Abstract
Background
Long noncoding RNAs (lncRNAs) have been revealed to function as competing endogenous RNAs (ceRNAs), which can seclude the common microRNAs (miRNAs) and hence prevent the miRNAs from binding to their ancestral gene. Nonetheless, the role of lncRNA-mediated ceRNAs in prostate cancer has not yet been elucidated.
Material/Methods
Using The Cancer Genome Atlas (TCGA) database, lncRNA, miRNA, and mRNA profiles from 499 prostate cancer tissues and 52 normal prostate tissues were analyzed with the R package “DESeq” to identify the differentially expressed RNAs. GO and KEGG pathway analyses were performed using “DAVID6.8” and R packages “Clusterprofile.” The ceRNA network in prostate cancer was constructed using miRDB, miRTarBase, and TargetScan databases. Survival analysis was performed with Kaplan-Meier analysis.
Results
A total of 376 lncRNAs, 33 miRNAs, and 687 mRNAs were identified as significant factors in tumorigenesis. Based on the hypothesis that the ceRNA network (lncRNA-miRNA-mRNA regulatory axis) is involved in prostate cancer and forms competitive interrelations between miRNA and mRNA or lncRNA, we constructed a ceRNA network that included 23 lncRNAs, 6 miRNAs, and 2 mRNAs that were differentially expressed in prostate cancer. Only 3 lncRNAs (LINC00308, LINC00355, and OSTN-AS1) had a significant association with survival (P<0.05). The 3 prostate cancer-specific lncRNA were validated in prostate cancer cell lines PC3 and DU145 using qRT-PCR.
Conclusions
We demonstrated the differential lncRNA expression profiles in prostate cancer, which provides new insights for future studies of the ceRNA network and its regulatory mechanisms in prostate cancer.
MeSH Keywords: Carcinogenesis; MicroRNAs; Prostatic Neoplasms; RNA, Long Noncoding
Background
The most common cancer in males worldwide is prostate cancer, accounting for 13% of cancer-related deaths. In 2016, prostate cancer resulted in 648 400 new cases and 80 900 deaths in developed countries [1]. Despite the rapid development of diagnosis and treatment of prostate cancer, poor therapeutic effect and high prevalence are still serious clinical challenges. Therefore, identification of new potential biomarkers and therapeutic targets is crucial to improving alternative therapies.
With the development of genome-wide analysis, up to 20 000 pseudogenes have been found in the human genome [2]. In general, pseudogenes are characterized by inaction in protein coding, such as long noncoding RNA (lncRNA). However, numerous studies suggest that they may execute important functions in carcinogenesis. Recently, a series of pseudogenes has been revealed to function as competing endogenous RNAs (ceRNAs), which can seclude the common microRNAs (miRNAs) and hence prevent the binding of miRNAs to their target genes [3,4], such as PTENP1, the pseudogene of PTEN tumor suppressor that includes a poly-A tail and shares a common 5′ and 3′UTR sequence with PTEN [5]. Zhang et al. showed that PTENP1 can act as a ceRNA to alter PTEN expression level by sponging miR-106b and miR-93 in gastric cancer [6]. In addition, Chen et al. suggested that the lncRNA ROR promotes radioresistance in hepatocellular carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression [7].
In the present study, the expression profiles of lncRNAs, miRNAs, and mRNAs were obtained from The Cancer Genome Atlas (TCGA) database. The lncRNA-miRNA-mRNA regulatory axis was positively correlated with prostate cancer. A ceRNA network was subsequently constructed using miRDB, miRTarBase, and TargetScan databases. Among the 23 lncRNAs, 6 miRNAs, and 2 mRNAs in the ceRNA network, 3 specific lncRNAs were found to have a strong association with the survival of prostate cancer patients. The results of this study help to describe the executive mechanisms of lncRNAs through the lncRNA-miRNA-mRNA network in prostate cancer, which may provide new insights for future research on prostate cancer.
Material and Methods
Patients and samples information
The RNA sequencing data from 499 prostate cancer tissues and 52 samples from non-tumorous prostate tissues were acquired from the TCGA database in 2018. The GDC Data Transfer Tool (https://gdc.cancer.gov/access-data/gdc-data-transfer-tool) was used to download the level 3 mRNASeq and miRNAseq gene expression data, as well as clinical information of prostate patients. The RNA sequencing data were generated from Illumina HiSeqRNASeq and Illumina HiSeqmiRNASeq platforms. This study meets the publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines). Ethics Committee approval was not required as the data were obtained from TCGA.
Analysis of differentially expressed RNA
The “DESeq” package [8] in R software was used to identify the differentially expressed mRNAs, lncRNAs, and miRNAs with thresholds of |log2FoldChange| >2, false discovery rate (FDR) or adjusted P value <0.01. In addition, mRNA and lncRNA annotation were performed with ENSEMBL to define and encode the differentially expressed RNAs (htps://www.ensembl.org/).
Go and KEGG functional enrichment analysis
In order to understand the potential biological processes and pathways of discriminatively expressed genes, we used the Annotate, Visualize, and Integrate Discovery Database (DAVID 6.8) (http://david.abcc.ncifcrf.gov/) [8] to perform Gene Ontology (GO) biological processes at the significant level (FDR <0.05). The KEGG Orthology-Based Annotation System 3.0 (KOBAS3.0) (kobas.cbi.pku.edu.cn/) was used to conduct KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis at the significance level of adjusted P value<0.05. The “Goplot” package in R was used to conduct the chord plot. The network was assembled and globally visualized using Cytoscape v 3.6.1 [9].
Construction of the lncRNA-miRNA-mRNA ceRNA network
Based on the hypothesis that lncRNAs can sponge the common miRNAs and thereby prevent the miRNAs from binding to their target gene [10], a ceRNA network was constructed. StarBase v2.0 database (http://starbase.sysu.edu.cn/) was used to modify the miRNAs sequences, and the lncRNA-miRNA interactions were predicted by the miRanda database (http://www.microrna.org/). miRDB (http://www.mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) [11] and TargetScan (http://www.targetscan.org/) were used to predict the miRNAs target mRNAs. Also, in the present study, the aberrant expression data of the predicted miRNA were combined to select the intersecting lncRNA and mRNA. Construction and visualization of the lncRNA-miRNA-mRNA ceRNA network were performed by Cytoscape v3.6.1. lncRNA, miRNA, and mRNA with |log2FoldChange| >2 and P<0.05 were analyzed.
Survival analysis
To determine the prognosis of TCGA prostate cancer patients in relation to differentially expressed RNA signatures, Kaplan-Meier survival curves of differentially expressed lncRNAs, microRNAs, and mRNAs were visualized using the “survival” package in R. Survival analysis was performed with log-rank test and P<0.05 was considered significant.
Cell culture
Normal myofibroblast stromal cell line WPMY1 and 2 prostate cancer cell lines (PC3 and DU145) were purchased from the American Type Culture Collection (USA). Cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (Gibco, MA, USA), 100 mg/mL streptomycin, and 100 U/mL penicillin in a humidified atmosphere of 95% air and 5% CO2 at a temperature of 37°C.
RNA extraction and the quantitative real-time PCR
Total RNA was extracted from cells with Trizol reagent (Invitrogen, Grand Island, CA, USA) according to the manufacturer’s instructions. RNA was first reverse transcribed into cDNA using the PrimeScript RT kit (Takara, Japan) according to the manufacturer’s protocol. qRT-PCR was then performed using an SYBR PrimeScript RT-PCR kit (Takara, Japan). The following primers were used:
LINC00308, 5′-CAGATAAGACTCTGTCTACCCT-3′ (forward),
5′-ACTGAATAAAGGAATGATGCGT-3′(reverse);
LINC00355 5′-ACAGAGCTGGTGGGAGCTGGGAAT-3′ (forward),
5′-AGTATCAATAGCTGAATAGAC-3′(reverse);
OSTN-AS1: 5′-CCTGCCTCAGCTTCCCAAGGAG-3′ (forward),
5′-GTTGGCAATAAAAAGAAGACAAT-3′.
U6 was used as endogenous control U6 (5′-CTCGCTTCGGCAGC ACA-3′ (forward), and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse)). All samples were run in triplicates on the ABI 7900HT Real-Time PCR system (Applied Biosystems, CA, USA). Calculation of relative expression levels was performed using the 2−ΔΔCt formula.
Statistical analysis
Statistical analyses were performed using SPSS 21.0 software (SPSS Inc., Chicago, IL, USA). Data are expressed as mean ±SD. The t test was used for two-group comparisons and a P value <0.05 was considered statistically significant.
Results
Patient characteristics
The detailed clinical and pathological features of the TCGA prostate cancer study population are shown in Table 1. All 499 patients were pathologically diagnosed with prostate cancer. The median age was 61 years (range: 41–78 years). Follow-up time was 23–5024 days. The majority of patients were lymph node-free (69.9%) and at high risk according to Gleason score histologic grade (95.2%).
Table 1.
Clinicopathological characteristics of 499 patients with prostate cancer.
| Characteristics | Subtype | Patinets n (%) |
|---|---|---|
| Age | >61 | 224 (44.9) |
| ≤61 | 275 (55.1) | |
| Race | Asian | 2 (0.4%) |
| Black or African American | 7 (1.4%) | |
| White | 147 (29.5%) | |
| Unknown | 343 (68.7%) | |
| Histological type | Acinar type | 484 (97.0%) |
| Other subtype | 15 (3.0%) | |
| Tumor stage | T2a | 13 (2.6%) |
| T2b | 10 (2.0%) | |
| T2c | 145 (29.0%) | |
| T3a | 158 (31.7%) | |
| T3b | 136 (27.3%) | |
| T4 | 10 (2.0%) | |
| Unknown | 7 (1.4%) | |
| Lymph node | N0 | 347 (69.6%) |
| N1 | 79 (15.8%) | |
| Unknown | 73 (14.6%) | |
| Patients cancer status | With tumor | 89 (17.8%) |
| Tumor free | 347 (69.6%) | |
| Unknown | 63 (12.6%) | |
| Survival status | Alive | 489 (98.0) |
| Dead | 10 (2.0%) |
Differentially expressed RNAs and their functional enrichment analysis
A total of 687 genes were identified as differentially expressed: 353 (51.38%) genes were downregulated and 334 (48.62%) were upregulated. The complete list of the differentially expressed genes is presented in Supplementary Table 1. The mRNA expression levels are visualized in the heatmap (Supplementary Figure 1). Using the same cut-off criteria of |log2FoldChange| >2 and adjusted P value <0.01, 376 lncRNAs (170 downregulated and 206 upregulated) and 33 miRNAs (11 downregulated and 22 upregulated) were identified as differentially expressed in prostate cancer tissues as compared to the normal tissues (Supplementary Figures 2, 3).
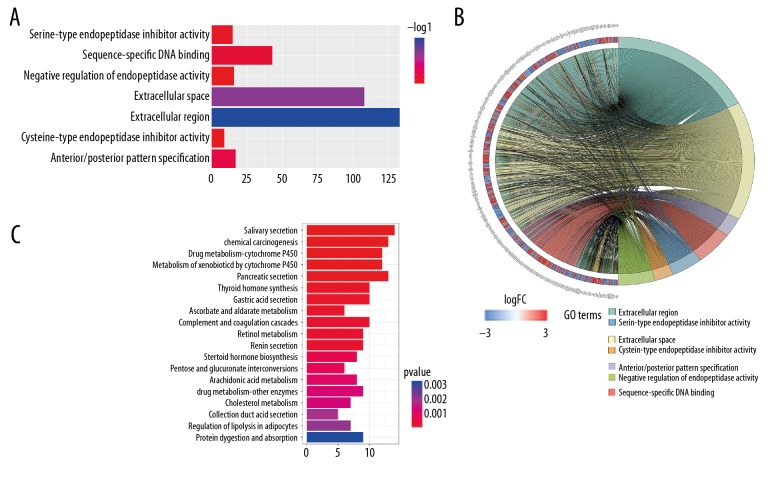
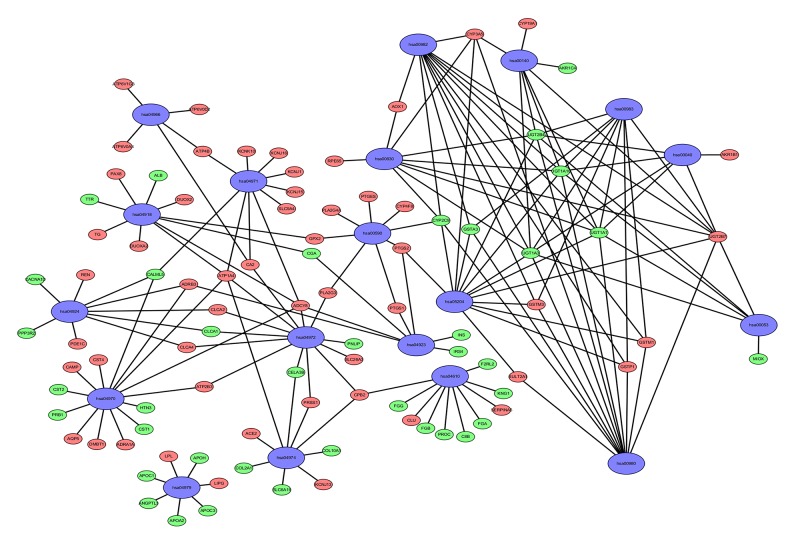
To understand the mechanism of oncogenesis underlying prostate cancer, functional enrichment characterization of these 687 mRNAs was performed by GO and KEGG analysis with DAVID6.8 and KOBAS3.0, respectively. Enrichment of GO analysis showed that 8 significant functions are involved in prostate cancer (FDR<0.01) (Figure 1A). Among them, 133 genes were enriched in the extracellular region and represent the lowest FDR, followed by extracellular space with 108 enriched genes. All enriched gene-function relationships are shown in the chord diagram (Figure 1B). In the KEGG pathway analysis, a total of 19 pathways were genetically enriched (Table 2); salivary secretion was the most important cancer-related pathway, which contains 14 genes (Figure 1C). All the expressions related to these KEGG pathway-enriched genes were visualized using Cytoscape software. In the network diagram, the red genes represent upregulation in this pathway and green genes represent the opposite expression (Figure 2).
Figure 1.
(A) Seven significant biological processes in GO analysis for differentially expressed mRNAs. (B) Chord diagram of all enriched genes in GO analysis. (C) Nineteen enrichment of KEGG pathways for differentially expressed mRNAs in prostate cancer.
Table 2.
KEGG pathways enriched by the differentially expressed mRNAs involved in ceRNA network.
| Pathway ID | Description | Adj. P-value | Number of DERNAs |
|---|---|---|---|
| hsa04970 | Salivary secretion | 0.00022 | 14 |
| hsa05204 | Chemical carcinogenesis | 0.00022 | 13 |
| hsa00982 | Drug metabolism – cytochrome P450 | 0.00022 | 12 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 0.00030 | 12 |
| hsa04972 | Pancreatic secretion | 0.00075 | 13 |
| hsa04918 | Thyroid hormone synthesis | 0.00591 | 10 |
| hsa04971 | Gastric acid secretion | 0.00591 | 10 |
| hsa00053 | Ascorbate and aldarate metabolism | 0.00593 | 6 |
| hsa04610 | Complement and coagulation cascades | 0.00593 | 10 |
| hsa00830 | Retinol metabolism | 0.00593 | 9 |
| hsa04924 | Renin secretion | 0.00593 | 9 |
| hsa00140 | Steroid hormone biosynthesis | 0.01369 | 8 |
| hsa00040 | Pentose and glucuronate interconversions | 0.01440 | 6 |
| hsa00590 | Arachidonic acid metabolism | 0.01642 | 8 |
| hsa00983 | Drug metabolism – other enzymes | 0.01823 | 9 |
| hsa04979 | Cholesterol metabolism | 0.01823 | 7 |
| hsa04966 | Collecting duct acid secretion | 0.02425 | 5 |
| hsa04923 | Regulation of lipolysis in adipocytes | 0.02566 | 7 |
| hsa04974 | Protein digestion and absorption | 0.03680 | 9 |
Figure 2.
The KEGG network of differentially expressed mRNAs in prostate cancer. Downregulated genes are represented by a green ellipse and upregulated genes are represented by a red ellipse.
Construction of ceRNA network in prostate cancer
To further explore the mechanisms of these differentially expressed genes in prostate cancer, a lncRNA-miRNA-mRNA (ceRNA) network was constructed based on the above data. Consequently, 6 miRNAs targeted 24 key lncRNAs were described in the ceRNAs network (Table 3). The miRNA-targeted mRNA was predicted through miRDB, miRTarBase, and TargetScan (Table 4). Only 2 differentially expressed mRNAs in the ceRNA network, PTGS2 and DUSP2, have previously been reported as tumor suppressor genes [10,12–14] (Figure 3). In the ceRNA network, 12 lncRNAs, 2 miRNAs, and 2 mRNAs are downregulated, while 12 lncRNAs and 4 miRNAs are upregulated (Table 5, Figure 4). Subsequently, we successfully constructed the dysregulated ceRNA network with differentially expressed RNAs, which included 24 lncRNAs, 6 miRNAs, and 2 mRNAs. Results indicate that differentially expressed lncRNAs indirectly interact with mRNAs through miRNAs in prostate cancer (Figure 5). To further identify the differentially expressed RNAs with prognostic significance, Kaplan-Meier survival analysis was used. As a result, 3 out of 24 differentially expressed lncRNAs (LINC00308, LINC00355, and OSTN-AS1) were significantly associated with overall survival (log-rank P<0.05) (Figure 6A). However, no differentially expressed miRNA and mRNA in this ceRNA network were found to be associated with prognosis.
Table 3.
miRNAs targeting cancer-specific lncRNAs in ceRNA network.
| LncRNA | miRNA |
|---|---|
| C5orf64 | miR-184; miR-122; miR-506 |
| LINC00308 | miR-137 |
| LINC00313 | miR-372; miR-187; miR-122 |
| LINC00336 | miR-506 |
| UCA1 | miR-184; miR-122; mir-506 |
| PCA3 | miR-137 |
| LINC00355 | miR-122; miR-506 |
| HCG22 | miR-122; miR-506 |
| XIST | miR-372; miR-137; miR-122; miR-506 |
| EMX2OS | miR-184; miR-506 |
| AL161645.1 | miR-184; miR-122 |
| NALCN-AS1 | miR-372; miR-506 |
| ERVH48-1 | miR-137; miR-184; miR-187 |
| OSTN-AS1 | miR-137; miR-506 |
| DSCAM-AS1 | miR-137; miR-122 |
| GPC5-AS1 | miR-372 |
| ZBTB20-AS3 | miR-122; miR-506 |
| AL356133.2 | miR-372 |
| HNF1A-AS1 | miR-372; miR-122 |
| AL353803.1 | miR-122 |
| ALDH1L1-AS2 | miR-372 |
| LNX1-AS2 | miR-506 |
| PCAT1 | miR-372; mir-122; mir-506 |
| ANO1-AS2 | mir-372 |
Table 4.
miRNAs targeted cancer-specific mRNAs in ceRNA network.
| miRNA | mRNA |
|---|---|
| miR-137 | PTGS2 |
| miR-122 | DUSP2 |
| miR-372 | DUSP2 |
Figure 3.

Venn diagram analysis of differentially expressed mRNAs in ceRNA network.
Table 5.
Differentially expressed RNAs involved in ceRNA network.
| RNAs | Regulation | Fold change | P-value | FDR |
|---|---|---|---|---|
| C5orf64(lncRNA) | Down-regulation | −3.036662036 | 1.46E-30 | 7.31E-29 |
| LINC00308(lncRNA) | Up-regulation | 3.235382205 | 1.26E-10 | 9.30E-10 |
| LINC00313(lncRNA) | Down-regulation | −2.476850161 | 1.82E-26 | 6.67E-25 |
| LINC00336(lncRNA) | Down-regulation | −2.276139635 | 4.31E-18 | 7.72E-17 |
| UCA1(lncRNA) | Down-regulation | −2.914678098 | 1.98E-47 | 2.4E-45 |
| PCA3(lncRNA) | Up-regulation | 3.331178572 | 5.63E-18 | 9.84E-17 |
| LINC00355(lncRNA) | Up-regulation | 3.435319383 | 9.65E-06 | 3.36E-05 |
| HCG22(lncRNA) | Down-regulation | −3.277663954 | 2.48E-37 | 1.94E-35 |
| XIST(lncRNA) | Down-regulation | −2.216949544 | 5.07E-11 | 3.99E-10 |
| EMX2OS(lncRNA) | Down-regulation | −5.978725669 | 2.84E-215 | 2.21E-211 |
| AL161645.1(lncRNA) | Down-regulation | −4.932391705 | 3.99E-151 | 6.2E-148 |
| NALCN-AS1(lncRNA) | Up-regulation | 2.337318012 | 9.23E-11 | 6.94E-10 |
| ERVH48-1(lncRNA) | Up-regulation | 2.606725757 | 5.73E-11 | 4.46E-10 |
| OSTN-AS1(lncRNA) | Up-regulation | 3.328039464 | 6.52E-07 | 2.79E-06 |
| DSCAM-AS1(lncRNA) | Up-regulation | 2.039530693 | 0.0001981 | 0.000546871 |
| GPC5-AS1(lncRNA) | Up-regulation | 4.291545148 | 1.31E-13 | 1.40E-12 |
| ZBTB20-AS3(lncRNA) | Up-regulation | 2.09029431 | 0.00131952 | 0.003071229 |
| AL356133.2(lncRNA) | Up-regulation | 4.946182739 | 5.14E-12 | 4.57E-11 |
| HNF1A-AS1(lncRNA) | Up-regulation | 2.202659839 | 1.29E-05 | 4.40E-05 |
| AL353803.1(lncRNA) | Down-regulation | −2.924748531 | 3.83E-31 | 2.03E-29 |
| ALDH1L1-AS2(lncRNA) | Down-regulation | −2.224445417 | 3.96E-39 | 3.31E-37 |
| LNX1-AS2(lncRNA) | Down-regulation | −2.482616679 | 3.81E-32 | 2.16E-30 |
| PCAT1(lncRNA) | Up-regulation | 2.659207972 | 3.07E-22 | 8.05E-21 |
| ANO1-AS2(lncRNA) | Down-regulation | −4.049375728 | 1.09E-50 | 1.81E-48 |
| miR-184(miRNA) | Down-regulation | −2.560858037 | 4.86E-27 | 6.04E-26 |
| miR-122(miRNA) | Up-regulation | 3.84488164 | 1.24E-07 | 3.63E-07 |
| miR-506(miRNA) | Up-regulation | 3.111522156 | 0.000277343 | 0.000532199 |
| miR-137(miRNA) | Up-regulation | 2.357657061 | 1.76E-05 | 4.00E-05 |
| miR-372(miRNA) | Up-regulation | 3.819509783 | 2.81E-08 | 8.84E-08 |
| miR-187(miRNA) | Down-regulation | −2.371598045 | 4.72E-33 | 9.78E-32 |
| PTGS2(mRNA) | Down-regulation | −3.018672047 | 6.74E-64 | 1.08E-61 |
| DUSP2(mRNA) | Down-regulation | −2.374155832 | 1.06E-51 | 1.22E-49 |
Figure 4.
A list of differentially expressed 24 lncRNAs, 6 miRNAs, and 2 mRNAs in prostate cancer.
Figure 5.

CeRNA network of prostate cancer. Downregulated genes are represented by a green ellipse and upregulated genes are represented by a red ellipse.
Figure 6.
Characteristics of LINC00308, LINC00355, and OSTN-AS1 in prostate cancer. (A) LINC00308, LINC00355, and OSTN-AS1 were significantly associated with survival (P<0.05). (B) The expressions of LINC00308, LINC00355, and OSTN-AS1 was significantly upregulated in prostate cancer tissues (P<0.05). (C) Expressions of LINC00308, LINC00355, and OSTN-AS1 were significantly upregulated in prostate cancer cell lines (* p<0.05, ** p<0.01).
Characteristics of differentially expressed lncRNAs in prostate cancer
Based on the results of lncRNAs related to survival, we re-examined the expression data of prostate cancer tissues and normal tissues. The results showed that the expression of LINC00308, LINC00355, and OSTN-AS1 was significantly higher in cancer tissues compared to the non-cancerous tissues (Figure 6B). In addition, we performed qRT-PCR in cell lines to examine the expression levels of LINC00308, LINC00355, and OSTN-AS1 in 2 prostate cancer cell lines, PC3 and DU145, as well as in the normal myofibroblast stromal cell line WPMY1. The results showed that LINC00308, LINC00355, and OSTN-AS1 are overexpressed in the prostate cancer cell line compared to the normal myofibroblast stromal cell line (Figure 6C).
Discussion
Prostate cancer is known to be a multifactorial disease with miscellaneous genetic factors. Traditional prognostic and predictive factors for prostate cancer, including tumor size, histologic grade, TNM stage, and number of lymph node involvement, may correlate with the clinical prognosis of patients. However, due to molecular heterogeneity, they seem to have limitations in distinguishing clinical outcomes among cancer risk subgroups [15,16]. Interestingly, we found 687 significantly differentially expressed mRNAs in prostate cancer. The functional enrichment analysis demonstrated that these genes were mainly enriched in the “extracellular region.” Several studies have revealed that DMBT1, ISG15, and EPPIN play an important role in the pathogenesis of prostate cancer [17–19]. The KEGG pathway analysis results showed that 14 genes were enriched in the salivary secretion pathway, including Cystatin, which has been shown to have a strong correlation with prostate cancer by modulating via the MAPK/Erk and androgen receptor pathways [20].
Many recent studies have revealed that various ncRNAs show great potential in the regulation of cancer. However, there have been few studies with large sample sizes (n=499) focusing on the identification of prostate cancer-related ncRNAs. Yang et al. revealed that lncRNA SNHG12 could be an oncogene in gastric carcinoma cell by targeting miRNA-199a/b-5p [21]. Wu et al. showed that lncRNA-PAGBC acts directly on tumor-suppressive microRNAs by activating the AKT/mTOR pathway, thus promoting tumorigenesis [22]. Because of the strong correlation between ncRNA expression and tumor status, lncRNAs and miRNAs may be valuable as diagnostic and prognostic biomarkers [23,24].
The expression of ncRNAs is low in the normal state but becomes increasingly upregulated in pathological states such as cancer. Hence, ncRNAs are also promising biomarker candidates for cancer [25] and numerous studies have focused on profiling RNA expression in relation to cancer state in search of potential biomarkers of cancer [26,27]. In the present study, we analyzed the public TCGA dataset based on human prostate cancer samples, which is extremely useful to find potential biomarkers. A total of 24 lncRNAs, 6 miRNAs, and 2 mRNAs were shown to harbor altered expression in the ceRNA network. One of the altered mRNAs was found to be PTGS2, also known as cyclooxygenase (COX-2). PTGS2 is an enzyme with a critical function in prostaglandin biosynthesis, and several reports have revealed its potential function in prostate cancer [28–30]. Our results confirm the function of PTGS2 in prostate cancer, and we also provide another potential mechanism: it may be regulated by XIST, PCA3, OSTN-AS1, DSCAM-AS1, LINC00308, and PCA3 in competing with mir-137. However, to the best our knowledge, the relationship between DUSP2 and prostate cancer has not been reported yet and our result suggest that DUSP2 may have a role in prostate carcinogenesis, but this remains to be elucidated mechanistically. Recently, there have been many reports that miRNAs can act as oncogenes and tumor suppressor genes to play important roles in proliferation, invasion, migration, apoptosis, EMT, and other malignant biological behaviors of tumor cells [31,32]. MiR-372, one of the differentially expressed miRNAs in our study, has been shown to inhibit prostate cancer cells migration and invasion by targeting p65 [33]. In addition, mir-184, which was also found to be differentially expressed in our study, has previously been reported as a potential signaling pathway in prostate cancer, acting through the mir-184/Bcl-2 axis [34]. Several new targets and potential mechanisms can be found in our ceRNA network studies, which may provide the basis for future research. Although lncRNAs have received much attention in recent years, the study of miRNAs is still very important because it plays a pivotal role in ceRNAs.
However, in our ceRNA network, only 3 lncRNAs could predict patient survival. LINC00308 has been reported to be overexpressed in the testes, but the function of LINC00308 has not been studied [35]. Moreover, LINC00355, which has been reported as deregulated in patients with sepsis, shows central properties in the molecular pathways associated with disease pathogenesis and gene expression regulatory loops that may be involved in poor disease outcomes [36]. So far, none of the 3 differentially expressed lncRNA (LINC00355, LINC00308, and OSTN-AS1) reported in our study has been shown to be associated with cancer by other researchers. Hence, in the present study, the expression of these 3 lncRNAs was evaluated in prostate cancer lines. These lncRNAs were not only significantly associated with poor survival, but also were overexpressed in vitro.
Conclusions
To conclude, we have identified 3 novel lncRNAs – LINC00308, LINC00355, and OSTN-AS1 – associated with prognosis of prostate cancer, which could be clinically useful as potential prognostic biomarkers for prostate cancer. However, the underlying molecular mechanisms of action remain to be further elucidated in functional studies.
Supplementary Files
Supplementary Table 1.
Differentially expressed mRNAs in prostate cancer.
| Gene | logFC | logCPM | PValue | FDR |
|---|---|---|---|---|
| SERPINA5 | −6.785068673 | 4.397738393 | 0 | 0 |
| MFSD2A | −5.966707923 | 3.456236811 | 0 | 0 |
| ACSL6 | −4.996876596 | 2.406174962 | 4.98E-299 | 2.97E-295 |
| MCF2 | −5.267342895 | 0.929684863 | 1.59E-262 | 7.09E-259 |
| EMX2 | −6.785444801 | 2.244788495 | 8.95E-260 | 3.20E-256 |
| HOXB8 | −6.244268143 | 1.047398016 | 4.88E-250 | 1.45E-246 |
| CLDN2 | −7.911204171 | 3.21684931 | 1.11E-247 | 2.82E-244 |
| AKR1B1 | −3.874547187 | 5.715503602 | 6.03E-236 | 1.35E-232 |
| SPINK2 | −7.409528496 | 2.678272683 | 7.41E-236 | 1.47E-232 |
| CYP19A1 | −5.406080831 | −1.222596337 | 2.05E-234 | 3.66E-231 |
| KLHL14 | −4.247424278 | 1.572359202 | 2.71E-206 | 4.41E-203 |
| SPINK13 | −5.528436256 | −0.392023032 | 2.65E-203 | 3.95E-200 |
| PATE2 | −5.418513576 | 0.297970005 | 1.42E-195 | 1.95E-192 |
| TMEM114 | −8.944978033 | 0.54167799 | 2.54E-188 | 3.24E-185 |
| NDRG4 | −3.348967314 | 2.992267305 | 3.41E-182 | 4.07E-179 |
| WNT9B | −4.877722819 | −0.703836945 | 1.10E-181 | 1.23E-178 |
| CRTAC1 | −4.292389971 | 2.665559252 | 1.29E-177 | 1.35E-174 |
| RASL10B | −3.465987979 | 2.057893571 | 1.17E-166 | 1.16E-163 |
| PIP | −7.351408595 | 6.495875509 | 3.33E-165 | 3.13E-162 |
| ANXA13 | −5.51532869 | 0.947383484 | 4.18E-165 | 3.74E-162 |
| PATE4 | −9.213661816 | 3.236652784 | 2.41E-157 | 2.05E-154 |
| CYSLTR2 | −3.722193167 | 1.029914851 | 9.93E-155 | 8.07E-152 |
| PAEP | −9.922475379 | 3.345532284 | 1.48E-151 | 1.15E-148 |
| AQP2 | −9.29515775 | 4.480800053 | 9.30E-151 | 6.93E-148 |
| CA2 | −4.440192491 | 2.780775663 | 5.54E-150 | 3.96E-147 |
| CRISP1 | −8.667915771 | 1.60380184 | 5.58E-149 | 3.84E-146 |
| MGAM | −4.692243468 | 0.243483116 | 7.50E-149 | 4.97E-146 |
| HOXB6 | −4.584092575 | 1.419092909 | 1.02E-146 | 6.53E-144 |
| SLC2A9 | −2.696283469 | 2.324688774 | 5.63E-140 | 3.47E-137 |
| SPINT3 | −7.357342601 | −1.907652893 | 5.44E-137 | 3.24E-134 |
| KCP | −3.487546788 | 0.465803728 | 4.69E-136 | 2.70E-133 |
| ANO1 | −2.868561416 | 5.461105992 | 9.81E-134 | 5.48E-131 |
| C3orf36 | −3.509127685 | 0.36085329 | 1.07E-131 | 5.80E-129 |
| ATP13A4 | −4.250554958 | −0.531092553 | 1.48E-130 | 7.79E-128 |
| SLC13A2 | −6.292561769 | 1.033617138 | 4.23E-129 | 2.16E-126 |
| KCNJ5 | −3.119719349 | 3.007097953 | 7.69E-129 | 3.82E-126 |
| PNMT | −4.667142036 | 1.022812758 | 2.83E-127 | 1.37E-124 |
| STAC2 | −4.416170051 | 3.417454375 | 8.66E-126 | 4.08E-123 |
| ABCG2 | −2.809960333 | 3.970415449 | 4.78E-124 | 2.19E-121 |
| FAM83A | −5.078067363 | −0.012191609 | 3.78E-123 | 1.69E-120 |
| SLC16A12 | −3.749170694 | 0.162005786 | 5.37E-121 | 2.34E-118 |
| SEMG2 | −9.505742358 | 10.32686946 | 8.68E-120 | 3.69E-117 |
| C1orf186 | −3.363933794 | 0.80475646 | 6.92E-119 | 2.88E-116 |
| PAQR8 | −2.568199676 | 3.692816808 | 4.02E-118 | 1.63E-115 |
| UGT2B7 | −5.583084346 | −0.640545992 | 7.63E-118 | 3.03E-115 |
| MRO | −3.090785129 | −0.234389189 | 1.54E-116 | 5.97E-114 |
| SLCO4C1 | −4.798092265 | −0.639044646 | 3.84E-116 | 1.46E-113 |
| ACE2 | −3.568169252 | 0.516117235 | 6.46E-114 | 2.41E-111 |
| FUT3 | −4.111844574 | 0.379086415 | 6.37E-113 | 2.33E-110 |
| PTGES | −3.039801329 | 4.655930611 | 4.92E-112 | 1.76E-109 |
| HS3ST5 | −4.914778132 | −1.407416204 | 2.03E-110 | 7.10E-108 |
| KIRREL3 | −3.012684881 | −0.860795205 | 2.44E-109 | 8.40E-107 |
| SEMG1 | −9.757312184 | 11.35548942 | 3.89E-108 | 1.31E-105 |
| DEFB129 | −6.866593977 | −2.315654961 | 2.00E-107 | 6.62E-105 |
| PAX2 | −4.064900582 | 0.376708484 | 2.81E-106 | 9.13E-104 |
| PIK3C2G | −4.380016398 | 0.469725146 | 1.07E-102 | 3.40E-100 |
| SLC46A2 | −4.113213363 | −1.409293159 | 3.17E-102 | 9.95E-100 |
| SNAP25 | −3.062100018 | 2.046817113 | 1.04E-96 | 3.21E-94 |
| LPL | −3.698585 | 3.897033258 | 1.73E-96 | 5.25E-94 |
| PLA2G4A | −2.306657328 | 3.506160074 | 5.45E-96 | 1.62E-93 |
| HOXB9 | −3.738394139 | 1.187071894 | 6.96E-96 | 2.04E-93 |
| POU3F3 | −7.820353855 | 1.441534386 | 1.02E-93 | 2.93E-91 |
| SLC28A3 | −4.429709489 | −0.22811352 | 4.13E-91 | 1.17E-88 |
| GSTM3 | −2.283273278 | 5.654161731 | 4.79E-91 | 1.34E-88 |
| FRMD3 | −2.232713406 | 2.362727796 | 5.80E-91 | 1.60E-88 |
| EVA1A | −2.609102352 | 0.046188518 | 6.32E-91 | 1.71E-88 |
| PRDM16 | −2.585618819 | 0.810572803 | 5.09E-90 | 1.36E-87 |
| FBP2 | −3.625501212 | −0.395808669 | 9.04E-90 | 2.38E-87 |
| GCNT4 | −2.419664765 | 1.308525087 | 2.55E-89 | 6.62E-87 |
| CLU | −2.575286197 | 9.076442775 | 1.20E-88 | 3.06E-86 |
| EDDM3A | −9.915750757 | 0.930978027 | 1.71E-88 | 4.30E-86 |
| KCNJ16 | −5.300617001 | 1.33688112 | 9.23E-88 | 2.29E-85 |
| KCNJ15 | −3.274002031 | 1.948780543 | 1.64E-87 | 4.02E-85 |
| PATE1 | −8.559529455 | 4.39114703 | 4.70E-87 | 1.14E-84 |
| KRT24 | −4.993860718 | −2.05900135 | 1.41E-86 | 3.36E-84 |
| SULT2A1 | −6.511620056 | −1.487548002 | 1.54E-84 | 3.58E-82 |
| NPFFR2 | −4.628734826 | −1.871185299 | 1.24E-82 | 2.84E-80 |
| GNAO1 | −2.487785948 | 3.631013922 | 4.77E-81 | 1.08E-78 |
| DEFB131A | −7.992232167 | −2.717399459 | 1.12E-79 | 2.51E-77 |
| PDK4 | −3.01435869 | 6.971020626 | 2.35E-79 | 5.19E-77 |
| UNC5B | −2.199076184 | 5.027632209 | 2.67E-79 | 5.83E-77 |
| CAMP | −3.53381043 | −1.328045181 | 9.63E-78 | 2.08E-75 |
| SLC31A2 | −2.111149911 | 0.324281459 | 2.44E-77 | 5.20E-75 |
| PON3 | −3.31897902 | −0.714040817 | 1.52E-76 | 3.20E-74 |
| ADCY8 | −4.727507179 | −2.378656903 | 2.96E-76 | 6.15E-74 |
| TMEM238L | −3.460883798 | 0.330286585 | 3.96E-76 | 8.14E-74 |
| EDDM3B | −9.095154357 | −1.282761718 | 1.53E-75 | 3.08E-73 |
| LHX1 | −6.152126939 | −2.569155343 | 2.06E-75 | 4.09E-73 |
| TMEM171 | −2.978213867 | −2.490124781 | 2.99E-73 | 5.88E-71 |
| ELSPBP1 | −5.964830301 | −2.613099778 | 9.37E-73 | 1.82E-70 |
| QPRT | −2.396260233 | 2.787898063 | 2.12E-72 | 4.07E-70 |
| FAM167A | −2.899049257 | 2.215708554 | 1.58E-71 | 3.00E-69 |
| RASAL1 | −3.075775921 | 0.258555744 | 1.90E-71 | 3.57E-69 |
| G0S2 | −2.281974698 | 2.740748523 | 1.01E-70 | 1.89E-68 |
| SLC26A3 | −4.713543952 | 4.255428037 | 4.42E-70 | 8.15E-68 |
| GLIS3 | −2.10224676 | 3.141701783 | 5.68E-69 | 1.04E-66 |
| ADTRP | −3.145256188 | 1.796113975 | 1.34E-67 | 2.38E-65 |
| PATE3 | −5.878495255 | −2.049038515 | 6.54E-67 | 1.12E-64 |
| PAX8 | −3.163424949 | 2.312640932 | 4.67E-66 | 7.95E-64 |
| PALM3 | −2.862055728 | 1.75017837 | 2.61E-65 | 4.36E-63 |
| SLPI | −3.550360086 | 5.950826934 | 3.38E-65 | 5.60E-63 |
| SCGB1D4 | −6.616957825 | −3.643904883 | 1.38E-64 | 2.27E-62 |
| CA14 | −2.628944503 | 1.565656785 | 1.41E-64 | 2.29E-62 |
| GRXCR1 | −7.152527699 | −1.442456622 | 4.35E-64 | 7.00E-62 |
| PTGS2 | −3.018672047 | 6.0997409 | 6.74E-64 | 1.08E-61 |
| DEFB125 | −7.823009219 | −3.088436223 | 1.21E-63 | 1.90E-61 |
| SP6 | −2.048223343 | 2.495992609 | 1.74E-63 | 2.70E-61 |
| LIPG | −2.797035036 | 2.502758674 | 2.57E-63 | 3.96E-61 |
| HOXB7 | −2.538554469 | 1.469159627 | 4.12E-63 | 6.24E-61 |
| AOX1 | −2.401455097 | 4.820050441 | 7.83E-63 | 1.18E-60 |
| SIM1 | −5.801689063 | −1.342000003 | 8.05E-63 | 1.20E-60 |
| TRIM9 | −2.21544454 | −0.284214026 | 1.28E-60 | 1.86E-58 |
| MUCL1 | −3.994926646 | 1.918418391 | 1.13E-59 | 1.61E-57 |
| TEDDM1 | −3.162014508 | −2.397478949 | 1.84E-59 | 2.62E-57 |
| GDPD2 | −2.614839978 | −2.578615619 | 1.32E-58 | 1.84E-56 |
| CYP4F8 | −4.234944216 | 5.957323259 | 4.62E-58 | 6.40E-56 |
| HSPA6 | −3.56230163 | 3.163452791 | 1.40E-57 | 1.93E-55 |
| PTGS1 | −2.488186912 | 4.442555664 | 1.50E-57 | 2.05E-55 |
| HOXB5 | −3.507475786 | −0.118278523 | 2.93E-57 | 3.94E-55 |
| ARC | −2.791928795 | 1.302315162 | 5.92E-57 | 7.79E-55 |
| C2orf88 | −2.342319604 | 2.417718221 | 7.27E-57 | 9.48E-55 |
| APOBEC3C | −2.037731259 | 4.968381179 | 1.06E-56 | 1.37E-54 |
| TFAP2B | −3.887211428 | −2.562766626 | 2.07E-56 | 2.66E-54 |
| PADI3 | −4.318296193 | 0.033579355 | 5.97E-56 | 7.63E-54 |
| CES5A | −5.399715292 | −2.273300712 | 6.88E-55 | 8.60E-53 |
| ATP1A4 | −2.650615554 | −0.8604139 | 1.12E-54 | 1.39E-52 |
| CCDC27 | −2.230774182 | −2.116580292 | 4.27E-54 | 5.27E-52 |
| WFDC9 | −7.370447463 | −3.418331864 | 1.29E-53 | 1.59E-51 |
| DCAF12L1 | −3.039717642 | −2.084125111 | 3.70E-53 | 4.50E-51 |
| SBSPON | −2.274088177 | 3.432870313 | 1.42E-52 | 1.70E-50 |
| DUSP2 | −2.374155832 | 4.249770546 | 1.06E-51 | 1.22E-49 |
| EPHA10 | 2.181110197 | 3.496632325 | 5.18E-51 | 5.87E-49 |
| GSTP1 | −2.053371722 | 6.920002831 | 1.40E-50 | 1.55E-48 |
| LMO3 | −2.170273005 | 3.259518865 | 5.16E-50 | 5.52E-48 |
| CXCR2 | −2.604659545 | −0.234591526 | 1.54E-49 | 1.62E-47 |
| LYVE1 | −2.250739542 | 1.531070033 | 6.82E-49 | 7.09E-47 |
| VWA5B2 | −2.355784274 | 1.103997608 | 1.52E-48 | 1.56E-46 |
| ANGPT1 | −2.047422403 | 3.901247666 | 1.86E-48 | 1.90E-46 |
| MUC6 | −4.984635603 | 7.087473698 | 2.08E-48 | 2.10E-46 |
| WFDC8 | −4.53427633 | −4.039274837 | 2.45E-48 | 2.46E-46 |
| DEFB127 | −6.707184857 | −3.923081354 | 6.20E-48 | 6.16E-46 |
| LCN15 | −3.320802889 | −0.939497584 | 6.94E-48 | 6.86E-46 |
| AL163195.3 | −3.923578432 | −4.064090252 | 7.69E-48 | 7.55E-46 |
| DEFB132 | −2.327019581 | 2.024711463 | 1.45E-46 | 1.39E-44 |
| TNMD | −3.2653763 | −1.061318979 | 2.36E-46 | 2.24E-44 |
| TMEM132C | −2.486194486 | 1.508679117 | 2.43E-46 | 2.30E-44 |
| HRASLS5 | −2.182632517 | 0.139647911 | 2.89E-46 | 2.72E-44 |
| SLC9A4 | −3.17506859 | −2.826706326 | 3.82E-46 | 3.58E-44 |
| SCN11A | −2.21835192 | −1.749469071 | 9.22E-46 | 8.54E-44 |
| DUOX1 | −2.137499241 | 4.097778228 | 1.46E-45 | 1.34E-43 |
| MUC15 | −2.661608288 | 1.132262497 | 2.45E-44 | 2.19E-42 |
| TRPM5 | −3.176120461 | −0.890607598 | 3.48E-44 | 3.10E-42 |
| SLC39A2 | −3.854360669 | 2.020647601 | 5.33E-44 | 4.72E-42 |
| TRPM3 | −2.29500896 | −0.137630395 | 6.04E-44 | 5.32E-42 |
| ATP10B | −2.893980809 | −2.015181875 | 3.56E-43 | 3.05E-41 |
| TRIM61 | −2.028327421 | −2.615002542 | 4.08E-43 | 3.47E-41 |
| HPN | 2.51071704 | 8.064534927 | 1.06E-42 | 8.91E-41 |
| KLHL4 | −2.039991565 | −0.923853849 | 1.86E-42 | 1.55E-40 |
| GATA3 | −2.202253741 | 3.782637479 | 2.30E-42 | 1.91E-40 |
| GPD1 | −2.398405613 | −0.219024849 | 3.92E-42 | 3.24E-40 |
| LCN9 | −5.438725778 | −3.834512629 | 4.84E-42 | 3.99E-40 |
| FAM110C | −2.187790425 | 3.123504141 | 6.00E-42 | 4.92E-40 |
| ADGRD2 | −2.978963237 | −1.102374977 | 6.30E-42 | 5.14E-40 |
| MAT1A | −2.847220497 | −1.240774796 | 1.50E-41 | 1.19E-39 |
| C20orf202 | −2.009368734 | −2.563236881 | 2.80E-41 | 2.20E-39 |
| HCAR2 | −2.004843097 | 1.194232598 | 6.33E-41 | 4.94E-39 |
| ASPA | −2.078527277 | 1.495235978 | 6.77E-41 | 5.26E-39 |
| KCNJ1 | −2.762883764 | −2.903446081 | 1.75E-40 | 1.33E-38 |
| STAC | −2.388338573 | 2.991906842 | 1.81E-40 | 1.36E-38 |
| GPX2 | −2.515514209 | 2.45210946 | 1.96E-40 | 1.47E-38 |
| DCHS2 | −2.148971591 | 0.710909297 | 5.37E-40 | 3.95E-38 |
| C10orf99 | −3.579716304 | −2.223557443 | 6.29E-40 | 4.59E-38 |
| PCP4L1 | −2.206007918 | 1.907712695 | 8.99E-40 | 6.45E-38 |
| DMRT2 | −2.991089671 | −2.296521924 | 1.56E-39 | 1.11E-37 |
| CLDN19 | −2.531145647 | −2.60827072 | 1.71E-39 | 1.22E-37 |
| IVL | −3.726131389 | −1.441202673 | 2.34E-39 | 1.66E-37 |
| SLC34A2 | −3.167340855 | 3.466038276 | 5.84E-39 | 4.05E-37 |
| SLC18A2 | −2.562912473 | 1.63627357 | 1.20E-38 | 8.23E-37 |
| NKX2-3 | 4.295926347 | 0.960315432 | 1.82E-38 | 1.24E-36 |
| PLCZ1 | −3.528460242 | −4.111131902 | 2.16E-38 | 1.46E-36 |
| SH2D1B | −2.070812969 | −2.247153696 | 2.30E-38 | 1.55E-36 |
| CCNI2 | −2.393120663 | −0.757806717 | 4.32E-38 | 2.89E-36 |
| OVCH2 | −2.474604409 | −1.229899903 | 4.36E-38 | 2.91E-36 |
| FOXQ1 | −2.113706442 | 2.340709388 | 1.77E-37 | 1.15E-35 |
| KCNS1 | −2.740616324 | 0.817741242 | 1.92E-37 | 1.24E-35 |
| REG3G | −5.362768922 | −3.996026989 | 2.04E-37 | 1.31E-35 |
| C21orf62 | −2.497748317 | 0.055828895 | 2.58E-37 | 1.65E-35 |
| LCN1 | −6.139941235 | −1.286853617 | 4.00E-37 | 2.53E-35 |
| EPHB1 | −2.071871109 | 1.406334339 | 4.28E-37 | 2.69E-35 |
| KY | −2.334715222 | −0.154030942 | 4.95E-37 | 3.09E-35 |
| SLC6A2 | −2.10678614 | −1.995090013 | 6.49E-37 | 4.03E-35 |
| KCNH5 | −3.179639439 | −2.263489674 | 1.54E-36 | 9.37E-35 |
| SIM2 | 2.246683676 | 6.39245743 | 2.10E-36 | 1.26E-34 |
| SIAH3 | −2.391041994 | −2.100220433 | 2.26E-36 | 1.36E-34 |
| TMEM213 | −2.938009836 | −0.973458402 | 3.77E-36 | 2.25E-34 |
| HOXB4 | −2.013128998 | 0.556074277 | 4.13E-36 | 2.44E-34 |
| WFDC2 | −2.278298566 | 5.691380285 | 6.46E-36 | 3.76E-34 |
| SYT8 | −2.62796781 | −0.04504407 | 9.39E-36 | 5.40E-34 |
| RAD21L1 | −3.931426302 | −4.029582937 | 1.14E-35 | 6.49E-34 |
| JPH4 | −2.032898156 | 3.896512339 | 1.52E-35 | 8.59E-34 |
| CLCA2 | −2.99069792 | 1.679389133 | 2.18E-35 | 1.22E-33 |
| GSTM1 | −3.348712796 | 3.902420778 | 2.36E-35 | 1.31E-33 |
| LY6D | −3.253162593 | 0.960481773 | 4.33E-35 | 2.40E-33 |
| LRCOL1 | −2.383609469 | −2.229173553 | 4.98E-35 | 2.74E-33 |
| KRT222 | −2.281961674 | −0.980864847 | 8.50E-35 | 4.62E-33 |
| CST4 | −3.656991558 | 1.448466198 | 9.21E-35 | 4.96E-33 |
| NEFM | −2.864624272 | 0.554358796 | 1.63E-34 | 8.52E-33 |
| DUOX2 | −2.585503889 | 2.635300446 | 1.67E-34 | 8.69E-33 |
| OPCML | −2.590292577 | −1.566109784 | 2.15E-34 | 1.11E-32 |
| DUOXA2 | −2.845461401 | −0.495623973 | 2.55E-34 | 1.29E-32 |
| PDE1C | −2.216681928 | 0.951037818 | 4.56E-34 | 2.27E-32 |
| MSLN | −2.673143906 | 2.312078872 | 9.59E-34 | 4.63E-32 |
| ATP6V1G3 | −3.407739337 | −2.880382936 | 1.15E-33 | 5.49E-32 |
| PRSS1 | −4.522834207 | −1.662313282 | 3.21E-33 | 1.50E-31 |
| FOXI2 | −2.306900792 | −2.155968961 | 3.52E-33 | 1.64E-31 |
| DCC | −2.459225535 | −0.956958564 | 4.76E-33 | 2.20E-31 |
| DUOXA1 | −2.155945912 | 2.560577531 | 2.07E-32 | 9.23E-31 |
| EMX1 | −2.835853169 | −3.197852638 | 2.31E-32 | 1.03E-30 |
| LVRN | −2.019593234 | −2.758757798 | 4.93E-32 | 2.12E-30 |
| CYP4B1 | −2.165077764 | 3.831392901 | 5.23E-32 | 2.24E-30 |
| TRH | −2.994128142 | −3.124637854 | 8.51E-32 | 3.59E-30 |
| HCAR3 | −2.221534692 | −1.323656559 | 9.86E-32 | 4.13E-30 |
| ITLN1 | −2.679097356 | −0.945384899 | 1.00E-31 | 4.18E-30 |
| SYT10 | −2.319711561 | −1.05820299 | 3.21E-31 | 1.29E-29 |
| ADRA1A | −2.022518109 | 2.151912159 | 4.83E-31 | 1.93E-29 |
| CDH8 | −2.263389081 | −0.939571117 | 5.72E-31 | 2.28E-29 |
| TMEM252 | −2.122053299 | 0.986298859 | 8.91E-31 | 3.52E-29 |
| AQP5 | −2.201067826 | −0.242796203 | 9.48E-31 | 3.74E-29 |
| PYY | −2.304539289 | −2.689358214 | 9.95E-31 | 3.91E-29 |
| HOXD4 | −2.010658856 | −1.667951124 | 1.07E-30 | 4.18E-29 |
| ASTL | −2.271544473 | −2.255063905 | 1.29E-30 | 4.99E-29 |
| FOLR1 | −2.267963855 | 0.443328675 | 1.60E-30 | 6.16E-29 |
| MGAT4C | −2.127179772 | −1.600917474 | 2.23E-30 | 8.48E-29 |
| LRRC3B | −2.226821286 | −3.265920448 | 2.37E-30 | 8.95E-29 |
| KCNF1 | −2.008639046 | −1.489871946 | 2.41E-30 | 9.07E-29 |
| CRNN | −4.196298519 | −3.602598059 | 3.11E-30 | 1.16E-28 |
| IL1RL1 | −2.164943372 | 0.580067069 | 4.59E-30 | 1.69E-28 |
| IGSF1 | −2.205150167 | 1.353299712 | 5.01E-30 | 1.84E-28 |
| CDO1 | −2.25897076 | 3.975578364 | 6.05E-30 | 2.21E-28 |
| UCN | 2.225028987 | 0.886371038 | 6.40E-30 | 2.32E-28 |
| AMACR | 3.271375874 | 7.272807932 | 1.43E-29 | 5.08E-28 |
| GLRA4 | −2.291078633 | −3.061966898 | 1.61E-29 | 5.68E-28 |
| CAPN6 | −2.167733352 | 3.083222877 | 1.67E-29 | 5.88E-28 |
| KCNJ13 | −2.677244912 | −2.098731552 | 2.33E-29 | 8.06E-28 |
| ROS1 | −2.664547966 | −1.41857366 | 2.56E-29 | 8.87E-28 |
| MEI4 | −2.157511068 | −3.504964788 | 2.79E-29 | 9.63E-28 |
| SLC45A2 | 5.233859283 | 3.020506268 | 3.11E-29 | 1.07E-27 |
| CYP4F22 | −2.48815109 | 1.361704428 | 4.42E-29 | 1.50E-27 |
| CLPSL1 | −3.884589239 | −3.179552875 | 7.45E-29 | 2.49E-27 |
| EVX2 | −2.201786126 | 0.147793705 | 1.24E-28 | 4.07E-27 |
| KRT16 | −2.392624596 | 2.060345166 | 1.80E-28 | 5.83E-27 |
| FAM83C | −2.840632484 | −1.823672187 | 2.44E-28 | 7.86E-27 |
| CHIA | −4.044693208 | −3.761939133 | 2.80E-28 | 8.97E-27 |
| CFAP65 | 2.17924865 | 1.923214285 | 4.94E-28 | 1.57E-26 |
| IL20 | −2.734032718 | −2.568179433 | 1.09E-27 | 3.38E-26 |
| HSD17B13 | −2.59308437 | 2.765460154 | 1.20E-27 | 3.67E-26 |
| ATP6V0D2 | −2.062948021 | −0.049347798 | 1.43E-27 | 4.33E-26 |
| FNDC10 | 2.189675352 | 3.1851198 | 1.43E-27 | 4.34E-26 |
| CLPSL2 | −2.9526799 | −3.435156672 | 4.32E-27 | 1.25E-25 |
| LGR6 | −2.077080456 | 2.417215605 | 6.96E-27 | 1.99E-25 |
| S100A14 | −2.094417081 | 2.768825991 | 7.06E-27 | 2.01E-25 |
| SFRP5 | −2.760385146 | 0.642123766 | 7.15E-27 | 2.04E-25 |
| NETO2 | 2.090951916 | 3.425360608 | 1.14E-26 | 3.22E-25 |
| LMX1B | 2.910958396 | 1.214627903 | 1.29E-26 | 3.60E-25 |
| GJB4 | −2.106346704 | −0.022277162 | 2.07E-26 | 5.69E-25 |
| SLITRK3 | −2.314417037 | 0.590359859 | 2.24E-26 | 6.13E-25 |
| CDH16 | −3.658293176 | −2.481108891 | 2.75E-26 | 7.49E-25 |
| ACTC1 | −2.549775894 | 5.199099142 | 3.86E-26 | 1.03E-24 |
| IL19 | −2.963272464 | −4.136314081 | 4.35E-26 | 1.16E-24 |
| LINGO2 | −2.353643152 | −1.61961467 | 2.36E-25 | 5.84E-24 |
| TGM5 | −2.041707954 | −1.130721222 | 2.51E-25 | 6.21E-24 |
| FAM83B | −2.016553537 | 0.635021108 | 3.30E-25 | 8.00E-24 |
| DNAH5 | 2.32012031 | 5.526858819 | 4.15E-25 | 1.00E-23 |
| CYP3A5 | −2.005756625 | 3.220640626 | 4.78E-25 | 1.15E-23 |
| KRT13 | −2.974414253 | 5.283076591 | 5.95E-25 | 1.42E-23 |
| LINC00694 | −2.00291498 | −2.423117021 | 7.37E-25 | 1.75E-23 |
| SCARA5 | −2.212531236 | 1.054207669 | 7.76E-25 | 1.84E-23 |
| PLA2G3 | −2.484369479 | −2.123775808 | 8.97E-25 | 2.12E-23 |
| SLIT1 | 2.991124143 | 4.235802648 | 9.13E-25 | 2.15E-23 |
| ADAMTS18 | −2.485445465 | −1.555485261 | 9.65E-25 | 2.26E-23 |
| ATP4B | −2.170350626 | −3.615853337 | 2.04E-24 | 4.61E-23 |
| MATN4 | −2.253033183 | −1.39482938 | 3.30E-24 | 7.32E-23 |
| C2orf72 | 2.352501555 | 6.112412268 | 3.37E-24 | 7.45E-23 |
| CBLN4 | −2.282088492 | −2.141588262 | 3.46E-24 | 7.64E-23 |
| HRASLS | −2.750037652 | −2.668890752 | 4.62E-24 | 1.01E-22 |
| PENK | −2.157694308 | 2.279458961 | 5.35E-24 | 1.17E-22 |
| CCDC198 | −3.361534597 | −3.550866101 | 6.39E-24 | 1.38E-22 |
| BTN1A1 | −2.058438498 | −3.223659882 | 8.65E-24 | 1.84E-22 |
| TG | −2.866055894 | 5.040539624 | 9.02E-24 | 1.91E-22 |
| SMR3B | −4.345403915 | −1.407810214 | 9.65E-24 | 2.04E-22 |
| C2CD4C | 2.365102335 | 1.292132594 | 1.03E-23 | 2.16E-22 |
| ATP6V0A4 | −2.469261417 | −0.171828266 | 1.44E-23 | 2.99E-22 |
| C1orf61 | −2.037014861 | −1.442631941 | 2.23E-23 | 4.54E-22 |
| SERPINB5 | −2.026028458 | 3.221299557 | 2.88E-23 | 5.82E-22 |
| OR7C1 | −2.022970619 | −2.050624749 | 3.09E-23 | 6.23E-22 |
| KBTBD13 | −2.109275337 | −3.695224421 | 3.30E-23 | 6.63E-22 |
| TH | −2.763714651 | −1.950206659 | 3.39E-23 | 6.81E-22 |
| MATK | 2.589971629 | 3.389770366 | 3.57E-23 | 7.14E-22 |
| ARHGAP19-SLIT1 | 3.000318492 | −2.097085148 | 4.54E-23 | 9.00E-22 |
| PAQR6 | 2.309347403 | 3.648588239 | 7.11E-23 | 1.37E-21 |
| CIDEC | −2.214864174 | −1.468234676 | 7.64E-23 | 1.47E-21 |
| FADS6 | −2.326944554 | −3.777201655 | 1.38E-22 | 2.59E-21 |
| DPP6 | −2.186696088 | 0.124736584 | 1.39E-22 | 2.61E-21 |
| WIF1 | −2.21507643 | 2.513039346 | 1.94E-22 | 3.57E-21 |
| CPB2 | −2.040650414 | −2.684359228 | 2.11E-22 | 3.86E-21 |
| APOC1 | 2.315598498 | 4.432027844 | 2.22E-22 | 4.06E-21 |
| NLRP12 | 2.474330308 | 1.187055602 | 2.27E-22 | 4.14E-21 |
| CPNE6 | −2.170399828 | 1.772299025 | 2.49E-22 | 4.53E-21 |
| HOXC6 | 2.328170608 | 3.774272822 | 2.79E-22 | 5.03E-21 |
| HOXC4 | 2.31331291 | 2.665774136 | 2.79E-22 | 5.03E-21 |
| ARSF | −2.138562635 | −2.870411011 | 2.97E-22 | 5.34E-21 |
| CERS1 | 2.553768773 | 1.469026165 | 3.80E-22 | 6.80E-21 |
| TRIM31 | −2.091716954 | 0.490285758 | 5.70E-22 | 1.00E-20 |
| ATP2B3 | −2.394530451 | −3.269639342 | 7.38E-22 | 1.29E-20 |
| ANKRD66 | 2.801025739 | −0.784999268 | 9.06E-22 | 1.57E-20 |
| MMP26 | 3.066474235 | 1.352487044 | 9.41E-22 | 1.62E-20 |
| KCNA4 | −2.346476072 | −2.699325651 | 1.18E-21 | 2.01E-20 |
| MUC21 | −3.114340581 | −2.477022768 | 1.22E-21 | 2.08E-20 |
| CLEC18B | 2.167387184 | −1.095435957 | 1.69E-21 | 2.84E-20 |
| TMEM196 | −2.596176551 | −3.648797811 | 2.57E-21 | 4.24E-20 |
| CCDC78 | 2.642490882 | 2.309466825 | 2.97E-21 | 4.87E-20 |
| EPGN | −2.120950779 | −2.692080174 | 3.34E-21 | 5.46E-20 |
| DEFB134 | −2.58614525 | −4.035042203 | 3.75E-21 | 6.09E-20 |
| GAS2L2 | −2.487311305 | −1.119961385 | 5.49E-21 | 8.72E-20 |
| CHP2 | −2.13935326 | 1.031772412 | 5.59E-21 | 8.87E-20 |
| FAM163A | −2.082472426 | −2.065990547 | 9.75E-21 | 1.52E-19 |
| TNFSF11 | −2.003738684 | −2.569011336 | 1.24E-20 | 1.90E-19 |
| KRT27 | −2.305026411 | −3.966447429 | 1.69E-20 | 2.55E-19 |
| SCGB1A1 | −2.475823685 | 3.909002352 | 2.53E-20 | 3.75E-19 |
| COLEC10 | −2.558276109 | −2.501754928 | 3.26E-20 | 4.77E-19 |
| APOF | 2.433781339 | 3.137119857 | 4.46E-20 | 6.45E-19 |
| PDIA2 | 4.280402557 | 2.013335255 | 4.70E-20 | 6.77E-19 |
| ADRB3 | −2.365978611 | −2.540179574 | 5.72E-20 | 8.14E-19 |
| FLG2 | −2.599596026 | −2.865814995 | 6.83E-20 | 9.67E-19 |
| SHISA8 | 2.993519423 | −0.189202204 | 6.86E-20 | 9.69E-19 |
| TMPRSS11A | −2.790567956 | −1.570310054 | 9.22E-20 | 1.29E-18 |
| RPE65 | −2.175383876 | −0.00527736 | 1.31E-19 | 1.80E-18 |
| HJURP | 2.068082168 | 2.187879526 | 1.34E-19 | 1.85E-18 |
| CCDC83 | 3.229819171 | −1.017747147 | 1.44E-19 | 1.96E-18 |
| ACSM1 | 2.868523218 | 6.613879148 | 1.57E-19 | 2.13E-18 |
| CLCA4 | −2.496145495 | 1.627115296 | 1.66E-19 | 2.25E-18 |
| C10orf82 | −2.135397958 | −0.911355049 | 2.24E-19 | 2.98E-18 |
| KCNK10 | −2.011525621 | −2.711656881 | 2.32E-19 | 3.08E-18 |
| TERT | 2.688569503 | −1.772060648 | 2.39E-19 | 3.16E-18 |
| EPPIN | −2.691805498 | −3.769965344 | 2.52E-19 | 3.32E-18 |
| DMBT1 | −2.236532941 | 0.1096238 | 3.62E-19 | 4.67E-18 |
| ONECUT2 | 2.681251387 | 3.221606201 | 3.64E-19 | 4.69E-18 |
| SFTPC | −2.25322321 | −3.281266307 | 5.39E-19 | 6.82E-18 |
| TWIST1 | 2.177091444 | 3.574725101 | 9.65E-19 | 1.19E-17 |
| FAM196B | −2.217220736 | −1.695478869 | 1.03E-18 | 1.27E-17 |
| PHGR1 | 3.386311578 | 1.820119662 | 1.36E-18 | 1.66E-17 |
| MOV10L1 | 2.27846602 | 1.581465328 | 1.41E-18 | 1.71E-17 |
| TBX4 | −2.17375547 | 2.268146306 | 1.58E-18 | 1.92E-17 |
| AMH | 3.230456823 | 1.135656782 | 1.79E-18 | 2.16E-17 |
| ZIC2 | 3.465784012 | 2.099443841 | 1.95E-18 | 2.35E-17 |
| REN | −2.010856028 | −1.82427371 | 2.05E-18 | 2.46E-17 |
| ARL14 | −2.041449796 | −2.836582438 | 2.77E-18 | 3.27E-17 |
| GPR149 | −3.565113736 | −2.173936635 | 2.97E-18 | 3.49E-17 |
| CACNA1D | 2.177413498 | 5.843737038 | 3.34E-18 | 3.89E-17 |
| NOX4 | 2.005839671 | 1.199904726 | 4.41E-18 | 5.07E-17 |
| SCGB3A1 | −2.174104533 | 2.495869638 | 7.08E-18 | 8.03E-17 |
| FOXN4 | 2.808885496 | −0.819378366 | 8.85E-18 | 9.88E-17 |
| DLX1 | 2.964944227 | 5.245627 | 1.43E-17 | 1.56E-16 |
| ARHGDIG | 2.722116583 | 1.951835142 | 1.80E-17 | 1.94E-16 |
| F2RL2 | 2.081512112 | 0.759279249 | 2.12E-17 | 2.26E-16 |
| LCN8 | −2.945514338 | −3.234638837 | 2.79E-17 | 2.94E-16 |
| OTX1 | 2.103182365 | 3.115942657 | 2.93E-17 | 3.08E-16 |
| KRT4 | −2.137873946 | 2.801389922 | 4.58E-17 | 4.75E-16 |
| ALB | 4.431655563 | 3.09043822 | 6.92E-17 | 7.03E-16 |
| TSPAN19 | 3.208847265 | 0.730279277 | 9.00E-17 | 9.05E-16 |
| GNG13 | 2.803827842 | 0.126341753 | 1.01E-16 | 1.01E-15 |
| DSG4 | −2.466048424 | −2.345854057 | 1.03E-16 | 1.03E-15 |
| EFNA2 | 3.057267493 | −1.886740535 | 1.45E-16 | 1.43E-15 |
| CCER2 | 2.000918092 | −0.3544635 | 1.48E-16 | 1.45E-15 |
| FAM163B | −2.321266796 | −2.413589392 | 1.63E-16 | 1.60E-15 |
| WFDC13 | −2.806598091 | −3.536010249 | 1.88E-16 | 1.83E-15 |
| ULBP1 | 2.15541201 | −1.028236929 | 1.89E-16 | 1.84E-15 |
| OR51E2 | 2.414972238 | 9.179968408 | 2.69E-16 | 2.59E-15 |
| TGM3 | 2.329959919 | 3.971772096 | 3.43E-16 | 3.25E-15 |
| MIOX | 2.023651528 | −0.851072617 | 6.30E-16 | 5.82E-15 |
| VGF | 2.505950604 | 1.28386305 | 6.83E-16 | 6.28E-15 |
| OR51S1 | 3.289112641 | −3.044498407 | 1.27E-15 | 1.14E-14 |
| KISS1R | 3.422260325 | −2.414859811 | 1.44E-15 | 1.28E-14 |
| HTR1E | −2.206015072 | −3.426655036 | 1.62E-15 | 1.42E-14 |
| GLYATL1B | 2.200776254 | −0.628848988 | 1.84E-15 | 1.60E-14 |
| KLK15 | 2.046781312 | 2.991115701 | 1.94E-15 | 1.67E-14 |
| THRSP | −2.470893848 | −0.92431372 | 2.09E-15 | 1.80E-14 |
| ANGPTL3 | 3.829532766 | 1.319655772 | 2.86E-15 | 2.43E-14 |
| TDRD1 | 2.968954599 | 4.312041873 | 4.32E-15 | 3.60E-14 |
| TNN | 2.314343045 | −0.352090036 | 4.70E-15 | 3.90E-14 |
| FOXD1 | 2.796864479 | 1.801714027 | 4.83E-15 | 4.00E-14 |
| DEFB126 | −2.714005591 | −1.975045093 | 6.47E-15 | 5.29E-14 |
| MYH6 | −2.531908454 | 0.826728283 | 1.06E-14 | 8.52E-14 |
| OR51H1 | 3.267819899 | −3.615043094 | 1.09E-14 | 8.69E-14 |
| SPON2 | 2.315586856 | 10.45792138 | 1.10E-14 | 8.78E-14 |
| ZMYND10 | 2.085925052 | 1.832701834 | 1.13E-14 | 9.04E-14 |
| ZP1 | 2.210730897 | 0.676381856 | 1.32E-14 | 1.04E-13 |
| KRTAP5-1 | 2.140411485 | −1.919084242 | 1.36E-14 | 1.07E-13 |
| SLC22A10 | 3.112889405 | 0.336771949 | 1.63E-14 | 1.27E-13 |
| NUTM2F | 2.599677343 | −2.440419577 | 1.81E-14 | 1.40E-13 |
| C11orf87 | −2.120433611 | −3.11909289 | 2.06E-14 | 1.59E-13 |
| SMIM28 | 2.307959645 | −1.425734172 | 2.32E-14 | 1.77E-13 |
| AC233992.2 | 2.188468656 | −2.988603347 | 2.55E-14 | 1.94E-13 |
| TRPC5 | −2.627768694 | −3.195666087 | 2.61E-14 | 1.99E-13 |
| ADGRG4 | −2.227928687 | −4.008538955 | 3.07E-14 | 2.32E-13 |
| DLX2 | 2.520792049 | 1.771274957 | 3.45E-14 | 2.59E-13 |
| AC013470.2 | 2.37626599 | −2.264634427 | 3.74E-14 | 2.80E-13 |
| 12-Sep | 2.32842503 | −1.614438692 | 3.83E-14 | 2.87E-13 |
| CHRM2 | −2.627807986 | −0.761918657 | 4.63E-14 | 3.42E-13 |
| PLPPR5 | −2.051535096 | −3.828879148 | 4.65E-14 | 3.44E-13 |
| KLB | 2.917388689 | 2.020757836 | 5.17E-14 | 3.79E-13 |
| KRT20 | 3.970669871 | −0.213705133 | 5.31E-14 | 3.90E-13 |
| SRARP | 3.763182115 | 4.202486163 | 5.51E-14 | 4.04E-13 |
| BAIAP2L2 | 2.226024178 | 2.628704883 | 5.96E-14 | 4.35E-13 |
| HIST1H4E | 2.159254954 | 1.043399573 | 7.18E-14 | 5.20E-13 |
| BRSK2 | 2.354199485 | 2.719801489 | 7.53E-14 | 5.44E-13 |
| AK5 | 2.022452828 | 3.960729447 | 8.23E-14 | 5.91E-13 |
| EPHA8 | 2.42213167 | −0.482075108 | 1.34E-13 | 9.40E-13 |
| HBQ1 | 3.058254424 | −2.360212262 | 1.94E-13 | 1.34E-12 |
| DLL3 | 2.036697223 | −0.709591972 | 2.50E-13 | 1.71E-12 |
| ATP8A2 | 2.805192641 | 3.941027197 | 2.73E-13 | 1.85E-12 |
| UGT3A1 | −2.300754877 | −2.195684518 | 2.90E-13 | 1.96E-12 |
| NKX6-1 | 3.539257475 | −0.069017009 | 3.14E-13 | 2.11E-12 |
| FOXB2 | 4.120786237 | −0.339067966 | 3.43E-13 | 2.29E-12 |
| OR2B6 | 2.070727347 | −1.791175567 | 3.50E-13 | 2.34E-12 |
| ZIC5 | 3.400800848 | 0.951706604 | 3.68E-13 | 2.45E-12 |
| B3GNT6 | 4.168735091 | 3.52191008 | 4.20E-13 | 2.78E-12 |
| FGL1 | 3.035621437 | 1.63122323 | 4.37E-13 | 2.88E-12 |
| TFF3 | 2.769530436 | 8.238324054 | 8.51E-13 | 5.41E-12 |
| CST2 | 3.537958116 | 4.052951673 | 9.02E-13 | 5.71E-12 |
| FOXL2 | 2.386028893 | 1.084381767 | 9.59E-13 | 6.06E-12 |
| TBX10 | 3.419608802 | 1.522870413 | 1.20E-12 | 7.46E-12 |
| RHCG | −2.008813569 | 0.619765959 | 1.42E-12 | 8.83E-12 |
| NLRP13 | 2.952695433 | −1.963847664 | 1.51E-12 | 9.33E-12 |
| ISG15 | 2.015582231 | 5.871285045 | 1.62E-12 | 9.94E-12 |
| FEZF1 | 2.983525183 | −2.371592675 | 1.62E-12 | 9.98E-12 |
| UPK1B | −2.479748949 | −1.685662964 | 1.77E-12 | 1.09E-11 |
| FABP5 | 2.55866377 | 5.395196065 | 1.92E-12 | 1.18E-11 |
| NR2E1 | 3.803357172 | −1.451927234 | 2.09E-12 | 1.27E-11 |
| SPINK1 | 4.492947788 | 4.716448182 | 2.20E-12 | 1.34E-11 |
| FUT6 | −2.34107223 | −0.394817546 | 2.42E-12 | 1.46E-11 |
| HOXC12 | 4.618237944 | −0.951730916 | 2.59E-12 | 1.55E-11 |
| OLFM4 | −2.001351853 | 7.271508686 | 2.61E-12 | 1.57E-11 |
| ANKRD30A | 6.088697199 | 0.552547891 | 2.75E-12 | 1.64E-11 |
| SPZ1 | 7.464030724 | 1.177592669 | 3.19E-12 | 1.90E-11 |
| HOXC13 | 3.203398538 | −1.939843545 | 4.23E-12 | 2.47E-11 |
| CLC | −2.056516349 | −3.77853227 | 4.31E-12 | 2.51E-11 |
| RPRML | 2.940731461 | −1.095714471 | 4.65E-12 | 2.70E-11 |
| OR51T1 | 2.54982834 | −2.792011523 | 5.76E-12 | 3.32E-11 |
| FOXI1 | −2.138580221 | 1.090697338 | 6.04E-12 | 3.47E-11 |
| NKAIN1 | 2.258465266 | 4.792143603 | 6.04E-12 | 3.47E-11 |
| DNAH8 | 2.464627376 | 5.112883427 | 7.54E-12 | 4.27E-11 |
| WFDC6 | −2.255684544 | −3.632961702 | 7.55E-12 | 4.28E-11 |
| COL10A1 | 2.453464464 | 3.270292451 | 7.57E-12 | 4.29E-11 |
| COMP | 2.16226434 | 5.850848074 | 8.07E-12 | 4.56E-11 |
| SPRR3 | −2.650341647 | −2.524440816 | 8.16E-12 | 4.61E-11 |
| ERG | 2.494207074 | 6.99904595 | 9.37E-12 | 5.25E-11 |
| SSTR1 | 2.060786454 | 4.37450598 | 1.04E-11 | 5.80E-11 |
| UGT2B4 | 3.221566911 | 2.827464057 | 1.08E-11 | 5.99E-11 |
| IL36RN | 3.03414543 | −1.504655432 | 1.26E-11 | 6.97E-11 |
| C19orf81 | 2.546159141 | 1.113739849 | 1.30E-11 | 7.16E-11 |
| VAX1 | 3.961513127 | −2.632789163 | 1.64E-11 | 8.92E-11 |
| LRRTM3 | −2.107441126 | −1.24681776 | 1.91E-11 | 1.03E-10 |
| FAM57B | 2.031437105 | −0.698387388 | 2.09E-11 | 1.13E-10 |
| GRIN3A | 2.638844764 | 4.65540954 | 2.23E-11 | 1.20E-10 |
| TUBB4A | 2.469186771 | 2.047892023 | 2.36E-11 | 1.26E-10 |
| ADAM29 | −2.02667846 | −3.775698756 | 2.84E-11 | 1.50E-10 |
| BMP5 | −2.18782084 | 2.684465934 | 4.71E-11 | 2.43E-10 |
| COL2A1 | 3.785158888 | 6.15210744 | 4.83E-11 | 2.48E-10 |
| KRT72 | 3.761702653 | −1.981764703 | 5.03E-11 | 2.58E-10 |
| BARX1 | 2.391463924 | −1.658459129 | 5.47E-11 | 2.80E-10 |
| ROPN1L | 2.092640047 | −0.046359297 | 5.55E-11 | 2.83E-10 |
| ARMC3 | 2.136397252 | 0.189599604 | 6.22E-11 | 3.16E-10 |
| PROC | 2.67020989 | 1.131805312 | 7.29E-11 | 3.67E-10 |
| KCNG3 | 2.132031411 | 2.045373485 | 9.35E-11 | 4.64E-10 |
| PCDHA1 | 2.795627976 | 0.375523362 | 1.20E-10 | 5.89E-10 |
| UNC5A | 2.709726989 | 3.92396467 | 1.24E-10 | 6.07E-10 |
| SUCNR1 | 3.081480451 | 1.536548908 | 1.41E-10 | 6.85E-10 |
| SMIM21 | 3.427582218 | −2.986773972 | 1.60E-10 | 7.73E-10 |
| ETV4 | 3.816023431 | 5.623026412 | 1.67E-10 | 8.03E-10 |
| SERPINA11 | 2.404577025 | 2.545674419 | 1.74E-10 | 8.34E-10 |
| GBX2 | 4.378429224 | −2.072125849 | 1.79E-10 | 8.57E-10 |
| CDC20B | 4.150567923 | 1.534510645 | 1.84E-10 | 8.79E-10 |
| GJA3 | 2.040848411 | 0.77159763 | 1.88E-10 | 8.94E-10 |
| C1QTNF9B | 2.025651594 | 0.385827111 | 1.89E-10 | 9.01E-10 |
| SMIM23 | 2.240012939 | −3.890952299 | 3.15E-10 | 1.46E-09 |
| CRYGD | −2.157881056 | −3.558055362 | 3.46E-10 | 1.60E-09 |
| GRPR | 2.082638062 | 1.422602298 | 4.01E-10 | 1.84E-09 |
| SLC22A31 | 2.15465431 | 1.682264922 | 4.15E-10 | 1.90E-09 |
| ZNF560 | 3.579830078 | 0.907262077 | 4.42E-10 | 2.02E-09 |
| PTPRR | 2.677564298 | 1.278570082 | 4.65E-10 | 2.11E-09 |
| SFTPA2 | 2.026666077 | 6.660829826 | 4.67E-10 | 2.12E-09 |
| HAO1 | 3.180714369 | −0.216881708 | 5.37E-10 | 2.43E-09 |
| CHIT1 | 2.124877061 | 3.520488959 | 6.25E-10 | 2.80E-09 |
| DMBX1 | 2.075728929 | −2.899045915 | 6.67E-10 | 2.98E-09 |
| UTS2B | 2.623610217 | 0.763054819 | 7.50E-10 | 3.33E-09 |
| PNMA5 | 5.583595807 | 1.256206222 | 7.52E-10 | 3.33E-09 |
| TTR | 2.263010905 | 1.534436019 | 7.85E-10 | 3.47E-09 |
| CER1 | −2.01557727 | −3.982760873 | 8.51E-10 | 3.74E-09 |
| GMNC | 2.222390659 | 1.731673014 | 9.70E-10 | 4.23E-09 |
| TGM4 | −2.299530484 | 9.058863089 | 1.06E-09 | 4.62E-09 |
| KRT73 | 2.784757012 | −3.396159935 | 1.60E-09 | 6.83E-09 |
| SLC24A2 | 2.263093544 | 0.036838795 | 1.70E-09 | 7.21E-09 |
| ANKRD30B | 3.073498165 | −1.41579624 | 1.91E-09 | 8.05E-09 |
| FEZF2 | 5.1526634 | −2.123364605 | 1.95E-09 | 8.20E-09 |
| NKX6-3 | 2.059995247 | −2.900165006 | 1.98E-09 | 8.34E-09 |
| OR52I1 | 2.176973868 | −3.001764047 | 2.05E-09 | 8.58E-09 |
| CAMKV | 2.881570973 | −2.038814296 | 2.86E-09 | 1.18E-08 |
| NOTUM | 2.308455321 | 0.786652156 | 4.24E-09 | 1.70E-08 |
| SFTA3 | −2.106019445 | −2.466203685 | 4.27E-09 | 1.71E-08 |
| POU4F1 | 3.693208616 | −2.095987674 | 4.39E-09 | 1.76E-08 |
| IL1F10 | 2.260744127 | −3.909896446 | 4.80E-09 | 1.91E-08 |
| PBOV1 | 2.085675747 | −2.154992386 | 7.36E-09 | 2.85E-08 |
| DIO1 | 2.26987907 | 0.891743028 | 7.38E-09 | 2.86E-08 |
| NETO1 | 3.568491117 | 0.942120675 | 8.02E-09 | 3.10E-08 |
| PPP3R2 | 2.570526002 | −2.677368344 | 8.10E-09 | 3.12E-08 |
| HNF1A | 2.906394479 | 0.190866031 | 8.41E-09 | 3.24E-08 |
| OR51A7 | 2.638046472 | −3.56624315 | 9.38E-09 | 3.59E-08 |
| GSTA3 | 2.806870513 | −1.42037873 | 9.39E-09 | 3.59E-08 |
| AC011604.2 | 2.143482743 | −3.108314603 | 9.51E-09 | 3.64E-08 |
| SMR3A | −2.635771323 | −4.144131086 | 1.06E-08 | 4.01E-08 |
| PANX3 | 2.326305366 | −3.160277377 | 1.42E-08 | 5.32E-08 |
| POTEB3 | 3.730685749 | −3.713452026 | 1.60E-08 | 5.94E-08 |
| APOA2 | 6.084002355 | −0.989674056 | 1.65E-08 | 6.13E-08 |
| HMX1 | 3.620329015 | −2.282249925 | 1.71E-08 | 6.34E-08 |
| KLK14 | 2.199159846 | 2.763344004 | 1.74E-08 | 6.44E-08 |
| TEX19 | 2.109864247 | −3.789954315 | 1.75E-08 | 6.48E-08 |
| FOXB1 | 2.548534069 | −3.326675239 | 1.83E-08 | 6.77E-08 |
| OR2T4 | −2.107781427 | −4.107614379 | 1.95E-08 | 7.16E-08 |
| DEFA6 | 8.471273836 | 1.646111486 | 2.42E-08 | 8.78E-08 |
| HIST2H2AA3 | 2.438596056 | −2.699403297 | 2.65E-08 | 9.60E-08 |
| CPN1 | 3.764667337 | −3.646951466 | 2.86E-08 | 1.03E-07 |
| SHD | 2.349865496 | −1.643837394 | 3.07E-08 | 1.10E-07 |
| SLC17A4 | 3.661486856 | 0.041233394 | 3.45E-08 | 1.23E-07 |
| AFP | 2.561513116 | −1.819167591 | 3.62E-08 | 1.29E-07 |
| A1CF | 2.968271138 | −0.721602281 | 3.74E-08 | 1.33E-07 |
| CENPVL3 | 2.108254917 | −3.236233695 | 3.84E-08 | 1.36E-07 |
| DEFA5 | 9.398510106 | 3.364489602 | 3.89E-08 | 1.38E-07 |
| ADAM2 | 2.268866369 | 1.975249434 | 3.95E-08 | 1.40E-07 |
| OR51F2 | 2.001274564 | −3.505887555 | 4.08E-08 | 1.44E-07 |
| VIL1 | 2.444441479 | −1.715247996 | 4.27E-08 | 1.51E-07 |
| GC | 5.399505393 | 0.791302383 | 4.74E-08 | 1.66E-07 |
| GLP1R | 2.638173244 | 0.005120757 | 5.23E-08 | 1.82E-07 |
| HABP2 | 2.053307035 | −0.144814994 | 6.05E-08 | 2.08E-07 |
| AMBN | 5.985888716 | −2.328310244 | 6.27E-08 | 2.16E-07 |
| HELT | 2.793749673 | −3.373803166 | 7.89E-08 | 2.68E-07 |
| ETV1 | 2.242859964 | 5.823683585 | 8.08E-08 | 2.74E-07 |
| CGA | 3.095258408 | 2.341413952 | 1.05E-07 | 3.52E-07 |
| SCGB2A2 | 4.164296891 | −2.821900282 | 1.26E-07 | 4.18E-07 |
| NXPH1 | 2.742113092 | −2.408586056 | 1.51E-07 | 4.94E-07 |
| PAX1 | 2.566319949 | 0.613275567 | 1.56E-07 | 5.11E-07 |
| FOXG1 | 4.413970069 | −2.305401919 | 1.66E-07 | 5.40E-07 |
| KRT75 | 2.558006156 | −1.069230467 | 1.66E-07 | 5.41E-07 |
| CD5L | 3.99795932 | −2.274198075 | 1.96E-07 | 6.32E-07 |
| INSM2 | 2.764301541 | −2.31599136 | 1.97E-07 | 6.34E-07 |
| TM4SF20 | 2.812654396 | −2.785614732 | 2.06E-07 | 6.62E-07 |
| SLCO1B7 | 2.01296759 | −3.7612757 | 2.07E-07 | 6.65E-07 |
| DYTN | 3.239601987 | −0.530752709 | 2.16E-07 | 6.93E-07 |
| POTEC | 3.432674516 | −3.289200505 | 2.22E-07 | 7.11E-07 |
| NAA11 | 2.650446317 | −2.486862885 | 2.26E-07 | 7.21E-07 |
| MYH13 | 2.066063048 | −2.482864861 | 2.50E-07 | 7.92E-07 |
| CDHR4 | 2.337589644 | −0.649813964 | 2.54E-07 | 8.03E-07 |
| AKAP14 | 2.517486668 | −2.274812756 | 2.62E-07 | 8.29E-07 |
| S100A7A | −2.151656547 | −4.15011295 | 2.68E-07 | 8.45E-07 |
| CACNA1I | 2.167388874 | −0.161792924 | 2.78E-07 | 8.76E-07 |
| MAGEA12 | 4.253847274 | −1.629199508 | 2.83E-07 | 8.91E-07 |
| MS4A15 | 2.679036167 | −1.548974492 | 3.54E-07 | 1.10E-06 |
| C6orf10 | 2.932100927 | −1.675723134 | 3.67E-07 | 1.14E-06 |
| ZBBX | 2.218935272 | −1.371954365 | 4.27E-07 | 1.31E-06 |
| NWD2 | 2.691936171 | −1.688128902 | 5.21E-07 | 1.58E-06 |
| APOBEC4 | 2.432607195 | −2.444614927 | 5.34E-07 | 1.62E-06 |
| DRGX | 5.581607276 | −0.548299892 | 5.36E-07 | 1.62E-06 |
| CAPSL | 2.141430537 | −0.478931097 | 5.42E-07 | 1.64E-06 |
| SNTN | 2.12725837 | −0.315435664 | 5.66E-07 | 1.70E-06 |
| KNG1 | 2.617105398 | −2.091853619 | 6.24E-07 | 1.87E-06 |
| PRSS48 | 3.821948787 | −2.662238738 | 6.31E-07 | 1.89E-06 |
| WDR38 | 2.180078528 | −0.64579903 | 7.41E-07 | 2.20E-06 |
| SCN1A | 3.512060368 | −0.40236363 | 8.22E-07 | 2.43E-06 |
| PNLIP | 2.328354606 | −1.788983069 | 9.04E-07 | 2.65E-06 |
| CST1 | 2.792052996 | 5.564777783 | 9.11E-07 | 2.68E-06 |
| AHSG | 3.199438746 | −2.773945144 | 1.06E-06 | 3.08E-06 |
| CRYBA4 | 2.772621752 | −1.320349749 | 1.06E-06 | 3.08E-06 |
| IAPP | 3.324462289 | −0.773721066 | 1.07E-06 | 3.11E-06 |
| ZNF716 | 3.052270573 | −4.06518223 | 1.16E-06 | 3.35E-06 |
| LIPF | 6.326103 | 2.912495858 | 1.35E-06 | 3.87E-06 |
| SERPINA6 | 2.943682115 | −1.793361739 | 1.43E-06 | 4.10E-06 |
| IRS4 | 2.223425105 | −0.100658114 | 1.47E-06 | 4.21E-06 |
| MAGEA4 | 6.312010735 | −1.437186342 | 1.48E-06 | 4.22E-06 |
| MAGEA8 | 2.386730522 | −2.00924606 | 2.03E-06 | 5.70E-06 |
| APOC3 | 4.606628396 | −2.654273715 | 2.09E-06 | 5.86E-06 |
| ZIC3 | 2.575224827 | −0.857692039 | 2.48E-06 | 6.86E-06 |
| FGB | 4.323384685 | −0.115174883 | 2.96E-06 | 8.13E-06 |
| UGT1A3 | 2.37472128 | 0.542246428 | 3.02E-06 | 8.29E-06 |
| UGT1A1 | 3.262526158 | −1.071235534 | 3.03E-06 | 8.30E-06 |
| MAGEA1 | 4.69097688 | −1.863592199 | 3.13E-06 | 8.56E-06 |
| HRG | 3.137868897 | −3.339954046 | 3.64E-06 | 9.89E-06 |
| PRAME | 2.341567845 | 0.717925667 | 3.89E-06 | 1.05E-05 |
| SCRT2 | 3.638442309 | −3.636567385 | 3.91E-06 | 1.06E-05 |
| CHAT | 3.19066275 | −1.980217619 | 4.36E-06 | 1.17E-05 |
| GAPDHS | 2.418577557 | −3.713135428 | 4.53E-06 | 1.21E-05 |
| BARHL1 | 3.015059095 | −3.539516674 | 4.66E-06 | 1.25E-05 |
| C8B | 2.984778011 | −4.131577497 | 5.40E-06 | 1.43E-05 |
| C1orf158 | 2.231593922 | −2.248366293 | 5.60E-06 | 1.48E-05 |
| TUBA4B | 2.083283692 | −1.021407683 | 5.88E-06 | 1.55E-05 |
| TAC3 | 2.369711799 | −1.769799158 | 6.35E-06 | 1.67E-05 |
| DCAF4L2 | 4.181333285 | −3.336994371 | 6.42E-06 | 1.68E-05 |
| MAGEA11 | −2.334344997 | −4.12452691 | 6.47E-06 | 1.70E-05 |
| OOSP2 | 3.211424198 | −4.128101138 | 6.88E-06 | 1.80E-05 |
| CALML5 | 2.96302721 | −0.373435971 | 6.97E-06 | 1.82E-05 |
| CELA3B | 2.509137069 | −3.052407547 | 7.67E-06 | 1.99E-05 |
| FGF3 | 3.877010188 | −3.428917251 | 7.80E-06 | 2.02E-05 |
| AKR1C4 | 2.494703252 | −3.934584544 | 8.50E-06 | 2.19E-05 |
| SULT1C3 | 2.186968852 | −1.355115719 | 1.01E-05 | 2.57E-05 |
| GPR52 | 2.388393967 | −3.786669653 | 1.09E-05 | 2.77E-05 |
| FAM92B | 2.05640783 | −1.500495388 | 1.13E-05 | 2.87E-05 |
| OR5B2 | 2.609194338 | −3.807269643 | 1.20E-05 | 3.03E-05 |
| FAM216B | 2.374779824 | −0.518608291 | 1.23E-05 | 3.11E-05 |
| CA6 | 2.513401867 | −2.76088127 | 1.31E-05 | 3.29E-05 |
| CTAG2 | 4.925601332 | −0.782603962 | 1.32E-05 | 3.31E-05 |
| TAS2R38 | 2.622407082 | −3.630778383 | 1.34E-05 | 3.36E-05 |
| GLRA3 | 3.223686569 | −0.188435203 | 1.51E-05 | 3.75E-05 |
| GUCA2A | 3.562863475 | −2.288541517 | 1.72E-05 | 4.24E-05 |
| LHX3 | 2.215818134 | −4.129011745 | 1.75E-05 | 4.30E-05 |
| TMEM212 | 2.577624226 | −3.145400789 | 1.88E-05 | 4.61E-05 |
| UCN3 | 2.272500127 | −1.200540549 | 2.21E-05 | 5.37E-05 |
| MAGEC1 | 4.686921382 | −1.392116181 | 2.23E-05 | 5.41E-05 |
| CT45A10 | 4.045321429 | −3.548760894 | 2.50E-05 | 6.03E-05 |
| AL035425.2 | 2.735879316 | −3.680989103 | 2.58E-05 | 6.19E-05 |
| CLCA1 | 3.776638394 | −1.530223149 | 2.64E-05 | 6.33E-05 |
| PDE6H | 2.153226877 | −3.916344045 | 2.81E-05 | 6.71E-05 |
| KRTAP13-2 | 2.243103436 | 0.468205823 | 2.93E-05 | 6.99E-05 |
| COX7B2 | 4.741214645 | −1.121111491 | 2.99E-05 | 7.10E-05 |
| MAGEA3 | 4.054216829 | −1.757379075 | 3.30E-05 | 7.81E-05 |
| MAGEA6 | 4.274899658 | −1.956846537 | 3.38E-05 | 7.99E-05 |
| IQCM | 2.579364621 | −4.047403881 | 3.50E-05 | 8.25E-05 |
| LYPD8 | 2.256064736 | −1.241702493 | 3.54E-05 | 8.34E-05 |
| NKX2-5 | 2.801832118 | −2.378705948 | 3.87E-05 | 9.07E-05 |
| REG3A | 3.736515651 | −3.332085299 | 4.10E-05 | 9.56E-05 |
| IGFBP1 | 2.14975531 | −3.545161931 | 4.49E-05 | 0.000104171 |
| MC4R | 3.25596124 | −2.085215369 | 4.57E-05 | 0.000105886 |
| APOH | 3.130989094 | −0.333603889 | 4.69E-05 | 0.00010841 |
| PRSS56 | 3.770792623 | −1.397434252 | 4.82E-05 | 0.000111245 |
| FMR1NB | 2.403293682 | −3.119019324 | 5.25E-05 | 0.000120751 |
| CARD18 | 3.618460498 | −3.641480606 | 5.95E-05 | 0.000135585 |
| C6orf15 | 3.15734794 | −3.509243731 | 6.39E-05 | 0.000144602 |
| ABCC12 | 2.447749702 | −0.858545305 | 6.48E-05 | 0.00014656 |
| SLC6A18 | 3.381785529 | −2.97339517 | 6.57E-05 | 0.000148554 |
| IFNK | 2.684220062 | −3.827516977 | 6.73E-05 | 0.000151888 |
| TRIM49 | 3.177581725 | −3.476981911 | 7.20E-05 | 0.00016148 |
| PRB1 | 3.326542308 | −3.729099839 | 7.35E-05 | 0.000164634 |
| TMEM207 | 2.647209716 | −4.101090504 | 7.69E-05 | 0.000171776 |
| INSL6 | 2.073304179 | −2.965377473 | 8.41E-05 | 0.000186712 |
| OBP2B | 2.109540767 | −1.09806609 | 8.91E-05 | 0.000197245 |
| SOX3 | 2.572786664 | −2.593667145 | 9.58E-05 | 0.000210846 |
| PIH1D3 | 2.051029477 | −3.248873998 | 0.000111697 | 0.000243629 |
| OR52E8 | 2.720510067 | −3.728113618 | 0.000130226 | 0.000281198 |
| IBSP | 2.064740445 | −2.938509758 | 0.000152455 | 0.000325344 |
| CRX | 2.116262219 | −2.812440876 | 0.000155787 | 0.000332099 |
| RAX | 2.56782095 | −2.046630028 | 0.000161204 | 0.000342911 |
| SLC6A19 | 2.216142084 | 2.683437613 | 0.000161511 | 0.000343482 |
| SLCO6A1 | 3.116278974 | −4.027389676 | 0.000171515 | 0.000363202 |
| FGF4 | 3.429130493 | −3.126190388 | 0.000186328 | 0.000392617 |
| VPREB1 | 3.522957645 | −3.50589044 | 0.000189054 | 0.000397891 |
| PRDM9 | 3.406136372 | −3.227166206 | 0.000197076 | 0.000413366 |
| LGALS16 | 3.375273982 | −3.96329701 | 0.000210268 | 0.000439385 |
| CSAG1 | 2.447454631 | −1.666596873 | 0.000211937 | 0.000442615 |
| FGA | 3.556112828 | −1.177897264 | 0.000231462 | 0.000480249 |
| TM4SF5 | 2.007153954 | −3.706723742 | 0.000236151 | 0.000489409 |
| BARHL2 | 2.496114201 | −3.403339289 | 0.000325784 | 0.000661897 |
| GIF | 2.108620219 | −3.831577628 | 0.000351243 | 0.000709509 |
| OR1N2 | 3.286097731 | −2.80552916 | 0.000522382 | 0.001028372 |
| LIN28B | 2.080144856 | −3.937283675 | 0.000578444 | 0.001130644 |
| OTOP1 | 2.945283928 | −3.891057771 | 0.000579885 | 0.001132841 |
| SLC18A3 | 2.169664988 | −1.026690515 | 0.000704233 | 0.001358103 |
| APCS | 2.769781173 | −3.531743743 | 0.000885631 | 0.001680557 |
| SP9 | 2.005599586 | −2.921939416 | 0.000947647 | 0.00178931 |
| PRB4 | 2.598407211 | −4.012142174 | 0.00097319 | 0.001834634 |
| CYP2C9 | 2.028397643 | −4.040000218 | 0.0011091 | 0.002073755 |
| TMPRSS11B | 3.351520815 | −2.537086097 | 0.001404034 | 0.002581258 |
| TPTE | 2.586293693 | −3.849215013 | 0.001431842 | 0.002628596 |
| DSCR8 | 2.143606791 | −3.733115111 | 0.001984814 | 0.003563974 |
| UGT1A10 | 2.29302094 | −1.689729261 | 0.002529475 | 0.004471915 |
| HTN3 | 2.485178564 | −4.015732583 | 0.002830284 | 0.00496153 |
| OR2T10 | 2.159593297 | −2.004107925 | 0.003677882 | 0.0063374 |
| FGG | 2.324040335 | −2.102893311 | 0.004863442 | 0.008221775 |
| MT4 | 2.334444629 | −3.227723792 | 0.005440693 | 0.009120681 |
| INS | 2.330844774 | −3.828167758 | 0.00568305 | 0.009495118 |
Differentially expressed mRNAs in prostate cancer. The horizontal axis shows sample names in TCGA. The right vertical axis means the mRNA names. Downregulated genes are green and upregulated genes are red.
Differentially expressed lncRNAs in prostate cancer. The horizontal axis shows sample names in TCGA. The right vertical axis means the lncRNA names. Downregulated genes are green and upregulated genes are red.
Differentially expressed miRNAs in prostate cancer. The horizontal axis shows sample names in TCGA. The right vertical axis means the miRNA names. Downregulated genes are green and upregulated genes are red.
Footnotes
Source of support: This work was supported by grants from the Qiqihar City Science and Technology Plan Project (SFGG-201435)
Conflicts of interests
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chan WL, Yuo CY, Yang WK, et al. Transcribed pseudogene psiPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma. Nucleic Acids Res. 2013;41:3734–47. doi: 10.1093/nar/gkt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–38. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotake Y, Nakagawa T, Kitagawa K, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: Origin and features. Brief Funct Genomic Proteomic. 2004;3:157–67. doi: 10.1093/bfgp/3.2.157. [DOI] [PubMed] [Google Scholar]
- 6.Zhang R, Guo Y, Ma Z, et al. Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer. Oncotarget. 2017;8:26079–89. doi: 10.18632/oncotarget.15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Shen Z, Zhi Y, et al. Long non-coding RNA ROR promotes radioresistance in hepatocellular carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophys. 2018;645:117–25. doi: 10.1016/j.abb.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 8.McCarter JP, Mitreva MD, Martin J, et al. Analysis and functional classification of transcripts from the nematode Meloidogyne incognita. Genome Biol. 2003;4:R26. doi: 10.1186/gb-2003-4-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Yang X, Wang T, et al. Knockdown delta-5-desaturase in breast cancer cells that overexpress COX-2 results in inhibition of growth, migration and invasion via a dihomo-gamma-linolenic acid peroxidation dependent mechanism. BMC Cancer. 2018;18:330. doi: 10.1186/s12885-018-4250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu SD, Tseng YT, Shrestha S, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Zhu GQ, Zhang K, et al. Cyclooxygenase-2 mediated synergistic effect of ursolic acid in combination with paclitaxel against human gastric carcinoma. Oncotarget. 2017;8:92770–77. doi: 10.18632/oncotarget.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong W, Li N, Pei X, Wu X. Differential expression of DUSP2 in left- and right-sided colon cancer is associated with poor prognosis in colorectal cancer. Oncol Lett. 2018;15:4207–14. doi: 10.3892/ol.2018.7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartmann S, Schuhmacher B, Rausch T, et al. Highly recurrent mutations of SGK1, DUSP2 and JUNB in nodular lymphocyte predominant Hodgkin lymphoma. Leukemia. 2016;30:844–53. doi: 10.1038/leu.2015.328. [DOI] [PubMed] [Google Scholar]
- 15.Bravaccini S, Puccetti M, Bocchini M, et al. PSMA expression: A potential ally for the pathologist in prostate cancer diagnosis. Sci Rep. 2018;8:4254. doi: 10.1038/s41598-018-22594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Z, Choi N, Wang R, et al. Simultaneous detection of dual prostate specific antigens using surface-enhanced raman scattering-based immunoassay for accurate diagnosis of prostate cancer. ACS Nano. 2017;11:4926–33. doi: 10.1021/acsnano.7b01536. [DOI] [PubMed] [Google Scholar]
- 17.Du J, Guan M, Fan J, Jiang H. Loss of DMBT1 expression in human prostate cancer and its correlation with clinical progressive features. Urology. 2011;77:509.e9–13. doi: 10.1016/j.urology.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Wood LM, Pan ZK, Seavey MM, et al. The ubiquitin-like protein, ISG15, is a novel tumor-associated antigen for cancer immunotherapy. Cancer Immunol Immunother. 2012;61:689–700. doi: 10.1007/s00262-011-1129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumi K, Zheng Y, Miyamoto H. Eppin expression in prostate cancer. Eur Urol. 2011;59:1071–72. doi: 10.1016/j.eururo.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 20.Wegiel B, Jiborn T, Abrahamson M, et al. Cystatin C is downregulated in prostate cancer and modulates invasion of prostate cancer cells via MAPK/Erk and androgen receptor pathways. PLoS One. 2009;4:e7953. doi: 10.1371/journal.pone.0007953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang BF, Cai W, Chen B. LncRNA SNHG12 regulated the proliferation of gastric carcinoma cell BGC-823 by targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci. 2018;22:1297–306. doi: 10.26355/eurrev_201803_14471. [DOI] [PubMed] [Google Scholar]
- 22.Wu XS, Wang F, Li HF, et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18:1837–53. doi: 10.15252/embr.201744147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485–91. doi: 10.1093/carcin/bgx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson DW, Dinger ME. Endogenous microRNA sponges: Evidence and controversy. Nat Rev Genet. 2016;17:272–83. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 25.Guil S, Esteller M. RNA-RNA interactions in gene regulation: The coding and noncoding players. Trends Biochem Sci. 2015;40:248–56. doi: 10.1016/j.tibs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Haas J, Mester S, Lai A, et al. Genomic structural variations lead to dysregulation of important coding and non-coding RNA species in dilated cardiomyopathy. EMBO Mol Med. 2018;10:107–20. doi: 10.15252/emmm.201707838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Zhao X, Liu Q, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med. 2017;23:1331–41. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng TM, Chin YT, Ho Y, et al. Resveratrol induces sumoylated COX-2-dependent anti-proliferation in human prostate cancer LNCaP cells. Food Chem Toxicol. 2018;112:67–75. doi: 10.1016/j.fct.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Ko CJ, Lan SW, Lu YC, et al. Inhibition of cyclooxygenase-2-mediated matriptase activation contributes to the suppression of prostate cancer cell motility and metastasis. Oncogene. 2017;36:4597–609. doi: 10.1038/onc.2017.82. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Liang J, Huang J. Downregulated microRNA-26a modulates prostate cancer cell proliferation and apoptosis by targeting COX-2. Oncol Lett. 2016;12:3397–402. doi: 10.3892/ol.2016.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uddin A, Chakraborty S. Role of miRNAs in lung cancer. J Cell Physiol. :2018. doi: 10.1002/jcp.26607. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Jiang X, Liang X, Jiang G. Molecular and cellular mechanisms of castration resistant prostate cancer. Oncol Lett. 2018;15:6063–76. doi: 10.3892/ol.2018.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong X, Qian X, Duan L, et al. microRNA-372 suppresses migration and invasion by targeting p65 in human prostate cancer cells. DNA Cell Biol. 2016;35:828–35. doi: 10.1089/dna.2015.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Wang X, Zhang J, et al. Artesunate suppresses the viability and mobility of prostate cancer cells through UCA1, the sponge of miR-184. Oncotarget. 2017;8:18260–70. doi: 10.18632/oncotarget.15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardiner K, Slavov D, Bechtel L, Davisson M. Annotation of human chromosome 21 for relevance to Down syndrome: Gene structure and expression analysis. Genomics. 2002;79:833–43. doi: 10.1006/geno.2002.6782. [DOI] [PubMed] [Google Scholar]
- 36.Pellegrina D, Severino P, Barbeiro HV, et al. Insights into the function of long noncoding RNAs in sepsis revealed by gene co-expression network analysis. Noncoding RNA. 2017;3(1) doi: 10.3390/ncrna3010005. pii: E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Differentially expressed mRNAs in prostate cancer.
| Gene | logFC | logCPM | PValue | FDR |
|---|---|---|---|---|
| SERPINA5 | −6.785068673 | 4.397738393 | 0 | 0 |
| MFSD2A | −5.966707923 | 3.456236811 | 0 | 0 |
| ACSL6 | −4.996876596 | 2.406174962 | 4.98E-299 | 2.97E-295 |
| MCF2 | −5.267342895 | 0.929684863 | 1.59E-262 | 7.09E-259 |
| EMX2 | −6.785444801 | 2.244788495 | 8.95E-260 | 3.20E-256 |
| HOXB8 | −6.244268143 | 1.047398016 | 4.88E-250 | 1.45E-246 |
| CLDN2 | −7.911204171 | 3.21684931 | 1.11E-247 | 2.82E-244 |
| AKR1B1 | −3.874547187 | 5.715503602 | 6.03E-236 | 1.35E-232 |
| SPINK2 | −7.409528496 | 2.678272683 | 7.41E-236 | 1.47E-232 |
| CYP19A1 | −5.406080831 | −1.222596337 | 2.05E-234 | 3.66E-231 |
| KLHL14 | −4.247424278 | 1.572359202 | 2.71E-206 | 4.41E-203 |
| SPINK13 | −5.528436256 | −0.392023032 | 2.65E-203 | 3.95E-200 |
| PATE2 | −5.418513576 | 0.297970005 | 1.42E-195 | 1.95E-192 |
| TMEM114 | −8.944978033 | 0.54167799 | 2.54E-188 | 3.24E-185 |
| NDRG4 | −3.348967314 | 2.992267305 | 3.41E-182 | 4.07E-179 |
| WNT9B | −4.877722819 | −0.703836945 | 1.10E-181 | 1.23E-178 |
| CRTAC1 | −4.292389971 | 2.665559252 | 1.29E-177 | 1.35E-174 |
| RASL10B | −3.465987979 | 2.057893571 | 1.17E-166 | 1.16E-163 |
| PIP | −7.351408595 | 6.495875509 | 3.33E-165 | 3.13E-162 |
| ANXA13 | −5.51532869 | 0.947383484 | 4.18E-165 | 3.74E-162 |
| PATE4 | −9.213661816 | 3.236652784 | 2.41E-157 | 2.05E-154 |
| CYSLTR2 | −3.722193167 | 1.029914851 | 9.93E-155 | 8.07E-152 |
| PAEP | −9.922475379 | 3.345532284 | 1.48E-151 | 1.15E-148 |
| AQP2 | −9.29515775 | 4.480800053 | 9.30E-151 | 6.93E-148 |
| CA2 | −4.440192491 | 2.780775663 | 5.54E-150 | 3.96E-147 |
| CRISP1 | −8.667915771 | 1.60380184 | 5.58E-149 | 3.84E-146 |
| MGAM | −4.692243468 | 0.243483116 | 7.50E-149 | 4.97E-146 |
| HOXB6 | −4.584092575 | 1.419092909 | 1.02E-146 | 6.53E-144 |
| SLC2A9 | −2.696283469 | 2.324688774 | 5.63E-140 | 3.47E-137 |
| SPINT3 | −7.357342601 | −1.907652893 | 5.44E-137 | 3.24E-134 |
| KCP | −3.487546788 | 0.465803728 | 4.69E-136 | 2.70E-133 |
| ANO1 | −2.868561416 | 5.461105992 | 9.81E-134 | 5.48E-131 |
| C3orf36 | −3.509127685 | 0.36085329 | 1.07E-131 | 5.80E-129 |
| ATP13A4 | −4.250554958 | −0.531092553 | 1.48E-130 | 7.79E-128 |
| SLC13A2 | −6.292561769 | 1.033617138 | 4.23E-129 | 2.16E-126 |
| KCNJ5 | −3.119719349 | 3.007097953 | 7.69E-129 | 3.82E-126 |
| PNMT | −4.667142036 | 1.022812758 | 2.83E-127 | 1.37E-124 |
| STAC2 | −4.416170051 | 3.417454375 | 8.66E-126 | 4.08E-123 |
| ABCG2 | −2.809960333 | 3.970415449 | 4.78E-124 | 2.19E-121 |
| FAM83A | −5.078067363 | −0.012191609 | 3.78E-123 | 1.69E-120 |
| SLC16A12 | −3.749170694 | 0.162005786 | 5.37E-121 | 2.34E-118 |
| SEMG2 | −9.505742358 | 10.32686946 | 8.68E-120 | 3.69E-117 |
| C1orf186 | −3.363933794 | 0.80475646 | 6.92E-119 | 2.88E-116 |
| PAQR8 | −2.568199676 | 3.692816808 | 4.02E-118 | 1.63E-115 |
| UGT2B7 | −5.583084346 | −0.640545992 | 7.63E-118 | 3.03E-115 |
| MRO | −3.090785129 | −0.234389189 | 1.54E-116 | 5.97E-114 |
| SLCO4C1 | −4.798092265 | −0.639044646 | 3.84E-116 | 1.46E-113 |
| ACE2 | −3.568169252 | 0.516117235 | 6.46E-114 | 2.41E-111 |
| FUT3 | −4.111844574 | 0.379086415 | 6.37E-113 | 2.33E-110 |
| PTGES | −3.039801329 | 4.655930611 | 4.92E-112 | 1.76E-109 |
| HS3ST5 | −4.914778132 | −1.407416204 | 2.03E-110 | 7.10E-108 |
| KIRREL3 | −3.012684881 | −0.860795205 | 2.44E-109 | 8.40E-107 |
| SEMG1 | −9.757312184 | 11.35548942 | 3.89E-108 | 1.31E-105 |
| DEFB129 | −6.866593977 | −2.315654961 | 2.00E-107 | 6.62E-105 |
| PAX2 | −4.064900582 | 0.376708484 | 2.81E-106 | 9.13E-104 |
| PIK3C2G | −4.380016398 | 0.469725146 | 1.07E-102 | 3.40E-100 |
| SLC46A2 | −4.113213363 | −1.409293159 | 3.17E-102 | 9.95E-100 |
| SNAP25 | −3.062100018 | 2.046817113 | 1.04E-96 | 3.21E-94 |
| LPL | −3.698585 | 3.897033258 | 1.73E-96 | 5.25E-94 |
| PLA2G4A | −2.306657328 | 3.506160074 | 5.45E-96 | 1.62E-93 |
| HOXB9 | −3.738394139 | 1.187071894 | 6.96E-96 | 2.04E-93 |
| POU3F3 | −7.820353855 | 1.441534386 | 1.02E-93 | 2.93E-91 |
| SLC28A3 | −4.429709489 | −0.22811352 | 4.13E-91 | 1.17E-88 |
| GSTM3 | −2.283273278 | 5.654161731 | 4.79E-91 | 1.34E-88 |
| FRMD3 | −2.232713406 | 2.362727796 | 5.80E-91 | 1.60E-88 |
| EVA1A | −2.609102352 | 0.046188518 | 6.32E-91 | 1.71E-88 |
| PRDM16 | −2.585618819 | 0.810572803 | 5.09E-90 | 1.36E-87 |
| FBP2 | −3.625501212 | −0.395808669 | 9.04E-90 | 2.38E-87 |
| GCNT4 | −2.419664765 | 1.308525087 | 2.55E-89 | 6.62E-87 |
| CLU | −2.575286197 | 9.076442775 | 1.20E-88 | 3.06E-86 |
| EDDM3A | −9.915750757 | 0.930978027 | 1.71E-88 | 4.30E-86 |
| KCNJ16 | −5.300617001 | 1.33688112 | 9.23E-88 | 2.29E-85 |
| KCNJ15 | −3.274002031 | 1.948780543 | 1.64E-87 | 4.02E-85 |
| PATE1 | −8.559529455 | 4.39114703 | 4.70E-87 | 1.14E-84 |
| KRT24 | −4.993860718 | −2.05900135 | 1.41E-86 | 3.36E-84 |
| SULT2A1 | −6.511620056 | −1.487548002 | 1.54E-84 | 3.58E-82 |
| NPFFR2 | −4.628734826 | −1.871185299 | 1.24E-82 | 2.84E-80 |
| GNAO1 | −2.487785948 | 3.631013922 | 4.77E-81 | 1.08E-78 |
| DEFB131A | −7.992232167 | −2.717399459 | 1.12E-79 | 2.51E-77 |
| PDK4 | −3.01435869 | 6.971020626 | 2.35E-79 | 5.19E-77 |
| UNC5B | −2.199076184 | 5.027632209 | 2.67E-79 | 5.83E-77 |
| CAMP | −3.53381043 | −1.328045181 | 9.63E-78 | 2.08E-75 |
| SLC31A2 | −2.111149911 | 0.324281459 | 2.44E-77 | 5.20E-75 |
| PON3 | −3.31897902 | −0.714040817 | 1.52E-76 | 3.20E-74 |
| ADCY8 | −4.727507179 | −2.378656903 | 2.96E-76 | 6.15E-74 |
| TMEM238L | −3.460883798 | 0.330286585 | 3.96E-76 | 8.14E-74 |
| EDDM3B | −9.095154357 | −1.282761718 | 1.53E-75 | 3.08E-73 |
| LHX1 | −6.152126939 | −2.569155343 | 2.06E-75 | 4.09E-73 |
| TMEM171 | −2.978213867 | −2.490124781 | 2.99E-73 | 5.88E-71 |
| ELSPBP1 | −5.964830301 | −2.613099778 | 9.37E-73 | 1.82E-70 |
| QPRT | −2.396260233 | 2.787898063 | 2.12E-72 | 4.07E-70 |
| FAM167A | −2.899049257 | 2.215708554 | 1.58E-71 | 3.00E-69 |
| RASAL1 | −3.075775921 | 0.258555744 | 1.90E-71 | 3.57E-69 |
| G0S2 | −2.281974698 | 2.740748523 | 1.01E-70 | 1.89E-68 |
| SLC26A3 | −4.713543952 | 4.255428037 | 4.42E-70 | 8.15E-68 |
| GLIS3 | −2.10224676 | 3.141701783 | 5.68E-69 | 1.04E-66 |
| ADTRP | −3.145256188 | 1.796113975 | 1.34E-67 | 2.38E-65 |
| PATE3 | −5.878495255 | −2.049038515 | 6.54E-67 | 1.12E-64 |
| PAX8 | −3.163424949 | 2.312640932 | 4.67E-66 | 7.95E-64 |
| PALM3 | −2.862055728 | 1.75017837 | 2.61E-65 | 4.36E-63 |
| SLPI | −3.550360086 | 5.950826934 | 3.38E-65 | 5.60E-63 |
| SCGB1D4 | −6.616957825 | −3.643904883 | 1.38E-64 | 2.27E-62 |
| CA14 | −2.628944503 | 1.565656785 | 1.41E-64 | 2.29E-62 |
| GRXCR1 | −7.152527699 | −1.442456622 | 4.35E-64 | 7.00E-62 |
| PTGS2 | −3.018672047 | 6.0997409 | 6.74E-64 | 1.08E-61 |
| DEFB125 | −7.823009219 | −3.088436223 | 1.21E-63 | 1.90E-61 |
| SP6 | −2.048223343 | 2.495992609 | 1.74E-63 | 2.70E-61 |
| LIPG | −2.797035036 | 2.502758674 | 2.57E-63 | 3.96E-61 |
| HOXB7 | −2.538554469 | 1.469159627 | 4.12E-63 | 6.24E-61 |
| AOX1 | −2.401455097 | 4.820050441 | 7.83E-63 | 1.18E-60 |
| SIM1 | −5.801689063 | −1.342000003 | 8.05E-63 | 1.20E-60 |
| TRIM9 | −2.21544454 | −0.284214026 | 1.28E-60 | 1.86E-58 |
| MUCL1 | −3.994926646 | 1.918418391 | 1.13E-59 | 1.61E-57 |
| TEDDM1 | −3.162014508 | −2.397478949 | 1.84E-59 | 2.62E-57 |
| GDPD2 | −2.614839978 | −2.578615619 | 1.32E-58 | 1.84E-56 |
| CYP4F8 | −4.234944216 | 5.957323259 | 4.62E-58 | 6.40E-56 |
| HSPA6 | −3.56230163 | 3.163452791 | 1.40E-57 | 1.93E-55 |
| PTGS1 | −2.488186912 | 4.442555664 | 1.50E-57 | 2.05E-55 |
| HOXB5 | −3.507475786 | −0.118278523 | 2.93E-57 | 3.94E-55 |
| ARC | −2.791928795 | 1.302315162 | 5.92E-57 | 7.79E-55 |
| C2orf88 | −2.342319604 | 2.417718221 | 7.27E-57 | 9.48E-55 |
| APOBEC3C | −2.037731259 | 4.968381179 | 1.06E-56 | 1.37E-54 |
| TFAP2B | −3.887211428 | −2.562766626 | 2.07E-56 | 2.66E-54 |
| PADI3 | −4.318296193 | 0.033579355 | 5.97E-56 | 7.63E-54 |
| CES5A | −5.399715292 | −2.273300712 | 6.88E-55 | 8.60E-53 |
| ATP1A4 | −2.650615554 | −0.8604139 | 1.12E-54 | 1.39E-52 |
| CCDC27 | −2.230774182 | −2.116580292 | 4.27E-54 | 5.27E-52 |
| WFDC9 | −7.370447463 | −3.418331864 | 1.29E-53 | 1.59E-51 |
| DCAF12L1 | −3.039717642 | −2.084125111 | 3.70E-53 | 4.50E-51 |
| SBSPON | −2.274088177 | 3.432870313 | 1.42E-52 | 1.70E-50 |
| DUSP2 | −2.374155832 | 4.249770546 | 1.06E-51 | 1.22E-49 |
| EPHA10 | 2.181110197 | 3.496632325 | 5.18E-51 | 5.87E-49 |
| GSTP1 | −2.053371722 | 6.920002831 | 1.40E-50 | 1.55E-48 |
| LMO3 | −2.170273005 | 3.259518865 | 5.16E-50 | 5.52E-48 |
| CXCR2 | −2.604659545 | −0.234591526 | 1.54E-49 | 1.62E-47 |
| LYVE1 | −2.250739542 | 1.531070033 | 6.82E-49 | 7.09E-47 |
| VWA5B2 | −2.355784274 | 1.103997608 | 1.52E-48 | 1.56E-46 |
| ANGPT1 | −2.047422403 | 3.901247666 | 1.86E-48 | 1.90E-46 |
| MUC6 | −4.984635603 | 7.087473698 | 2.08E-48 | 2.10E-46 |
| WFDC8 | −4.53427633 | −4.039274837 | 2.45E-48 | 2.46E-46 |
| DEFB127 | −6.707184857 | −3.923081354 | 6.20E-48 | 6.16E-46 |
| LCN15 | −3.320802889 | −0.939497584 | 6.94E-48 | 6.86E-46 |
| AL163195.3 | −3.923578432 | −4.064090252 | 7.69E-48 | 7.55E-46 |
| DEFB132 | −2.327019581 | 2.024711463 | 1.45E-46 | 1.39E-44 |
| TNMD | −3.2653763 | −1.061318979 | 2.36E-46 | 2.24E-44 |
| TMEM132C | −2.486194486 | 1.508679117 | 2.43E-46 | 2.30E-44 |
| HRASLS5 | −2.182632517 | 0.139647911 | 2.89E-46 | 2.72E-44 |
| SLC9A4 | −3.17506859 | −2.826706326 | 3.82E-46 | 3.58E-44 |
| SCN11A | −2.21835192 | −1.749469071 | 9.22E-46 | 8.54E-44 |
| DUOX1 | −2.137499241 | 4.097778228 | 1.46E-45 | 1.34E-43 |
| MUC15 | −2.661608288 | 1.132262497 | 2.45E-44 | 2.19E-42 |
| TRPM5 | −3.176120461 | −0.890607598 | 3.48E-44 | 3.10E-42 |
| SLC39A2 | −3.854360669 | 2.020647601 | 5.33E-44 | 4.72E-42 |
| TRPM3 | −2.29500896 | −0.137630395 | 6.04E-44 | 5.32E-42 |
| ATP10B | −2.893980809 | −2.015181875 | 3.56E-43 | 3.05E-41 |
| TRIM61 | −2.028327421 | −2.615002542 | 4.08E-43 | 3.47E-41 |
| HPN | 2.51071704 | 8.064534927 | 1.06E-42 | 8.91E-41 |
| KLHL4 | −2.039991565 | −0.923853849 | 1.86E-42 | 1.55E-40 |
| GATA3 | −2.202253741 | 3.782637479 | 2.30E-42 | 1.91E-40 |
| GPD1 | −2.398405613 | −0.219024849 | 3.92E-42 | 3.24E-40 |
| LCN9 | −5.438725778 | −3.834512629 | 4.84E-42 | 3.99E-40 |
| FAM110C | −2.187790425 | 3.123504141 | 6.00E-42 | 4.92E-40 |
| ADGRD2 | −2.978963237 | −1.102374977 | 6.30E-42 | 5.14E-40 |
| MAT1A | −2.847220497 | −1.240774796 | 1.50E-41 | 1.19E-39 |
| C20orf202 | −2.009368734 | −2.563236881 | 2.80E-41 | 2.20E-39 |
| HCAR2 | −2.004843097 | 1.194232598 | 6.33E-41 | 4.94E-39 |
| ASPA | −2.078527277 | 1.495235978 | 6.77E-41 | 5.26E-39 |
| KCNJ1 | −2.762883764 | −2.903446081 | 1.75E-40 | 1.33E-38 |
| STAC | −2.388338573 | 2.991906842 | 1.81E-40 | 1.36E-38 |
| GPX2 | −2.515514209 | 2.45210946 | 1.96E-40 | 1.47E-38 |
| DCHS2 | −2.148971591 | 0.710909297 | 5.37E-40 | 3.95E-38 |
| C10orf99 | −3.579716304 | −2.223557443 | 6.29E-40 | 4.59E-38 |
| PCP4L1 | −2.206007918 | 1.907712695 | 8.99E-40 | 6.45E-38 |
| DMRT2 | −2.991089671 | −2.296521924 | 1.56E-39 | 1.11E-37 |
| CLDN19 | −2.531145647 | −2.60827072 | 1.71E-39 | 1.22E-37 |
| IVL | −3.726131389 | −1.441202673 | 2.34E-39 | 1.66E-37 |
| SLC34A2 | −3.167340855 | 3.466038276 | 5.84E-39 | 4.05E-37 |
| SLC18A2 | −2.562912473 | 1.63627357 | 1.20E-38 | 8.23E-37 |
| NKX2-3 | 4.295926347 | 0.960315432 | 1.82E-38 | 1.24E-36 |
| PLCZ1 | −3.528460242 | −4.111131902 | 2.16E-38 | 1.46E-36 |
| SH2D1B | −2.070812969 | −2.247153696 | 2.30E-38 | 1.55E-36 |
| CCNI2 | −2.393120663 | −0.757806717 | 4.32E-38 | 2.89E-36 |
| OVCH2 | −2.474604409 | −1.229899903 | 4.36E-38 | 2.91E-36 |
| FOXQ1 | −2.113706442 | 2.340709388 | 1.77E-37 | 1.15E-35 |
| KCNS1 | −2.740616324 | 0.817741242 | 1.92E-37 | 1.24E-35 |
| REG3G | −5.362768922 | −3.996026989 | 2.04E-37 | 1.31E-35 |
| C21orf62 | −2.497748317 | 0.055828895 | 2.58E-37 | 1.65E-35 |
| LCN1 | −6.139941235 | −1.286853617 | 4.00E-37 | 2.53E-35 |
| EPHB1 | −2.071871109 | 1.406334339 | 4.28E-37 | 2.69E-35 |
| KY | −2.334715222 | −0.154030942 | 4.95E-37 | 3.09E-35 |
| SLC6A2 | −2.10678614 | −1.995090013 | 6.49E-37 | 4.03E-35 |
| KCNH5 | −3.179639439 | −2.263489674 | 1.54E-36 | 9.37E-35 |
| SIM2 | 2.246683676 | 6.39245743 | 2.10E-36 | 1.26E-34 |
| SIAH3 | −2.391041994 | −2.100220433 | 2.26E-36 | 1.36E-34 |
| TMEM213 | −2.938009836 | −0.973458402 | 3.77E-36 | 2.25E-34 |
| HOXB4 | −2.013128998 | 0.556074277 | 4.13E-36 | 2.44E-34 |
| WFDC2 | −2.278298566 | 5.691380285 | 6.46E-36 | 3.76E-34 |
| SYT8 | −2.62796781 | −0.04504407 | 9.39E-36 | 5.40E-34 |
| RAD21L1 | −3.931426302 | −4.029582937 | 1.14E-35 | 6.49E-34 |
| JPH4 | −2.032898156 | 3.896512339 | 1.52E-35 | 8.59E-34 |
| CLCA2 | −2.99069792 | 1.679389133 | 2.18E-35 | 1.22E-33 |
| GSTM1 | −3.348712796 | 3.902420778 | 2.36E-35 | 1.31E-33 |
| LY6D | −3.253162593 | 0.960481773 | 4.33E-35 | 2.40E-33 |
| LRCOL1 | −2.383609469 | −2.229173553 | 4.98E-35 | 2.74E-33 |
| KRT222 | −2.281961674 | −0.980864847 | 8.50E-35 | 4.62E-33 |
| CST4 | −3.656991558 | 1.448466198 | 9.21E-35 | 4.96E-33 |
| NEFM | −2.864624272 | 0.554358796 | 1.63E-34 | 8.52E-33 |
| DUOX2 | −2.585503889 | 2.635300446 | 1.67E-34 | 8.69E-33 |
| OPCML | −2.590292577 | −1.566109784 | 2.15E-34 | 1.11E-32 |
| DUOXA2 | −2.845461401 | −0.495623973 | 2.55E-34 | 1.29E-32 |
| PDE1C | −2.216681928 | 0.951037818 | 4.56E-34 | 2.27E-32 |
| MSLN | −2.673143906 | 2.312078872 | 9.59E-34 | 4.63E-32 |
| ATP6V1G3 | −3.407739337 | −2.880382936 | 1.15E-33 | 5.49E-32 |
| PRSS1 | −4.522834207 | −1.662313282 | 3.21E-33 | 1.50E-31 |
| FOXI2 | −2.306900792 | −2.155968961 | 3.52E-33 | 1.64E-31 |
| DCC | −2.459225535 | −0.956958564 | 4.76E-33 | 2.20E-31 |
| DUOXA1 | −2.155945912 | 2.560577531 | 2.07E-32 | 9.23E-31 |
| EMX1 | −2.835853169 | −3.197852638 | 2.31E-32 | 1.03E-30 |
| LVRN | −2.019593234 | −2.758757798 | 4.93E-32 | 2.12E-30 |
| CYP4B1 | −2.165077764 | 3.831392901 | 5.23E-32 | 2.24E-30 |
| TRH | −2.994128142 | −3.124637854 | 8.51E-32 | 3.59E-30 |
| HCAR3 | −2.221534692 | −1.323656559 | 9.86E-32 | 4.13E-30 |
| ITLN1 | −2.679097356 | −0.945384899 | 1.00E-31 | 4.18E-30 |
| SYT10 | −2.319711561 | −1.05820299 | 3.21E-31 | 1.29E-29 |
| ADRA1A | −2.022518109 | 2.151912159 | 4.83E-31 | 1.93E-29 |
| CDH8 | −2.263389081 | −0.939571117 | 5.72E-31 | 2.28E-29 |
| TMEM252 | −2.122053299 | 0.986298859 | 8.91E-31 | 3.52E-29 |
| AQP5 | −2.201067826 | −0.242796203 | 9.48E-31 | 3.74E-29 |
| PYY | −2.304539289 | −2.689358214 | 9.95E-31 | 3.91E-29 |
| HOXD4 | −2.010658856 | −1.667951124 | 1.07E-30 | 4.18E-29 |
| ASTL | −2.271544473 | −2.255063905 | 1.29E-30 | 4.99E-29 |
| FOLR1 | −2.267963855 | 0.443328675 | 1.60E-30 | 6.16E-29 |
| MGAT4C | −2.127179772 | −1.600917474 | 2.23E-30 | 8.48E-29 |
| LRRC3B | −2.226821286 | −3.265920448 | 2.37E-30 | 8.95E-29 |
| KCNF1 | −2.008639046 | −1.489871946 | 2.41E-30 | 9.07E-29 |
| CRNN | −4.196298519 | −3.602598059 | 3.11E-30 | 1.16E-28 |
| IL1RL1 | −2.164943372 | 0.580067069 | 4.59E-30 | 1.69E-28 |
| IGSF1 | −2.205150167 | 1.353299712 | 5.01E-30 | 1.84E-28 |
| CDO1 | −2.25897076 | 3.975578364 | 6.05E-30 | 2.21E-28 |
| UCN | 2.225028987 | 0.886371038 | 6.40E-30 | 2.32E-28 |
| AMACR | 3.271375874 | 7.272807932 | 1.43E-29 | 5.08E-28 |
| GLRA4 | −2.291078633 | −3.061966898 | 1.61E-29 | 5.68E-28 |
| CAPN6 | −2.167733352 | 3.083222877 | 1.67E-29 | 5.88E-28 |
| KCNJ13 | −2.677244912 | −2.098731552 | 2.33E-29 | 8.06E-28 |
| ROS1 | −2.664547966 | −1.41857366 | 2.56E-29 | 8.87E-28 |
| MEI4 | −2.157511068 | −3.504964788 | 2.79E-29 | 9.63E-28 |
| SLC45A2 | 5.233859283 | 3.020506268 | 3.11E-29 | 1.07E-27 |
| CYP4F22 | −2.48815109 | 1.361704428 | 4.42E-29 | 1.50E-27 |
| CLPSL1 | −3.884589239 | −3.179552875 | 7.45E-29 | 2.49E-27 |
| EVX2 | −2.201786126 | 0.147793705 | 1.24E-28 | 4.07E-27 |
| KRT16 | −2.392624596 | 2.060345166 | 1.80E-28 | 5.83E-27 |
| FAM83C | −2.840632484 | −1.823672187 | 2.44E-28 | 7.86E-27 |
| CHIA | −4.044693208 | −3.761939133 | 2.80E-28 | 8.97E-27 |
| CFAP65 | 2.17924865 | 1.923214285 | 4.94E-28 | 1.57E-26 |
| IL20 | −2.734032718 | −2.568179433 | 1.09E-27 | 3.38E-26 |
| HSD17B13 | −2.59308437 | 2.765460154 | 1.20E-27 | 3.67E-26 |
| ATP6V0D2 | −2.062948021 | −0.049347798 | 1.43E-27 | 4.33E-26 |
| FNDC10 | 2.189675352 | 3.1851198 | 1.43E-27 | 4.34E-26 |
| CLPSL2 | −2.9526799 | −3.435156672 | 4.32E-27 | 1.25E-25 |
| LGR6 | −2.077080456 | 2.417215605 | 6.96E-27 | 1.99E-25 |
| S100A14 | −2.094417081 | 2.768825991 | 7.06E-27 | 2.01E-25 |
| SFRP5 | −2.760385146 | 0.642123766 | 7.15E-27 | 2.04E-25 |
| NETO2 | 2.090951916 | 3.425360608 | 1.14E-26 | 3.22E-25 |
| LMX1B | 2.910958396 | 1.214627903 | 1.29E-26 | 3.60E-25 |
| GJB4 | −2.106346704 | −0.022277162 | 2.07E-26 | 5.69E-25 |
| SLITRK3 | −2.314417037 | 0.590359859 | 2.24E-26 | 6.13E-25 |
| CDH16 | −3.658293176 | −2.481108891 | 2.75E-26 | 7.49E-25 |
| ACTC1 | −2.549775894 | 5.199099142 | 3.86E-26 | 1.03E-24 |
| IL19 | −2.963272464 | −4.136314081 | 4.35E-26 | 1.16E-24 |
| LINGO2 | −2.353643152 | −1.61961467 | 2.36E-25 | 5.84E-24 |
| TGM5 | −2.041707954 | −1.130721222 | 2.51E-25 | 6.21E-24 |
| FAM83B | −2.016553537 | 0.635021108 | 3.30E-25 | 8.00E-24 |
| DNAH5 | 2.32012031 | 5.526858819 | 4.15E-25 | 1.00E-23 |
| CYP3A5 | −2.005756625 | 3.220640626 | 4.78E-25 | 1.15E-23 |
| KRT13 | −2.974414253 | 5.283076591 | 5.95E-25 | 1.42E-23 |
| LINC00694 | −2.00291498 | −2.423117021 | 7.37E-25 | 1.75E-23 |
| SCARA5 | −2.212531236 | 1.054207669 | 7.76E-25 | 1.84E-23 |
| PLA2G3 | −2.484369479 | −2.123775808 | 8.97E-25 | 2.12E-23 |
| SLIT1 | 2.991124143 | 4.235802648 | 9.13E-25 | 2.15E-23 |
| ADAMTS18 | −2.485445465 | −1.555485261 | 9.65E-25 | 2.26E-23 |
| ATP4B | −2.170350626 | −3.615853337 | 2.04E-24 | 4.61E-23 |
| MATN4 | −2.253033183 | −1.39482938 | 3.30E-24 | 7.32E-23 |
| C2orf72 | 2.352501555 | 6.112412268 | 3.37E-24 | 7.45E-23 |
| CBLN4 | −2.282088492 | −2.141588262 | 3.46E-24 | 7.64E-23 |
| HRASLS | −2.750037652 | −2.668890752 | 4.62E-24 | 1.01E-22 |
| PENK | −2.157694308 | 2.279458961 | 5.35E-24 | 1.17E-22 |
| CCDC198 | −3.361534597 | −3.550866101 | 6.39E-24 | 1.38E-22 |
| BTN1A1 | −2.058438498 | −3.223659882 | 8.65E-24 | 1.84E-22 |
| TG | −2.866055894 | 5.040539624 | 9.02E-24 | 1.91E-22 |
| SMR3B | −4.345403915 | −1.407810214 | 9.65E-24 | 2.04E-22 |
| C2CD4C | 2.365102335 | 1.292132594 | 1.03E-23 | 2.16E-22 |
| ATP6V0A4 | −2.469261417 | −0.171828266 | 1.44E-23 | 2.99E-22 |
| C1orf61 | −2.037014861 | −1.442631941 | 2.23E-23 | 4.54E-22 |
| SERPINB5 | −2.026028458 | 3.221299557 | 2.88E-23 | 5.82E-22 |
| OR7C1 | −2.022970619 | −2.050624749 | 3.09E-23 | 6.23E-22 |
| KBTBD13 | −2.109275337 | −3.695224421 | 3.30E-23 | 6.63E-22 |
| TH | −2.763714651 | −1.950206659 | 3.39E-23 | 6.81E-22 |
| MATK | 2.589971629 | 3.389770366 | 3.57E-23 | 7.14E-22 |
| ARHGAP19-SLIT1 | 3.000318492 | −2.097085148 | 4.54E-23 | 9.00E-22 |
| PAQR6 | 2.309347403 | 3.648588239 | 7.11E-23 | 1.37E-21 |
| CIDEC | −2.214864174 | −1.468234676 | 7.64E-23 | 1.47E-21 |
| FADS6 | −2.326944554 | −3.777201655 | 1.38E-22 | 2.59E-21 |
| DPP6 | −2.186696088 | 0.124736584 | 1.39E-22 | 2.61E-21 |
| WIF1 | −2.21507643 | 2.513039346 | 1.94E-22 | 3.57E-21 |
| CPB2 | −2.040650414 | −2.684359228 | 2.11E-22 | 3.86E-21 |
| APOC1 | 2.315598498 | 4.432027844 | 2.22E-22 | 4.06E-21 |
| NLRP12 | 2.474330308 | 1.187055602 | 2.27E-22 | 4.14E-21 |
| CPNE6 | −2.170399828 | 1.772299025 | 2.49E-22 | 4.53E-21 |
| HOXC6 | 2.328170608 | 3.774272822 | 2.79E-22 | 5.03E-21 |
| HOXC4 | 2.31331291 | 2.665774136 | 2.79E-22 | 5.03E-21 |
| ARSF | −2.138562635 | −2.870411011 | 2.97E-22 | 5.34E-21 |
| CERS1 | 2.553768773 | 1.469026165 | 3.80E-22 | 6.80E-21 |
| TRIM31 | −2.091716954 | 0.490285758 | 5.70E-22 | 1.00E-20 |
| ATP2B3 | −2.394530451 | −3.269639342 | 7.38E-22 | 1.29E-20 |
| ANKRD66 | 2.801025739 | −0.784999268 | 9.06E-22 | 1.57E-20 |
| MMP26 | 3.066474235 | 1.352487044 | 9.41E-22 | 1.62E-20 |
| KCNA4 | −2.346476072 | −2.699325651 | 1.18E-21 | 2.01E-20 |
| MUC21 | −3.114340581 | −2.477022768 | 1.22E-21 | 2.08E-20 |
| CLEC18B | 2.167387184 | −1.095435957 | 1.69E-21 | 2.84E-20 |
| TMEM196 | −2.596176551 | −3.648797811 | 2.57E-21 | 4.24E-20 |
| CCDC78 | 2.642490882 | 2.309466825 | 2.97E-21 | 4.87E-20 |
| EPGN | −2.120950779 | −2.692080174 | 3.34E-21 | 5.46E-20 |
| DEFB134 | −2.58614525 | −4.035042203 | 3.75E-21 | 6.09E-20 |
| GAS2L2 | −2.487311305 | −1.119961385 | 5.49E-21 | 8.72E-20 |
| CHP2 | −2.13935326 | 1.031772412 | 5.59E-21 | 8.87E-20 |
| FAM163A | −2.082472426 | −2.065990547 | 9.75E-21 | 1.52E-19 |
| TNFSF11 | −2.003738684 | −2.569011336 | 1.24E-20 | 1.90E-19 |
| KRT27 | −2.305026411 | −3.966447429 | 1.69E-20 | 2.55E-19 |
| SCGB1A1 | −2.475823685 | 3.909002352 | 2.53E-20 | 3.75E-19 |
| COLEC10 | −2.558276109 | −2.501754928 | 3.26E-20 | 4.77E-19 |
| APOF | 2.433781339 | 3.137119857 | 4.46E-20 | 6.45E-19 |
| PDIA2 | 4.280402557 | 2.013335255 | 4.70E-20 | 6.77E-19 |
| ADRB3 | −2.365978611 | −2.540179574 | 5.72E-20 | 8.14E-19 |
| FLG2 | −2.599596026 | −2.865814995 | 6.83E-20 | 9.67E-19 |
| SHISA8 | 2.993519423 | −0.189202204 | 6.86E-20 | 9.69E-19 |
| TMPRSS11A | −2.790567956 | −1.570310054 | 9.22E-20 | 1.29E-18 |
| RPE65 | −2.175383876 | −0.00527736 | 1.31E-19 | 1.80E-18 |
| HJURP | 2.068082168 | 2.187879526 | 1.34E-19 | 1.85E-18 |
| CCDC83 | 3.229819171 | −1.017747147 | 1.44E-19 | 1.96E-18 |
| ACSM1 | 2.868523218 | 6.613879148 | 1.57E-19 | 2.13E-18 |
| CLCA4 | −2.496145495 | 1.627115296 | 1.66E-19 | 2.25E-18 |
| C10orf82 | −2.135397958 | −0.911355049 | 2.24E-19 | 2.98E-18 |
| KCNK10 | −2.011525621 | −2.711656881 | 2.32E-19 | 3.08E-18 |
| TERT | 2.688569503 | −1.772060648 | 2.39E-19 | 3.16E-18 |
| EPPIN | −2.691805498 | −3.769965344 | 2.52E-19 | 3.32E-18 |
| DMBT1 | −2.236532941 | 0.1096238 | 3.62E-19 | 4.67E-18 |
| ONECUT2 | 2.681251387 | 3.221606201 | 3.64E-19 | 4.69E-18 |
| SFTPC | −2.25322321 | −3.281266307 | 5.39E-19 | 6.82E-18 |
| TWIST1 | 2.177091444 | 3.574725101 | 9.65E-19 | 1.19E-17 |
| FAM196B | −2.217220736 | −1.695478869 | 1.03E-18 | 1.27E-17 |
| PHGR1 | 3.386311578 | 1.820119662 | 1.36E-18 | 1.66E-17 |
| MOV10L1 | 2.27846602 | 1.581465328 | 1.41E-18 | 1.71E-17 |
| TBX4 | −2.17375547 | 2.268146306 | 1.58E-18 | 1.92E-17 |
| AMH | 3.230456823 | 1.135656782 | 1.79E-18 | 2.16E-17 |
| ZIC2 | 3.465784012 | 2.099443841 | 1.95E-18 | 2.35E-17 |
| REN | −2.010856028 | −1.82427371 | 2.05E-18 | 2.46E-17 |
| ARL14 | −2.041449796 | −2.836582438 | 2.77E-18 | 3.27E-17 |
| GPR149 | −3.565113736 | −2.173936635 | 2.97E-18 | 3.49E-17 |
| CACNA1D | 2.177413498 | 5.843737038 | 3.34E-18 | 3.89E-17 |
| NOX4 | 2.005839671 | 1.199904726 | 4.41E-18 | 5.07E-17 |
| SCGB3A1 | −2.174104533 | 2.495869638 | 7.08E-18 | 8.03E-17 |
| FOXN4 | 2.808885496 | −0.819378366 | 8.85E-18 | 9.88E-17 |
| DLX1 | 2.964944227 | 5.245627 | 1.43E-17 | 1.56E-16 |
| ARHGDIG | 2.722116583 | 1.951835142 | 1.80E-17 | 1.94E-16 |
| F2RL2 | 2.081512112 | 0.759279249 | 2.12E-17 | 2.26E-16 |
| LCN8 | −2.945514338 | −3.234638837 | 2.79E-17 | 2.94E-16 |
| OTX1 | 2.103182365 | 3.115942657 | 2.93E-17 | 3.08E-16 |
| KRT4 | −2.137873946 | 2.801389922 | 4.58E-17 | 4.75E-16 |
| ALB | 4.431655563 | 3.09043822 | 6.92E-17 | 7.03E-16 |
| TSPAN19 | 3.208847265 | 0.730279277 | 9.00E-17 | 9.05E-16 |
| GNG13 | 2.803827842 | 0.126341753 | 1.01E-16 | 1.01E-15 |
| DSG4 | −2.466048424 | −2.345854057 | 1.03E-16 | 1.03E-15 |
| EFNA2 | 3.057267493 | −1.886740535 | 1.45E-16 | 1.43E-15 |
| CCER2 | 2.000918092 | −0.3544635 | 1.48E-16 | 1.45E-15 |
| FAM163B | −2.321266796 | −2.413589392 | 1.63E-16 | 1.60E-15 |
| WFDC13 | −2.806598091 | −3.536010249 | 1.88E-16 | 1.83E-15 |
| ULBP1 | 2.15541201 | −1.028236929 | 1.89E-16 | 1.84E-15 |
| OR51E2 | 2.414972238 | 9.179968408 | 2.69E-16 | 2.59E-15 |
| TGM3 | 2.329959919 | 3.971772096 | 3.43E-16 | 3.25E-15 |
| MIOX | 2.023651528 | −0.851072617 | 6.30E-16 | 5.82E-15 |
| VGF | 2.505950604 | 1.28386305 | 6.83E-16 | 6.28E-15 |
| OR51S1 | 3.289112641 | −3.044498407 | 1.27E-15 | 1.14E-14 |
| KISS1R | 3.422260325 | −2.414859811 | 1.44E-15 | 1.28E-14 |
| HTR1E | −2.206015072 | −3.426655036 | 1.62E-15 | 1.42E-14 |
| GLYATL1B | 2.200776254 | −0.628848988 | 1.84E-15 | 1.60E-14 |
| KLK15 | 2.046781312 | 2.991115701 | 1.94E-15 | 1.67E-14 |
| THRSP | −2.470893848 | −0.92431372 | 2.09E-15 | 1.80E-14 |
| ANGPTL3 | 3.829532766 | 1.319655772 | 2.86E-15 | 2.43E-14 |
| TDRD1 | 2.968954599 | 4.312041873 | 4.32E-15 | 3.60E-14 |
| TNN | 2.314343045 | −0.352090036 | 4.70E-15 | 3.90E-14 |
| FOXD1 | 2.796864479 | 1.801714027 | 4.83E-15 | 4.00E-14 |
| DEFB126 | −2.714005591 | −1.975045093 | 6.47E-15 | 5.29E-14 |
| MYH6 | −2.531908454 | 0.826728283 | 1.06E-14 | 8.52E-14 |
| OR51H1 | 3.267819899 | −3.615043094 | 1.09E-14 | 8.69E-14 |
| SPON2 | 2.315586856 | 10.45792138 | 1.10E-14 | 8.78E-14 |
| ZMYND10 | 2.085925052 | 1.832701834 | 1.13E-14 | 9.04E-14 |
| ZP1 | 2.210730897 | 0.676381856 | 1.32E-14 | 1.04E-13 |
| KRTAP5-1 | 2.140411485 | −1.919084242 | 1.36E-14 | 1.07E-13 |
| SLC22A10 | 3.112889405 | 0.336771949 | 1.63E-14 | 1.27E-13 |
| NUTM2F | 2.599677343 | −2.440419577 | 1.81E-14 | 1.40E-13 |
| C11orf87 | −2.120433611 | −3.11909289 | 2.06E-14 | 1.59E-13 |
| SMIM28 | 2.307959645 | −1.425734172 | 2.32E-14 | 1.77E-13 |
| AC233992.2 | 2.188468656 | −2.988603347 | 2.55E-14 | 1.94E-13 |
| TRPC5 | −2.627768694 | −3.195666087 | 2.61E-14 | 1.99E-13 |
| ADGRG4 | −2.227928687 | −4.008538955 | 3.07E-14 | 2.32E-13 |
| DLX2 | 2.520792049 | 1.771274957 | 3.45E-14 | 2.59E-13 |
| AC013470.2 | 2.37626599 | −2.264634427 | 3.74E-14 | 2.80E-13 |
| 12-Sep | 2.32842503 | −1.614438692 | 3.83E-14 | 2.87E-13 |
| CHRM2 | −2.627807986 | −0.761918657 | 4.63E-14 | 3.42E-13 |
| PLPPR5 | −2.051535096 | −3.828879148 | 4.65E-14 | 3.44E-13 |
| KLB | 2.917388689 | 2.020757836 | 5.17E-14 | 3.79E-13 |
| KRT20 | 3.970669871 | −0.213705133 | 5.31E-14 | 3.90E-13 |
| SRARP | 3.763182115 | 4.202486163 | 5.51E-14 | 4.04E-13 |
| BAIAP2L2 | 2.226024178 | 2.628704883 | 5.96E-14 | 4.35E-13 |
| HIST1H4E | 2.159254954 | 1.043399573 | 7.18E-14 | 5.20E-13 |
| BRSK2 | 2.354199485 | 2.719801489 | 7.53E-14 | 5.44E-13 |
| AK5 | 2.022452828 | 3.960729447 | 8.23E-14 | 5.91E-13 |
| EPHA8 | 2.42213167 | −0.482075108 | 1.34E-13 | 9.40E-13 |
| HBQ1 | 3.058254424 | −2.360212262 | 1.94E-13 | 1.34E-12 |
| DLL3 | 2.036697223 | −0.709591972 | 2.50E-13 | 1.71E-12 |
| ATP8A2 | 2.805192641 | 3.941027197 | 2.73E-13 | 1.85E-12 |
| UGT3A1 | −2.300754877 | −2.195684518 | 2.90E-13 | 1.96E-12 |
| NKX6-1 | 3.539257475 | −0.069017009 | 3.14E-13 | 2.11E-12 |
| FOXB2 | 4.120786237 | −0.339067966 | 3.43E-13 | 2.29E-12 |
| OR2B6 | 2.070727347 | −1.791175567 | 3.50E-13 | 2.34E-12 |
| ZIC5 | 3.400800848 | 0.951706604 | 3.68E-13 | 2.45E-12 |
| B3GNT6 | 4.168735091 | 3.52191008 | 4.20E-13 | 2.78E-12 |
| FGL1 | 3.035621437 | 1.63122323 | 4.37E-13 | 2.88E-12 |
| TFF3 | 2.769530436 | 8.238324054 | 8.51E-13 | 5.41E-12 |
| CST2 | 3.537958116 | 4.052951673 | 9.02E-13 | 5.71E-12 |
| FOXL2 | 2.386028893 | 1.084381767 | 9.59E-13 | 6.06E-12 |
| TBX10 | 3.419608802 | 1.522870413 | 1.20E-12 | 7.46E-12 |
| RHCG | −2.008813569 | 0.619765959 | 1.42E-12 | 8.83E-12 |
| NLRP13 | 2.952695433 | −1.963847664 | 1.51E-12 | 9.33E-12 |
| ISG15 | 2.015582231 | 5.871285045 | 1.62E-12 | 9.94E-12 |
| FEZF1 | 2.983525183 | −2.371592675 | 1.62E-12 | 9.98E-12 |
| UPK1B | −2.479748949 | −1.685662964 | 1.77E-12 | 1.09E-11 |
| FABP5 | 2.55866377 | 5.395196065 | 1.92E-12 | 1.18E-11 |
| NR2E1 | 3.803357172 | −1.451927234 | 2.09E-12 | 1.27E-11 |
| SPINK1 | 4.492947788 | 4.716448182 | 2.20E-12 | 1.34E-11 |
| FUT6 | −2.34107223 | −0.394817546 | 2.42E-12 | 1.46E-11 |
| HOXC12 | 4.618237944 | −0.951730916 | 2.59E-12 | 1.55E-11 |
| OLFM4 | −2.001351853 | 7.271508686 | 2.61E-12 | 1.57E-11 |
| ANKRD30A | 6.088697199 | 0.552547891 | 2.75E-12 | 1.64E-11 |
| SPZ1 | 7.464030724 | 1.177592669 | 3.19E-12 | 1.90E-11 |
| HOXC13 | 3.203398538 | −1.939843545 | 4.23E-12 | 2.47E-11 |
| CLC | −2.056516349 | −3.77853227 | 4.31E-12 | 2.51E-11 |
| RPRML | 2.940731461 | −1.095714471 | 4.65E-12 | 2.70E-11 |
| OR51T1 | 2.54982834 | −2.792011523 | 5.76E-12 | 3.32E-11 |
| FOXI1 | −2.138580221 | 1.090697338 | 6.04E-12 | 3.47E-11 |
| NKAIN1 | 2.258465266 | 4.792143603 | 6.04E-12 | 3.47E-11 |
| DNAH8 | 2.464627376 | 5.112883427 | 7.54E-12 | 4.27E-11 |
| WFDC6 | −2.255684544 | −3.632961702 | 7.55E-12 | 4.28E-11 |
| COL10A1 | 2.453464464 | 3.270292451 | 7.57E-12 | 4.29E-11 |
| COMP | 2.16226434 | 5.850848074 | 8.07E-12 | 4.56E-11 |
| SPRR3 | −2.650341647 | −2.524440816 | 8.16E-12 | 4.61E-11 |
| ERG | 2.494207074 | 6.99904595 | 9.37E-12 | 5.25E-11 |
| SSTR1 | 2.060786454 | 4.37450598 | 1.04E-11 | 5.80E-11 |
| UGT2B4 | 3.221566911 | 2.827464057 | 1.08E-11 | 5.99E-11 |
| IL36RN | 3.03414543 | −1.504655432 | 1.26E-11 | 6.97E-11 |
| C19orf81 | 2.546159141 | 1.113739849 | 1.30E-11 | 7.16E-11 |
| VAX1 | 3.961513127 | −2.632789163 | 1.64E-11 | 8.92E-11 |
| LRRTM3 | −2.107441126 | −1.24681776 | 1.91E-11 | 1.03E-10 |
| FAM57B | 2.031437105 | −0.698387388 | 2.09E-11 | 1.13E-10 |
| GRIN3A | 2.638844764 | 4.65540954 | 2.23E-11 | 1.20E-10 |
| TUBB4A | 2.469186771 | 2.047892023 | 2.36E-11 | 1.26E-10 |
| ADAM29 | −2.02667846 | −3.775698756 | 2.84E-11 | 1.50E-10 |
| BMP5 | −2.18782084 | 2.684465934 | 4.71E-11 | 2.43E-10 |
| COL2A1 | 3.785158888 | 6.15210744 | 4.83E-11 | 2.48E-10 |
| KRT72 | 3.761702653 | −1.981764703 | 5.03E-11 | 2.58E-10 |
| BARX1 | 2.391463924 | −1.658459129 | 5.47E-11 | 2.80E-10 |
| ROPN1L | 2.092640047 | −0.046359297 | 5.55E-11 | 2.83E-10 |
| ARMC3 | 2.136397252 | 0.189599604 | 6.22E-11 | 3.16E-10 |
| PROC | 2.67020989 | 1.131805312 | 7.29E-11 | 3.67E-10 |
| KCNG3 | 2.132031411 | 2.045373485 | 9.35E-11 | 4.64E-10 |
| PCDHA1 | 2.795627976 | 0.375523362 | 1.20E-10 | 5.89E-10 |
| UNC5A | 2.709726989 | 3.92396467 | 1.24E-10 | 6.07E-10 |
| SUCNR1 | 3.081480451 | 1.536548908 | 1.41E-10 | 6.85E-10 |
| SMIM21 | 3.427582218 | −2.986773972 | 1.60E-10 | 7.73E-10 |
| ETV4 | 3.816023431 | 5.623026412 | 1.67E-10 | 8.03E-10 |
| SERPINA11 | 2.404577025 | 2.545674419 | 1.74E-10 | 8.34E-10 |
| GBX2 | 4.378429224 | −2.072125849 | 1.79E-10 | 8.57E-10 |
| CDC20B | 4.150567923 | 1.534510645 | 1.84E-10 | 8.79E-10 |
| GJA3 | 2.040848411 | 0.77159763 | 1.88E-10 | 8.94E-10 |
| C1QTNF9B | 2.025651594 | 0.385827111 | 1.89E-10 | 9.01E-10 |
| SMIM23 | 2.240012939 | −3.890952299 | 3.15E-10 | 1.46E-09 |
| CRYGD | −2.157881056 | −3.558055362 | 3.46E-10 | 1.60E-09 |
| GRPR | 2.082638062 | 1.422602298 | 4.01E-10 | 1.84E-09 |
| SLC22A31 | 2.15465431 | 1.682264922 | 4.15E-10 | 1.90E-09 |
| ZNF560 | 3.579830078 | 0.907262077 | 4.42E-10 | 2.02E-09 |
| PTPRR | 2.677564298 | 1.278570082 | 4.65E-10 | 2.11E-09 |
| SFTPA2 | 2.026666077 | 6.660829826 | 4.67E-10 | 2.12E-09 |
| HAO1 | 3.180714369 | −0.216881708 | 5.37E-10 | 2.43E-09 |
| CHIT1 | 2.124877061 | 3.520488959 | 6.25E-10 | 2.80E-09 |
| DMBX1 | 2.075728929 | −2.899045915 | 6.67E-10 | 2.98E-09 |
| UTS2B | 2.623610217 | 0.763054819 | 7.50E-10 | 3.33E-09 |
| PNMA5 | 5.583595807 | 1.256206222 | 7.52E-10 | 3.33E-09 |
| TTR | 2.263010905 | 1.534436019 | 7.85E-10 | 3.47E-09 |
| CER1 | −2.01557727 | −3.982760873 | 8.51E-10 | 3.74E-09 |
| GMNC | 2.222390659 | 1.731673014 | 9.70E-10 | 4.23E-09 |
| TGM4 | −2.299530484 | 9.058863089 | 1.06E-09 | 4.62E-09 |
| KRT73 | 2.784757012 | −3.396159935 | 1.60E-09 | 6.83E-09 |
| SLC24A2 | 2.263093544 | 0.036838795 | 1.70E-09 | 7.21E-09 |
| ANKRD30B | 3.073498165 | −1.41579624 | 1.91E-09 | 8.05E-09 |
| FEZF2 | 5.1526634 | −2.123364605 | 1.95E-09 | 8.20E-09 |
| NKX6-3 | 2.059995247 | −2.900165006 | 1.98E-09 | 8.34E-09 |
| OR52I1 | 2.176973868 | −3.001764047 | 2.05E-09 | 8.58E-09 |
| CAMKV | 2.881570973 | −2.038814296 | 2.86E-09 | 1.18E-08 |
| NOTUM | 2.308455321 | 0.786652156 | 4.24E-09 | 1.70E-08 |
| SFTA3 | −2.106019445 | −2.466203685 | 4.27E-09 | 1.71E-08 |
| POU4F1 | 3.693208616 | −2.095987674 | 4.39E-09 | 1.76E-08 |
| IL1F10 | 2.260744127 | −3.909896446 | 4.80E-09 | 1.91E-08 |
| PBOV1 | 2.085675747 | −2.154992386 | 7.36E-09 | 2.85E-08 |
| DIO1 | 2.26987907 | 0.891743028 | 7.38E-09 | 2.86E-08 |
| NETO1 | 3.568491117 | 0.942120675 | 8.02E-09 | 3.10E-08 |
| PPP3R2 | 2.570526002 | −2.677368344 | 8.10E-09 | 3.12E-08 |
| HNF1A | 2.906394479 | 0.190866031 | 8.41E-09 | 3.24E-08 |
| OR51A7 | 2.638046472 | −3.56624315 | 9.38E-09 | 3.59E-08 |
| GSTA3 | 2.806870513 | −1.42037873 | 9.39E-09 | 3.59E-08 |
| AC011604.2 | 2.143482743 | −3.108314603 | 9.51E-09 | 3.64E-08 |
| SMR3A | −2.635771323 | −4.144131086 | 1.06E-08 | 4.01E-08 |
| PANX3 | 2.326305366 | −3.160277377 | 1.42E-08 | 5.32E-08 |
| POTEB3 | 3.730685749 | −3.713452026 | 1.60E-08 | 5.94E-08 |
| APOA2 | 6.084002355 | −0.989674056 | 1.65E-08 | 6.13E-08 |
| HMX1 | 3.620329015 | −2.282249925 | 1.71E-08 | 6.34E-08 |
| KLK14 | 2.199159846 | 2.763344004 | 1.74E-08 | 6.44E-08 |
| TEX19 | 2.109864247 | −3.789954315 | 1.75E-08 | 6.48E-08 |
| FOXB1 | 2.548534069 | −3.326675239 | 1.83E-08 | 6.77E-08 |
| OR2T4 | −2.107781427 | −4.107614379 | 1.95E-08 | 7.16E-08 |
| DEFA6 | 8.471273836 | 1.646111486 | 2.42E-08 | 8.78E-08 |
| HIST2H2AA3 | 2.438596056 | −2.699403297 | 2.65E-08 | 9.60E-08 |
| CPN1 | 3.764667337 | −3.646951466 | 2.86E-08 | 1.03E-07 |
| SHD | 2.349865496 | −1.643837394 | 3.07E-08 | 1.10E-07 |
| SLC17A4 | 3.661486856 | 0.041233394 | 3.45E-08 | 1.23E-07 |
| AFP | 2.561513116 | −1.819167591 | 3.62E-08 | 1.29E-07 |
| A1CF | 2.968271138 | −0.721602281 | 3.74E-08 | 1.33E-07 |
| CENPVL3 | 2.108254917 | −3.236233695 | 3.84E-08 | 1.36E-07 |
| DEFA5 | 9.398510106 | 3.364489602 | 3.89E-08 | 1.38E-07 |
| ADAM2 | 2.268866369 | 1.975249434 | 3.95E-08 | 1.40E-07 |
| OR51F2 | 2.001274564 | −3.505887555 | 4.08E-08 | 1.44E-07 |
| VIL1 | 2.444441479 | −1.715247996 | 4.27E-08 | 1.51E-07 |
| GC | 5.399505393 | 0.791302383 | 4.74E-08 | 1.66E-07 |
| GLP1R | 2.638173244 | 0.005120757 | 5.23E-08 | 1.82E-07 |
| HABP2 | 2.053307035 | −0.144814994 | 6.05E-08 | 2.08E-07 |
| AMBN | 5.985888716 | −2.328310244 | 6.27E-08 | 2.16E-07 |
| HELT | 2.793749673 | −3.373803166 | 7.89E-08 | 2.68E-07 |
| ETV1 | 2.242859964 | 5.823683585 | 8.08E-08 | 2.74E-07 |
| CGA | 3.095258408 | 2.341413952 | 1.05E-07 | 3.52E-07 |
| SCGB2A2 | 4.164296891 | −2.821900282 | 1.26E-07 | 4.18E-07 |
| NXPH1 | 2.742113092 | −2.408586056 | 1.51E-07 | 4.94E-07 |
| PAX1 | 2.566319949 | 0.613275567 | 1.56E-07 | 5.11E-07 |
| FOXG1 | 4.413970069 | −2.305401919 | 1.66E-07 | 5.40E-07 |
| KRT75 | 2.558006156 | −1.069230467 | 1.66E-07 | 5.41E-07 |
| CD5L | 3.99795932 | −2.274198075 | 1.96E-07 | 6.32E-07 |
| INSM2 | 2.764301541 | −2.31599136 | 1.97E-07 | 6.34E-07 |
| TM4SF20 | 2.812654396 | −2.785614732 | 2.06E-07 | 6.62E-07 |
| SLCO1B7 | 2.01296759 | −3.7612757 | 2.07E-07 | 6.65E-07 |
| DYTN | 3.239601987 | −0.530752709 | 2.16E-07 | 6.93E-07 |
| POTEC | 3.432674516 | −3.289200505 | 2.22E-07 | 7.11E-07 |
| NAA11 | 2.650446317 | −2.486862885 | 2.26E-07 | 7.21E-07 |
| MYH13 | 2.066063048 | −2.482864861 | 2.50E-07 | 7.92E-07 |
| CDHR4 | 2.337589644 | −0.649813964 | 2.54E-07 | 8.03E-07 |
| AKAP14 | 2.517486668 | −2.274812756 | 2.62E-07 | 8.29E-07 |
| S100A7A | −2.151656547 | −4.15011295 | 2.68E-07 | 8.45E-07 |
| CACNA1I | 2.167388874 | −0.161792924 | 2.78E-07 | 8.76E-07 |
| MAGEA12 | 4.253847274 | −1.629199508 | 2.83E-07 | 8.91E-07 |
| MS4A15 | 2.679036167 | −1.548974492 | 3.54E-07 | 1.10E-06 |
| C6orf10 | 2.932100927 | −1.675723134 | 3.67E-07 | 1.14E-06 |
| ZBBX | 2.218935272 | −1.371954365 | 4.27E-07 | 1.31E-06 |
| NWD2 | 2.691936171 | −1.688128902 | 5.21E-07 | 1.58E-06 |
| APOBEC4 | 2.432607195 | −2.444614927 | 5.34E-07 | 1.62E-06 |
| DRGX | 5.581607276 | −0.548299892 | 5.36E-07 | 1.62E-06 |
| CAPSL | 2.141430537 | −0.478931097 | 5.42E-07 | 1.64E-06 |
| SNTN | 2.12725837 | −0.315435664 | 5.66E-07 | 1.70E-06 |
| KNG1 | 2.617105398 | −2.091853619 | 6.24E-07 | 1.87E-06 |
| PRSS48 | 3.821948787 | −2.662238738 | 6.31E-07 | 1.89E-06 |
| WDR38 | 2.180078528 | −0.64579903 | 7.41E-07 | 2.20E-06 |
| SCN1A | 3.512060368 | −0.40236363 | 8.22E-07 | 2.43E-06 |
| PNLIP | 2.328354606 | −1.788983069 | 9.04E-07 | 2.65E-06 |
| CST1 | 2.792052996 | 5.564777783 | 9.11E-07 | 2.68E-06 |
| AHSG | 3.199438746 | −2.773945144 | 1.06E-06 | 3.08E-06 |
| CRYBA4 | 2.772621752 | −1.320349749 | 1.06E-06 | 3.08E-06 |
| IAPP | 3.324462289 | −0.773721066 | 1.07E-06 | 3.11E-06 |
| ZNF716 | 3.052270573 | −4.06518223 | 1.16E-06 | 3.35E-06 |
| LIPF | 6.326103 | 2.912495858 | 1.35E-06 | 3.87E-06 |
| SERPINA6 | 2.943682115 | −1.793361739 | 1.43E-06 | 4.10E-06 |
| IRS4 | 2.223425105 | −0.100658114 | 1.47E-06 | 4.21E-06 |
| MAGEA4 | 6.312010735 | −1.437186342 | 1.48E-06 | 4.22E-06 |
| MAGEA8 | 2.386730522 | −2.00924606 | 2.03E-06 | 5.70E-06 |
| APOC3 | 4.606628396 | −2.654273715 | 2.09E-06 | 5.86E-06 |
| ZIC3 | 2.575224827 | −0.857692039 | 2.48E-06 | 6.86E-06 |
| FGB | 4.323384685 | −0.115174883 | 2.96E-06 | 8.13E-06 |
| UGT1A3 | 2.37472128 | 0.542246428 | 3.02E-06 | 8.29E-06 |
| UGT1A1 | 3.262526158 | −1.071235534 | 3.03E-06 | 8.30E-06 |
| MAGEA1 | 4.69097688 | −1.863592199 | 3.13E-06 | 8.56E-06 |
| HRG | 3.137868897 | −3.339954046 | 3.64E-06 | 9.89E-06 |
| PRAME | 2.341567845 | 0.717925667 | 3.89E-06 | 1.05E-05 |
| SCRT2 | 3.638442309 | −3.636567385 | 3.91E-06 | 1.06E-05 |
| CHAT | 3.19066275 | −1.980217619 | 4.36E-06 | 1.17E-05 |
| GAPDHS | 2.418577557 | −3.713135428 | 4.53E-06 | 1.21E-05 |
| BARHL1 | 3.015059095 | −3.539516674 | 4.66E-06 | 1.25E-05 |
| C8B | 2.984778011 | −4.131577497 | 5.40E-06 | 1.43E-05 |
| C1orf158 | 2.231593922 | −2.248366293 | 5.60E-06 | 1.48E-05 |
| TUBA4B | 2.083283692 | −1.021407683 | 5.88E-06 | 1.55E-05 |
| TAC3 | 2.369711799 | −1.769799158 | 6.35E-06 | 1.67E-05 |
| DCAF4L2 | 4.181333285 | −3.336994371 | 6.42E-06 | 1.68E-05 |
| MAGEA11 | −2.334344997 | −4.12452691 | 6.47E-06 | 1.70E-05 |
| OOSP2 | 3.211424198 | −4.128101138 | 6.88E-06 | 1.80E-05 |
| CALML5 | 2.96302721 | −0.373435971 | 6.97E-06 | 1.82E-05 |
| CELA3B | 2.509137069 | −3.052407547 | 7.67E-06 | 1.99E-05 |
| FGF3 | 3.877010188 | −3.428917251 | 7.80E-06 | 2.02E-05 |
| AKR1C4 | 2.494703252 | −3.934584544 | 8.50E-06 | 2.19E-05 |
| SULT1C3 | 2.186968852 | −1.355115719 | 1.01E-05 | 2.57E-05 |
| GPR52 | 2.388393967 | −3.786669653 | 1.09E-05 | 2.77E-05 |
| FAM92B | 2.05640783 | −1.500495388 | 1.13E-05 | 2.87E-05 |
| OR5B2 | 2.609194338 | −3.807269643 | 1.20E-05 | 3.03E-05 |
| FAM216B | 2.374779824 | −0.518608291 | 1.23E-05 | 3.11E-05 |
| CA6 | 2.513401867 | −2.76088127 | 1.31E-05 | 3.29E-05 |
| CTAG2 | 4.925601332 | −0.782603962 | 1.32E-05 | 3.31E-05 |
| TAS2R38 | 2.622407082 | −3.630778383 | 1.34E-05 | 3.36E-05 |
| GLRA3 | 3.223686569 | −0.188435203 | 1.51E-05 | 3.75E-05 |
| GUCA2A | 3.562863475 | −2.288541517 | 1.72E-05 | 4.24E-05 |
| LHX3 | 2.215818134 | −4.129011745 | 1.75E-05 | 4.30E-05 |
| TMEM212 | 2.577624226 | −3.145400789 | 1.88E-05 | 4.61E-05 |
| UCN3 | 2.272500127 | −1.200540549 | 2.21E-05 | 5.37E-05 |
| MAGEC1 | 4.686921382 | −1.392116181 | 2.23E-05 | 5.41E-05 |
| CT45A10 | 4.045321429 | −3.548760894 | 2.50E-05 | 6.03E-05 |
| AL035425.2 | 2.735879316 | −3.680989103 | 2.58E-05 | 6.19E-05 |
| CLCA1 | 3.776638394 | −1.530223149 | 2.64E-05 | 6.33E-05 |
| PDE6H | 2.153226877 | −3.916344045 | 2.81E-05 | 6.71E-05 |
| KRTAP13-2 | 2.243103436 | 0.468205823 | 2.93E-05 | 6.99E-05 |
| COX7B2 | 4.741214645 | −1.121111491 | 2.99E-05 | 7.10E-05 |
| MAGEA3 | 4.054216829 | −1.757379075 | 3.30E-05 | 7.81E-05 |
| MAGEA6 | 4.274899658 | −1.956846537 | 3.38E-05 | 7.99E-05 |
| IQCM | 2.579364621 | −4.047403881 | 3.50E-05 | 8.25E-05 |
| LYPD8 | 2.256064736 | −1.241702493 | 3.54E-05 | 8.34E-05 |
| NKX2-5 | 2.801832118 | −2.378705948 | 3.87E-05 | 9.07E-05 |
| REG3A | 3.736515651 | −3.332085299 | 4.10E-05 | 9.56E-05 |
| IGFBP1 | 2.14975531 | −3.545161931 | 4.49E-05 | 0.000104171 |
| MC4R | 3.25596124 | −2.085215369 | 4.57E-05 | 0.000105886 |
| APOH | 3.130989094 | −0.333603889 | 4.69E-05 | 0.00010841 |
| PRSS56 | 3.770792623 | −1.397434252 | 4.82E-05 | 0.000111245 |
| FMR1NB | 2.403293682 | −3.119019324 | 5.25E-05 | 0.000120751 |
| CARD18 | 3.618460498 | −3.641480606 | 5.95E-05 | 0.000135585 |
| C6orf15 | 3.15734794 | −3.509243731 | 6.39E-05 | 0.000144602 |
| ABCC12 | 2.447749702 | −0.858545305 | 6.48E-05 | 0.00014656 |
| SLC6A18 | 3.381785529 | −2.97339517 | 6.57E-05 | 0.000148554 |
| IFNK | 2.684220062 | −3.827516977 | 6.73E-05 | 0.000151888 |
| TRIM49 | 3.177581725 | −3.476981911 | 7.20E-05 | 0.00016148 |
| PRB1 | 3.326542308 | −3.729099839 | 7.35E-05 | 0.000164634 |
| TMEM207 | 2.647209716 | −4.101090504 | 7.69E-05 | 0.000171776 |
| INSL6 | 2.073304179 | −2.965377473 | 8.41E-05 | 0.000186712 |
| OBP2B | 2.109540767 | −1.09806609 | 8.91E-05 | 0.000197245 |
| SOX3 | 2.572786664 | −2.593667145 | 9.58E-05 | 0.000210846 |
| PIH1D3 | 2.051029477 | −3.248873998 | 0.000111697 | 0.000243629 |
| OR52E8 | 2.720510067 | −3.728113618 | 0.000130226 | 0.000281198 |
| IBSP | 2.064740445 | −2.938509758 | 0.000152455 | 0.000325344 |
| CRX | 2.116262219 | −2.812440876 | 0.000155787 | 0.000332099 |
| RAX | 2.56782095 | −2.046630028 | 0.000161204 | 0.000342911 |
| SLC6A19 | 2.216142084 | 2.683437613 | 0.000161511 | 0.000343482 |
| SLCO6A1 | 3.116278974 | −4.027389676 | 0.000171515 | 0.000363202 |
| FGF4 | 3.429130493 | −3.126190388 | 0.000186328 | 0.000392617 |
| VPREB1 | 3.522957645 | −3.50589044 | 0.000189054 | 0.000397891 |
| PRDM9 | 3.406136372 | −3.227166206 | 0.000197076 | 0.000413366 |
| LGALS16 | 3.375273982 | −3.96329701 | 0.000210268 | 0.000439385 |
| CSAG1 | 2.447454631 | −1.666596873 | 0.000211937 | 0.000442615 |
| FGA | 3.556112828 | −1.177897264 | 0.000231462 | 0.000480249 |
| TM4SF5 | 2.007153954 | −3.706723742 | 0.000236151 | 0.000489409 |
| BARHL2 | 2.496114201 | −3.403339289 | 0.000325784 | 0.000661897 |
| GIF | 2.108620219 | −3.831577628 | 0.000351243 | 0.000709509 |
| OR1N2 | 3.286097731 | −2.80552916 | 0.000522382 | 0.001028372 |
| LIN28B | 2.080144856 | −3.937283675 | 0.000578444 | 0.001130644 |
| OTOP1 | 2.945283928 | −3.891057771 | 0.000579885 | 0.001132841 |
| SLC18A3 | 2.169664988 | −1.026690515 | 0.000704233 | 0.001358103 |
| APCS | 2.769781173 | −3.531743743 | 0.000885631 | 0.001680557 |
| SP9 | 2.005599586 | −2.921939416 | 0.000947647 | 0.00178931 |
| PRB4 | 2.598407211 | −4.012142174 | 0.00097319 | 0.001834634 |
| CYP2C9 | 2.028397643 | −4.040000218 | 0.0011091 | 0.002073755 |
| TMPRSS11B | 3.351520815 | −2.537086097 | 0.001404034 | 0.002581258 |
| TPTE | 2.586293693 | −3.849215013 | 0.001431842 | 0.002628596 |
| DSCR8 | 2.143606791 | −3.733115111 | 0.001984814 | 0.003563974 |
| UGT1A10 | 2.29302094 | −1.689729261 | 0.002529475 | 0.004471915 |
| HTN3 | 2.485178564 | −4.015732583 | 0.002830284 | 0.00496153 |
| OR2T10 | 2.159593297 | −2.004107925 | 0.003677882 | 0.0063374 |
| FGG | 2.324040335 | −2.102893311 | 0.004863442 | 0.008221775 |
| MT4 | 2.334444629 | −3.227723792 | 0.005440693 | 0.009120681 |
| INS | 2.330844774 | −3.828167758 | 0.00568305 | 0.009495118 |
Differentially expressed mRNAs in prostate cancer. The horizontal axis shows sample names in TCGA. The right vertical axis means the mRNA names. Downregulated genes are green and upregulated genes are red.
Differentially expressed lncRNAs in prostate cancer. The horizontal axis shows sample names in TCGA. The right vertical axis means the lncRNA names. Downregulated genes are green and upregulated genes are red.
Differentially expressed miRNAs in prostate cancer. The horizontal axis shows sample names in TCGA. The right vertical axis means the miRNA names. Downregulated genes are green and upregulated genes are red.