Abstract
The incidence of HPV-related oropharyngeal squamous cell carcinoma (OPSCC) has increased more than 200% in the past 20 years. Recent genetic sequencing efforts have elucidated relevant genes in head and neck cancer, but HPV-related tumors have consistently shown few DNA mutations. In this study, we sought to analyze alternative splicing events (ASE) that could alter gene function independent of mutations. To identify ASE unique to HPV-related tumors, RNA sequencing was performed on 46 HPV-positive OPSCC and 25 normal tissue samples. A novel algorithm using outlier statistics on RNA-sequencing junction expression identified 109 splicing events, which were confirmed in a validation set from the Cancer Genome Atlas (TCGA). Because the most common type of splicing event identified was an alternative start site (39%), MBD-seq genome-wide CpG methylation data were analyzed for methylation alterations at promoter regions. ASE in six genes showed significant negative correlation between promoter methylation and expression of an alternative transcriptional start site, including AKT3. The novel AKT3 transcriptional variant and methylation changes were confirmed using qRT-PCR and qMSP methods. In vitro silencing of the novel AKT3 variant resulted in significant growth inhibition of multiple head and neck cell lines, an effect not observed with wild type AKT3 knockdown. Analysis of ASE in HPV-related OPSCC identified multiple alterations likely involved in carcinogenesis, including a novel, functionally active transcriptional variant of AKT3. Our data indicate that ASEs represent a significant mechanism of oncogenesis with untapped potential for understanding complex genetic changes that result in development of cancer.
Keywords: AKT3, alternative splice expression, HPV, oropharynx squamous cell carcinoma, outlier analysis
Introduction
In recent years, major efforts fueled by quickly advancing technologies have been dedicated to sequencing cancer genomes through the Cancer Genome Atlas (TCGA) and other independent groups.(1–3) These studies have advanced the understanding of many cancer types, especially through identification of mutated genes that result in oncogenesis. For head and neck squamous cell carcinoma (HNSCC), DNA sequencing studies and TCGA have identified new genes and pathways that are frequently mutated including NOTCH1, CCND1 and PIK3CA in addition to genes that have been known to be relevant such as TP53 and EGFR.(1) However in HPV-related HNSCC, while some genetic alterations have been identified in the PI3K/AKT3 pathway, overall fewer actionable genetic alterations have been identified in HPV-related HNSCC, which has been significantly increasing in incidence.(4),(3)
In HPV-related tumors, the presence of fewer of oncogenic genetic alterations limits our ability to understand the underlying biology of this disease.(1) Patients with HPV-related disease have improved prognosis; however improved understanding of network alterations in HPV-related tumors is still needed to identify poor prognosis subgroups or those that may benefit from de-escalated therapy.(5),(6) Improved understanding of the disease biology could also help to open doors for development of new therapeutics.
In the case of HPV-related OPSCC, a better understanding of the disease biology necessitates exploration of changes that play a role at the post-transcriptional level, such as splice variants or epigenetic alterations. These changes may significantly modify gene and protein function independent of DNA sequence alterations. Alternative splicing is an important mechanism for post-transcriptional change that allows for significant expansion of protein diversity and function. It is estimated that up to 95% of multi-exonic genes in the human genome undergo some form of alternative splicing in normal tissues.(7)
Alternative splicing has been a proposed mechanism of oncogenesis for many years. For instance, alternatively spliced forms of BCL2L1 (apoptotic regulator) and TNR6 (Fas receptor) have been shown to be either pro or anti-apoptotic in human cancers, based on inclusion of specific exons.(8) Alternative splicing events have also been identified in head and neck cancer using gene expression arrays.(9) With the advent of RNA-sequencing (RNA-seq) technology, systematic analysis of alternative splicing events is now possible with the ability to detect novel splice variants that are not dependent on existing probes for identification. Instead, RNA-seq identifies junctions, or specific connections between exons, throughout the whole genome.(10) However, currently existing algorithms for identification of splice variants using RNA-seq data are not well suited for analysis of heterogeneous cancer sample populations.(11) Previously, we demonstrated that applying outlier statistics to exon specific microarray data was well suited for identifying cancer-specific splice variants in HNSCC.(9) RNA-seq offers an opportunity to directly discover novel exon junctions spanning distinct gene isoforms. Therefore, we adapted these outlier statistics to RNA-sequencing data to directly discover novel exon junctions occurring within a heterogeneous tumor population.(12)
Through use of a novel algorithm for systematic identification of tumor-specific alternative splicing events and correlation with DNA methylation data, we identified over 100 alternative splicing alterations in HPV-related OPSCC, including a novel splice variant of AKT3. This novel variant expresses a unique first exon which may be functionally active in promoting growth in head and neck cancer.
Materials & Methods
Patient samples
Primary tumor tissue samples were obtained from a test cohort of 46 patients with HPV-related oropharyngeal squamous cell carcinoma, as previously described.(13) For comparison, normal oropharynx mucosal tissue from uvulopalatopharyngoplasty (UPPP) surgical specimens were obtained from 25 cancer unaffected controls. All tissue samples were collected from the Johns Hopkins Tissue Core under an approved IRB protocol (#NA_00036235) and written informed consent was obtained from each patient prior to collection of samples in accordance to ethical guidelines according to the Belmont Report.
RNA preparation and RNA-seq analysis
RNA was extracted from 0.35 mm thick frozen tissue sections from tissue samples above and RNA sequencing was performed, as previously described.(13) Samples were required to achieve an RNA Integrity Number (RIN) of at least 7.0. Barcoded, stranded RNA-seq libraries from ribosomal RNA depleted total RNA (400 ng) was prepared using the Illumina TruSeq stranded total RNA Seq Gold kit (San Diego, CA). Sequencing was performed using the HiSeq 2500 platform sequencer (Illumina) and the TruSeq Cluster Kit, resulting in approximately 80 million 100×100 paired end reads per sample. Next, the RNA sequencing data were normalized based on the version 2 protocols developed by TCGA.(1) Alignment was performed using MapSplice2 version 2.0.1.9 to the GRCh37/hg19 genome assembly. Junction data from sequence alignment was extracted for further analysis. Gene expression values were quantified from RNA sequencing data using RSEM version 1.2.9 and upper quartile normalization according to the TCGA RSEM v2 normalization pipeline.(1,13)
Splice variant identification and outlier statistics
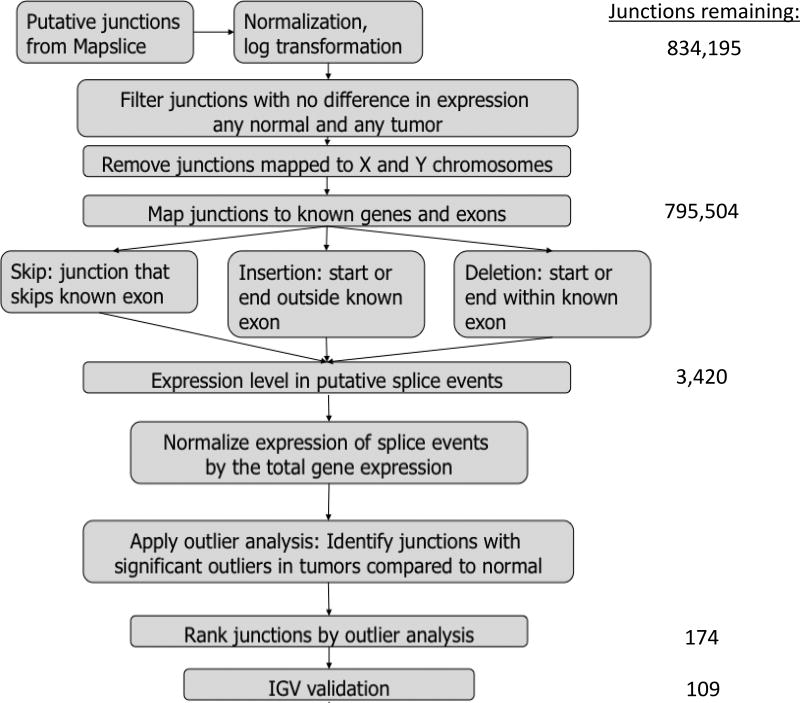
For identification of splice variants unique to tumor samples, an algorithm utilizing outlier analysis was applied to putative junctions identified from MapSplice alignment output (Figure 1), utilizing R version 3.2.1. Junction expression was compiled across all samples, using a value of 0 for samples that did not have the putative junction. Values were then normalized as RPM (reads per million) and log transformed. Junctions were removed if there was no difference in expression between any tumor and any normal, as well as if junctions were located on X or Y chromosomes. Junctions were then mapped to known genes and exons based on hg19 genome assembly, and selected as putative splice variants if they were identified either as a skip (junction that skips a known exon), insertion (start or end outside a known exon) or deletion (start or end within a known exon) package org.Hs.eg.db, version 3.4.0.(14) The junction expression for selected junctions were normalized by dividing by total gene expression (using RSEM(15)) and gene alignments were determined by genome assembly or manual curation if overlapping genes were present. Outlier analysis was performed to identify the number of tumors with outlier expression in comparison to the distribution of expression in normal tissue, using functions from the OGSA Bioconductor package(12,16). Next the number of outliers occurring in tumors was compared to normal using a Fisher’s exact test. P-values were FDR adjusted with Benjamini–Hochberg method and adjusted p-values below 0.05 were used to determine junctions with significantly higher outliers in tumors compared to normal samples (source code within Supplemental data).
Figure 1.
Schematic of algorithm for identification of significant alternative splicing events through RNA-seq analysis. Initially, from MapSplice output 834,195 raw junctions, representing splice variant isoforms, were present within sequenced cohort. These junctions were normalized and filtered to exclude junctions without variation or those on X & Y chromosomes. Then potential splice variants were identified based on possible alternative splicing patterns to identify 3,420 potential candidates. Outlier statistics were applied to identify candidates with differential expression in tumors, and of these 109 splice variants were confirmed.
IGV confirmation
Putative junctions identified as significant through outlier statistics were then visualized using the Integrative Genome Viewer (IGV, Broad Institute, version 2.3).(17) From RNA sequencing data, BAM files were loaded into IGV to directly visualize compiled RNA read data. Read data was visualized at start and end of each junction and compared between normal and tumor samples. Putative junctions were confirmed if the overall gene expression was observed in both normal and tumor tissue, and a unique wild type splicing event was identifiable in normal tissue. Junctions were then categorized as either alternative start site, canonical skipping, insertion, deletion, intron retention, alternative end site, intronic short segment (junction confined within one intron), or non-coding (junction start and end site occur within non-coding region).
MBD-Seq DNA methylation analysis
Genome-wide DNA methylation analysis was carried out using MBD-seq similar to what was described previously.(18,19) Briefly, DNA was sonicated, end-repaired, and ligated to SOLiD P1 and P2 sequencing adaptors lacking 5' phosphate groups, using the NEBNext DNA Library Prep Set for SOLiD according to the manufacturer's recommended protocol (NEB). Libraries were then nick-translated with Platinum Taq polymerase and divided into two fractions: an enriched methylated fraction that was subjected to isolation and elution of CpG-methylated library fragments by using MBD2-MBD-bound magnetic beads as described previously,(19) and a total input fraction that was left unenriched. These fractions were then amplified using 4–6 cycles for the total input, and 10–12 cycles for the enriched methylated fractions according to the NEBNext DNA Library Prep Set for SOLiD kit (NEB). The resulting libraries were subjected to emulsion PCR, bead enrichment, and sequencing on a SOLiD sequencer to generate on average ~25–50 million 50 bp single-end reads per sample according to the manufacturer's protocols (Life Tech). The resulting color-space reads were aligned using Bioscope software (Applied Biosystems). MACS(20) peak identification software was used to identify regions of methylation by comparing the enriched versus total input libraries for each sample with a default cutoff of p < 10−5.
The methylation status of each 100 bp segment across the genome for each sample in the cohort described above was determined as an indicator of presence of any intersection of the segment with regions of DNA methylation as identified by MACS peak calling.(20) For each of 43 splice variants associated with an alternative start site junction, methylation was analyzed for the 2000 bp region surrounding the start of the junction, the end of the junction, and the canonical transcriptional start site. The differential methylation was tested in each of 100 bp segment within these regions of interest number by comparison of numbers of samples with and without methylation signal in the segment between tumors and normal samples using Fisher’s exact test. All the methods are available as ‘differential.coverage’ R package.(21,22) The same methods were also used to compare methylation status between tumors expressing the alternative splicing event and tumors without the ASE. ASEs with a significant Fisher test when comparing tumors with and without ASE expression (p<0.05) in any interrogated region were identified as significant. ASEs where methylation was positively correlated with splice expression were excluded.
Validation with TCGA RNA-seq data
To perform validation within TCGA, raw fastq files for the RNA-Seq data was obtained for 44 HPV-positive oropharynx tumors and 16 normal samples within the TCGA using cghub(1). These data were realigned to the genome using MapSplice2 and above described methods to obtain junction expression in non-canonical junctions, not available from the level 3 TCGA junction expression data. Gene expression was determined using publicly available RSEM data. Splice variant identification and outlier statistics were then applied to these normalized junction data from TCGA using the same methods (Figure 1) to identify significant splice variants with HPV-positive TCGA tumors.
Additional TCGA data was obtained for analysis of identified unique AKT3 junction in a larger TCGA data set including both HPV-positive and HPV-negative tumors (499 tumors and 44 normal samples). These AKT3 junction data were then normalized using RSEM data expression for further analysis. Splice variant expression was compared between tumors and normal tissue and between HPV-positive and negative tumors using student’s t-test. Tumors were identified as harboring AKT3 splice variant based on outlier statistics as described above.
Validation in biologic samples
For validation of gene expression of both wild type AKT3 (AKT3 wt) and variant AKT3 (AKT3 var), primer and probe sets were designed specifically to span the junction between the canonical first and second exon (AKT3 wt) as well as the unique tumor first exon and second exon (AKT3 var). Primers and probes were designed using PrimerQuest tools (Integrated DNA Technologies, sequences in Supplemental Table 1). Touchdown PCR was used to confirm accurate PCR product of appropriate length for primers used. Quantitative RT-PCR was performed on cDNA generated from extracted primary tissue RNA from the test cohort to determine gene expression
To validate methylation of primary tissue, quantitative methylation specific PCR (qMSP) was performed on bisulfite converted DNA using an Epitect kit (Qiagen).(23,24) Primers specific for methylated bisulfite converted DNA were designed for the region where methylation was noted to be significantly altered between tumors with and without the unique splice variant on MBD-Seq and where a CpG island was identified. Primer sets were verified using bisulfite sequencing and used to confirm DNA methylation in selected tumor and normal samples..
AKT3 siRNA transfection
Cell lines used in this study, SKN3 (Japanese Collection of Research Bioresources), SCC61(25) and UM-SCC-47,(26) were fingerprinted and authenticated within 9 months of experimental studies using short tandem repeat analysis through Johns Hopkins Genetic Resources Core Facility (Supplemental Files). UM-SCC-47 was obtained in 2011 from Dr. Thomas Carey at University of Michigan, at which time mycoplasma testing was performed. SCC61 and SKN3 were obtained from Dr. Ralph Weichselbaum from University of Chicago and the Japanese Collection of Research Bioresources, respectively, in 2009, at which time mycoplasma testing was performed.(27) Experiments were carried out between passage 18–25 for each of the three cell lines. SKN3 and SCC61 were cultured in RPMI media with 10% FBS (fetal bovine serum) and DMEM/F12 media with 10% FBS, respectively. Transient knockdown of wild type AKT3 and variant AKT3 was achieved using siRNA transfection. Two unique custom designed siRNA were obtained from Dharmacon for specific targeting of novel first exon identified by tumor-specific splice junction using ON-TARGETplus (Supplemental Table 1). The siRNA for knockdown of overall AKT3 gene expression was obtained from Dharmacon, utilizing ON-TARGETplus SMARTpool for AKT3 (L-003002-00), and a scrambled ON-TARGETplus non-targeting pool siRNA (D-001810-10) was used as a control. Cells were plated in 96 well plates, with 5 replicates per experiment. Transfection was performed in reduced-serum media (Gibco Opti-MEM) for 16 hours, using lipofectamine reagent (ThermoFisher Scientific). Growth was assessed on day of transfection, and at 24, 48 and 72 hours after transfection using alamarBlue assay (Bio-Rad) for evaluation of cell viability at each time point.
RNA was isolated from cell pellets at 48 hours after transfection using Qiazol reagent (Qiagen) and RNeasy extraction kit (Qiagen). Reverse transcription was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems).
Western blot protein analysis
Protein analysis was performed on cell lysates of siRNA transfected cell lines. Cell lysates were collected at 48 hours after transfection. Media was aspirated and cells were rinsed twice with ice-cold PBS. Next, ice-cold RIPA buffer (50 mM Tris pH 7.4, 0.5% sodium deoxycholate, 150mM sodium chloride, 2mM EDTA, 0.1% SDS, 1% NP-40, 50mM NaF containing protease and phosphatase inhibitor cocktails) was applied to cells and incubated for 5 minutes on ice. Lysates were sonicated on ice and then centrifuged at 15 min at max speed at 4C, and supernatant was collected for further analysis. Protein concentrations were measured using Protein Assay Kit (DC™ Protein Assay, BIO-RAD). Equal amounts of denatured protein were loaded for western blot assay using Mini-PROTEAN TGX gels (Bio-Rad).
Rabbit primary antibodies used for western blot analysis were obtained from Cell Signaling Technology. These included pan-AKT (9272), phospho-AKT S473 (4060), phospho-AKT T308 (9275S), phospho-S6 (4858) and GAPDH (SC25778) all at 1:2000 dilutions. S6 antibody (2317) was used at 1:1000 dilution due to high signal level. HRP-conjugated goat Anti-Rabbit Ig (7074) was used for secondary antibody. ECL (Pierce™ ECL Western Blotting Substrate, Thermo Scientific™) reagent was used for western blot development.
Gene set analysis
Utilizing RNA sequencing data from the study cohort of 46 tumors and 25 normal samples, gene set analysis was performed on AKT/PI3K pathways in Hallmark and canonical pathway gene sets from MSigDB.(28,29) Wilcoxon gene set statistics were performed on differential expression statistics from LIMMA VOOM(30) and data was compared within these gene sets between tumors and normal samples, and between tumors harboring AKT3 splice variant and tumors without the variant. Benjamini-Hochberg correction was applied to p-values from gene set analysis to correct for multiple comparisons.
Results
Clinical cohort
RNA sequencing and subsequent analysis was performed on primary tumor tissue obtained from a cohort of 46 patients with HPV-related oropharyngeal squamous cell carcinoma (OPSCC). Clinical characteristics are summarized in Table 1, with detailed data in Supplemental Table 2. The median age at the time of diagnosis was 55 (range 25–75). A majority of the cohort was male (89.4%) and Caucasian (95.7%). With regard to smoking history, 37% of patients were never smokers, 37% of patients who were former smokers, and 26% of patients were current smokers at the time of diagnosis and specimen collection.
Table 1.
Patient and control clinical characteristics
| Patient Characteristics |
HPV+ OPSCC (N=46) N (%) |
UPPP controls (N=25) N (%) |
|---|---|---|
| Age | ||
| Median (range) | 55 (35–75) | 27 (18–51) |
|
| ||
| Male gender | 41 (89.1) | 10 (40) |
|
| ||
| Caucasian | 44 (95.6) | 14 (56) |
|
| ||
| Smoking status | ||
| Never | 17 (36.9) | 19 (76) |
| Prior | 17 (36.9) | 4 (16) |
| Current | 12 (26.1) | 2 (8) |
|
| ||
| T stage | ||
| T0 | 2 (4.3) | - |
| T1 | 17 (36.9) | |
| T2 | 18 (39.1) | |
| T3 | 8 (17.4) | |
| T4 | 1 (2.2) | |
|
| ||
| N stage | ||
| 0 | 1 (2.2) | - |
| 1 | 6 (13.0) | |
| 2A | 12 (26.1) | |
| 2B | 20 (43.5) | |
| 2C | 5 (10.9) | |
| 3 | 2 (4.3) | |
|
| ||
| Overall stage | ||
| I | 0 | - |
| II | 0 | |
| III | 7 (15.2) | |
| IV | 39 (84.8) | |
RNA-Seq ASE pipeline and IGV confirmation
A total of 834,195 putative junctions were identified by MapSplice2 alignment results for all samples. An algorithm utilizing outlier statistics (Figure 1, as described in methods) was applied to identify potential alternative splicing events (ASEs) by comparing junction expression levels in tumors compared to normal tissue. Filtering criterion left a total of 3420 junctions for analysis. Junctions were then normalized based on gene expression within each sample, using RSEM calculations to estimate gene expression. Outlier analysis found 174 junctions with significant outlier expression relative to the distribution the normalized junction expression values.
Next, each statistically significant junction was visualized in the Integrated Genome Viewer (IGV) to confirm the presence of a true ASE when comparing tumors and normal tissue. Of junctions identified, 109 (62.2%) of significant junctions were confirmed after visualization, involving 99 unique genes (Supplemental Table 3).
TCGA Validation
In order to validate junctions identified in this cohort, comparisons were performed using publicly available RNA-Seq data from TCGA. Junction and gene expression data from 44 HPV-positive HNSCC primary tumors and 16 normal samples were used for comparison. Using junction data available in TCGA, only 249,577 junctions were available for comparison due to increased filtering applied to pipelines. Therefore, RNA sequencing data was realigned using the same MapSplice2 pipeline. Using this pipeline, 108 (99%) of statistically significant junctions from the validation cohort were also identified in the TCGA data set. When junction expression levels were compared, 44 (41%) of these ASEs showed significant differential expression in tumors compared to normal tissue using t-test comparisons.
Characterization of ASEs
After validation, the 109 ASEs were characterized based on location of junctions and configuration of surrounding reads as visualized in IGV. Junctions were then characterized as alternative splicing events into the following categories: alternative start site (39%), insertion (21%), canonical exon skipping (9%), intron retention (9%), deletion (8%), alternative end site (7%), or non-coding (3%). When compared to gene isoforms documented in RefSeq, 29.4% of junctions were found and considered to be canonical, while 70.6% were novel splicing events. Within the 46 OPSCC tumors in the discovery set, the number of ASEs identified within individual tumors ranged from 9–77 (median = 47 ASEs per tumor, Supplemental Figure 1). These ASEs were frequently expressed across multiple tumors, and the prevalence of a given individual ASE among the tumor cohort was between 30.4% to 82.6% of tumors harboring the same ASE. (Supplemental Table 3) ASEs with a frequency of less than 30% among tumors were not identified as significant. In addition, 24.8% (n=27) of ASEs were significantly overexpressed in more than half of tumors within the cohort. Lastly, there was no association between the number of splice variants identified within a tumor and the patient smoking status, either as a current smoker (p=0.524) or never smoker (p=0.601).
MBD-Seq correlation
Given high prevalence of alternative start sites identified, we hypothesized that methylation could play a role in regulating expression of the identified alternative start site events. Therefore, methyl-CpG binding domain protein enrichment sequencing (MBD-Seq) was performed on the same tumor and normal tissue cohort. For each of the 43 ASEs which correlated with an alternative start site, the methylation level of three regions surrounding the start of each junction, the end of each junction, and the canonical transcriptional start site region were evaluated. When methylation of these regions was compared between tumors harboring the ASE of interest and tumors without the ASE, six genes showed significant negative correlation between level of methylation in these regions and expression of the ASE: AKT3, ELOVL1, KBTBD12, SNHG6, SYCP2, and ZNF331.
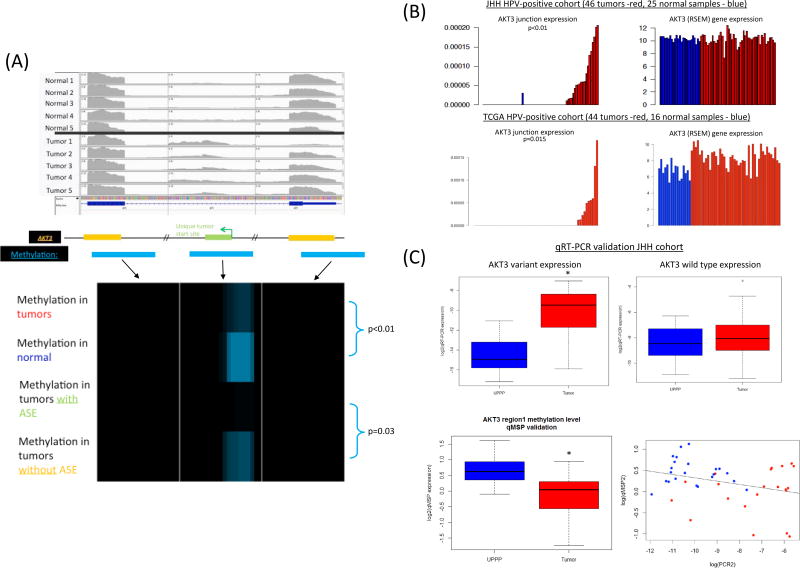
Given the important role of the AKT pathway in HNSCC, we chose to focus further study of the AKT3 alternative start site splice variant. In our cohort, differential methylation was present in the region of the alternative start site which was unique to tumors. In this region, methylation was significantly higher in normal tissues compared to tumors (p<0.01), and in addition methylation was significantly higher in tumors without the ASE compared to tumors expressing the ASE (p=0.03) (Figure 2A). Methylation was not able to be validated in TCGA as methylation probes in the Illumina 450K array performed in TCGA did not cover the region of this AKT3 alternative start site. The expression of the junction correlating with the AKT3 alternative start site was unique to tumors with significantly higher expression of the alternative AKT3 splice junction (Figure 2B) in both our cohort (p<0.01) and the TCGA HPV+ tumor cohort (p=0.015). These differences in AKT3 splice variant expression between tumors and normal tissues were not fully explained by AKT3 gene expression based on RSEM RNA seq data. While in the TCGA cohort normal tissue showed lower overall AKT3 gene expression compared to tumors, these changes in expression do not correlate with AKT3 splice variant expression (Figure 2B). In addition, AKT3 splice variant expression was not associated with patient smoking status, either never smokers (p=0.544) or current smokers (p=0.976).
Figure 2.
A. IGV visualization of AKT3 alternative start site in JHU cohort data (top) showing a unique non-canonical start site with expression unique to tumors. Using MBD-seq data, level of methylation was evaluated for three regions surrounding the novel exon and the two adjacent exons. The frequency of methylation in each 100 bp region is shown for the following category of samples: tumors, normal samples, tumors with alternative splicing event (ASE) and tumors without the ASE; with blue shade showing higher rate of methylation. In the region of the alternative start site ASE, decreased methylation is observed in tumors (p<0.01). In particular methylation in tumors expressing the ASE of interest had significantly decreased methylation compared to tumors without expression of the ASE (p=0.03).
B. Unique AKT3 junction expression (left) and overall AKT3 gene expression through RNA Seq analysis is shown. Expression of the AKT3 junction and splice variant is identified exclusively in tumors among both the JHU patient cohort including 46 tumors and 25 normal samples (top) and HPV+tumors in TCGA including 44 tumors and 16 normal samples (bottom). The corresponding overall AKT3 gene expression (calculated using RSEM methods) show that these differences in AKT3 splice variant expression are not fully explained by overall gene expression.
C. Biologic validation of AKT3 splice variant expression and methylation in primary tumor tissues. Quantitative PCR for identification of the AKT3 splice variant shows significantly increased expression in tumors compared to normal tissue (top left, p<0.0001), while wild type AKT3 gene expression is similar between normal UPPP tissue and tumors (top right, p=0.356). Quantitative Methylation specific PCR (qMSP) revealed significantly higher methylation in normal tissue compared to tumors at the alternative start site of AKT3 (bottom left, p<0.001), and there was a significant negative correlation between methylation in this region and expression of the splice variant (bottom right, p<0.01).
Biologic validation of AKT3 ASE (qRT-PCR) and methylation (qMSP)
Additional validation was performed in primary tissues through qRT-PCR to confirm the AKT3 alternative start site that was identified through the genome-wide studies. In this analysis, qRT-PCR data confirmed expression of the AKT3 variant significantly higher in tumors compared to normal tissue (Figure 2C, p<0.001). Interestingly, the qRT-PCR assay was more sensitive for identification of the splice variant and identified presence of the splice variant in a majority (73%) of tumor samples compared to 43% of tumors based on the RNA-Seq algorithm.
Next, methylation was also biologically validated through qMSP methods. First, the region of differential methylation was analyzed using bisulfite sequencing which confirmed methylation in this region. Based on bisulfite sequencing results, qMSP primer and probe sets were designed. Consistent with computational analysis of the MBD-seq data, the methylation in this region was significantly higher in normal tissue when compared to tumors (Figure 2C, p<0.001). A significant negative correlation was identified between expression of the AKT3 splice variant based on qRT-PCR and level of methylation based on qMSP (p<0.01, Figure 2C).
In addition, within TCGA the presence of the AKT3 splice variant was analyzed within a larger data set (499 tumors, 44 normal samples) including both HPV-positive and HPV-negative tumors. AKT3 splice variant was identified almost exclusively in tumors (25.4%) and only one normal sample (2.2%) with significantly higher expression in tumors (p<0.001). There was no difference seen between prevalence of the splice variant in HPV-positive (28.3%, n=92 tumors) and HPV-negative tumors (24.8%, n=407 tumors, p=0.742).
Biological significance of alternative first exon in AKT3 correlating to AKT1 E17K mutational hotspot
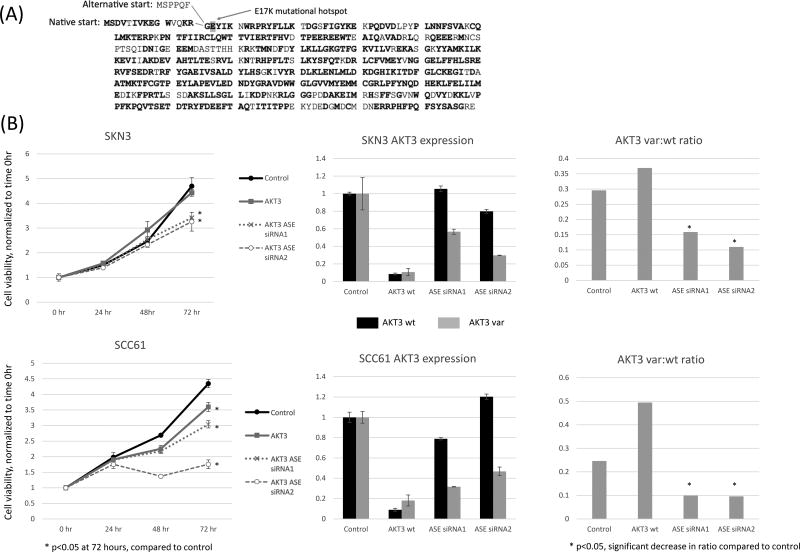
To determine the potential biological significance of an alternative start site or first exon in AKT3, a literature search was performed to identify known alterations in the start site region of AKT. Within COSMIC databases, no hotspot mutations have been identified in AKT3.(31) However, in AKT1, a gene with high sequence homology with AKT3, a hot spot mutation has been identified in the beginning of the gene near the PH domain.(32) This E17K mutation was shown to be transforming with increased activation of AKT1, and this mutation was identified in a subset of solid tumors including breast, colorectal and ovarian cancers.
The transforming properties of a mutation in this region offer indirect evidence that alterations at the 5’ end of AKT family genes could potentially result in oncogenic activation. Further analysis shows high protein sequence homology between AKT1 and AKT3, where the glutamic acid (E) site of the E17K mutational hotspot is conserved between AKT1 and AKT3 (Figure 3A). Furthermore, the E17K mutational hotspot mutation is located at the beginning of the second exon. The novel alternative start site isoform identified in this study results in a shortened first exon, which could result in a conformational change surrounding the E17K mutational hotspot. Based on these findings, we hypothesized that the novel AKT3 variant isoform identified by our algorithms had potential functional and transformative properties compared to wild type AKT3.
Figure 3.
A. Diagram showing AKT3 native and alternative start site predicted protein sequences. Amino acids with homology with AKT1 are shown in bold, and E17K AKT1 mutational hotspot is highlighted.
B. In adherent cell culture, silencing of the whole AKT3 gene using pooled siRNA (left panel, AKT3) and the AKT3 alternative splicing event (ASE) variant by independent two siRNA s(AKT3 ASE siRNA1 & siRNA2) is shown in two independent cell lines. Growth is normalized to time zero and measured over 72 hours. Significant growth inhibition was seen with specific silencing of the ASE of interest in both cell lines compared to silencing of the whole AKT3 gene. Gene expression from qRT-PCR shows decreased expression of both wild type (wt) AKT3 and and ASE AKT3 (var) when treated with pooled siRNA for AKT3. The ASE siRNA2 and siRNA3 show specific inhibition of the alternatively spliced AKT3 with minimal inhibition of the native gene. Specifically, the right panel shows the ratio of variant AKT3 to wild type expression, showing that this ratio was significantly decreased by both ASE siRNA1 and siRNA2 in both cell lines.
AKT3 siRNA knockdown
Specific siRNA constructs were designed for silencing of the overall AKT3 gene as well as silencing the tumor-specific isoform of AKT3 with alternative first exon. First, using sequence-specific primers and probes, expression of wild type and variant AKT3 was assessed in a panel of 17 HNSCC cell lines and 3 immortalized keratinocyte lines (Supplemental Figure 2). Using this data, three adherent cell lines were selected for siRNA knockdown functional growth assays: two cell lines with expression of the splice variant (SKN3, HPV-negative and SCC61, HPV-negative) and one cell line without native expression of the splice variant (UM-SCC-47, HPV-positive).
Using assessment of cell viability over 72 hours, significant growth inhibition was seen in both SKN3 and SCC61 after treatment with two independent ASE specific siRNAs (Figure 3B). Notably, this growth inhibition was significant compared to both control (scramble siRNA) as well as knockdown of overall AKT3 gene expression (pooled siRNA for the whole AKT3 gene). In cell lines without native expression of the splice variant (UM-SCC-47), minimal growth inhibition was seen with treatment using ASE specific siRNA (data not shown). Gene expression showed that siRNA was specific for inhibition of the AKT3 ASE variant, with only mild inhibition of the wild type AKT3 gene expression in both SCC61 and SKN3. Some variation in wild type AKT3 gene expression was seen after treatment with siRNAs targeted against the splice variant, however the overall ratio of variant expression to wild type AKT3 expression was significantly inhibited by the siRNAs in both cell lines (Figure 3B). As expected, siRNA designed to target the whole AKT3 gene decreased both the wild type and variant expression of AKT3 (Figure 3B).
AKT3 spice variant and PI3K/AKT pathways
In order to elucidate possible mechanisms of growth inhibition seen through silencing of the AKT3 splice variant, western blot protein analysis of the canonical phosphorylated AKT1 signaling pathway was performed. No consistent differences were seen in activation of pAKT1 or downstream AKT1 signaling pathway including S6 phosphorylation with silencing of the AKT3 splice variant, despite consistent growth inhibition [Supplemental Figure 3]. We were therefore unable to demonstrate an effect of AKT3 splice variant knockdown on pAKT1 or downstream AKT1 signaling as defined by changes in phospho-S6, despite inhibitory growth effects. We conclude that this indicates that the growth effects of AKT3 splice variants are at least in part mediated by pAKT1 and pAKT1 target independent mechanisms.
Additional bioinformatics analysis was performed to evaluate the potential role of the AKT3 splice variant within known pathways.Analysis of mutations and gene amplifications in TCGA tumors was utilized to understand potential relationship between presence of AKT3 splice variant and downstream AKT pathway activation. There was no correlation between presence of AKT3 splice variant and presence or absence of the most frequent 50 genetic alterations along the PI3K/AKT pathway including amplifications and mutations (Supplemental figure 4). No alterations were noted to occur more frequently or be mutually exclusive with the presence of the AKT3 splice variant.
Gene set analyses were also performed based on RNA-seq data available from the HPV-positive OPSCC cohort from this study to compare gene expression in gene sets from MSigDB in the PI3K/AKT pathways. First, when gene sets were compared between tumors and normal tissue, as expected, multiple gene sets showed significant differences including pathways involved in PI3K signaling, RELA-p65, PI3K/AKT/MTOR signaling pathway, NF-kappa B, and HIF-1 alpha. When analysis was restricted to tumors, gene set related to RELA-p65 was noted to be significantly different in tumors with the AKT3 splice variant compared to those without (p=0.013).
Discussion
TCGA and other recent sequencing efforts have been a tremendous resource in understanding the genetic landscape of cancer.(33) New mutational analyses have led to understanding cancer mutations that occur across organ systems to allow for the design of specific basket trials, where therapies can be rationally targeted towards specific mutations.(34) Alternative splicing is another mechanism by which alternative isoforms may have significant impact on gene expression and function, independent of genetic mutations.(8) In this study, a novel algorithm is utilized to systematically identify alternative splicing events unique to tumors to show that alternative splicing can represent an important functional mechanism of oncogenesis in HPV-related oropharyngeal cancer that has not been previously appreciated.
Alternative splicing has long been recognized as an important method of increasing protein diversity in eukaryotic cells. Within the human genome, it is estimated that more than 95% of multi-exonic genes undergo some level of alternative splicing, and this process has been shown to play a role in carcinogenesis in both solid and liquid tumors.(7,8) While splice variation has been studied in the past, the availability of high throughput RNA sequencing now allows for systematic transcriptome wide analysis of splicing variation, which has not been previously reported in head and neck cancer.
Several algorithms have been developed for analysis of differential splicing to identify significant differential gene isoforms.(35–37) Utilization of outlier statistics has been shown to better capture significant events in cancer tissue, (12,38) and our adaptation of this algorithm to RNA-sequencing data identified novel splicing events that have not been previously described. Of the validated splicing events identified, 70% were non-canonical. These may represent previously unreported gene isoforms or aberrant splicing events occurring within an altered cancer genome. While some of these identified splicing events may represent passenger alterations, many may be functionally active, such as the AKT3 variant described.
In addition, expression of some ASEs correlated with changes in alternative start sites showed significant negative correlation with methylation change, suggesting that methylation may play a role in regulating these splicing events. Epigenetic changes traditionally are recognized to inhibit gene expression through promoter hypermethylation, but additional roles for DNA methylation are increasingly being recognized.(39,40) There has been some evidence in the literature for regulation of RNA splicing through DNA methylation.(41) While only 15% of alternative start site ASEs in this study showed correlated epigenetic alterations, these data suggest that methylation changes may be one method by which aberrant splicing is regulated within the cancer genome. Another possibility is that an alternative start site splicing variant could be a mechanism by which cancer cells can overcome hypermethylation at a canonical promoter site. Further investigation is needed to clarify and confirm the relationship between promotor hypermethylation and alternative start site splicing events observed in this study.
Through described methods, a functionally relevant novel splice variant of AKT3 was identified within HPV-positive OPSCC primary tumors. The AKT pathway is known to play an important role in the pathogenesis of HNSCC. Genes within this pathway axis are mutated in HNSCC, including receptor tyrosine kinases (EGFR, FGFR), PIK3R1, and PIK3CA, which was more frequently mutated in HPV-positive tumors.(1) The replacement of the first exon occurs at a known mutational hotspot in the AKT gene family (AKT1), and growth inhibition was associated with specific silencing of this novel splice variant. Notably, this growth inhibition was dependent on the ratio of AKT3 variant expression compared to wild-type expression. Knockdown of the AKT3 variant alone produced significant growth inhibition, however knockdown of the whole AKT3 gene including both variant and wild-type did not produce significant growth inhibition compared to controls. Protein studies showed that this growth inhibition does not occur through the canonical AKT1 pathway, and further studies are required to elucidate the mechanism of action of this alternative AKT3 splice variant that are beyond the scope of the current study endeavor.
This study is limited by some factors inherent to clinical samples and RNA preparation methods, limited validation and need for additional functional studies. In this study, primary patient tumor samples were compared against non-matched cancer unaffected controls. Other studies including TCGA(1) utilize adjacent normal tissue for controls; however in head and neck cancer patients there is potential for field cancerization resulting in molecular changes in normal adjacent tissue.(42) Data from normal tissue within this study showed strong uniformity, even with variations in age or smoking status, and cancer specific variants were not identified among normal samples. Within this cohort, patients with prior or current smoking status did not exhibit higher rates of ASEs within tumors, and smoking status did not correlate with the presence of the identified AKT3 splice variant, suggesting that smoking status plays a smaller role in splice variant changes than is seen with DNA level genetic alterations. Additionally, ASEs identified within this algorithm were validated in the TCGA data set. In addition, there are some limitations to RNA-sequencing methods utilized. However, multi-platform analyses have shown strong concordance between library prep platforms.(43) Short read RNA sequencing libraries have some limitations in inferring gene isoform variations, but long read technologies are currently more expensive with high rates of sequencing errors.(44,45)
In addition, the performance of described techniques for identification of differential splicing events in heterogeneous tumor samples need to be computationally benchmarked against other algorithms described in the literature with simulated datasets.(11) While the outlier analysis is effective at prioritizing individual junctions associated with differential gene isoform usage, it cannot integrate complex changes that occur over an entire gene. Therefore, in the time that we have completed this analysis of the characterization of splice variants in HPV-positive HNSCC, we have continued to refine techniques for identifying ASE variant expression, and have piloted additional in silico techniques including multivariate, non-parametric variability statistics for differential ASE analysis in RNA-Seq data that capture alternative splicing behavior in hetereogenous primary cancer samples, called Splice Expression Variability Analysis (manuscript in submission).
Utilizing described methods, we were able to successfully identify more than 100 tumor specific splice variant candidates within HPV-positive head and neck cancer, including a splice variant of AKT3 which may have functional relevance within these tumors. The investigations in this study suggest a functional role for this AKT3 splice variant with growth inhibition observed within cell culture, as well as involvement of well know downstream mediators of the AKT pathway, including S6, as well as a significant association of AKT3 splice variant expression with RELA-p65 gene set enrichment, indicating NF-kB alteration consistent with nuclear activity of AKT affecting NF-kB transcription. (46) However, additional functional studies beyond the scope of the goals of this study are needed to define the pathway mechanism of the proposed functionally active isoform of AKT3 identified in this study, as these mechanisms likely involve pathways outside of canonical, AKT1 reported pathways that have been well characterized. After this, further animal work could confirm the functional significance of this splice variant alteration. Furthermore, additional studies are needed to better elucidate mechanisms behind the relationship between methylation and regulation of alternative start site splice variants.
Lastly, this splice variant was identified within HPV-positive tumors, but is also identified in HPV-negative tumors, which is consistent with in vitro data which showed impact on growth in HPV-negative cell lines. While the identified splice variant may not mechanistically be related HPV, it represents a post translational genetic alteration that has high prevalence in HPV-positive tumors (28–43%), and therefore maintains relevance in the biologic understanding and treatment of this disease. Based on qRT-PCR data, this prevalence may even be higher (more than 70% of tumors in this study) than is estimated by RNA-sequencing analysis methods which are dependent on read depth and stringent outlier statistics.
This current study highlights the potential of RNA-sequencing analysis to identify functionally relevant splicing events in primary tumor tissues. While splice variants have been identified to some extent in head and neck cancer,(9,47) this represents the first report of a functionally active splice variant in HPV-related OPSCC. Future studies may further explore the role of AKT3 and other splice variants identified in this study to further elucidate the biological pathways that are activated in HPV-related disease independent of DNA level genetic mutations and alterations. These data also suggested that at least some alternative splicing events may be epigenetically regulated. Finally, the described methods for identification of significant splicing events can be applied to other cancer types to expand our understanding of the complex perturbations that can occur within cancer genomes.
Supplementary Material
Acknowledgments
Funding support: This work was supported by NIH R01DE023347 (J. Califano), NIH 2T32DC000027-26 (T. Guo), NIH P50 DE 019032 (J. Califano), NIH R21DE025398 (D.A. Gaykalova), NIH P30 CA006973 (A.V. Favorov), R01CA183965 (S. Yegnasubramanian and S.J. Wheelan), U01CA196390 (S. Yegnasubramanian and S.J. Wheelan) and Russian Foundation for Basic Research 14-04-01872 (A.V. Favorov).
We thank the members of the Sidney Kimmel Comprehensive Cancer Center (SKCCC) Experimental and Computational Genomics Core for performing RNA-seq and MBD-seq.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:612–9. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia A, Burtness B. Human Papillomavirus-Associated Oropharyngeal Cancer: Defining Risk Groups and Clinical Trials. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:3243–50. doi: 10.1200/JCO.2015.61.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fakhry C, Zhang Q, Nguyen-Tan PF, Rosenthal D, El-Naggar A, Garden AS, et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:3365–73. doi: 10.1200/JCO.2014.55.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34:1–14. doi: 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- 9.Li R, Ochs MF, Ahn SM, Hennessey P, Tan M, Soudry E, et al. Expression microarray analysis reveals alternative splicing of LAMA3 and DST genes in head and neck squamous cell carcinoma. PloS one. 2014;9:e91263. doi: 10.1371/journal.pone.0091263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afsari B, Geman D, Fertig EJ. Learning dysregulated pathways in cancers from differential variability analysis. Cancer Inform. 2014;13:61–7. doi: 10.4137/CIN.S14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochs MF, Farrar JE, Considine M, Wei Y, Meshinchi S, Arceci RJ. Outlier Analysis and Top Scoring Pair for Integrated Data Analysis and Biomarker Discovery. IEEE/ACM Trans Comput Biol Bioinform. 2014;11:520–32. doi: 10.1109/TCBB.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo T, Gaykalova DA, Considine M, Wheelan S, Pallavajjala A, Bishop JA, et al. Characterization of functionally active gene fusions in human papillomavirus related oropharyngeal squamous cell carcinoma. International journal of cancer Journal international du cancer. 2016;139:373–82. doi: 10.1002/ijc.30081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson M. org.Hs.eg.db: Genome wide annotation for Human. R package version 340: R package version 3.4.0. 2016 [Google Scholar]
- 15.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochs MF. OGSA: Outlier Gene Set Analysis. R package version 120: Bioconductor. 2015 [Google Scholar]
- 17.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinclair SH, Yegnasubramanian S, Dumler JS. Global DNA methylation changes and differential gene expression in Anaplasma phagocytophilum-infected human neutrophils. Clin Epigenetics. 2015;7:77. doi: 10.1186/s13148-015-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yegnasubramanian S, Wu Z, Haffner MC, Esopi D, Aryee MJ, Badrinath R, et al. Chromosome-wide mapping of DNA methylation patterns in normal and malignant prostate cells reveals pervasive methylation of gene-associated and conserved intergenic sequences. BMC Genomics. 2011;12:313. doi: 10.1186/1471-2164-12-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favorov A. differential.coverage. R package. 2015 < https://github.com/favorov/differential.coverage>.
- 22.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2016 < https://www.r-project.org/] >.
- 23.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaykalova DA, Vatapalli R, Wei Y, Tsai HL, Wang H, Zhang C, et al. Outlier Analysis Defines Zinc Finger Gene Family DNA Methylation in Tumors and Saliva of Head and Neck Cancer Patients. PloS one. 2015;10:e0142148. doi: 10.1371/journal.pone.0142148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res. 2011;17:7248–64. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, et al. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget. 2014;5:8906–23. doi: 10.18632/oncotarget.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatakeyama H, Cheng H, Wirth P, Counsell A, Marcrom SR, Wood CB, et al. Regulation of heparin-binding EGF-like growth factor by miR-212 and acquired cetuximab-resistance in head and neck squamous cell carcinoma. PloS one. 2010;5:e12702. doi: 10.1371/journal.pone.0012702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–44. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magliacane G, Grassini G, Bartocci P, Francaviglia I, Dal Cin E, Barbieri G, et al. Rapid targeted somatic mutation analysis of solid tumors in routine clinical diagnostics. Oncotarget. 2015;6:30592–603. doi: 10.18632/oncotarget.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartley SW, Mullikin JC. Detection and visualization of differential splicing in RNA-Seq data with JunctionSeq. Nucleic Acids Res. 2016;44:e127. doi: 10.1093/nar/gkw501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, Huang Y, Du Y, Orellana CF, Singh D, Johnson AR, et al. DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res. 2013;41:e39. doi: 10.1093/nar/gks1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Z, Ihle NT, Rejto PA, Zarrinkar PP. Outlier analysis of functional genomic profiles enriches for oncology targets and enables precision medicine. BMC Genomics. 2016;17:455. doi: 10.1186/s12864-016-2807-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 40.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 41.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lydiatt WM, Anderson PE, Bazzana T, Casale M, Hughes CJ, Huvos AG, et al. Molecular support for field cancerization in the head and neck. Cancer. 1998;82:1376–80. [PubMed] [Google Scholar]
- 43.Li S, Tighe SW, Nicolet CM, Grove D, Levy S, Farmerie W, et al. Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat Biotechnol. 2014;32:915–25. doi: 10.1038/nbt.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Au KF, Underwood JG, Lee L, Wong WH. Improving PacBio long read accuracy by short read alignment. PloS one. 2012;7:e46679. doi: 10.1371/journal.pone.0046679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho H, Davis J, Li X, Smith KS, Battle A, Montgomery SB. High-resolution transcriptome analysis with long-read RNA sequencing. PloS one. 2014;9:e108095. doi: 10.1371/journal.pone.0108095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madrid LV, Mayo MW, Reuther JY, Baldwin AS., Jr Akt stimulates the transactivation potential of the RelA/p65 Subunit of NF-kappa B through utilization of the Ikappa B kinase and activation of the mitogen-activated protein kinase p38. The Journal of biological chemistry. 2001;276:18934–40. doi: 10.1074/jbc.M101103200. [DOI] [PubMed] [Google Scholar]
- 47.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.