reaction buffer, MgCl2 (2 μM), DNTP mix (0.2 μM), 2.5 U of HotStart DNA polymerase (Promega, Madison, Wisconsin, USA) and ultrapure water in 50 μl reactions was amplified at 94 °C for 1 min, 30 cycles of 94 °C for 1 min, 56 °C for 1 min and 72 °C for 1 min, and a final extension of 72 °C for 2 min. PCR products (460 bp) were purified with 20% PEG and sequenced. At 5 days post-infection, the supernatant of Vero cells infected with Viola virus passage 5 was centrifuged at 159,000× g for 75 min using 3 ml of 25% sucrose cushion in polyallomer ultracentrifuge tubes (Beckman Coulter, Atlanta, Georgia, USA). The pellet was resuspended in PBS-1X and prepared for transmission electron microscopy (TEM) by negative staining [15] and observed in a JEOL JEM-1010 TEM at 100 kV.
Results
Specimens of sand flies captured in RAPELD systems at North Pantanal and Chapada dos Guimarães National Park
In total, 106 sand flies captured in the rainy (n = 86; 81.2%), transitional (n = 3; 2.8%) and dry (n = 17; 16.0%) periods, 38 (35.8%) in the CGNP and 68 (64.2%) at North Pantanal, comprised three pools of Lutzomyia sp., one of Lutzomyia (Lutzomyia) longipalpis, two of Nyssomyia whitmani, one of Evandromyia (Aldamyia) evandroi, one of Ev. (Ald.) carmelinoi, one of Lu. (Tricholateralis) sherlocki, one of Ev. (Eva.) wilsoni and two of Brumptomyia sp. (Table 1).
Table 1.
Viral sequences obtained by high throughput sequencing from sand flies pools captured in the rainy, intermediate and dry period at North Pantanal and Chapada dos Guimarães National Park, Mato Grosso, Brazil
| Place | Period | Pool | Species (n) | Reads | Contigs | Viral hits | Genome | Size (nt) | Identity (%)a | E value | GenBank ID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| North Pantanal | Rainy | 1 | Evandromyia (Aldamyia) evandroi (5) | 983,899,176 | 11,747 | – | – | – | – | – | – |
| 2 | Lutzomyia sp. (25) | 849,154.470 | 61,862 | – | – | – | – | – | – | ||
| 3 | Ev. (Eva.) wilsoni (11) | 819,202,516 | 43,993 | – | – | – | – | – | – | ||
| 4 | Evandromyia (Aldamyia) carmelinoi (1) | 1,179,702,220 | 91,115 | – | – | – | – | – | – | ||
| 5 | Lu. (Tricholateralis) sherlocki (3) | 449,698,864 | 28,893 | – | – | – | – | – | – | ||
| 6 | Brumptomyia sp. (15) | 785,799,190 | 73,656 | Vesiculovirus | ssRNA- | 69 | 44.9 | 3.83e-12 | – | ||
| Transitional | 7 | Lu. longipalpis (3) | 645,865,104 | 13,763 | Phlebovirus (Seg L) | ssRNA- | 6341 | 61 | 0 | MF289183 | |
| Phlebovirus (Seg M) | ssRNA- | 4403 | 46 | 0 | MF289182 | ||||||
| Phlebovirus (Seg S) | ssRNA- | 1747 | 53 | 2e-79 | MH119632 | ||||||
| Dry | 8 | Lutzomyia sp. (5) | 1,115,860,120 | 94,199 | Vesiculovirus | ssRNA- | 72 | 40.3 | 4.10e-11 | – | |
| Flavivirus | ssRNA+ | 48 | 47.9 | 5.88e-10 | – | ||||||
| CGNP | Rainy | 9 | Lutzomyia sp. (22) | 999,786,678 | 28,460 | – | – | – | – | – | – |
| 10 | Nyssomyia whitmani (3) | 787,599,818 | 37,990 | – | – | – | – | – | – | ||
| 11 | Brumptomyia sp. (1) | – | – | – | – | – | – | – | – | ||
| Dry | 12 | Nyssomyia whitmani (12) | 692,307,934 | 30,666 | – | – | – | – | – | – |
aIdentity refers to the closest phlebovirus species Chagres virus indicated by BLASTx (GenBank)
Abbreviations: CGNP Chapada dos Guimarães National Park, n number, nt nucleotides, Seg segment
Specimens of Lu. longipalpis were identified using the HTS data against a database of ND4 NADH dehydrogenase genes from eukaryotes. To avoid misidentification, only contigs > 300 nt were used in this analysis (data not shown).
Sequencing data analysis
Illumina sequencing generated 9,308,876,090 reads and 516,344 contigs from 12 pools. BLASTx analysis of the contigs against a RefSeq viral database resulted in 1198(0.23%) viral hits.
A substantial amount of the assembled reads had no significant similarity to any of the sequences deposited in the viral RefSeq database. BLASTx (nr) generated hits with less than 100 bp with some viral families. Three pools of sand flies captured in North Pantanal in the three climatic periods presented contigs of 43–57 nt with 40–47% of identity with members of the families Rhabdoviridae (genus Vesiculovirus) and Flaviviridae (genus Flavivirus) (Table 1).
Characterization of a new species of Phlebovirus
One pool containing extracts of three specimens of Lu. longipalpis females and males captured in the transitional period in Pirizal, North Pantanal presented the genome of a tripartite segmented virus belonging to phlebotomus fever serogroup, genus Phlebovirus, family Phenuiviridae, order Bunyavirales.
The partial genome sequence of this virus showed a low similarity with other species classified in the same genus and therefore, represents a putative new viral species that was named Viola phlebovirus.
After the identification through HTS, this virus was isolated in Vero cells in the fourth passage and confirmed by RT-PCR using L segment specific oligonucleotides and nucleotide sequencing, and also by visualizing the viral particles in the supernatant after TEM (Fig. 2).
Fig. 2.
RT-PCR for a region of L segment (460 bp) at passage 4 (a) and transmission electron microscopy of Viola phlebovirus in Vero cells at passage 5 (b)
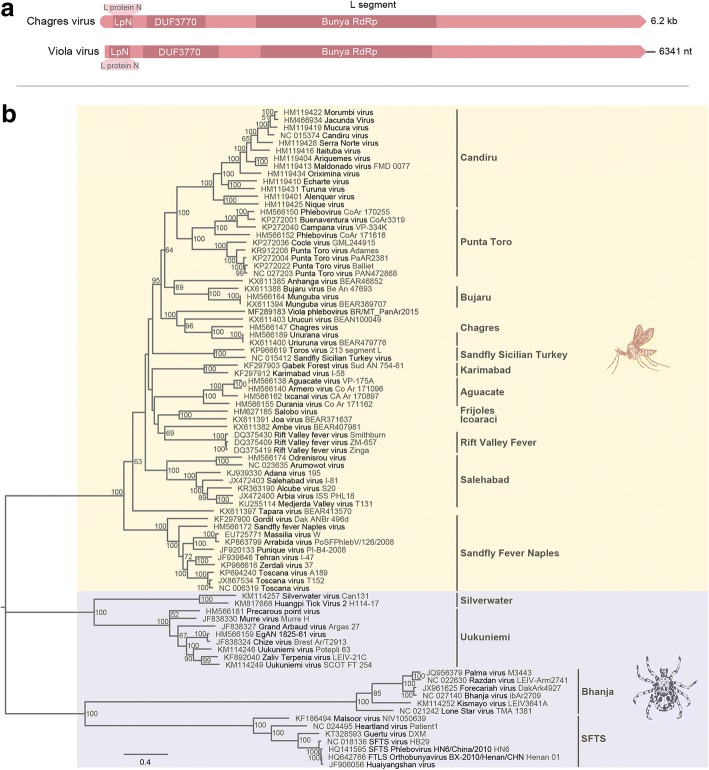
The L segment presents 6341 nucleotides (nt) in length and encodes the RNA-dependent RNA polymerase (RdRp) (2082 amino acids, aa) (Fig. 3a). The main RdRp domain of Viola phlebovirus has 662 aa, the L protein N domain has 85 aa and the subdomain DUF 3770 has 297 aa. BLASTp analysis revealed 61% identity with the closest phlebovirus, Chagres virus.
Fig. 3.
a Genomic organization of Viola phlebovirus L segment; and b phylogenetic tree based on amino acid sequences of phleboviruses by maximum likelihood method Jones-Taylor-Thornton model. Phlebotomus fever and Uukuniemi serogroups are marked in orange and purple, respectively. Bars indicate serocomplexes
The M segment is 4442 nt in length and encodes a polyprotein (4423 nt, 1467 aa) processed into: (i) NSm protein (41–259 aa; 219 aa of length), characteristic of phlebotomus fever serogroup; (ii) the G1 glycoprotein (315–840 aa; 526 aa of length); and (iii) the G2 glycoprotein (845–1324 aa; 480 aa of length) (Fig. 4a). Transmembrane domains were identified in the initial portion of G1 (741–763 aa), at the final region of G2 (1315–1337 aa) and a third between the end of G1 and beginning of G2 (828–850 aa). These domains are classical for phleboviruses, responsible for the anchoring of these glycoproteins to viral envelope [16]. BLASTp analysis revealed 43% of identity with the closest phlebovirus, Uriurana virus.
Fig. 4.
a Genomic organization of Viola phlebovirus M segment; and b phylogenetic tree based on amino acid sequences of phleboviruses by maximum likelihood method Jones-Taylor-Thornton model. Phlebotomus fever and Uukuniemi serogroups are marked in orange and purple, respectively. Bars indicate serocomplexes
The partial S segment is 1732 nt in length and encodes the nucleoprotein (N) and a nonstructural (NSs) protein in an ambisense coding strategy (Fig. 5a). The N protein (42–779 nt; 245 aa) presents the characteristic Tenui N domain of pheboviruses (pfam05733), involved in nucleocapsid formation. The NSs protein (1732–929 nt; 267 aa) codified by ambissense strategy presents the NSs domain (pfam11073), which is a major determinant of virulence by antagonizing interferon beta gene expression. BLASTp analysis revealed 49% and 37% identity (N and NSs, respectively) with the closest phlebovirus, Uriurana virus.
Fig. 5.
a Genomic organization of Viola phlebovirus S segment; and b phylogenetic tree based on amino acid sequences of phleboviruses by maximum likelihood method Jones-Taylor-Thornton model. Phlebotomus fever and Uukuniemi serogroup are marked in orange and purple, respectively. Bars indicate serocomplexes
Phylogenetic analysis of the aligned amino acid sequence of the RdRp, NSm/glycoproteins and nucleoprotein suggested that Viola phlebovirus is a distinct member of the family Phenuiviridae, genus Phlebovirus, phlebotomus fever serogroup, clustering with Urucuri virus, Changres virus and Uriurana virus (Figs. 3b, 4b, 5b).
As previously shown, the pairwise comparisons of the RdRp and nucleoprotein (the two most conserved virus proteins) showed similarity of 61% and 49%, respectively (Additional file 1: Figure S1). Taken together, these results confirm that Viola phlebovirus is indeed a novel phlebovirus.
Discussion
Viruses are the most diverse microorganisms. Metagenomic analysis has contributed to the description of viral diversity in several ecosystems and hosts, allowing the discovery of several previously unknown viral species and, allowing more precise and complete description of viral evolution. Despite the large number of arboviruses described in sand flies in the Amazon, studies involving viral detection in those arthropods are lacking in Midwestern Brazil. In this study, a new species of Phlebovirus belonging to phlebotomus fever serogroup was identified in Lu. longipalpis captured at North Pantanal.
In Brazil, sand flies have been associated with transmission of arboviruses only in the Amazon region [6, 17, 18]. Among these, the most relevant to public health are those belonging to the family Phenuiviridae, genus Phlebovirus, phlebotomus fever serogroup, associated with acute mild febrile illness in humans [19]. Despite their public health importance, genomic diversity of viruses associated with sand flies remains largely underestimated.
Previously, Bunyaviridae was a large family of negative or ambissense tripartite single stranded RNA viruses, primarily classified by serological methods. Recently, this family was reclassified into a new order, Bunyavirales, which contains now nine viral families, including Phenuiviridae, where the genus Phlebovirus was allocated [20]. Diversity and evolution of these viruses is largely associated to plasticity of their RNA genomic segments, resulting in point mutations, recombination and reassortment events [21]. These characteristics have contributed to the unfeasibility of serological tests to classify novel phleboviruses against all other known members of this viral order.
Within phleboviruses, S segment presents ambissense strategy, encoding two ORFs: the NSs protein associated with the ability of the virus to replicate in mammalian cells, and the N protein. M and L segments are negative sense segments encoding the envelope glycoproteins and NSm protein and the RdRp, respectively [22, 23]. Since the Uukuniemi serogroup does not encode NSm protein, the finding of NSm in the M segment of Viola phlebovirus indicates this virus belongs to phlebotomus fever serogroup, which includes arboviruses. NSm is a virulence factor associated with the inhibition of apoptosis in infected cells [24].
Viola virus was more closely related to phleboviruses from phlebotomus fever serogroup isolated from sand flies (Uriurana virus) and rodents Proechimys guyannensis (Urucuri virus) in the Amazon and in the Utinga Forest, State of Pará, respectively [25], and also isolated from humans with febrile illness in Panamá (Chagres virus) [26].
In addition, viral isolation of Viola virus in mammalian cell lines (Vero cells) and the presence of NSs and NSm proteins indicate that it is not an insect-specific virus (ISV) and might represent a virus that infect vertebrates.
Conclusions
This detection and isolation of a putative new Phlebovirus (Viola phlebovirus) in Lu. longipalpis and its initial characterization contributes to our knowledge about viral diversity in arthropods from sylvatic areas of North Pantanal. Phylogeny revealed proximity with viruses causing disease in humans, rodents and isolated from sand flies belonging to phlebotomus fever serogroup. Isolation of Viola virus in mammalian cells indicates this virus is not an ISV and represents a novel species with no known vertebrate host. Therefore, future studies involving Viola phlebovirus may address its importance for public or veterinary health. This is the first study reporting a new species of virus infecting arthropods through HTS and able to replicate in Vero cells in Mato Grosso, Midwestern Brazil.
Additional file
Figure S1. Pairwise amino acid comparision of Viola phlebovirus M segment sequences with all other M segment sequences of members of the genus Phlebovirus available in the GenBank database. (PNG 62 kb)
Acknowledgments
We thank Fabricio Campos for his important assistance with the PCR protocol, Ana Lucia Ribeiro, Maria Cristina F. Bezerra and Chico Bio for their fundamental assistance in the collections, as well as COMCERRADO and Pirizal RAPELD modules for access to those areas. To Bruno Cidade for his help with the maps, Marcio Nunes (IEC) for his assistance with phylogenetic tree and Nicholas Komar (CDC) for reviewing the manuscript. MSC and AZLP received Master scholarships from CNPq and CAPES, respectively.
Funding
This work was supported by Rede Pro-Centro-Oeste of viral biodiversity - CNPQ process 407817/2013-1.
Availability of data and materials
The nucleic acid sequences of segments L, M and S of the Viola virus are available in GenBank (accession numbers MF289182, MF289183 and MH119632). Other data used in the present study are available from the corresponding author upon request.
Author’s contributions
MSC: developed the study. AZLP: helped in all experimental procedures. FLM: experiment design, data analysis and writing. AP and JSVR: field work and identification. BMR: experimental design, financial support, writing the manuscript. RDS: coordinator, advisor, experimental design and development, writing the manuscript. All authors read and approved the final manuscript.
Abbreviations
- HTS
high throughput sequencing
- TEM
transmission electron microscopy
- RNA
ribonucleic acid
- RdRp
RNA dependent RNA polymerase
- RT-PCR
reverse transcription polymerase chain reaction
- RAPELD
Rapid Inventary Long Term Ecological Research
- aa
amino acid
- G
glycoprotein
- L
large
- M
medium
- S
small
- MT
Mato Grosso
- CGNP
Chapada dos Guimarães National Park
- CDC
Centers for Diseases Control and Prevention
- NCBI
National Center for Biotechnology Information
- cDNA
complementary deoxyribonucleic acid
- dscDNA
double-stranded cDNA
- SegLF/SegLR
segment L forward and reverse primers
- SiSBio
System of Biodiversity Authorization and Information
- ICMBio
Chico Mendes Institute for the Conservation of Biodiversity
- IBAMA
Brazilian Institute of the Environment and Renewable Natural Resources
- PEG
polyethyleneglycol
- BLAST
basic local alignment search tool
- RefSeq
reference sequence
- Ev/Eva
Evandromyia
- Ald
Aldamyia
- Lu.
Lutzomyia; nt: nucleotide
- nr
non-redundant
- N
nucleoprotein
- ORF
open reading frame
- ISV
insect-specific virus
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2985-3) contains supplementary material, which is available to authorized users.
Contributor Information
Michellen S. de Carvalho, Email: michellen.scarvalho@gmail.com
Andressa Z. de Lara Pinto, Email: andressazelenski@gmail.com
Aquirya Pinheiro, Email: aquiryapinhiero@hotmail.com.
Jorge S. V. Rodrigues, Email: senatorevargas@yahoo.com.br
Fernando L. Melo, Email: flucasmelo@gmail.com
Leonardo Assis da Silva, Email: leocbq@gmail.com.
Bergmann M. Ribeiro, Email: bergmann.ribeiro@gmail.com
Renata Dezengrini-Slhessarenko, Email: renatadezengrini@yahoo.com.br.
References
- 1.Hall-Mendelin S, Allcock R, Kresoje N, van den Hurk AF, Warrilow D. Detection of arboviruses and other micro-organisms in experimentally infected mosquitoes using massively parallel sequencing. PLoS One. 2013;8:e58026. doi: 10.1371/journal.pone.0058026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pauvolid-Correa A, Solberg O, Couto-Lima D, Nogueira RM, Langevin S, Komar N. Novel viruses isolated from mosquitoes in Pantanal, Brazil. Genome Announc. 2016;4:e01195-16. [DOI] [PMC free article] [PubMed]
- 3.Karabatsos N. International catalogue of arthropod-borne viruses. 3rd edition. San Antonio, Texas: American Society of Tropical Medicine and Hygiene for The Subcommittee on Information Exchange of the American Committee on Arthropod-borne Viruses; 1985.
- 4.Lima-Camara TN. Emerging arboviruses and public health challenges in Brazil. Rev Saude Publica. 2016;50: 10.1590/S1518-8787.2016050006791. [DOI] [PMC free article] [PubMed]
- 5.Rodrigues SG, da Rosa T, Vasconcelos PF, Travassos da Rosa ES, Tesh RB. Characterization of two new phleboviruses associated with human illness from the Amazon region of Brazil. An overview of arbovirology in Brazil and neighbouring countries. Belem: Instituto Evandro Chagas; 1998. pp. 100–6.
- 6.Brito VN, Almeida Ado B, Nakazato L, Duarte R, Souza Cde O, Sousa VR. Phlebotomine fauna, natural infection rate and feeding habits of Lutzomyia cruzi in Jaciara, state of Mato Grosso, Brazil. Mem Inst Oswaldo Cruz. 2014;109:899–904. [DOI] [PMC free article] [PubMed]
- 7.Shimabukuro PHF, Andrade AJ, Galati EAB. Checklist of American sand flies (Diptera, Psychodidae, Phlebotominae): genera, species, and their distribution. ZooKeys. 2017;660:67–106. doi: 10.3897/zookeys.660.10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Missawa NA, Dias ES. Phlebotomine sand flies (Diptera: Psychodidae) in the municipality of Varzea Grande: an area of transmission of visceral leishmaniasis in the state of Mato Grosso, Brazil. Mem Inst Oswaldo Cruz. 2007;102:913–918. doi: 10.1590/S0074-02762007000800004. [DOI] [PubMed] [Google Scholar]
- 9.Girard P. Hydrology of surface and ground waters in the Pantanal floodplains. In: Junk WJ, da Silva CJ, Nunes da Cunha C, editors. The Pantanal: ecology, biodiversity and sustainable management of a large neotropical seasonal wetland. Sofia: Pensoft: Publishers; 2011. p. 103–26.
- 10.Pinto JRR, ATd OF. Perfil florístico e estrutura da comunidade arbórea de uma floresta de vale no Parque Nacional da Chapada dos Guimarães, Mato Grosso, Brasil. Rev Bras Bot. 1999;22:53–67. doi: 10.1590/S0100-84041999000100008. [DOI] [Google Scholar]
- 11.Young D, Duncan M. Guide to the identification and geographic distribution of Lutzomyia Sand Flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae) Gainesville, Florida, USA: Associated Publishers; 1994. [Google Scholar]
- 12.Stang A, Korn K, Wildner O, Uberla K. Characterization of virus isolates by particle-associated nucleic acid PCR. J Clin Microbiol. 2005;43:716–720. doi: 10.1128/JCM.43.2.716-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kluge M, Campos FS, Tavares M, de Amorim DB, Valdez FP, Giongo A, et al. Metagenomic survey of viral diversity obtained from feces of Subantarctic and South American fur seals. PLoS One. 2016;11:e0151921. doi: 10.1371/journal.pone.0151921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.AZd LP, Santos CM, de Melo FL, Ribeiro ALM, Morais RB, Dezengrini Slhessarenko R. Novel viruses in salivary glands of mosquitoes from sylvatic Cerrado, Midwestern Brazil. PLoS One. 2017;12:e0187429. doi: 10.1371/journal.pone.0187429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner S, Horne RW. A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta. 1959;34:103–110. doi: 10.1016/0006-3002(59)90237-9. [DOI] [PubMed] [Google Scholar]
- 16.Halldorsson S, Behrens AJ, Harlos K, Huiskonen JT, Elliott RM, Crispin M, et al. Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion. Proc Natl Acad Sci USA. 2016;113:7154–9. [DOI] [PMC free article] [PubMed]
- 17.Vasconcelos PF, Rosa A, Degallier N, Rosa JF, Pinheiro FP. Clinical and ecoepidemiological situation of human arboviruses in Brazilian Amazonia. Ciênc Cult (Säo Paulo) 1992;44:117–124. [Google Scholar]
- 18.Tesh RB. The genus Phlebovirus and its vectors. Annu Rev Entomol. 1988;33:169–181. doi: 10.1146/annurev.en.33.010188.001125. [DOI] [PubMed] [Google Scholar]
- 19.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 20.Briese T, Alkhovsky S, Beer M, Calisher CH, Charrel R, Ebihara H, et al. Create a new order, Bunyavirales, to accommodate nine families (eight new, one renamed) comprising thirteen genera. 2016. https://data.ictvonline.org/proposals/2016.030a-vM.A.v6.Bunyavirales.pdf. Accessed 13 Feb 2018.
- 21.Liang G, Gao X, Gould EA. Factors responsible for the emergence of arboviruses; strategies, challenges and limitations for their control. Emerg Microbes Infect. 2015;4:e18. doi: 10.1038/emi.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calisher CH. History, classification, and taxonomy of viruses in the family Bunyaviridae. In: Elliott RM, editor. The Bunyaviridae. Boston, MA: Springer; 1996. pp. 1–17. [Google Scholar]
- 23.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus taxonomy: VIIIth report of the International Committee on Taxonomy of Viruses: Academic Press; 2005.
- 24.Colon-Ramos DA, Irusta PM, Gan EC, Olson MR, Song J, Morimoto RI, et al. Inhibition of translation and induction of apoptosis by Bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol Bio Cell. 2003;14:4162–4172. doi: 10.1091/mbc.e03-03-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunes-Neto JP, Souza WM, Acrani GO, Romeiro MF, Fumagalli M, Vieira LC, et al. Characterization of the Bujaru, frijoles and Tapara antigenic complexes into the sandfly fever group and two unclassified phleboviruses from Brazil. J Gen Virol. 2017;98:585–594. doi: 10.1099/jgv.0.000724. [DOI] [PubMed] [Google Scholar]
- 26.Peralta PH, Shelokov A, Brody JA. Chagres virus: a new human isolate from Panama. Am J Trop Med Hyg. 1965;14:146–151. doi: 10.4269/ajtmh.1965.14.146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Pairwise amino acid comparision of Viola phlebovirus M segment sequences with all other M segment sequences of members of the genus Phlebovirus available in the GenBank database. (PNG 62 kb)
Data Availability Statement
The nucleic acid sequences of segments L, M and S of the Viola virus are available in GenBank (accession numbers MF289182, MF289183 and MH119632). Other data used in the present study are available from the corresponding author upon request.